Abstract
Purpose: To develop a pharmacokinetic modelfree framework to analyze the dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) data for assessment of response of brain metastases to radiation therapy.
Methods: Twenty patients with 45 analyzable brain metastases had MRI scans prior to whole brain radiation therapy (WBRT) and at the end of the 2-week therapy. The volumetric DCE images covering the whole brain were acquired on a 3T scanner with approximately 5 s temporal resolution and a total scan time of about 3 min. DCE curves from all voxels of the 45 brain metastases were normalized and then temporally aligned. A DCE matrix that is constructed from the aligned DCE curves of all voxels of the 45 lesions obtained prior to WBRT is processed by principal component analysis to generate the principal components (PCs). Then, the projection coefficient maps prior to and at the end of WBRT are created for each lesion. Next, a pattern recognition technique, based upon fuzzy-c-means clustering, is used to delineate the tumor subvolumes relating to the value of the significant projection coefficients. The relationship between changes in different tumor subvolumes and treatment response was evaluated to differentiate responsive from stable and progressive tumors. Performance of the PC-defined tumor subvolume was also evaluated by receiver operating characteristic (ROC) analysis in prediction of nonresponsive lesions and compared with physiological-defined tumor subvolumes.
Results: The projection coefficient maps of the first three PCs contain almost all response-related information in DCE curves of brain metastases. The first projection coefficient, related to the area under DCE curves, is the major component to determine response while the third one has a complimentary role. In ROC analysis, the area under curve of 0.88 ± 0.05 and 0.86 ± 0.06 were achieved for the PC-defined and physiological-defined tumor subvolume in response assessment.
Conclusions: The PC-defined subvolume of a brain metastasis could predict tumor response to therapy similar to the physiological-defined one, while the former is determined more rapidly for clinical decision-making support.
Keywords: DCE-MRI, therapy assessment, tumor subvolumes, brain metastases
INTRODUCTION
Dynamic contrast enhanced magnetic resonance imaging (DCE-MRI, see Table 1) is a noninvasive imaging tool extensively used in oncology for diagnosis and therapy assessment,1 and also as an imaging biomarker for development of antiangiogenic drugs.2 The DCE-MRI technique is based upon rapid acquisition of a series of T1-weighted images from a tissue of interest before, during, and after intravenous bolus injection of a gadolinium-based contrast agent (CA).3 As the CA perfuses into the tissue under investigation, T1 values of the tissue water decrease to an extent which depends upon the CA concentration. Hence, the characteristic signal intensity time course, relating to the CA concentration, in a region of interest or a voxel of the tissue can be subsequently analyzed by a pharmacokinetic (PK) model. By fitting the DCE-MRI data to a PK model, a set of volumetric maps of physiological parameters can be obtained, for example, tissue perfusion, microvascular permeability, and extravascular extracellular volume.4 Longitudinal changes in maps of the physiological parameters of a tissue of interest from pre- to post-treatment could aid in for assessment and prediction of treatment response and outcome as well as for drug development.5, 6
Table 1.
Frequently used abbreviation.
| Abbreviation | Description |
|---|---|
| 2W | Week 2 |
| AIF | Arterial Input Function |
| AUC | Area Under Curve |
| cPC | Projection Coefficient |
| cPDF | Pooled Histogram |
| DCE-MRI | Dynamic Contrast Enhanced Magnetic Resonance Imaging |
| FCM | Fuzzy-C-Means Clustering |
| GTV | Gross Tumor Volume |
| H | Histogram: Interchangeably used with Probability Density Function |
| P | Progressive |
| PCA | Principle Component Analysis |
| Probability Density Function: interchangeably used with Histogram | |
| PK | Pharmacokinetic |
| Pre-RT | Prior to Radiation Therapy |
| R | Responsive lesions |
| rCBV | Regional Cerebral Blood Volume |
| ROC | Receiver Operating Characteristic |
| ROI | Region of Interest |
| S | Stable lesions |
| SV | Sub-volume |
| WBRT | Whole Brain Radiation Therapy |
The physiological parameter maps derived from the PK models, although useful and meaningful for diagnosis and therapy assessment, involve in a series of uncertainties.7 Foremost, the DCE-MRI data often cannot be fitted to a selected PK model perfectly, and hence may be a source of errors affecting the accuracy of the physiological parameters. In addition, the “physiological parameters” derived from the PK models may not accurately reflect the underlying physiology, due to oversimplification in the models and lack of physiological validation of the models. For example, interpretation of Ktrans derived from the Toft model depends upon flow limited, permeability limited, or mixed. Also, computing the physiological parameters by fitting the DCE-MRI data into the PK models is a time consuming process, which is inadequate for real-time decision making support in adaptive radiation therapy, especially to handle the number of patients and the clinical workload. Hence, a modelfree approach to analyze the DCE-MRI data, e.g., the methods based on factor analysis,8, 9 independent component analysis (ICA),10 and principal component analysis (PCA),11, 12, 13, 14 could potentially facilitate the development of the real-time decision-making supportive tools in diagnosis and therapy assessment. PCA has shown the potential to be a very robust and fast technique in analyzing the DCE-MRI data,11, 12, 13, 14, 15, 16 and especially in prostate12, 13, 14 and breast15, 16 cancer. Eyal et al.12 applied PCA to dynamic intensity-scaled (IS) and enhancement-scaled (ES) datasets to develop and evaluate a method for image processing of the dynamic contrast enhanced MRI of the prostate cancer. They found that the first IS-eigenvector captured the major part of the signal variance, and the next two IS-eigenvectors captured the signal changes due to the tissue contrast-enhancement; whereas the remaining eigenvectors captured noise fluctuations. When they were applying the approach to distinguish benign lesions from malignant prostate cancers, the high values of 2nd and 3rd eigenvectors were able to differentiate the two.13 Although the model-based approach achieved the similar results, the PCA-based methods were fast and robust for analyzing the DCE data. Furthermore, the PCA-based approach has been applied to the DCE images of breast cancer, in which the 2nd and 3rd eigenvectors were found to be indicators of the wash-in and wash-out of the contrast agent.15 However, although these works have shown promising results for diagnostic purposes, to the best of our knowledge, no standard approach has been proposed so far to incorporate PCA into an automatic supportive tool for decision-making in therapy assessment.
A previous study17 has shown that an early change in the subvolume of a brain metastatic tumor with high-rCBV and high-Ktrans is a better predictor for postradiation therapy response than a tumor volumetric change and a change in the mean tumor rCBV observed in the same time interval. In this previous work, the physiological imaging-defined tumor subvolume, as a response-predictor, is determined through a two-step process: (1) deriving the physiological parameters by fitting the DCE MRI data to a PK model and (2) delineating the subvolume by analysis of the physiological parameters in the tumor using a fuzzy-c-means (FCM)-based technique. Hence, in this work our main goal is to develop a general framework to derive a response-predictor from DCE-MRI data without using the PK modeling and to have a semiautomated or fully automated tool for response assessment and therapy guidance in radiation therapy of brain metastases, the most common form of intracranial tumors exceeding the number of primary brain tumors by at least ten times and occurring in approximately 25% of all patients with cancers.18 In our proposed approach, we transfer the DCE curves into an N-dimensional feature space using principal component analysis, then, identify the most response-related features using a FCM-based technique similar to what is proposed in Ref. 17, and finally combine the features to define the tumor subvolumes. We evaluate whether our modelfree approach could provide a similar or even better metric, in defining the subvolume of a tumor compared to the subvolume defined by the physiological parameters obtained from the PK model.
MATERIALS AND METHOD
Patients
Twenty patients (11 women and 9 men, ages 41–76 yr), diagnosed with brain metastases and enrolled in an institutional review board (IRB)-approved prospective MRI study and previously described in Ref. 17, were also included in this study (Table 2). The histology consisted of melanoma (11), nonsmall cell lung cancer (6), renal cell carcinoma (1), breast cancer (1), and head & neck squamous cell carcinoma (1). All patients received standard WBRT with a total dose of 30 Gy in 10 fractions (13 patients) or 37.5 Gy in 15 fractions (7 patients). If a patient had less than three brain metastatic lesions, all lesions were included. If a patient had more than three lesions, only the three largest lesions were included. If a patient had more than three lesions larger than 1 cm3, up to three lesions greater than 1 cm3 were also included. As a total, 45 lesions with a median volume of 1.65 cm3 and a range of 0.1–17.6 cm3 were analyzed.
Table 2.
Patient characteristics information.
| Total accumulated | Concurrent drug | No. of | Volume | |||
|---|---|---|---|---|---|---|
| Pt. #. | G/Age (Y) | Histology | dose/Fx (Gy) | treatment | lesions | range (cm3) |
| 1 | Female/54 | Breast Cancer | 37.5/2.5 | None | 3 | 4.23–11.78 |
| 2 | Male/63 | Renal Cell Carcinoma | 30/3 | Bortezomib | 2 | 13.23–14.67 |
| 3 | Male/41 | Melanoma | 37.5/2.5 | Bortezomib | 3 | 0.15–1.24 |
| 4 | Female/60 | NSC Lung Cancer | 37.5/2.5 | None | 1 | 0.52 |
| 5 | Female/52 | Melanoma | 37.5/2.5 | Bortezomib | 1 | 2.74 |
| 6 | Female/45 | Melanoma | 30/3 | Bortezomib | 1 | 2.07 |
| 7 | Male/49 | Melanoma | 30/3 | Bortezomib | 2 | 0.17–4.09 |
| 8 | Female/51 | NSC Lung Cancer | 30/3 | Bortezomib | 3 | 0.50–4.55 |
| 9 | Male/61 | Melanoma | 37.5/2.5 | Bortezomib | 4 | 6.64–17.67 |
| 10 | Male/52 | NSC Lung Cancer | 30/3 | None | 1 | 0.479 |
| 11 | Female/55 | Melanoma | 30/3 | Bortezomib | 2 | 0.42–0.54 |
| 12 | Male/76 | Melanoma | 30/3 | Bortezomib | 1 | 0.68 |
| 13 | Female/46 | Melanoma | 30/3 | Bortezomib | 6 | 1.25–1.95 |
| 14 | Female/57 | Melanoma | 30/3 | Bortezomib | 2 | 0.94–1.58 |
| 15 | Female/64 | NSC Lung Cancer | 37.5/2.5 | None | 1 | 0.108 |
| 16 | Male/60 | Melanoma | 30/3 | Bortezomib | 3 | 0.18–1.31 |
| 17 | Female/74 | Melanoma | 30/3 | Bortezomib | 4 | 0.69–5.81 |
| 18 | Male/43 | Head & Neck SCC | 30/3 | None | 1 | 0.60 |
| 19 | Male/58 | NSC Lung Cancer | 30/3 | None | 3 | 2.38–10.69 |
| 20 | Female/66 | NSC Lung Cancer | 37.5/2.5 | None | 1 | 0.95 |
Abbreviation: Pt. #. = patient number; G = Gender; Y = year; NSC Lung Cancer = non-small cell Lung Cancer; and Head & Neck SCC = Head and Neck squamous cell carcinoma.
Imaging acquisition
All patients had MRI scans on a Philips 3T scanner prior to radiation therapy (Pre-RT), 2 weeks after the start of RT (2W), and 1 month after the completion of treatment (1M Post-RT). MRI scans included pre and post-Gd-DTPA volumetric T1 weighted images, multislice 2D T2 weighted images, and volumetric DCE T1 weighted images. The 3D volumetric DCE-images were acquired during bolus injection of a standard dose (0.1 mmol/kg) of Gd-DTPA with an injection rate of 2 cc/s. The DCE-images were acquired in the sagittal plane with an image matrix of 128 × 128 × 80, a field-of-view of 240 × 240 × 160 (mm), a voxel size of 2 × 2 × 2 (mm3), a flip angle(α) of 200, and TE/TR of 1.04/5.14 ms and a temporal resolution of 4-6 s. Using an in-house software package, both anatomical and DCE-MR images were coregistered and resampled to have a voxel size of 0.9375 × 0.9375 × 3 (mm3). Each lesion of interest was manually contoured by a physician on the post-Gd T1 weighed images obtained preRT, 2W and 1M post-RT.
Pharmacokinetic modelfree framework
Our proposed PK modelfree framework consists of two phases: development phase and usage phase. In the development phase, a sample of the DCE data from brain metastases is processed and analyzed to develop the model and the predictive metric. In the usage phase, we determine if the predictive metric could be extracted rapidly from the DCE data of a new patient scan.
Development phase
A flowchart of the development phase, shown in Fig. 1, includes preprocessing, modeling of DCE curves of a sample of brain metastases (including PCA and feature classification) to obtain a single metric, and evaluating the metric for response assessment. Each step is described in Subsections 2D1, 2F in detail.
Figure 1.
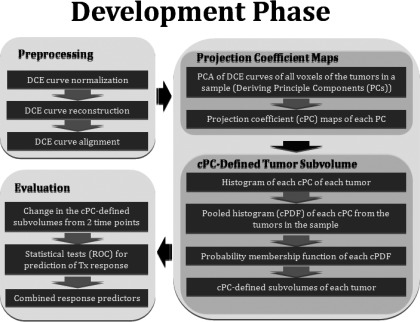
The flowchart of our proposed PK model free approach based upon tumor DCE data for supporting decision-making of assessment of therapy response. cPC: Projection coefficient map; cPDF: Pooled histogram corresponding to a cPC.
Preprocessing
DCE curves normalization
The dynamic curve at each voxel represents the temporal changes in signal intensity after the contrast injection. We calculate the signal intensity change ΔS from pre (baseline) to postcontrast as following:19
| (1) |
where S(t) and S0 represent signal intensities of a DCE curve at times t and 0 (the time of contrast injection), respectively. Note that ΔS(t)/S0 is proportional to ΔR1·T10, the change in relaxation rate caused by the contrast agent weighted by the initial spin-lattice relaxation time, as long as TR · R1 is much smaller than one. To account for the individual hemodynamic response to contrast, we normalize ΔS at each voxel using the peak of the arterial input function, AIFmax, obtained during the same scan as
| (2) |
An arterial input function can be determined from a region of interest (ROI) in a large artery (e.g., carotid artery for our application) manually, semiautomatically, or automatically. In the current work, a region of interest containing brain and neck is initially contoured. Then, DCE curves within the contour are averaged to determine the peak enhancement, Tmax, in the tissue. Assuming that the arterial input function reaches the enhancement peak prior to tissue, the first 20 voxels with the maximum enhancement in ΔS(Tmax − Δt), one time frame before Tmax, within the contour are thresholded, and then the corresponding DCE curves are averaged to be considered as an arterial input function.
DCE curve reconstruction
The DCE curves in each scan may not be acquired with the exactly same temporal resolutions and time durations. Hence, we standardize the DCE curves in such a way that all curves have the same temporal resolution and length. We use the spline curve-fitting method20 to reconstruct each DCE curve, and then resample them to have a temporal resolution of 4 s and a total length of 120 s, respectively.
DCE curve alignment
The DCE curves from voxels within the tumor volumes of all patients need to be temporally aligned for further processing. We use the arterial input function (AIF) obtained from each patient scan to align the DCE curves of voxels in the tumor volumes. First, we fit the Gamma variate function21 to each AIF as follows:
| (3) |
All AIFs are then aligned at t0 that is resigned to be time 0. Using the resultant time shifts, the DCE curves from each scan are adjusted accordingly. This makes all DCE curves aligned based on the start of enhancement in the arterial input function.
Projection coefficient map from Karhumen-Loeve expansion of DCE curves
Our primary goal is to extract response-predictive features rapidly and directly from the DCE curves. Hence, we expand the DCE curves using a set of basis functions, by which the coefficients of the projection vectors for each DCE curve are a unique representation in a new space. First, in the development stage, using DCE data from a sample of brain metastases (e.g., pretherapy DCE data), we construct matrix C (N × T) in which each row represents a DCE curve from one voxel in the tumors. N is the total number of voxels in all tumors and T is the number of time points in each curve. Next, we apply PCA to C to obtain a complete set of a total of T orthonormal principal components (PCi). Then, we perform the Karhumen-Loeve transformation of each DCE curve, ΔSN, in each voxel of the tumor to
| (4) |
where ai is the projection coefficient (cPC) corresponding to the ith principal component. Hence, each DCE curve in a tumor volume is represented uniquely by {αi} in a T-dimensional coefficient space. However, Eq. 4 can be truncated at the first M principal components which contain 99% of energy of the original DCE curves. We will show that M is noticeably smaller than T. Decomposing DCE curves of a new tumor to the first M PCs is much faster than fitting them to a PK model.
Projection coefficient defined tumor subvolumes
Probability density function of a projection coefficient in a tumor
Each PC depicts a feature of the tumor DCE curve. Each voxel in a tumor has a unique projection coefficient on each PC. For each PC, the projection coefficients of the voxels in a tumor, which can be presented as a volumetric map of a lesion, have a distinct role in predicting the treatment response and outcome. The distribution of the projection coefficients in a large tumor is heterogeneous, similar to the physiological parameters. Hence, similar to what has been done previously for the physiological parameters,17 we analyze the distribution patterns of a projection coefficient, ai, in the lesions, and subsequent changes during treatment. A probability density function (PDF) or histogram of ai of a lesion is generated using a nonparametric PDF estimator. The PDF consists of 150 evenly spaced points to cover the range of ai for all the lesions of interest. A value of the PDF at a point x, H(ai = x), of a lesion is calculated as
| (5) |
where ni is the number of voxels within |ai-x| < ɛ, and ɛ is a smooth factor of H and set as , where σ denotes a standard deviation of the ai distribution in the tumor. For each lesion, PDFs are calculated for scans at baseline [e.g., pretherapy as HPre(x)] and after starting therapy [e.g., at week-2 during therapy as H2W(x)]. After HPre(x) is normalized to have an area under the PDF curve equal to one (), the HPre(x)s of all the lesions are summed to generate a pooled PDF (cPDF), in which each lesion has an equal contribution regardless of its size. Figure 2 shows an example of a typical a1 PDF for a lesion with a volume of 17.6 cm3 at pretherapy and the pretherapy a1 pooled PDF, respectively.
Figure 2.
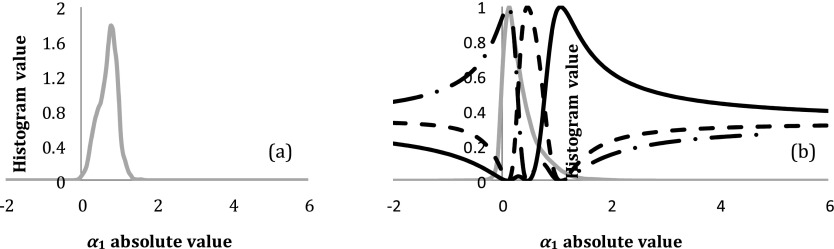
(a) An example of the pre-RT a1 histogram of a brain metastasis with a tumor volume of 17.6 cm3. (b) The pooled PDF (light gray) of the pre-RT a1 (cPDF1) from all the lesions and the three probability membership functions determined by FCM clustering. The cPDF1 is partitioned into three classes: representing low (dotted-dashed black), intermediate (dashed black), and high (solid black) a1 classes. a1: coefficient projection of the DCE curves on the 1st principal component.
Probabilistic membership functions of projection coefficients
Previous studies have suggested that the rCBV (or Ktrans) distribution in a brain tumor is abnormal compared to normal cerebral tissue, as elevated rCBV in a subvolume of the tumor and low rCBV in another one.22, 23, 24 A renormalization of tumor vasculature, such as decreasing the elevated rCBV and increasing the low one, could be an indicator of a tumor response to treatment.25 The DCE-derived physiological parameters (e.g., rCBV and Ktrans) and projection coefficients, (a1, a2…, aT), are two representations of the DCE curves. Therefore, it is reasonable to assume that ai in the brain metastases could also distribute abnormally in contrast to normal tissue, and changes during treatment could predict tumor response to therapy. Hence, similar to what has been done for rCBV previously,17 we classify the pooled distribution of Hpre(ai) to three classes as high, intermediate, and low ai classes using FCM clustering analysis by minimizing the objective function Jm:
| (6) |
where cj is a prototype vector of the jth class, Pj(ai) is a probabilistic membership of a ai value belonging to the jth class, and m is a fuzzy exponent and chosen as 2. The probabilistic membership function, Pj(ai), describes that a voxel having a projection coefficient ai has a probability P belonging to a class j, which is a new representation of a ai value of a tumor voxel (mathematically transfers the data from the ai space into a new space).
Projection coefficient defined tumor subvolume
Our primary interest is to test if a change in a subvolume of the tumor defined by high, intermediate, or low ai values is related to tumor treatment response. We define a subvolume (SV) of a tumor with low, intermediate, or high ai using the probabilistic membership function Pj(ai), and calculate a percentage change in the SV from pretherapy to after starting treatment (e.g., 2 weeks) as follows:
| (7) |
where GTV denotes the gross tumor volume. We will test whether a change in each of the first M cPC defined tumor subvolumes during RT is associated with post-RT tumor response in a group of patients, which will be described in Sec. 2F.
Tumor subvolume defined by combined projection coefficients
The overall aim of developing a prediction model for a clinical decision support system is to find a combination of factors that accurately anticipate an individual patient's outcome.26 Hence, we would like to test if combining different cPCs could improve prediction for tumor response compared to using one cPC. To do so, first a joint histogram of (a1, a2…, aM) of a lesion is computed, e.g., H(a1 = x1, a2 = x2, …, aM = xM). Then, a joint probability function, P({ai}, {βi}), is defined as follows:
| (8) |
where βi is the weighting factor of each coefficient and j ∈ {low, intermediate, or high}. Applying the joint probability function to Eq. 7, a percentage change in a subvolume of a tumor defined by {ai} classes from pretherapy to after starting treatment [e.g., 2 weeks (2W)] is given by
| (9) |
The weighting factor {βi} is selected based upon the best prediction of response from a developmental dataset and is evaluated by an independent dataset. We will demonstrate the principal in Sec. 3C.
Usage phase
For a new patient scan, first, we perform preprocessing of the DCE curves and then compute the projection coefficient maps of the first M or selected principal components. Next, we compute the histograms or a joint histogram of the selected coefficients within the tumor. Then, using the probability membership function obtained in the development step, we calculate the cPC-defined tumor subvolumes by Eqs. 8 or 9. Finally, a change of the subvolume from pretherapy to during or post-therapy is determined (Fig. 3).
Figure 3.
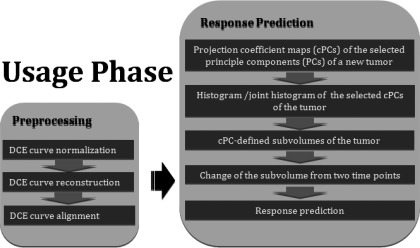
A flowchart of the procedure required for a new patient scan.
Evaluation
Endpoint
A percentage change in the gross tumor volume (GTV) from pre to post-RT was used as an endpoint for response assessment. Several patients did not have 3 or 6 months post-treatment imaging followups. For the patients in whom 3 and 6 months post-RT images were available, there were good correlations in the GTV changes between 1 and 3 months post-RT and between 3 and 6 months post-RT. Also, previous studies indicate that brain metastases exhibit little pseudoresponse and pseudoprogression one month after RT.27 Therefore, we used a percentage change in the GTV from pre-RT to 1 month post-RT, , as a measure of tumor response to therapy. From pre-RT to 1M post-RT, 16 tumors had a decrease in the GTV at least 25%, defined as responsive, 11 tumors had an increase at least 25%, defined as progressive, and the remaining 18 were defined as stable. We noticed that there were heterogeneous responses of multiple lesions from a single patient. Thus, each lesion was considered independently.
Parameter selection and evaluation
First, we ranked the priority of the candidate parameters and subvolumes by testing if the changes in significantly differentiated responsive tumors from combined stable and progressive ones using Mann-Whitney U Test. Considering M principal components and two independent subvolumes (only two of the three are independent) for each component, a p-value < (0.05/2M) with Bonferroni correction was considered as a significant cutoff to select the parameters. The conventional metrics, such as a percentage change in the GTV from pre-RT to 2W () and a change in the mean rCBV values of a tumor from pre-RT to 2W (), were not considered as the candidate parameters in our model, and thus not used for multiple comparison justification. Next, we performed univariate analysis to evaluate sensitivity and specificity of the selected significant metrics identified in the previous test for predicting responsive tumors using Receiver Operating Characteristic analysis (software package ROCKIT).28 Also, we compared these newly developed metrics with the conventional metrics including a percentage change in the GTV from pre-RT to 2W, , and a change in the mean rCBV values of a tumor from pre-RT to 2W, , for predicting post-treatment response. The significant difference of the area under ROC curves (AUC) between the metrics was compared by t-test, for which the standard error and the difference between the two AUCs were calculated by the method proposed by DeLong et al.29 To create the tumor subvolume defined by combining more than one cPC, we used the maximum AUC to determine the {βi}.
RESULTS
Principal components
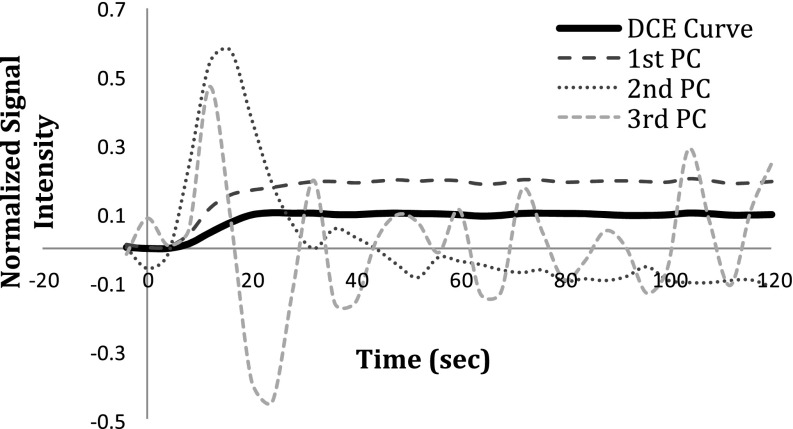
PCA revealed that the first three PCs comprised more than 99.99% of the energy of the DCE curves of brain metastases, of which the first component contributed approximately 99.8%, while the second and third components had 0.08% and 0.02% contributions, respectively (see Fig. 4). The first component (the predominant component) is highly related to the area under each DCE curve with the cross correlation coefficient of 0.99. Figure 5 shows the heterogeneous distributions of the first three cPC maps of a brain metastasis, indicating that an average cPC in the tumor would not be sensitive to a heterogeneous change. Therefore, the analysis described in Sec. 2 was applied to the histograms of the first three cPCs in a tumor to determine the subvolume of the tumor with a given class.
Figure 4.
An example of a typical AIFmax-normalized DCE curve of a voxel in a brain metastatic lesion and the first three principle components (PCs) resulted after applying PCA to the DCE-matrix. The coefficient map achieved by projecting the DCE curve onto the first component (the predominant component) is related to the area under the DCE curve (see the text for more details).
Figure 5.
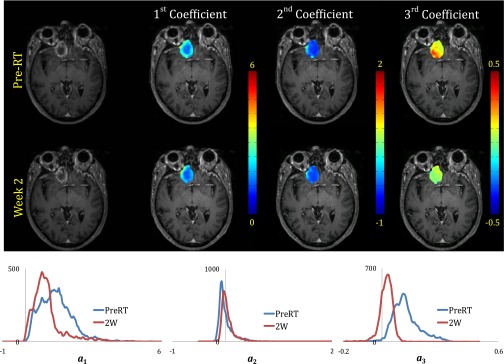
Distribution patterns and histograms of the first three projection coefficient maps for a responsive brain metastasis (from patient #19) overlaid on T1-weighted images at pre-RT and week 2 after start of whole brain radiation therapy.
Association of the cPC-defined tumor subvolumes with response
Associations of the changes in the first three cPC-defined brain metastases subvolumes with high, intermediate, or low coefficients from pre-RT to 2W with the tumor response to treatment are given in Table 3. Since we only compare the first three components, a p-value < 0.01 was considered as significance to select the candidate parameters. Median changes of −63.7 (range:-95.5 to 123), −13 (range: −98 to 198.4), and 44 (range: −42 to 444.3) were observed in the subvolume of the tumor with high-a1 (the most dominant component) from pre-RT to 2W for the responsive, stable, and progressive lesions, respectively. We found that for the responsive group, a percentage decrease in the high-a1 subvolumes of the tumors from pre-RT to 2W differed significantly from the group combining progressive and stable tumors (p < 0.0017), Table 3. We observed a similar but weaker trend for the high-a2 subvolume (p < 0.07). Furthermore, a percentage decrease in the low-a3 subvolume of the tumor was associated with tumor response (p < 0.01). A percentage decrease in the subvolumes defined by combining the high-a1 and low-a3 classes from pre-RT to 2W revealed that adding a3 improved the statistical significance for differentiating the responsive tumor from the group of stable and progressive lesions compared to either coefficient alone but a2 did not add discriminatory information. As a postanalysis, the comparisons between other lesion groups are also given in Table 3.
Table 3.
Differences between responsive, stable and progressive brain metastases using cPC defined tumor subvolume, physiological defined tumor subvolume and conventional metrics
| Group of lesions |
||||||
|---|---|---|---|---|---|---|
| Analysis | Post Analysis | |||||
| R vs. {S & P} | R vs. S | S vs. P | R vs. P | |||
| Metric | p-value | |||||
| Projection Coefficient Defined Tumor Subvolumes | j = low | 0.5937 | 0.8766 | 0.7024 | 0.3878 | |
| j = intermediate | 0.0773 | 0.2477 | 0.3339 | 0.0457 | ||
| j= high | 0.0017** | 0.0199* | 0.1321 | 0.0015** | ||
| j = low | 0.4843 | 0.7431 | 0.7702 | 0.3359 | ||
| j = intermediate | 0.5774 | 0.8766 | 0.2002 | 0.1596 | ||
| j= high | 0.0661 | 0.3979 | 0.0561 | 0.0096** | ||
| j= low | 0.0094** | 0.0068** | 0.2002 | 0.1323 | ||
| j = intermediate | 0.4133 | 0.7693 | 0.2909 | 0.2083 | ||
| j = high | 0.8403 | 0.1522 | 0.0143 | 0.1088 | ||
| ^ | 0.0005 | 0.0018 | 0.3568 | 0.0053 | ||
| Physiological Defined Tumor Subvolumes | 0.0057** | 0.0338* | 0.3568 | 0.0072** | ||
| 0.4992 | 0.6663 | 0.0162* | 0.0406* | |||
| 0.0015** | 0.0199* | 0.0687 | 0.0012** | |||
| Conventional Metrics | 0.0066** | 0.0049** | 0.2336 | 0.1088 | ||
| 0.8775 | 0.5233 | 0.1704 | 0.5704 | |||
| 0.0124* | 0.1086 | 0.0653 | 0.0039** | |||
Abbreviations: GTV = gross tumor volume; R = responders; S = stables; P = Progressive; cPC = projection Coefficient; ^ The optimum value of β3 is 0.3, see the results of the ROC analysis.*: P<0.05;**: P<0.01. The candidate subvolume for each principle component is highlighted.
For comparison, the results of the rCBV-Ktrans analysis that were reported previously17 and other conventional metrics are included in Table 3. In summary, we found that the percentage decrease in the high-rCBV subvolumes of the brain metastases from pre-RT to 2W of the responsive group differed significantly from the progressive group (p < 0.0072) and from a group combining progressive and stable tumors (p < 0.0057), but not from the stable group (p = 0.033) (Table 3). The percentage decrease in the brain metastases subvolumes defined by both the high rCBV and high Ktrans classes from pre-RT to 2W differentiated the three groups with improved statistical significances, compared to using either variable alone. Specifically, the responsive group significantly differed from the progressive group (p = 0.0012) and from the group combining the progressive and stable tumors (p = 0.0015). In this case, median changes of −47.8 (range: −88.5 to 213.84), −2 (range: −90 to 959.42), and 46 (range: −22 to 254.5) were observed in the subvolume of the tumor with high-rCBV and Ktrans from pre-RT to 2W for the responsive, stable, and progressive lesions, respectively. The statistical analysis regarding the conventional metrics, such as changes in the mean of the tumor rCBV and Ktrans and the GTV from pre-RT to 2W are also given in Table 3. These data show that both physiological-defined and cPC-defined subvolumes of a brain metastasis achieve a similar level of statistical significance in differentiation of responsive, stable, and progressive brain metastatic lesions.
Predictive values of the cPC-defined tumor subvolumes
We explored the predictive value of the decrease in the subvolumes of the brain metastases defined by the cPCs from pre-RT to 2W for predicting responsive tumors post-RT, and compared their performance with the decrease in subvolumes of the tumors defined by the high rCBV and high Ktrans and two conventional metrics. The ROC analysis showed that the AUCs were 0.83 ± 0.06(±SEM), 0.77 ± 0.07, 0.80 ± 0.07, 0.70 ± 0.08, 0.67 ± 0.08, and 0.56 ± 0.09 for , , , , , and , respectively (Fig. 6), indicating the high-a1 defined subvolume of the tumor had the best performance among the tested variables for predicting responsive tumor. The subvolumes defined by the high-a1 and low-a3 classes with the weighting factor = 0.3, determined by empirical evaluation of the AUCs (right panel of Fig. 6), resulted in the largest AUC, 0.88 ± 0.05. The subvolumes defined by the high-rCBV and high-Ktrans classes with the weighting factor = 0.6 resulted in the AUC of 0.86 ± 0.06.
Figure 6.
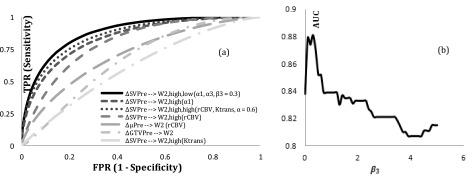
(a) ROC curves of the metrics listed in Table 3 for predicting responsive tumors; (b) AUC vs β3 in Eq. 9. FPR: False Positive Rate TPR: True Positive Rate; AUC: Area Under Curve.
The statistical analysis of the pairwise ROC curves revealed that was a predictor slightly but not significantly better than , and (p = 0.1 and p = 0.4, respectively). However, it was a predictor significantly better than and (p = 0.0463 and p = 0.02), respectively. Finally, the predictive value of was slightly but not significantly better than (p < 0.4).
Probability membership function maps
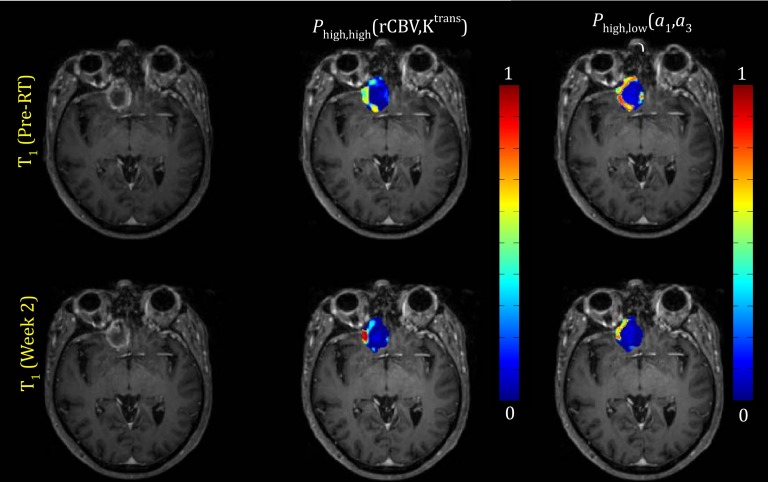
Examples of maps of the first coefficient maps and the corresponding histograms, and the probability functions belonging to the classes of high-rCBV and high-Ktrans and the classes of high-a1 and low-a3, of a responsive lesion pre-RT and at 2W are shown in Figs. 57, respectively. For this lesion, the voxel probability functions belonging to the high rCBV-Ktrans class decreased about 47% from pre-RT to 2W. A similar pattern was also observed for the probability functions belonging to the class of high-a1 and low-a3.
Figure 7.
Top row: Pre-RT T1 weighted image (left), probability function map of the class with the high-rCBV and high Ktrans pre-RT (middle) and probability function map of the class with the high-a1 and low-a3 pre-RT (right). Bottom row: the similar images at 2W. This lesion is responsive. The images are from patient #19 in Table 3.
DISCUSSION AND CONCLUSION
In this paper, we proposed a general framework based on PCA and a pattern recognition technique for directly delineating the response-driven subvolume of brain metastases from the DCE-MRI data. We compared the predictive values of the PC-defined tumor subvolume with the physiological-parameter-defined one [based upon regional cerebral volume (rCBV) and Ktrans] in the patients treated with whole brain radiation therapy. We found that the two approaches could predict the tumor response to therapy similarly while the PC-defined subvolume can be delineated more rapidly, which is required for supporting clinic decision making. In overall, our findings indicate that the projection coefficient maps from the first three PCs may contain almost all response-related information of the DCE curves. Our further investigation revealed that the first coefficient that is related to the area under the DCE curve is the main factor to determine the response (AUC = 0.83), while the third component could have complimentary information (AUC = 0.88). Our approach had the potential to be an effective tool for supporting real-time decision making.
Our proposed approach to analyze the DCE-MRI data has several advantages compared with the other model-free techniques which are based on factor analysis,8, 9 ICA,10 or extracting characteristic features, such as time to peak or maximum enhancement, from the DCE curves.30 Foremost, for each tumor type or body site, a set of PCs used in calculation of the coefficient maps can be achieved offline and are available before a new patient is scanned. Also, we address the heterogeneous distributions of the principal component coefficients within a tumor by using fuzzy clustering analysis to determine the probability membership functions of classes that the principal component coefficients belong to. Again, this relationship is established offline. Hence, for a new patient scan, computations for the DCE curves only involve in preprocessing the DCE curves, projecting them onto a couple of predetermined principal components, calculating the histograms of the two projection coefficients in the tumor, and computing the metric given in Eq. 9. Computing a couple of principal component coefficients is a much faster process than fitting PK modeling. Our model can be updated when the new patients’ data are accumulated, including PCs, the probability membership functions, and the predictive statistics given in Fig. 6a. Also, realigning and reconstructing all DCE curves from all tumors in the DCE-matrix to determine the PCs has the potential to overcome the interscan variation in the DCE time series. In addition, the set of projection coefficients obtained for each curve is a complete representation of the curve in an N-dimensional feature space wherein the data reduction is performed with the best approximation and without noticeable concerns regarding the information loss. As shown, almost all response-related information is derived from the first three components. However, for the approaches based on feature extraction, there is no guarantee that the computed features incorporate all relevant information for therapy assessment.
One very important note regarding the proposed methodology is that it is critical to use the arterial input function to normalize the DCE curves. Our investigations showed the enhancement peak of AIF is not linearly correlated with the enhancement peaks of the DCE curves of other tissues, such as normal white matter, gray matter, and even veins, indicating intrapatient variation on the peak enhancement. Also, the enhancement peak of AIF manifests great interpatient variation. It is worthwhile to point out that the projection coefficients (e.g., also first three components) and subsequent delineated subvolume determined by our approach have great tolerances on the temporal resolution and total acquisition time of the DCE curves, while the image acquisition parameters may affect the accuracy of the physiological parameters derived from the PK models.31 Hence, we may be able to estimate the projection coefficient maps of a tumor accurately using a lower temporal resolution, by which a high spatial resolution can be achieved to delineate the tumor heterogeneity. However, it is worthwhile to mention that due to the temporal resolution variation in the data, we had to use a spline-fitting to standardize the DCE curves in the DCE matrix. But, any manipulation of the original datasets may introduce errors and bias, and may affect the final results. Hence, an optimized and standardized protocol in terms of spatial and temporal resolution could decrease the errors in the dataset and improve the statistical power of the evaluation of response. Also, using a standard acquisition protocol could avoid the additional preprocessing of the data in the PK model-free approach, which is important using response assessment during the course of radiation therapy for adaptation treatment guidance. In the adaptation treatment setting, it requires to develop fully automated, fast, and robust algorithms to compute the quantitative response metric. Further development of such an algorithm and comparing its performance with the PK-model based metric are ongoing. In addition, in our calculation, we neglected T10 in normalization of the DCE data. We recognize that T10 can vary within the tumor volume. However, the influence of T10 on identifying a robust biomarker for therapy assessment needs to be further verified.
In summary, in this paper, we proposed a general framework for directly analyzing the DCE-MRI data to delineate the response-driven subvolume of a brain metastatic tumor. However, this approach needs to be further validated using an independent and larger dataset. Also, it could be extended and recalibrated to other tumor types, e.g., glioblastoma, for early assessment of tumor response to therapy.
ACKNOWLEDGMENT
This work is supported in part by the National Institutes of Health (NIH) Grant Nos. RO1 NS064973 and R21 CA113699. The authors report no conflicts of interest in conducting the research.
This work is in part presented in the 21th annual meeting and exhibition of International Society for Magnetic Resonance in Medicine (ISMRM 2013).
References
- Jackson A., Buckley D. L., and Parker G. J. M., Contrast-Enhanced Magnetic Resonance Imaging in Oncology (Springer, Berlin,2005). [Google Scholar]
- Jackson A., O’Connor J. P. B., Parker G. J. M., and Jayson G. C., “Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging,” Clin. Cancer Res. 13, 3449–3459 (2007). 10.1158/1078-0432.CCR-07-0238 [DOI] [PubMed] [Google Scholar]
- Yankeelov T. E. and Gore J. C., “Dynamic contrast enhanced magnetic resonance imaging in oncology: Theory, data acquisition, analysis, and examples,” Curr. Med. Imaging Rev. 3, 91–107 (2007). 10.2174/157340507780619179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofts P. S. et al. , “Estimating kinetic parameters from dynamic contrast enhanced T1 -weighted MRI of a diffusible tracer: Standardized quantities and symbols,” J. Magn. Reson. Imaging 10, 223–232 (1999). [DOI] [PubMed] [Google Scholar]
- Hayes C., Padhani A. R., and Leach M. O., “Assessing changes in tumor vascular function using dynamic contrast-enhanced magnetic resonance imaging,” NMR Biomed. 15, 154–163 (2002). 10.1002/nbm.756 [DOI] [PubMed] [Google Scholar]
- Brasch R. and Turetschek K., “MRI characterization and grading angiogenesis using a macromolecular contrast media: Status report,” Eur. J. Radiol. 34, 148–155 (2000). 10.1016/S0720-048X(00)00195-9 [DOI] [PubMed] [Google Scholar]
- Eyal E. and Degani H., “Model-based and model-free parametric analysis of breast dynamic-contrast-enhanced MRI,” NMR Biomed. 22, 40–53 (2009). 10.1002/nbm.1221 [DOI] [PubMed] [Google Scholar]
- Frouin F., Bazin J. P., Paola M. D., Jolivet O., and Paola R. D., “FAMIS: A software package for functional feature extraction from biomedical multidimensional images,” Comput. Med. Imaging Graph. 16, 81–91 (1992). 10.1016/0895-6111(92)90121-O [DOI] [PubMed] [Google Scholar]
- Zagdanski A. M., Sigal R., Bosq J., Bazin J. P., Vanel D., and Paola R. D., “Factor analysis of medical image sequences in MR of head and neck tumors,” Am. J. Neuroradiol. 15, 1359–1368 (1994). [PMC free article] [PubMed] [Google Scholar]
- Yoo S. S., Gil C. B., Han J. Y., and Hee Kim H., “Independent component analysis for the examination of dynamic contrast-enhanced breast magnetic resonance imaging data: Preliminary study,” Invest. Radiol. 37, 647–654 (2002). 10.1097/00004424-200212000-00002 [DOI] [PubMed] [Google Scholar]
- Twellmann T., Saalbach A., Gerstung O., Leach M. O., and Nattkemper T. W., “Image fusion for dynamic contrast enhanced magnetic resonance imaging,” Biomed. Eng. Online 3, 35 (2004). 10.1186/1475-925X-3-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal E., Bloch N. B., Rofsky N. M., Furman-Haran E., Genega E. M., Lenkinski R. E., and Degani H., “Principal component analysis of dynamic contrast enhanced MRI in human prostate cancer,” Invest. Radiol. 45, 174–181 (2010). 10.1097/RLI.0b013e3181d0a02f [DOI] [PubMed] [Google Scholar]
- Bloch B., Eyal E., Rofsky N. M., Furman-Haran E., Degani H., Genega E. M., Dewolf W. C., Bubley G. J., and Lenkinski R. E., “Computer-aided diagnosis of prostate cancer: Clinical utility of integrating model-free and kinetic-based analysis of high spatial resolution dynamic contrast enhanced 3 tesla MRI,” Proc. Intl. Soc. Mag. Reson. Med. 16 (2008). [Google Scholar]
- Bruwer M. J., MacGregor J. F., and Noseworthy M. D., “Dynamic contrast-enhanced MRI diagnostics in oncology via principal component analysis,” J. Chemom. 22, 708–716 (2008). 10.1002/cem.1143 [DOI] [Google Scholar]
- Eyal E., Badikhi D., Furman-Haran E., Kelcz F., Kirshenbaum K. J., and Degani H., “Principal component analysis of breast DCE-MRI adjusted with a model-based method,” J. Magn. Reson. Imaging 30, 989–998 (2009). 10.1002/jmri.21950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal E. et al. , “Combination of model-free and model-based analysis of dynamic contrast enhanced MRI for breast cancer diagnosis,” Proc. SPIE 6916, 69161B-1 (2008). 10.1117/12.770192 [DOI] [Google Scholar]
- Farjam R., Tsien C. I., Feng F. Y., Gomez-Hassan D., Hayman J. A., Lawrence T. S., and Cao Y., “Physiological imaging-defined response-driven subvolume of a tumor,” Int. J. Radiat. Oncol., Biol., Phys. 85, 1383–1390 (2013) 10.1016/j.ijrobp.2012.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre J. Y., Krol G., Thaler H. T., and Posner J. B., “Distribution of brain metastases,” Arch Neurol. 45, 741–744 (1988). 10.1001/archneur.1988.00520310047016 [DOI] [PubMed] [Google Scholar]
- Jackson A., Buckley D. L., and Parker G. J. M., Dynamic Contrast-Enhanced Magnetic Resonance Imaging in Oncology (Springer-Verlag, Berlin/Heidelberg, 2005), pp. 69–79. [Google Scholar]
- Spath H., One Dimensional Spline Interpolation Algorithms (A K Peters Ltd., Wellesley, MA, 1995), p. 416. [Google Scholar]
- Cao Y., Shen Z., Chenevert T. L., and Ewing J. R., “Estimate of vascular permeability and cerebral blood volume using Gd-DTPA contrast enhancement and dynamic T2*-weighted MRI,” J. Magn. Reson. Imaging 24, 288–296 (2006). 10.1002/jmri.20634 [DOI] [PubMed] [Google Scholar]
- Law M., Yang S., Babb J. S., Knopp E. A., Golfinos J. G., Zagzag D., and Johnson G., “Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with Glioma grade,” Am. J. Neuroradiol. 25, 746–755 (2007). [PMC free article] [PubMed] [Google Scholar]
- Law M. et al. , “Gliomas: Predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR Imaging,” Radiology 247, 490–498 (2008). 10.1148/radiol.2472070898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. et al. , “The extent and severity of vascular leakage as evidence of tumor aggressiveness in high-grade gliomas,” Cancer Res. 66, 8912–8917 (2006). 10.1158/0008-5472.CAN-05-4328 [DOI] [PubMed] [Google Scholar]
- Cao Y., Tsien C. I., Nagesh V., Junck L., Ten Haken R., Ross B. D., Chenevert T. L., and Lawrence T. S., “Survival prediction in high-grade gliomas by perfusion MRI prior to and during early stage of RT,” Int. J. Radiat. Oncol., Biol., Phys. 64, 876–885 (2006). 10.1016/j.ijrobp.2005.09.001 [DOI] [PubMed] [Google Scholar]
- Lambin P., Van Stiphout R. G., Starmans M. H., Rios-Velazquez E., Nalbantov G., Aerts H. J., Roelofs E., Van Elmpt W., Boutros P. C., Granone P., Valentini V., Begg A. C., De Ruysscher D., and Dekker A., “Predicting outcomes in radiation oncology—Multifactorial decision support systems,” Nat. Rev. Clin. Oncol. 10, 27–40 (2013). 10.1038/nrclinonc.2012.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber P. E. et al. , “Transient enlargement of contrast uptake on MRI after linear accelerator (linac) stereotactic radiosurgery for brain metastases,” Int. J. Radiat. Oncol., Biol., Phys. 49, 1339–1349 (2001). 10.1016/S0360-3016(00)01511-X [DOI] [PubMed] [Google Scholar]
- Eng J., ROC analysis: Web-based calculator for ROC curves (Johns Hopkins University, Baltimore, 2012). [Google Scholar]
- DeLong E. R., DeLong D. M., and Clarke-Pearson D. L., “Comparing the area under two or more correlated receiver operating characteristic curve: A nonparametric approach,” Biometrics 44, 837–845 (1988). 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- Tuncbilek N., Kaplan M., Altaner S., Atakan I. H., Süt N., Inci O., and Demir M. K., “Value of dynamic contrast-enhanced MRI and correlation with tumor angiogenesis in bladder cancer,” AJR, Am. J. Roentgenol. 192, 949–955 (2009). 10.2214/AJR.08.1332 [DOI] [PubMed] [Google Scholar]
- Cao Y., Li D., Shen Z., and Normolle D., “Sensitivity of quantitative metrics derived from DCE MRI and a pharmacokinetic model to image quality and acquisition parameters,” Acad. Radiol. 17, 468–478 (2010). 10.1016/j.acra.2009.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]