Mutations in CSF3R have been recently defined as the common genetic event in patients with myeloid neoplasms, including the rare entity known as chronic neutrophilic leukemia (CNL),1, 2, 3 becoming a potentially useful biomarker for diagnosing and therapy target.4 CSF3R encodes the transmembrane receptor for granulocyte colony-stimulating factor (G-CSF; CSF3), which provides the proliferative and survival signal for granulocytes and also contributes to their differentiation and function.5 Although there are several studies on massive next-generation sequencing of myeloid disorders, not a single comprehensive study has been reported in CNL. Here, we used whole-exome sequencing (WES) and RNA sequencing (RNA-seq) to identify new candidate genes to the disease pathogenesis of an index CNL patient.
A 66-year-old man was diagnosed with CNL, according to the 2008 World Health Organization (WHO) classification. At diagnosis, the patient presented peripheral blood leukocytosis (66 × 109/l), segmented neutrophils and band forms were 91.5% of the white blood cells counts (WBCs), immature granulocytes were <10% of WBCs and myeloblasts were <1%. The aspirate showed a hypercellular bone marrow (BM) with neutrophilic granulocytes increased in number and percentage and myeloblasts 0.5% of WBCs. No dysplastic features were observed in the myeloid lineages. His Zubrod Performance Status (ECOG) was 1. A GTG-banding chromosome analysis revealed a normal karyotype (46,XY[20]), and molecular biology studies were negative for BCR-ABL1 transcripts and JAK2 V617F mutation. The patient was treated with hydroxyurea but, unfortunately, died 7 months after the diagnosis due to an intensification of the disease.
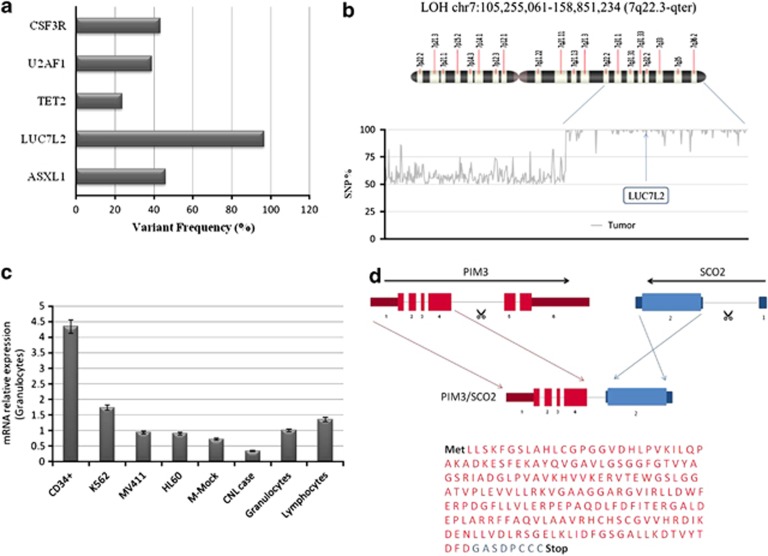
To improve our understanding of the genes involved in the pathogenesis of CNL, WES was performed on matched tumor and oral mucosa cell (germline) samples from the patient. Candidate somatic mutations were identified using RUbioSeq software.6 The bioinformatics analysis and the filtering steps to identify the coding variants are detailed in the Supplementary Material. In total, we found 1437 candidate variants; among them, 797 were somatic mutations (412 were intronic, intergenic, affecting non-conding-RNA or untranscribed regions and 385 were exonic). From the 385 exonic variants, we selected only those variants within coding regions that, after passing sequencing depth and quality filters, were, frameshift, stop gain/loss and non-synonymous amino acid changes predicted to produce a deleterious effect in the protein structure, resulting in 56 single-nucleototide variants (SNVs) and small insertions/deletions (indels). We selected 24 for further validation by Haloplex/Ion Torrent. In addition to the CSF3R p.Thr618Ile mutation, we validated mutations in U2AF1, TET2, LUC7L2 and ASXL1 (Figure 1a and Table 1).
Figure 1.
Molecular characterization of CNL. (a) In addition to the CSF3R T618I mutation, WES revealed mutations in U2AF1, TET2, LUC7L2 and ASXL1. LUC7L2 mutation was found in homozygosis; (b) LOH of 53.2 Mb in chromosome 7q including the locus of LUC7L2; (c) Downregulation of LUC7L2 expression in CNL patient, compared with other normal and leukemic myeloid cells; (d) PIM3–SCO2 fusion gene result of an intrachromosomal inversion of approximately 0.6 Mb in chromosome 22.
Table 1. Description of validated mutations in the CNL patient.
| Chrom | Position | Gene name | Zygosity | Ref | Variant | Var freq | Coverage | AA change | Type | Provean | SIFT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | 31024563 | ASXL1 | Het | C | T | 46.12 | 2433 | Q1350* | Stop gain | NA | NA |
| 7 | 139060825 | LUC7L2 | Hom | C | T | 96.55 | 261 | R93* | Stop gain | NA | NA |
| 4 | 106157539 | TET2 | Het | CG | C | 23.81 | 126 | NA | Frameshift | NA | NA |
| 21 | 44514777 | U2AF1 | Het | T | C | 38.96 | 2477 | Q157R | Single AA change | Deleterious | Damaging |
| 1 | 36933434 | CSF3R | Het | G | A | 43.23 | 1203 | T618I | Single AA change | Deleterious | Damaging |
Abbreviations: AA, amino acid; CNL, chronic neutrophilic leukemia; Het, heterozygotic; Hom, homozygotic; NA, not applicable.
The current study, first, confirms the observations by Maxon et al.1 and Pardanani et al.2 regarding the association between CNL and CSF3R mutations. Second, it presents a complete picture of the mutational profiling of CNL, certainly more complex than expected from these previous reports. In fact, we found and validated mutations affecting both splicing machinery and epigenetic genes. Kosmider et al.,3 very recently, showed that CSF3R somatic mutations can be identified in ∼4% of chronic myelomonocytic leukemias. These mutations, which affect distinct residues in CSF3R as compared with CNL, are frequently associated with mutations in the ASXL1 gene and have a poor prognostic impact on overall and acute myeloid leukemia (AML)-free survival. Together, these data indicate that CNL genome had a combination of few mutations with a pattern of cooperation with a strong biological relationship among genes and categories, similar to AML.7 Along this line of cooperating mutations on epigenetic genes, we also found in our CNL patient mutated copies of ASXL1 and TET2 genes.
The variant allelic frequency of LUC7L2 mutation was found to be high (more than 95%). We previously described mutations of this gene in myelodysplastic syndrome (MDS).8 As this gene is located in the 7q region, a frequently deleted chromosomal region in myeloid leukemias,9 we decided to investigate whether a critical deletion or a loss of heterozygosity (LOH) affecting this genomic region was also present in the patient. To study this phenomenon, we interrogate our WES data for the LOH across the whole genome of the sample. Interestingly, we found an LOH of 53.2 Mb in chromosome 7q including the locus of the LUC7L2 gene. Allele frequencies of each SNP along chromosome 7 are shown in Figure 1b and Supplementary Figure 4. As no del(7q) was detected with metaphase cytogenetics, our study demonstrates for the first time mutations in LUC7L2 accompanied by a copy-neutral LOH (uniparental disomy) in 7q in a patient with an aggressive CNL phenotype. To further evaluate the biological consequences of this homozygous mutation, we explored LUC7L2 expression in the BM cells of the patient and in some myeloid leukemia cell lines (Figure 1c). By using real-time PCR, we observed a downregulation in the expression of LUC7L2 in the patient cells compared with normal granulocytes, as well as a general downregulation in myeloid leukemia cell lines.
To determine the functional consequences of the mutations in LUC7L2 and U2AF1, genes involved in the splicing machinery, in the proper splicing process, we performed RNA-seq in our index patient as well as in CD34+ cells from a normal control BM. Although no clear genome-wide increase in intron retention was observed in the patient, as previously reported by us10 in some MDS cases, we found an altered pattern of splicing in the mRNA species transcribed from the RUNX1 gene. At the 3′ splice site of RUNX1 intron 5, the un-spliced reads were almost three times more frequent in the mutated patient than in the normal control (Supplementary Figure 1). Because of the large numbers of diverse mutations in the splicing machinery, larger studies will be needed to fully evaluate the impact of these mutations in splicing.
In relation with the effects of an aberrant RNA splicing due to the presence of two mutations in the genes responsible for these processes, we used RNA-seq data to investigate the presence of aberrant fusion transcripts in our CNL patient. In fact, we identified a chimeric transcript involving the PIM3 and SCO2 genes (both were located on 22q13.33), which was the result of an intrachromosomal inversion of approximately 0.6 Mb in chromosome 22 (Figure 1d). The PIM3 oncogene belongs to the Ser/Thr protein kinase family and PIM subfamily. This gene is overexpressed in hematological and epithelial tumors and is associated with MYC co-expression. It has a role in the regulation of signal–transduction cascades, contributing to both cell proliferation and survival, and provides a selective advantage in tumorigenesis.11 Interestingly, the inhibition of PIM kinases by pim kinase inhibitors in Myc-induced lymphoma resulted in cell death.12 Functional studies are needed to elucidate the role of this fusion gene in leukemogenesis.
Regarding the global expression profile of CNL, we obtained 2022 genes upregulated and 1884 genes downregulated in CNL compared with the control (FDR=0.05). Functional classification of genes differently expressed between CNL cells and CD34+, according to the Ingenuity Systems Analysis (Ingenuity Systems, www.ingenuity.com), revealed an enrichment of categories like cell signaling, cell death and survival, as well as gene expression (B-H P-value <0.05; Supplementary Figure 2). These findings reconfirmed the significant pathophysiology of such several synergetic unique genetic defects in the general CNL cohort, as well as in our index case.
The oncogenic CSF3R mutations T618I strongly activate the JAK/STAT pathway and are sensitive to inhibitors of SRC family TNK2 and JAK kinases and may provide a new avenue for therapy.4 In contrast, pim kinase inhibition could be a viable treatment strategy in certain human leukemias that rely on the PIM3 kinase expression.12 In addition, epigenetic modifiers provide new targets for therapeutic intervention, and targeting these enzymatic activities is currently being explored from a therapeutic standpoint in several types of leukemia.13, 14 Although Pardanani et al.,2 considering 35 cases of clinically suspected CNL, did not found alteration in the survival on the basis of the presence or absence of CSF3R mutations, our reported patient had a rapid disease progression and died 7 months after diagnosis, which was probably explained by the profoundly aberrant landscape of gene mutations and rearrangements with functional effects on the biology of the tumor cells.
In summary, our study provides, for the first time, a massive molecular and expression data, revealing a large amount of genomic alterations in CNL. In this complex scenario, a combination of new target therapies may be considered as reasonable options for the therapeutic management of this aggressive and rare subtype of leukemia.
Acknowledgments
We thank all the co-workers in our laboratory for their excellent technical assistance. We especially thank the help of Bartlomiej Przychodzen from Taussig Cancer Institute, Cleveland Clinic. We also thank all the co-workers in our laboratory for their excellent technical assistance. This work was supported by the grants INTRASALUD project PI12/00425 and Red Temática de Investigación Cooperativa en Cáncer (RTICC) RD12/0036/0037 to JCC from Instituto de Salud Carlos III. JM is recipient of a La Caixa International PhD Fellowship.
Author contributions
JM designed and performed the research, analyzed and interpreted data, and wrote the manuscript. FA performed the targeted next-generation sequencing and sequence data analysis. GG-L and DGP performed the analysis of sequence data for whole-exome sequencing. HM, IG and JPM performed the LOH analysis. AD and OG performed and analyzed the RNA-seq. SA analyzed and interpreted the data. MT reviewed the pathologic data and confirmed the diagnosis. JCC designed, directed the research and revised the manuscript, which all authors have approved.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Accession codes
Next-generation sequences have been deposited in the NCBI Sequence Read Archive (SRA), pending accession details.
Supplementary Material
References
- Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368:1781–1790. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardanani A, Lasho TL, Laborde RR, Elliott M, Hanson CA, Knudson RA, et al. CSF3R T618I is a highly prevalent and specific mutation in chronic neutrophilic leukemia. Leukemia. 2013;27:1870–1873. doi: 10.1038/leu.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider O, Itzykson R, Chesnais V, Lasho T, Laborde R, Knudson R, et al. Mutation of the colony-stimulating factor-3 receptor gene is a rare event with poor prognosis in chronic myelomonocytic leukemia. Leukemia. 2013;27:1946–1949. doi: 10.1038/leu.2013.182. [DOI] [PubMed] [Google Scholar]
- Gotlib J, Maxson JE, George TI, Tyner JW. The new genetics of chronic neutrophilic leukemia and atypical CML: implications for diagnosis and treatment. Blood. 2013;122:1707–1711. doi: 10.1182/blood-2013-05-500959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D. The granulocyte-macrophage colony stimulating factors. Cell. 1985;43:5–6. doi: 10.1016/0092-8674(85)90004-2. [DOI] [PubMed] [Google Scholar]
- Rubio-Camarillo M, Gomez-Lopez G, Fernandez JM, Valencia A, Pisano DG. RUbioSeq: a suite of parallelized pipelines to automate exome variation and bisulfite-seq analyses. Bioinformatics. 2013;29:1687–1689. doi: 10.1093/bioinformatics/btt203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerez A, Sugimoto Y, Makishima H, Verma A, Jankowska AM, Przychodzen B, et al. Loss of heterozygosity in 7q myeloid disorders: clinical associations and genomic pathogenesis. Blood. 119:6109–6117. doi: 10.1182/blood-2011-12-397620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang YY, Nakamoto Y, Li YY, Baba T, Kaneko S, et al. Accelerated hepatocellular carcinoma development in mice expressing the Pim-3 transgene selectively in the liver. Oncogene. 29:2228–2237. doi: 10.1038/onc.2009.504. [DOI] [PubMed] [Google Scholar]
- Forshell LP, Li Y, Forshell TZ, Rudelius M, Nilsson L, Keller U, et al. The direct Myc target Pim3 cooperates with other Pim kinases in supporting viability of Myc-induced B-cell lymphomas. Oncotarget. 2:448–460. doi: 10.18632/oncotarget.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LI, Frye SV. Targeting chromatin readers. Clin Pharmacol Ther. 2013;93:312–314. doi: 10.1038/clpt.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge S. Target watch: Drugging the epigenome. Nat Rev Drug Discov. 2013;12:92–93. doi: 10.1038/nrd3943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.