Abstract
Metaphase cytogenetics (MC) has a major role in the risk stratification of patients with myelodysplastic syndromes (MDSs) and can affect the choice of therapies. Azacitidine (AZA) has changed the outcome of patients with MDS or acute myeloid leukemia (AML) unfit for intensive chemotherapy. Identification of patients without the benefit of AZA would allow AZA combination or other drugs in first-line treatments. New whole-genome scanning technologies such as single nucleotide polymorphism microarray (SNP-A)-based molecular karyotyping (MK) improve the risk stratification in MDS and AML. Maintenance of genomic integrity is less than three megabases (Mbs) total disruption of the genome correlated with better overall survival (OS) in patients with lower-risk MDS. In this SNP-A study, we aimed at defining a cutoff value for total genomic copy number (CN) alterations (TGA) influencing the median OS in a cohort of 51 higher-risk MDS/AML patients treated with AZA. We observed that the relative risk of worse OS increased >100 Mb of TGA, as detected by SNP-A-based MK (8 and 15 months respectively, P=0.02). Our data suggest that precise measurement of TGA could provide predictive information in poor and very poor revised International Prognostic Scoring system (IPSS-R) patients treated with AZA.
Keywords: SNP array, myelodysplastic syndromes, azacitidine, predictive factor
Introduction
The myelodysplastic syndromes (MDSs) represent an heterogeneous group of clonal hematopoietic stem-cell disorders with a propensity to progress to acute myeloid leukemia (AML). Azacitidine (AZA) is an hypomethylating agent, providing about 50% of responses in MDS and AML with low blast count.1 AZA has changed the outcome of patients with MDS or AML with multilineage dysplasia (AML-MLD) unfit for intensive chemotherapy. Unfortunately, many patients are primary refractory to AZA. Identification of patients without the benefit of AZA would allow AZA combination or other drugs in first-line treatments. Performans status ⩾2, intermediate- and poor-risk IPSS cytogenetic groups, presence of circulating blasts and RBC transfusion dependency ⩾4 units per 8 weeks independently predicted poorer overall survival (OS) in a series of 282 high-risk MDS patients treated with AZA.2 Similarly, the revised International Prognostic Scoring system (IPSS-R) was recently shown to have a strong prognostic value for survival.3 Metaphase cytogenetics (MC) has a major role in the risk stratification of MDS patients.4 To complement MC, cytogenetic laboratories recently started implementation of new whole-genome scanning technologies such as comparative genomic hybridization arrays or single nucleotide polymorphism microarray (SNP-A)-based molecular karyotyping (MK). Indeed, MK is a valuable tool for identification of previously cryptic defects, such as submicroscopic copy number (CN) changes, and CN-neutral loss of heterozygosity, analogous to segmental uniparental disomy (UPD).5 By adding genomic complexity, MK was shown to improve the risk stratification in MDS and AML, with inferior survival seen in patients with additional SNP-A defects.6, 7 However, the clinical relevance of MK is not fully established. In particular, little is known regarding correlations between the full length of total genomic imbalance and OS. Starczynowski et al.8 have shown that maintenance of genomic integrity, defined as <3 Mb total disruption of the genome as detected using comparative genomic hybridization arrays, correlated with better OS in patients with lower-risk MDS. In this SNP-A study, we aimed at defining a cutoff value for genomic unbalanced defects influencing the median OS in a cohort of 51 higher-risk patients treated with AZA.
Patients and methods
Patients
Fifty-one consecutive higher-risk MDS and AML patients with bone marrow blast of 20–30 %, classified as RAEB-t according to the FAB classification, received at least one cycle of AZA. All patients had signed informed consent in accordance with the Declaration of Helsinki. All data were collected on the reference date of 31 December 2012.
Treatment
Patients were treated at six centers. The general plan for AZA treatment was subcutaneous administration according to the approved FDA/EMEA schedule (75 mg/m2 per day for 7 days, every 28 days) for at least four cycles1 or a reduced AZA schedule (75 mg/m2 for 5 days every 4 weeks). All patients who responded after four to six cycles of AZA were to continue treatment until disease progression.
Response criteria
Responses were evaluated after four to six treatment cycles by blood counts and marrow aspiration. Complete remission (CR), partial remission, marrow CR, stable disease, hematological improvement and progression were defined according to the IWG 2006 criteria.9 Responses were analyzed on an intent-to-treat basis, and patients who received fewer than four cycles of AZA without documented responses or progression were considered to be treatment failures.
Cytogenetics
MC abnormalities were classified according to the International System for Human Cytogenetic Nomenclature criteria. Cytogenetic risk was evaluated according to the IPSS-R classification.10 DNA samples, obtained before the start of AZA treatment, were available for the entire cohort. DNAs were processed and hybridized to Genome-Wide Human SNP 6.0 arrays (Affymetrix, High Wycombe, UK) as per the manufacturer's instructions (Affymetrix). Genotyping was performed using the Birdseed version 2 genotype-calling algorithm, and data were analyzed with the GeneChip Genotyping Console version 3.0.2 software (Affymetrix). We selected as lesions segments with a minimum genomic size of 200 kb and with >20 SNP/CN-altered markers; considering that inherited genetic background could have a role in individual patient's response to treatment and survival, the segments overlapping with CN variations (CNVs) cataloged in CNVs' databases were taken into account. Then, for each patient, we calculated the size of total genomic material loss (TGL) by measuring and adding up the sizes of all single genomic deletions found with Genotyping Console. The same was performed for genomic gains (TGG) and segmental UPDs. The total genomic alteration size (TGA) was defined by the association of TGL, TGG and total UPD. Results were expressed in Mb. MK was performed using the Copy Number Analyzer for GeneChip (CNAG version 3.3.0.1) algorithm.
Statistical analysis
The Smoothing spline Cox model analysis was used to define megabase cutoffs. OS was calculated from the date of the initial AZA treatment until death from any cause. Survival curves were estimated according to the Kaplan–Meier product-limit method and were compared with the log-rank test. The overall response rate (ORR) was defined as the total rate of CR, marrow CR, partial remission and stable disease with hematological improvement. Statistical tests were considered significant when the two-tailed P-value was <0.05. Confidence intervals were computed with 95% coverage. All statistical analyses were performed with SAS software (SAS, version 9.2, SAS institute, Cary, NC, USA).
Results and discussion
In this study, we evaluated the predictive value of MK for survival in 51 MDS or WHO-AML-MLD patients (median age 73 (42–88); female (n=23), male (n=28)) treated with AZA in six French hospitals. Diagnosis at the time of AZA treatment was RAEB-2 in 20 patients (IPSS int-1 in 4, int-2 in 10, high in 6) and WHO-AML-MLD in 31 (IPSS int-2 in 14, high in 17). IPSS-R cytogenetic groups were good (n=23), intermediate (n=9), poor (n=7) and very poor (n=12). The IPSS-R risk score was found to be low in one patient, intermediate in 8%, high in 50% and very high in 40%. The median baseline hemoglobin level was 9.4 g/dl (range: 6.9–13.4), the median baseline platelet count was 54 G/l (range: 5–473), and the median baseline neutrophil count was 0.8 G/l (range: 0–58.8). The median number of previous treatments was 1 (range: 0–3) (Table 1).
Table 1. Patient characteristics.
| n=51 | |
|---|---|
| Age (median) | 73 (42–88) |
| Sex ratio M/W | 28/23 |
| Number of AZA cycles (range) | 7 (1–42) |
| Hemoglobin, g/dl | |
| <8 | 10 (20%) |
| 8–10 | 20 (39%) |
| >10 | 21 (41%) |
| Absolute neutrophils count, x103/l | |
| <0.8 | 32 (63%) |
| >0.8 | 19 (37%) |
| Platelet count, × 103/l | |
| <50 | 22 (43%) |
| 50–100 | 18 (35%) |
| >100 | 11 (22%) |
| BM blasts | |
| <10% | 0 (0%) |
| >10% | 51 (100%) |
| >20% | 31 (61%) |
| Cytogenetic group | |
| Very good | 0 |
| Good | 23 (45%) |
| Intermediate | 9 (18%) |
| Poor | 8 (15%) |
| Very poor | 11 (22%) |
| IPSS-R classification | |
| Very low | 0 |
| Low | 1 (2%) |
| Intermediate | 4 (8%) |
| High | 25 (49%) |
| Very high | 21 (41%) |
Abbreviations: AZA, azacitidine; BM, bone marrow; IPSS-R, revised International Prognostic Scoring system.
The median number of cycles was 7 (range 1–42). The ORR was 55%, which included CR in 5 (10%), partial remission in 10 (20%) and stable disease with hematological improvement in 13 patients (25%). Thirteen patients (25%) received fewer than four cycles of AZA without documented responses or progression. According to the IPSS-R cytogenetic classification, we observed a similar response rate across all the subgroups (ORR of 61%, 56%, 57% and 33% in the good, intermediate, poor and very poor subgroups, respectively; P=0.13). Similarly, the ORR was not significantly different among the subgroups of the IPSS-R classification (58% and 47% in high and very high-risk subgroups, respectively; P=0.49).
The median follow-up was 10 months. The median OS of the cohort was 11 months (3–53). The median OS was 13 and 9 months for high and very high IPSS-R patients, respectively (P=0.40).
All patients had genomic modifications detected by the SNP-A analysis. MK data showed genomic gains in 100% of the patients (TGG median size 7.5 Mb, min: 1.8, max: 221), genomic losses in 84% (TGL median size: 1.6 Mb, min: 0.3, max: 446) and the presence of UPDs in 82% (total UPD median size: 7 Mb, min: 1.3, max: 138). The median size for TGA was 86 Mb (min: 3.3, max: 470).
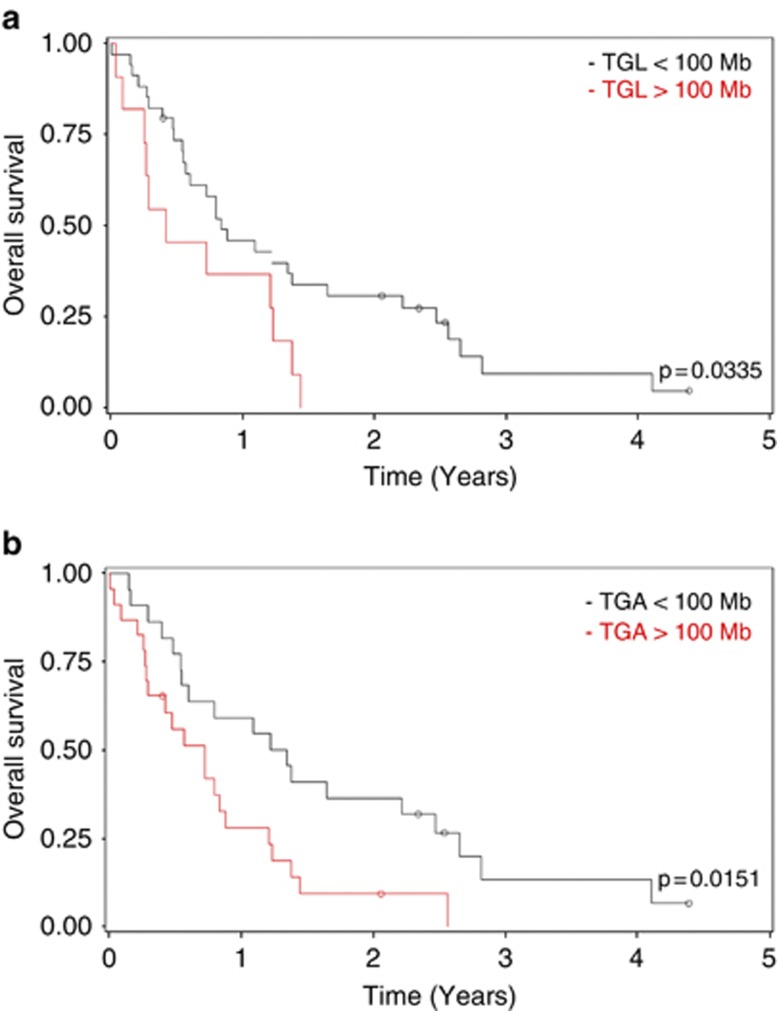
Starczynowski et al.8 have shown that genome alteration involving <3 Mb detectable by comparative genomic hybridization arrays correlated with better OS in patients with lower-risk MDS. In using the smoothing spline Cox model analysis, we observed that relative risk of the worse median OS increased above 100 Mb of TGL or TGA in high- and very high-risk MDS/AML. Patients with TGL or TGA (Figure 1) <100 or >100 Mb had significantly different OS (median 5 versus 10 months, P=0.03, and median 8 versus 15 months, P=0.02 (Figures 1a and b, respectively). Nevertheless, the 100-Mb TGL/TGA cutoff value had no impact on the ORR (TGL: 43% and 64%, respectively, P=0.14) and TGA (45% and 58%, respectively, P=0.48).
Figure 1.
Correlation between OS and TGL (a) and TGA (b) in high and very high IPSS-R risk AZA treated MDS/AML patients.
An increase in relative risk was found in intermediate, high and very high IPSS-R risk subgroups. Our search for an impact of TGL and TGA on the two RAEB-2 and AML subgroups, separately, was not conclusive, given the small size of these groups of patients.
Using the same statistical approach, we could not define a cutoff value for TGG influencing median OS. This is in agreement with data from Breems et al.11 showing that extra copies of one or more chromosomes (for example, trisomies), as detected by MC, did not seem to affect survival in AML. Conversely, extensive TGL involving >100 Mb correlated with poorer OS, consistent with the role of monosomal karyotypes (MSK) as strong indicator of poor prognosis in AML.12, 13 MSK was defined recently as the presence of two or more autosomal monosomies or a single monosomy associated with a structural abnormality. In comparison to complex aberrant karyotype, AML with MSK appears at the far end of the unfavorable prognostic spectrum;12 moreover, in a large retrospective study, combining MSK with poor-risk complex karyotypes four or more abnormalities identified the largest proportion of patients with very poor risk.13
As mentioned above, we took into account inherited CNVs or regions of apparent homozygosity for TGA measurement. Indeed, somatically acquired genomic abnormalities within the MDS/AML clone(s) are assumed to be linked to the pathogenesis of MDS and AML; however, one could hypothesize that inherited polymorphisms and CNVs may contribute to individual patient's response to treatments and survival. In other words, a combined study of MC and MK may help to define and distinguish cytogenetic patterns that can be attributed to ‘common features' of the pathology versus patterns that can be attributed to ‘personal or individual features' that are not common but could affect the response or resistance of some individuals to therapy and modulate prognosis. As is shown in Figure 1, TGA, which includes TGL, TGG and total segmental UPDs' size, is more predictive of shorter OS than TGL. Several studies have reported on the high frequency of segmental UPDs in MDS and AML samples.5, 14, 15 In our cohort, 82% of patients presented with UPDs, including 18 of 19 cases with normal karyotype. As for TGG, we could not define a cutoff value for total segmental UPDs' size influencing the median OS. UPDs result in CN-loss of heterozygosity and are therefore undetectable by MC. As whole-genome scanning technologies have been mainly used until now in research setting, whether UPDs per se have important diagnosis and prognosis information should be looked for prospectively on larger cohort of newly diagnosed patients.
Acquired UPDs have been instrumental in the detection of gene mutations, microdeletions or epigenetically suppressed regions in cancer cells. They point towards mutations that only have a pathogenic effect in a homozygous form. Similarly, germline UPDs theoretically may have a role in carcinogenesis by leading, for example, to duplication of susceptibility alleles or to gain or loss of imprinting through the duplication of methylated or unmethylated alleles. Hence, a multiplatform combining MC, MK and deep sequencing technology would be necessary to conduct a comprehensive cytogenetic profiling of our patients.
Conclusion
In conclusion, we show that TGL and/or TGA >100 Mb are relevant predictive markers for survival in a cohort of MDS and AML-MLD patients treated with AZA. To what extent do these data depend on the AZA treatment needs to go in depth, as MK was shown to affect patients' survival in MDS/AML even when therapeutic choices were not considered.6, 7 Therefore, additional studies are necessary to confirm our results. Many patients are refractory to AZA. Identification of patients without the benefit of AZA would allow to propose AZA combination or other drugs in first-line treatments. In this perspective, results described in our study will be helpful.
Acknowledgments
This work was supported by COST Action BM0801 (EuGESMA, European Genomic and Epigenetic Study on MDS and AML). SR received research funding from Celgene.
The authors declare no conflict of interest.
Footnotes
Author Contributions
TC, JMK, GG, DR, NM, JG, JGF and JPC collected the data; CM performed the SNP-A analysis; CM and NM performed the statistical analysis; TC and SR designed the study, TC, CM, PA and SR wrote the manuscript.
References
- Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzykson R, Thepot S, Quesnel B, Dreyfus F, Beyne-Rauzy O, Turlure P, et al. Prognostic factors for response and overall survival in 282 patients with higher-risk myelodysplastic syndromes treated with azacitidine. Blood. 2011;117:403–411. doi: 10.1182/blood-2010-06-289280. [DOI] [PubMed] [Google Scholar]
- Lamarque M, Raynaud S, Itzykson R, Thepot S, Quesnel B, Dreyfus F, et al. The revised IPSS is a powerful tool to evaluate the outcome of MDS patients treated with azacitidine: the GFM experience. Blood. 2012;120:5084–5085. doi: 10.1182/blood-2012-09-453555. [DOI] [PubMed] [Google Scholar]
- Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondek LP, Tiu R, O'Keefe CL, Sekeres MA, Theil KS, Maciejewski JP. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111:1534–1542. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiu RV, Gondek LP, O'Keefe CL, Elson P, Huh J, Mohamedali A, et al. Prognostic impact of SNP array karyotyping in myelodysplastic syndromes and related myeloid malignancies. Blood. 2011;117:4552–4560. doi: 10.1182/blood-2010-07-295857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiu RV, Gondek LP, O'Keefe CL, Huh J, Sekeres MA, Elson P, et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol. 2009;27:5219–5226. doi: 10.1200/JCO.2009.21.9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starczynowski DT, Vercauteren S, Telenius A, Sung S, Tohyama K, Brooks-Wilson A, et al. High-resolution whole genome tiling path array CGH analysis of CD34+ cells from patients with low-risk myelodysplastic syndromes reveals cryptic copy number alterations and predicts overall and leukemia-free survival. Blood. 2008;112:3412–3424. doi: 10.1182/blood-2007-11-122028. [DOI] [PubMed] [Google Scholar]
- Cheson BD, Greenberg PL, Bennett JM, Lowenberg B, Wijermans PW, Nimer SD, et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood. 2006;108:419–425. doi: 10.1182/blood-2005-10-4149. [DOI] [PubMed] [Google Scholar]
- Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, et al. Revised International Prognostic Scoring System (IPSS-R) for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breems DA, Van Putten WL, De Greef GE, Van Zelderen-Bhola SL, Gerssen-Schoorl KB, Mellink CH, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26:4791–4797. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
- Breems DA, Lowenberg B. Acute myeloid leukemia with monosomal karyotype at the far end of the unfavorable prognostic spectrum. Haematologica. 2011;96:491–493. doi: 10.3324/haematol.2011.043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach C, Alpermann T, Schnittger S, Kern W, Chromik J, Schmid C, et al. Prognostic value of monosomal karyotype in comparison to complex aberrant karyotype in acute myeloid leukemia: a study on 824 cases with aberrant karyotype. Blood. 2012;119:2122–2125. doi: 10.1182/blood-2011-10-385781. [DOI] [PubMed] [Google Scholar]
- Gondek LP, Tiu R, Haddad AS, O'Keefe CL, Sekeres MA, Theil KS, et al. Single nucleotide polymorphism arrays complement metaphase cytogenetics in detection of new chromosomal lesions in MDS. Leukemia. 2007;21:2058–2061. doi: 10.1038/sj.leu.2404745. [DOI] [PubMed] [Google Scholar]
- Heinrichs S, Kulkarni RV, Bueso-Ramos CE, Levine RL, Loh ML, Li C, et al. Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics. Leukemia. 2009;23:1605–1613. doi: 10.1038/leu.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]