Abstract
Computer-aided drug discovery/design methods have played a major role in the development of therapeutically important small molecules for over three decades. These methods are broadly classified as either structure-based or ligand-based methods. Structure-based methods are in principle analogous to high-throughput screening in that both target and ligand structure information is imperative. Structure-based approaches include ligand docking, pharmacophore, and ligand design methods. The article discusses theory behind the most important methods and recent successful applications. Ligand-based methods use only ligand information for predicting activity depending on its similarity/dissimilarity to previously known active ligands. We review widely used ligand-based methods such as ligand-based pharmacophores, molecular descriptors, and quantitative structure-activity relationships. In addition, important tools such as target/ligand data bases, homology modeling, ligand fingerprint methods, etc., necessary for successful implementation of various computer-aided drug discovery/design methods in a drug discovery campaign are discussed. Finally, computational methods for toxicity prediction and optimization for favorable physiologic properties are discussed with successful examples from literature.
I. Introduction
On October 5, 1981, Fortune magazine published a cover article entitled the “Next Industrial Revolution: Designing Drugs by Computer at Merck” (Van Drie, 2007). Some have credited this as being the start of intense interest in the potential for computer-aided drug design (CADD). Although progress was being made in CADD, the potential for high-throughput screening (HTS) had begun to take precedence as a means for finding novel therapeutics. This brute force approach relies on automation to screen high numbers of molecules in search of those that elicit the desired biologic response. The method has the advantage of requiring minimal compound design or prior knowledge, and technologies required to screen large libraries have become more efficient. However, although traditional HTS often results in multiple hit compounds, some of which are capable of being modified into a lead and later a novel therapeutic, the hit rate for HTS is often extremely low. This low hit rate has limited the usage of HTS to research programs capable of screening large compound libraries. In the past decade, CADD has reemerged as a way to significantly decrease the number of compounds necessary to screen while retaining the same level of lead compound discovery. Many compounds predicted to be inactive can be skipped, and those predicted to be active can be prioritized. This reduces the cost and workload of a full HTS screen without compromising lead discovery. Additionally, traditional HTS assays often require extensive development and validation before they can be used. Because CADD requires significantly less preparation time, experimenters can perform CADD studies while the traditional HTS assay is being prepared. The fact that both of these tools can be used in parallel provides an additional benefit for CADD in a drug discovery project.
For example, researchers at Pharmacia (now part of Pfizer) used CADD tools to screen for inhibitors of tyrosine phosphatase-1B, an enzyme implicated in diabetes. Their virtual screen yielded 365 compounds, 127 of which showed effective inhibition, a hit rate of nearly 35%. Simultaneously, this group performed a traditional HTS against the same target. Of the 400,000 compounds tested, 81 showed inhibition, producing a hit rate of only 0.021%. This comparative case effectively displays the power of CADD (Doman et al., 2002). CADD has already been used in the discovery of compounds that have passed clinical trials and become novel therapeutics in the treatment of a variety of diseases. Some of the earliest examples of approved drugs that owe their discovery in large part to the tools of CADD include the following: carbonic anhydrase inhibitor dorzolamide, approved in 1995 (Vijayakrishnan 2009); the angiotensin-converting enzyme (ACE) inhibitor captopril, approved in 1981 as an antihypertensive drug (Talele et al., 2010); three therapeutics for the treatment of human immunodeficiency virus (HIV): saquinavir (approved in 1995), ritonavir, and indinavir (both approved in 1996) (Van Drie 2007); and tirofiban, a fibrinogen antagonist approved in 1998 (Hartman et al., 1992).
One of the most striking examples of the possibilities presented from CADD occurred in 2003 with the search for novel transforming growth factor-β1 receptor kinase inhibitors. One group at Eli Lilly used a traditional HTS to identify a lead compound that was subsequently improved by examination of structure-activity relationship using in vitro assays (Sawyer et al., 2003), whereas a group at Biogen Idec used a CADD approach involving virtual HTS based on the structural interactions between a weak inhibitor and transforming growth factor-β1 receptor kinase (Singh et al., 2003a). Upon the virtual screening of compounds, the group at Biogen Idec identified 87 hits, the best hit being identical in structure to the lead compound discovered through the traditional HTS approach at Eli Lilly (Shekhar 2008). In this situation, CADD, a method involving reduced cost and workload, was capable of producing the same lead as a full-scale HTS (Fig. 1) (Sawyer et al., 2003).
Fig. 1.
Identical lead compounds are discovered in a traditional high-throughput screen and structure-based virtual high-throughput screen. I, X-ray crystal structures of 1 and 18 bound to the ATP-binding site of the TβR-I kinase domain discovered using traditional high-throughput screening. Compound 1, shown as the thinner wire-frame is the original hit from the HTS and is identical to that which was discovered using virtual screening. Compound 18 is a higher affinity compound after lead optimization. II, X-ray crystal structure of compound HTS466284 bound to the TβRI active site. This compound is identical to compound 1 in I but was discovered using structure-based virtual high-throughput screening.
A. Position of Computer-Aided Drug Design in the Drug Discovery Pipeline
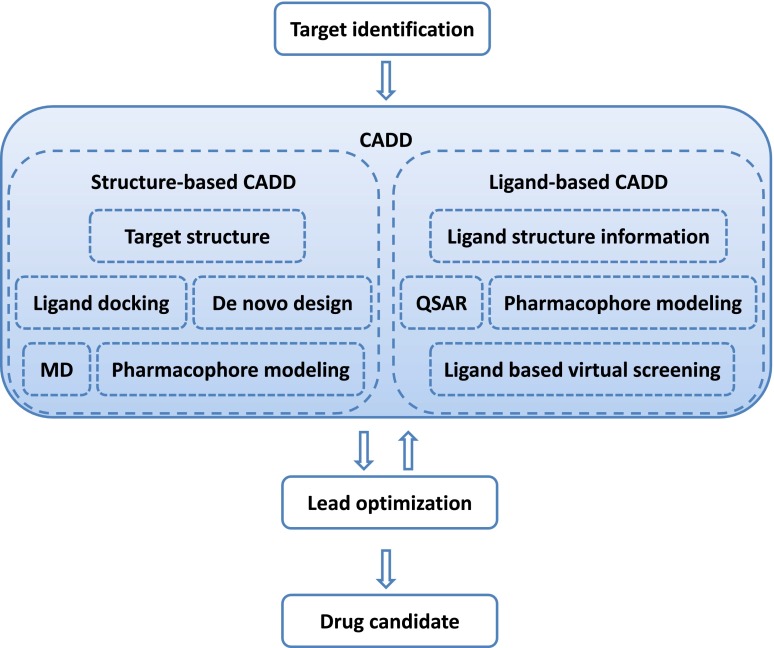
CADD is capable of increasing the hit rate of novel drug compounds because it uses a much more targeted search than traditional HTS and combinatorial chemistry. It not only aims to explain the molecular basis of therapeutic activity but also to predict possible derivatives that would improve activity. In a drug discovery campaign, CADD is usually used for three major purposes: (1) filter large compound libraries into smaller sets of predicted active compounds that can be tested experimentally; (2) guide the optimization of lead compounds, whether to increase its affinity or optimize drug metabolism and pharmacokinetics (DMPK) properties including absorption, distribution, metabolism, excretion, and the potential for toxicity (ADMET); (3) design novel compounds, either by "growing" starting molecules one functional group at a time or by piecing together fragments into novel chemotypes. Figure 2 illustrates the position of CADD in drug discovery pipeline.
Fig. 2.
CADD in drug discovery/design pipeline. A therapeutic target is identified against which a drug has to be developed. Depending on the availability of structure information, a structure-based approach or a ligand-based approach is used. A successful CADD campaign will allow identification of multiple lead compounds. Lead identification is often followed by several cycles of lead optimization and subsequent lead identification using CADD. Lead compounds are tested in vivo to identify drug candidates .
CADD can be classified into two general categories: structure-based and ligand-based. Structure-based CADD relies on the knowledge of the target protein structure to calculate interaction energies for all compounds tested, whereas ligand-based CADD exploits the knowledge of known active and inactive molecules through chemical similarity searches or construction of predictive, quantitative structure-activity relation (QSAR) models (Kalyaanamoorthy and Chen, 2011). Structure-based CADD is generally preferred where high-resolution structural data of the target protein are available, i.e., for soluble proteins that can readily be crystallized. Ligand-based CADD is generally preferred when no or little structural information is available, often for membrane protein targets. The central goal of structure-based CADD is to design compounds that bind tightly to the target, i.e., with large reduction in free energy, improved DMPK/ADMET properties, and are target specific, i.e., have reduced off-target effects (Jorgensen, 2010). A successful application of these methods will result in a compound that has been validated in vitro and in vivo and its binding location has been confirmed, ideally through a cocrystal structure.
One of the most common uses in CADD is the screening of virtual compound libraries, also known as virtual high-throughput screening (vHTS). This allows experimentalists to focus resources on testing compounds likely to have any activity of interest. In this way, a researcher can identify an equal number of hits while screening significantly less compounds, because compounds predicted to be inactive with high confidence may be skipped. Avoiding a large population of inactive compounds saves money and time, because the size of the experimental HTS is significantly reduced without sacrificing a large degree of hits. Ripphausen et al. (2010) note that the first mention of vHTS was in 1997 (Horvath, 1997) and chart an increasing rate of publication for the application of vHTS between 1997 and 2010. They also found that the largest fraction of hits has been obtained for G-protein-coupled receptors (GPCRs) followed by kinases (Ripphausen et al., 2010).
vHTS comes in many forms, including chemical similarity searches by fingerprints or topology, selecting compounds by predicted biologic activity through QSAR models or pharmacophore mapping, and virtual docking of compounds into target of interest, known as structure-based docking (Enyedy and Egan, 2008). These methods allow the ranking of “hits” from the virtual compound library for acquisition. The ranking can reflect a property of interest such as percent similarity to a query compound or predicted biologic activity, or in the case of docking, the lowest energy scoring poses for each ligand bound to the target of interest (Joffe, 1991). Often initial hits are rescored and ranked using higher level computational techniques that are too time consuming to be applied to full-scale vHTS. It is important to note that vHTS does not aim to identify a drug compound that is ready for clinical testing, but rather to find leads with chemotypes that have not previously been associated with a target. This is not unlike a traditional HTS where a compound is generally considered a hit if its activity is close to 10 µM. Through iterative rounds of chemical synthesis and in vitro testing, a compound is first developed into a “lead” with higher affinity, some understanding of its structure-activity-relation, and initial tests for DMPK/ADMET properties. Only after further iterative rounds of lead-to-drug optimization and in vivo testing does a compound reach a clinically appropriate potency and acceptable DMPK/ADMET properties (Jorgensen, 2004). For example, the literature survey performed by Ripphausen et al. (2010) revealed that a majority of successful vHTS applications identified a small number of hits that are usually active in the micromolar range, and hits with low nanomolar potency are only rarely identified.
The cost benefit of using computational tools in the lead optimization phase of drug development is substantial. Development of new drugs can cost anywhere in the range of 400 million to 2 billion dollars, with synthesis and testing of lead analogs being a large contributor to that sum (Basak, 2012). Therefore, it is beneficial to apply computational tools in hit-to-lead optimization to cover a wider chemical space while reducing the number of compounds that must be synthesized and tested in vitro. The computational optimization of a hit compound can involve a structure-based analysis of docking poses and energy profiles for hit analogs, ligand-based screening for compounds with similar chemical structure or improved predicted biologic activity, or prediction of favorable DMPK/ADMET properties. The comparably low cost of CADD compared with chemical synthesis and biologic characterization of compounds make these methods attractive to focus, reduce, and diversify the chemical space that is explored (Enyedy and Egan, 2008).
De novo drug design is another tool in CADD methods, but rather than screening libraries of previously synthesized compounds, it involves the design of novel compounds. A structure generator is needed to sample the space of chemicals. Given the size of the search space (more than 1060 molecules) (Bohacek et al., 1996) heuristics are used to focus these algorithms on molecules that are predicted to be highly active, readily synthesizable, devoid of undesirable properties, often derived from a starting scaffold with demonstrated activity, etc. Additionally, effective sampling strategies are used while dealing with large search spaces such as evolutionary algorithms, metropolis search, or simulated annealing (Schneider et al., 2009). The construction algorithms are generally defined as either linking or growing techniques. Linking algorithms involve docking of small fragments or functional groups such as rings, acetyl groups, esters, etc., to particular binding sites followed by linking fragments from adjacent sites. Growing algorithms, on the other hand, begin from a single fragment placed in the binding site to which fragments are added, removed, and changed to improve activity. Similar to vHTS, the role of de novo drug design is not to design the single compound with nanomolar activity and acceptable DMPK/ADMET properties but rather to design a lead compound that can be subsequently improved.
B. Ligand Databases for Computer-Aided Drug Design
Virtual HTS uses high-performance computing to screen large chemical data bases and prioritize compounds for synthesis. Current hardware and algorithms allow structure-based screening of up to 100,000 molecules per day using parallel processing clusters (Agarwal and Fishwick, 2010). To perform a virtual screen, however, a virtual library must be available for screening. Virtual libraries can be acquired in a variety of sizes and designs including general libraries that can be used to screen against any target, focused libraries that are designed for a family of related targets, and targeted libraries that are specifically designed for a single target (Takahashi et al., 2011).
General libraries can be constructed using a variety of computational and combinatorial tools. Early systems used molecular formula as the only constraint for structure generation, resulting in all possible structures for a predetermined limit in the number of atoms. As comprehensive computational enumeration of all chemical space is and will remain infeasible, additional restrictions are applied. Typically, chemical entities difficult to synthesize or known/expected to cause unfavorable DMPK/ADMET properties are excluded. Fink et al. proposed a generation method for the construction of virtual libraries that involved the use of connected graphs populated with C, N, O, and F atoms and pruned based on molecular structure constraints and the removal of unstable structures. The final data base proposed with this method is called the GDB (Generated a DataBase) and contains 26.4 million chemical structures that have been used for vHTS (Fink et al., 2005; Fink and Reymond, 2007). A more recent variation of this data base called GDB-13 includes atoms C, N, O, S, and Cl (F is not included in this variation to accelerate computation) and contains 970 million compounds (Blum and Reymond, 2009).
Most frequently, vHTS focuses on drug-like molecules that have been synthesized or can be easily derived from already available starting material. For this purpose several small molecule data bases are available that provide a variety of information including known/available chemical compounds, drugs, carbohydrates, enzymes, reactants, and natural products (Ortholand and Ganesan, 2004; Song et al., 2009). Some widely used data bases are listed in Table 1.
TABLE 1.
Widely used chemical compound repositories along with content information about class of compounds they host and the size of repositories
| Database | Type | Size |
|---|---|---|
| PubChem (Wheeler et al., 2006) | Biologic activities of small molecules | ∼40,000,000 |
| Accelrys Available Chemicals Directory (ACD) (Accelrys, 2012) | Consolidated catalog from major chemical suppliers | ∼7,000,000 |
| PDBeChem (Dimitropoulos, 2006) | Ligands and small molecules referred in PDB | 14,572 |
| Zinc (Irwin and Shoichet, 2005) | Annotated commercially available compounds | ∼21,000,000 |
| LIGAND (Goto et al., 2002) | Chemical compounds with target and reactions data | 16,838 |
| DrugBank (Wishart et al., 2006) | Detailed drug data with comprehensive drug target information | 6711 |
| ChemDB (Chen et al., 2005, 2007) | Annotated commercially available molecules | ∼5,000,000 |
| WOMBAT Data base (World of Molecular BioAcTivity) (Ekins et al., 2007; Hristozov et al., 2007) | Bioactivity data for compounds reported in medicinal chemistry journals | 331,872 |
| MDDR (MDL Drug Data Report) (Hristozov et al., 2007) | Drugs under development or released; descriptions of therapeutic | 180,000 |
| 3D MIND (Mandal et al., 2009). | Molecules with target interaction and tumor cell line screen data | 100,000 |
1. Preparation of Ligand Libraries for Computer-Aided Drug Design.
Ligand libraries are often constructed by enriching ligands for drug likeness or certain desirable physiochemical properties suitable for the target of interest. Even with rapid docking algorithms, docking millions of compounds requires considerable resources, and time can be saved through the elimination of non-drug like, unstable, or unfavorable compounds. Drug likeness is commonly evaluated using Lipinski’s rule of five (Lipinski et al., 2001), which states that in general, an orally active drug should have no more than one violation of the following criteria: (1) maximum of five hydrogen bond donors, (2) no more than 10 oxygen and nitrogen atoms; (3) molecular mass of less than 500 Da; and (4) an octanol-water partition coefficient of not greater than five. If two or more of the conditions are violated, poor adsorption can be expected. Similarly, polar molecular surface is also used as a determinant for oral absorption and brain penetration (Kelder et al., 1999). It is a common practice to filter molecules based on predicted DMPK/ADMET properties before initializing a vHTS campaign. Ligand-based methods to predict DMPK/ADMET properties use statistical and learning approaches, molecular descriptors, and experimental data to model biologic processes such as oral bioavailability, intestinal absorption/permeability, half-life time, distribution in human blood plasma, etc.
Compound libraries are often enriched for a particular target or family of targets. Physiochemical filters derived from observed ligand-target complexes are used for enriching a library with compounds that satisfy specific geometric or physicochemical constraints. Such libraries are prepared by searching for ligands that are similar to known active ligands (Orry et al., 2006; Harris et al., 2011). Several target-specific libraries exist in Cambridge Structure Data base like the kinase-biased, GPCR-biased, ion channel-biased sets, etc. In addition, a small molecule library requires preparations such as conformational sampling and assigning proper stereo isometric and protonation state (Cavasotto and Phatak, 2011; Anderson, 2012). Molecules are flexible in solvent environment and hence representation of conformational flexibility is an important aspect of molecular recognition. Often conformations of protein and ligand are precomputed using simulation or knowledge-based methods (Liwo et al., 2008; Foloppe and Chen, 2009).
2. Representation of Small Molecules as “SMILES”.
Development and efficient use of ligand data bases require universally applicable methods for the virtual representation of small molecules. SMILES (Simplified Molecular Input Line System) (Wiswesser, 1985) was developed as an unambiguous and reproducible method for computationally representing molecules. It was developed as an improvement over the Wiswesser Line Notation (Wiswesser, 1954), which had a cumbersome set of rules, but was a preferred method due to the representation of molecular structure as a linear string of symbols that could be efficiently read and stored by computer systems.
Commonly, SMILES does not explicitly encode hydrogen atoms (hydrogen-suppressed graph) and conventionally assumes that hydrogens make up the remainder of an atom’s lowest normal valence. All non-hydrogen atoms are represented by their atomic symbols enclosed in square brackets. Atoms may also be listed without square brackets, implying the presence of hydrogens. Formal charges are specifically assigned as + or − followed by an optional digit inside the appropriate brackets. Aromatic atoms are specified using the lowercase atomic symbols. Single bonds, double bonds, triple bonds, and aromatic bonds are denoted by “-,” “=,” “#,” and “:,” respectively. Branched systems are specified by enclosing them in parentheses. Cyclic structures are represented by breaking a ring at a single or aromatic bond and numbering the atoms on either side of the break with a number. For example, cyclohexane is represented with the SMILES string C1CCCCC1. Disconnected compounds are separated by a period, and ionic bonds are considered disconnected structures with complimentary formal charges (Weininger, 1988).
SMILES algorithms are capable of detecting most aromatic compounds with an extended version of Huckel’s rule (all atoms in the ring must be sp2 hybridized and the number of available π electrons must satisfy 4N + 2) (Weininger and Stermitz, 1984). Therefore, aromaticity does not necessarily need to be defined beforehand. However, tautomeric structures must be explicitly specified as separate SMILES strings. There are no SMILES definitions for tautomeric bonds or mobile hydrogens. SMILES was designed to have good human readability as a molecular file format. However, there are usually many different but equally valid SMILES descriptions for the same structure. It is most commonly used for storage and retrieval of compounds across multiple computer platforms.
SMARTS (SMILES ARbitrary Target Specification) is an extension of SMILES that allows for variability within the represented molecular structures. This provides substructure search functionality to SMILES. In addition to the SMILES naming conventions, SMARTS includes logical operators, such as "AND" (&), "OR" (,), and "NOT" (!), and special atomic and bond symbols that provide a level of flexibility to chemical names. For example, in SMARTS notation, [C,N] represents an atom that can be either an aliphatic carbon or an aliphatic nitrogen, and the symbol "∼" will match any bond type (Daylight Chemical Information Systems, 2008).
3. Small Molecule Representations for Modern Search Engines: InChIKey.
InChI (International Chemical Identifier) was released in 2005 as an open source structure representation algorithm that is meant to unify searches across multiple chemical data bases using modern internet search engines. It is maintained by the InChI Trust (http://www.inchi-trust.org) and currently supports chemical elements up to 112 (InChITRUST, 2013). The purpose of InChI and the hash-key version InChIKey is to provide a nonproprietary machine-readable code unique for all chemical structures that can be indexed by major search engines such as Google without any alteration. By use of this protocol, researchers can search for chemicals in a routine and straightforward manner. Before the development of INChI, chemical searches spanning multiple data bases using typical search engines were unreliable. Different systems have their own proprietary identification method for indexing chemicals, SMILES-based searches are also insufficient, because different data bases have adopted their own unique SMILES.
InChI is made up of several layers that represent different classes of structural information. The first two layers contain only general information, including the chemical formula and connections. More specific conformational information such as stereochemistry, tautomerism, and isotopic information is represented in additional optional layers. Bonds between atoms can be partitioned into up to three sublayers depending on the level of specification desired. These layers represent all bonds to nonbridging hydrogen atoms, immobile hydrogen atoms, and mobile hydrogen atoms, respectively. The InChI algorithm includes six normalization rules that apply qualities such as variable protonation and identification of tautomeric patterns and resonances to achieve a unique and consistent chemical representation (InChITRUST, 2013).
InChIKey is a hash-key version of InChI that generates two blocks using a truncated SHA-256 cryptographic hash function. This allows the keys to contain a fixed length of 27 characters with high collision resistance (minimal chance of two different molecules having the same hash key). Use of InChIKeys to search multiple data base with typical search engines was tested, and the incidence of false-positive hits was low (Southan, 2013). Publically available web applets are available that allow chemists to draw molecules and automatically search the web using an automatically calculated InChIKey (http://www.chemspider.com/StructureSearch.aspx).
C. Target Data Bases for Computer-Aided Drug Discovery/Design
The knowledge of the structure of the target protein is required for structure-based CADD. The Protein Data Bank (PDB) (2013), established in 1971 at the Brookhaven National Laboratory, and the Cambridge Crystallographic Data Center, are among the most commonly used data bases for protein structure. PDB currently houses more than 81,000 protein structures, the majority of which have been determined using X-ray crystallography and a smaller set determined using NMR spectroscopy. When an experimentally determined structure of a protein is not available, it is often possible to create a comparative model based on the experimental structure of a related protein. Most frequently the relation is based in evolution that introduced the term "homology model." The Swiss-Model server is one of the most widely used web-based tools for homology modeling (Arnold et al., 2006). Initially, static protein structures were used for all structure-based design methods. However, proteins are not static structures but rather exist as ensembles of different conformational states. The protein fluctuates through this ensemble depending on the relative free energies of each of these states, spending more time in conformations of lower free energy. Ligands are thought to interact with some conformations but not others, thus stabilizing conformational populations in the ensemble. Therefore, docking compounds into a static protein structure can be misleading, as the chosen conformation may not be representative of the conformation capable of binding the ligand. Recently, it has become state of the art to use additional computational tools such as molecular dynamics and molecular mechanics to simulate and evaluate a protein’s conformational space. Conformational sampling provides a collection of snapshots that can be used in place of a single structure that reflect the breadth of fluctuations the ligand may encounter in vivo. This approach was proven to be invaluable in CADD by Schames et al. (2004) in the 2004 identification of novel HIV integrase inhibitors (Durrant and McCammon, 2010). Some methods, such as ROSETTALIGAND (Meiler and Baker, 2006), are capable of incorporating protein flexibility during the actual docking procedure, omitting the need for snapshot ensembles.
The collection of events that occurs when a ligand binds a receptor extends far beyond the noncovalent interactions between ligand and protein. Desolvation of ligand and binding pocket, shifts in the ligand and protein conformational ensembles, and reordering of water molecules in the binding site all contribute to binding free energies. Consideration of water molecules as an integral part of binding sites is necessary for key mechanistic steps and binding (Levitt and Park, 1993; Ball, 2008). These water molecules shift the free energy change of ligand binding by either facilitating certain noncovalent interactions between the ligand and protein or by being displaced into more favorable direct interactions between the ligand and protein, causing an overall change in free energy upon binding (Ladbury, 1996; Li and Lazaridis, 2007). Improvements in computational resources allow inclusion of better representations of physiochemical interactions in computational methods to increase their accuracies (de Beer et al., 2010).
D. Benchmarking Techniques of Computer-Aided Drug Design
Effective benchmarks are essential for assessment of performance and accuracy of CADD algorithms. Design of the benchmark in terms of number and type of target proteins, size, and composition of active and inactive chemicals, and selection of quality measures play a key role when comparing new CADD methods with existing ones. Scientific benchmarks usually involve screening a library of compounds that include a subset of known actives combined with known inactive compounds and then evaluating the number of known actives that were identified by the CADD technique used (Stumpfe et al., 2012).
Performance is commonly reported by correlating predicted activities with experimentally observed activities through the use of receiver operating characteristic curves. These curves plot the number of true positive predictions on the y-axis versus the false-positive predictions on the x-axis. A random predictor would result in a plot of a line with a slope of 1, whereas curves with high initial slopes above this line represent increasing performance scores for the method tested (Cleves and Jain, 2006; Hristozov et al., 2007). Receiver operating characteristic curves are therefore analyzed by determining the area under the curve, positive predictive value—the ratio of true positives in a subset selected in a vHTS screen, or enrichment—a benchmark that normalizes positive predictive value by the background ratio of positives in the dataset.
For structure-based CADD, it is now common also to include decoy molecules that further test a technique’s ability to discern actives from inactives at high resolution. Irwin et al. (2008) created the Directory of Useful Decoys (DUD) dataset designed for high-resolution benchmarking. It includes experimental data for approximately 3000 ligands covering up to 40 different targets and a set of carefully chosen decoys (Huang et al., 2006). These decoys were designed to resemble positive ligands physically but not topologically (Irwin, 2008). These decoys, however, are not experimentally validated and are only postulated to be “inactive” against the targets. Good and Oprea (2008) developed clustered versions of DUD with added datasets from sources such as WOMBAT to avoid challenges in enrichment comparisons between methods due to different parameters and limited diversity (Good and Oprea, 2008).
The present review covers various established structure-based and ligand-based CADD methods followed by a section on CADD methods in ADMET profile prediction. The applications of various methods discussed in the manuscript are illustrated with recent studies. We prioritize studies that concluded in compounds that were at least tested in vivo and often entered clinical trials.
II. Structure-Based Computer-Aided Drug Design
Structure-based computer-aided drug design (SB-CADD) relies on the ability to determine and analyze 3D structures of biologic molecules. The core hypothesis of this approach is that a molecule’s ability to interact with a specific protein and exert a desired biologic effect depends on its ability to favorably interact with a particular binding site on that protein. Molecules that share those favorable interactions will exert similar biologic effects. Therefore, novel compounds can be elucidated through the careful analysis of a protein’s binding site. Structural information about the target is a prerequisite for any SB-CADD project. Scientists have been using a target protein’s structure to aid in drug discovery since the early 1980s (NIH-structure based). Since then, SB-CADD has become a commonly used drug discovery technique thanks to advances in genomics and proteomics that have led to the discovery of a large number of candidate drug targets (Bambini and Rappuoli, 2009; Lundstrom, 2011). Extensive use of biophysical techniques such as X-ray crystallography and NMR spectroscopy has led to the elucidation of a number of 3D structures of human and pathogenic proteins. For example, the PDB has over 81,000 protein structures, whereas data bases such as PDBBIND (Wang et al., 2004) and protein ligand data base house 5,671 and 129 (as of 2003) ligand-protein cocrystal structures, respectively. Drug discovery campaigns leveraging target structure information have sped up the discovery process and have led to the development of several clinical drugs. A prerequisite for the drug discovery process is the ability to rapidly determine potential binders to the target of biologic interest. Computational methods in drug discovery allow rapid screening of a large compound library and determination of potential binders through modeling/simulation and visualization techniques.
A. Preparation of a Target Structure
A target structure experimentally determined through X-ray crystallography or NMR techniques and deposited in the PDB is the ideal starting point for docking. Structural genomics has accelerated the rate at which target structures are being determined. In the absence of experimentally determined structures, several successful virtual screening campaigns have been reported based on comparative models of target proteins (Becker et al., 2006; Warner et al., 2006; Budzik et al., 2010). Efforts have also been made to incorporate information about binding properties of known ligands back into comparative modeling process (Evers et al., 2003; Evers and Klebe, 2004).
Success of virtual screening is dependent upon the amount and quality of structural information known about both the target and the small molecules being docked. The first step is to evaluate the target for the presence of an appropriate binding pocket (Hajduk et al., 2005; Fauman et al., 2011). This is usually done through the analysis of known target-ligand cocrystal structures or using in silico methods to identify novel binding sites (Laurie and Jackson, 2006).
1. Comparative Modeling.
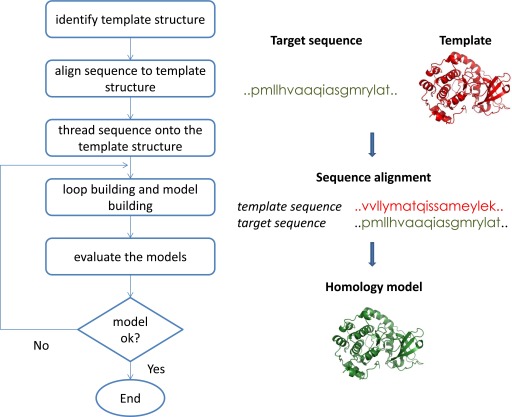
Advances in biophysical techniques. such as X-ray crystallography and NMR techniques. have led to increasing availability of protein structures. This has allowed use of structural information to guide drug discovery. In the absence of experimental structures, computational methods are used to predict the 3D structure of target proteins. Comparative modeling is used to predict target structure based on a template with a similar sequence, leveraging that protein structure is better conserved than sequence, i.e., proteins with similar sequences have similar structures. Homology modeling is a specific type of comparative modeling in which the template and target proteins share the same evolutionary origin. Comparative modeling involves the following steps: (1) identification of related proteins to serve as template structures, (2) sequence alignment of the target and template proteins, (3) copying coordinates for confidently aligned regions, (4) constructing missing atom coordinates of target structure, and (5) model refinement and evaluation. Figure 3 illustrates the steps involved in comparative modeling. Several computer programs and web servers exist that automate the comparative modeling process e.g., PSIPRED (Buchan et al., 2010) and MODELER (Marti-Renom et al., 2000).
Fig. 3.
Steps in homology model building process.
a. Template identification and alignment.
In the first step, the target sequence is used as a query for the identification of template structures in the PDB. Templates with high sequence similarity can be determined by a straightforward PDB-BLAST search (Altschul et al., 1990). More sophisticated fold recognition methods are available if PDB-BLAST does not yield any hits (Kelley and Sternberg, 2009; Soding and Remmert, 2011). Search for template structure is followed by sequence alignment using methods like ClustalW (Thompson et al., 1994), which is a multiple sequence alignment tool. For closely related protein structures, structurally conserved regions are identified and used to build the comparative model. Construction and evaluation of multiple comparative models from multiple good-scoring sequence alignments improve the quality of the comparative model (Chivian and Baker, 2006; Misura et al., 2006). It has been demonstrated that combination of multiple templates can improve comparative models by leveraging well-determined regions that are mutually exclusive (Rai and Fiser, 2006). Template selection is key for successful homology modeling. Careful consideration should be given to alignment length, sequence identity, resolution of template structure, and consistency of secondary structure between target and templates.
b. Model building.
Gaps or insertions in the original sequence alignment occur most frequently outside secondary structure elements and lead to chain breaks (gaps and insertions) and missing residues (gaps) in the initial target protein model. Modeling these missing regions involves connecting the anchor residues, which are the N- or C-terminal residues of protein segments on either side of the missing region. Two broad classes of loop-modeling methods exist: (1) knowledge-based methods and (2) de novo methods. Knowledge-based methods use loops from protein structures that have approximately the same anchors as found in target models. Loops from such structures are applied to the target structure. De novo methods generate a large number of loop conformations and use energy functions to judge the quality of predicted loops (Hillisch et al., 2004). Both methods, however, solve the “loop closure” problem, i.e., identifying low-energy loop conformations from a large conformational sample space that justify the structural restraint of connecting the two anchor points. Cyclic coordinate descent (Canutescu and Dunbrack, 2003) and kinematic closure (KIC) (Mandell et al., 2009) algorithms optimally search for conformations that satisfy constraints for loop closure in a target structure. Cyclic coordinate descent iteratively changes dihedral angles one at a time such that a distance constraint between anchor residues is satisfied (Canutescu and Dunbrack, 2003). The KIC algorithm derives from kinematic methods that allow geometric analysis of possible conformations of a system of rigid objects connected by flexible joints. The KIC algorithm generates a Fourier polynomial in N variables for a system of N rotatable bonds by analyzing bond lengths and bond angles constraints (Coutsias and Seok, 2004). Atom coordinates of the loop are then determined using the polynomial equation.
The loop modeling step can be affected by two classes of errors: scoring function errors and insufficient sampling. The former arises when nonnative conformations are assigned better scores. Confidence in scoring can be improved by scoring with different functions, assuming that true native conformation will likely be best ranked across multiple scoring methods. Insufficient sampling arises when near native conformations are not sampled. Sufficient sampling can be achieved by running multiple independent simulations to establish convergence.
The next step in comparative modeling is prediction of side-chain conformations. A statistical clustering of observed side-chain conformations in PDB, called a rotamer library, is used in most side-chain construction methods (Krivov et al., 2009). Methods such as dead-end elimination (Desmet et al., 1992) implemented in SCRWL (Dunbrack and Karplus, 1993; Dunbrack and Karplus, 1994; Bower et al., 1997) and Monte Carlo searches (Rohl et al., 2004) are used for side-chain conformation sampling. Dead-end elimination imposes conditions to identify rotamers that cannot be members of global minimum energy conformation. For example, the algorithm prunes a rotamer a if a second rotamer b exists, such that lowest energy conformation containing a is greater than highest energy conformations containing b. The SCRWL algorithm evaluates steric interactions between side chains through the use of a backbone-dependent rotamer library that expresses frequency of rotamers as a function of dihedral angles φ and ψ. Monte Carlo algorithms search the side-chain conformational space stochastically using the Metropolis criterion to guide the search into energetic minima.
Binding pockets in homology models or even crystal structures are often not amenable for ligand docking because of insufficient accuracy. Ligand information has been used to improve comparative models. Tanrikulu et al. (2009) and Tanrikulu and Schneider (2008) used a pseudoreceptor modeling method to improve a homology model of human histamine H4 receptor. Pseudoreceptor methods map binding pockets around one or more reference ligands by capturing their shape and interactions with the target. Conformation snapshots of the homology model were obtained by MD simulation, and pocket-forming coordinates were extracted. Binding pockets of MD frames that matched pseudoreceptor were prioritized for virtual screening. Hits from virtual screening were tested experimentally, and two compounds with diverse chemotypes exhibited pKi > 4 (Tanrikulu and Schneider, 2008; Tanrikulu et al., 2009). Katritich et al. (2010) used a combined homology modeling and ligand-guided backbone ensemble receptor optimization algorithm (LiBERO) for prediction of a protein-ligand complex in CASP experiments. The approach was identified as the best in that it identified 40% of the 70 contacts that the antagonist ZM241385 makes with adenosine A2a receptor (PDB:3EML). In LiBERO, framework multiple models are generated and normal mode analysis is used to generate backbone conformation ensembles. Conformers are selected according to docking performance through an iterative process of model building and docking (Katritch et al., 2010). Ligand information-assisted homology modeling is contingent on (1) availability of high-affinity ligands and (2) availability of structurally close homologs to ensure good quality initial homology model.
c. Model refinement and evaluation.
Atomic models are refined by introducing ideal bond geometries and by removing unfavorable contacts introduced by the initial modeling process. Refinement involves minimizing models using techniques such as molecular dynamics (Raval et al., 2012), Monte Carlo Metropolis minimization (Misura and Baker, 2005), or genetic algorithms (Xiang, 2006). For example, the ROSETTA refinement protocol fixes bond lengths and angles at ideal values and removes steric clashes in an initial low-resolution step. ROSETTA then minimizes energy as a function of backbone torsional angles φ, ψ, and ω using a Monte Carlo minimization strategy (Misura and Baker, 2005). Molecular dynamics-based refinement techniques have been used widely as refinement strategy in drug design-oriented homology models (Serrano et al., 2006; Li et al., 2008).
Model evaluation involves comparison of observed structural features with experimentally determined protein structures. Melo and Sali (2007) applied a genetic algorithm that used 21 input model features like sequence alignment scores, measures of protein packing, and geometric descriptors to assess folds of models. Critical Assessment of Techniques for Protein Structure Prediction (CASP) (Cozzetto et al., 2009) is a worldwide competition in which many groups participate for an objective assessment of methods in the area of protein structure prediction. Models are numerically assessed and ranked by estimating similarity between a model and corresponding experimental structure. Some evaluation methods used in CASP are full model root mean square deviation, global distance test-total scores (GDT-TS), and alignment accuracy (AL0 score). GDT-TS is the average maximum number of residues in a predicted model that deviate from corresponding residues in the target by no more than a specified distance, whereas AL0 represents the percentage of correctly aligned residues (Cozzetto et al., 2009).
d. Model data bases.
SWISS-MODEL (Kiefer et al., 2009) and MODBASE (Pieper et al., 2009) data bases store annotated comparative protein structure models. SWISS-MODEL repository contains annotated 3D protein models generated by homology modeling of all sequences in SWISS-PROT (Kiefer et al., 2009). As of May 2012, SWISS-MODEL contained 3.2 million entries for 2.2 million unique sequences in UNIPROT data base. MODBASE is organized into datasets of models for specific projects, which include datasets of 9 archaeal genomes, 13 bacterial genomes, and 18 eukaryotic genomes. Together with other datasets, MODBASE currently houses 5.2 million subdomain models across 1.6 million unique protein sequences (Pieper et al., 2009).
e. Example application in computer-aided drug design.
Park et al. (2009) used homology model of Cdc25A phosphatases, a drug target for cancer therapy, to identify novel inhibitors. The crystal structure of protein Cdc25B served as a template to generate structural models of Cdc25A. Docking of a library of 85,000 compounds led to the discovery of structurally diverse compounds with IC50 values ranging from 0.8 to 15 µM.
2. Binding Site Detection and Characterization.
Protein-ligand interaction is a prerequisite for drug activity. Often possible binding sites for small molecules are known from cocrystal structures of the target or a closely related protein with a natural or nonnatural ligand. In the absence of a cocrystal structure, mutational studies can pinpoint ligand binding sites. However, the ability to identify putative high-affinity binding sites on proteins is important if the binding site is unknown or if new binding sites are to be identified, e.g., for allosteric molecules. Computational methods like POCKET, SURFNET, Q-SITEFINDER, etc. (Laurie and Jackson, 2006; Henrich et al., 2010) are often used for binding site identification. Computational methods for identifying and characterizing binding sites can be divided into three general classes: (1) geometric algorithms to find shape concave invaginations in the target, (2) methods based on energetic consideration, and (3) methods considering dynamics of protein structures.
a. Geometric method.
Geometric algorithms identify binding sites through the detection of cavities on a protein’s surface. These algorithms frequently use grids to describe molecular surface or 3D structure of protein. The boundary of a pocket is determined by rolling a “spherical probe” over the grid surface. A pocket is identified if there is a period of noninteraction i.e., probe does not touch any target atoms, between periods of contact with protein. This technique is used by POCKET (Levitt and Banaszak, 1992) and LIGSITE (Hendlich et al., 1997). SURFNET (Laskowski, 1995) places spheres between all pairs of target atoms and then reduces the radius of spheres until each sphere contains only a pair of atoms. The program thus accumulates spheres in pockets, both inside the target and on the surface. The SPHGEN program (Desjarlais et al., 1988) generates overlapping spheres to describe the 3D shape of binding pocket. The algorithm creates a negative image of invaginations for target surface. Spheres are calculated all over the entire surface such that each sphere touches the molecular surface at two points. The overlapping dense representation of spheres is then filtered to include only the largest sphere associated with each target surface atom. The main disadvantages of geometric-based methods include that geometric descriptors are method dependent and subjective, the target protein is typically rigid, and the methods are often tied to a generalized concept of a binding pocket and may miss unorthodox binding sites within channels or on protein-protein interaction interfaces (Laurie and Jackson, 2006).
b. Example application in computer-aided drug design.
Trypanosoma brucei is the causative agent of human trypanosomiasis in Africa (Smithson et al., 2010). A binding pocket identified by LIGSITE was used for identifying inhibitors of ornithine decarboxylase, which is a molecular target for treatment of African trypanosomiasis (Smithson et al., 2010). SPHGEN was used to identify putative binding sites in BCL6 (Cerchietti et al., 2010), a therapeutic target for B cell lymphomas. Docking of a library of 1,000,000 commercially available compounds into the identified sites led to successful identification of inhibitors of BCL6 (Cerchietti et al., 2010).
c. Energy-based approaches.
Energy-based approaches calculate van der Waals, electrostatic, hydrogen-binding, hydrophobic, and solvent interactions of probes that could result in energetically favored binding. Simple energy-based methods tend to be as fast as geometric methods but are more sensitive and specific. The Q-SITEFINDER (Laurie and Jackson, 2005) algorithm calculates the Van der Waals interaction energy for aliphatic carbon probes on a grid and retains pockets with favorable interactions. The GRID (Reynolds et al., 1989; Wade et al., 1993) algorithm samples the potential on a 3D grid to determine favorable binding positions for different probes. GRID determines interaction energy as a sum of Lennard-Jones, Coulombic, and hydrogen-bond terms. Other algorithms like POCKETPICKER (Weisel et al., 2007) and FLAPSITE (Henrich et al., 2010) use similar approaches but different metrics to evaluate the quality of a putative binding site. For example, POCKETPICKER defines “buriedness” indices in its binding site elucidation. A serious limitation of these methods is that they result in many different energy minima on the surface of the protein, including many false-positives (Laurie and Jackson, 2006). These shortcomings can be addressed in part by including the solvation term in the scoring potential as is done in CS-Map algorithm (Kortvelyesi et al., 2003). More complex tools distinguish solvent accessible from solvent inaccessible surfaces. Kim et al. (2008) present a method for defining the topology of the protein as a Voronoi diagram of spheres and its use to elucidate binding pocket locations.
d. Example application in computer-aided drug design.
Segers et al. (2007) applied Q-SITEFINDER and POCKETFINDER to identify the binding site for the C2 domain of coagulation factor V whose interaction with platelet membrane is necessary for coagulation. Excessive coagulation caused by high thrombin production could be controlled by small molecule inhibitors of factor V. Docking of 300,000 compounds into the predicted sites identified four inhibitors with IC50 < 10 µM. Novel putative drug binding regions were identified in Avian Influenza Neuraminidase H5N1 using computational solvent mapping (Landon et al., 2008). Virtual screening of the binding site with a library of compounds led to the discovery of novel small-molecule inhibitor of H5N1 (An et al., 2009).
e. Pocket matching.
Methods like Catalytic Site Atlas (Porter et al., 2004), AFT (Arakaki et al., 2004), SURFACE (Ferre et al., 2004), POCKET-SURFER (Chikhi et al., 2010), and PATCH-SURFER (Sael and Kihara, 2012) detect similar pockets based on reference ligand binding sites. Catalytic Site Atlas contains annotated descriptors of enzyme active site residues as well as equivalent sites in related proteins found by sequence alignment. Query made by PDB code returns annotated catalytic residues highlighted on amino acid sequence and on the structure via RasMol (Sayle and Milner-White, 1995). SURFACE is a repository of annotated protein functional sites with sequence and structure-derived information about function or interactions. The comparison algorithm explores all combinations of similar/identical residues in a sequence-independent way between query protein and data base structures. Pocket-surfer and patch-surfer describe property of binding pockets. Pocket-surfer captures global similarity of pockets, whereas Patch-surfer evaluates and compares binding pocketed in small circular patches. These methods describe patches using four properties, the surface shape, visibility, the hydrophobicity, and the electrostatic potential.
f. Molecular dynamics-based detection.
The dynamic nature of biomolecules sometimes makes it insufficient to use a single static structure to predict putative binding sites. Multiple conformations of target are often used to account for structural dynamics of target. Classic molecular dynamics (MD) simulations can be used for obtaining an ensemble of target conformations beginning with a single structure. The MD method uses principles of Newtonian mechanics to calculate a trajectory of conformations of a protein as a function of time. The trajectory is calculated for a specific number of atoms in small time steps, typically 1–10 fs (Vangunsteren and Berendsen, 1990). Classic MD methods tend to get trapped in local energy minima. To overcome this, several advanced MD algorithms such as targeted-MD (Schlitter et al., 1994), SWARM-MD (Huber and van Gunsteren, 1998), conformational flooding simulations (Grubmuller, 1995), temperature accelerated MD simulations (Abrams and Vanden-Eijnden, 2010), and replica exchange MD (Sugita and Okamoto, 1999) have been implemented for traversing multiple-minima energy surface of proteins.
g. Example application in computer-aided drug design.
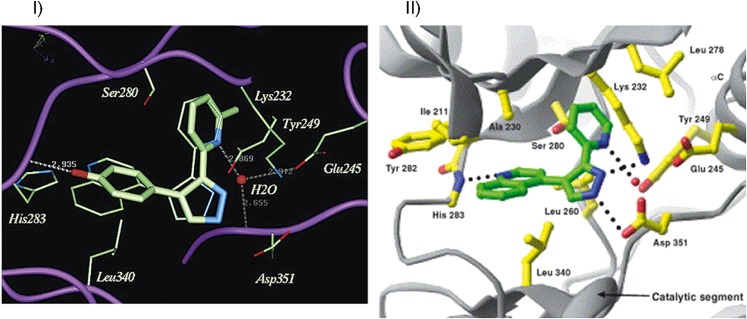
MD simulations elucidated a novel binding trench in HIV integrase (IN), which led to development of raltegravir, a drug used to treat HIV infection. MD simulations of 5CITEP, a known inhibitor of IN, showed that the inhibitor underwent various movements including entry into a novel binding trench (shown in Fig. 4) that went undetected with a static crystal structure (Schames et al., 2004). The discovery of this trench led to the development of raltegravir, by Merck (Summa et al., 2008). Frembgen-Kesner and Elcock (2006) reproduced a cryptic drug binding site in an explicit-solvent MD simulations of unliganded p38 mitogen-activated protein kinase (p38 MAPK) protein, a target in the treatment of inflammatory diseases.
Fig. 4.
Discovery of novel binding trench in HIV-1 IN. Ligand in green is similar to the crystal structure binding pose while the one in yellow is in the novel trench. Adapted from Schames et al. (2004).
B. Representing Small Molecules and Target Protein for Docking Simulations
There are three basic methods to represent target and ligand structures in silico: atomic, surface, and grid representations (Halperin et al., 2002; Kitchen et al., 2004). Atomic representation of the surface of the target is usually used when scoring and ranking is based on potential energy functions. An example is DARWIN, which uses CHARMM force-field to calculate energy (Taylor and Burnett, 2000). Surface methods represent the topography of molecules using geometric features. The surface is represented as a network of smooth convex, concave, and saddle shape surfaces. These features are generated by mapping part of van der Waals surface of atoms that is accessible to probe a sphere (Connolly, 1983). Docking is then guided by a complementary alignment of ligand and binding site surfaces. Earliest implementation of DOCK (Kuntz et al., 1982) used a set of nonoverlapping spheres to represent invaginations of target surface and the surface of the ligand (method described earlier in detail for SPHGEN). Geometric matching begins by systematically pairing one ligand sphere a1 with one receptor sphere b1. This is followed by pairing a second set of spheres, a2 and b2. The move is accepted if the change in atomic distances is less than an empirically determined cut-off value. The cut-off value specifies the maximum allowed deviation between ligand and receptor internal distance. The pairing step is repeated for a third pair of atoms with the same internal distance checks as above. A minimum of four assignable pairs is essential for determining orientation, otherwise the match is rejected. For the grid representation, the target is encoded as physicochemical features of its surface. A grid method described by Katchalskikatzir et al. (1992) digitizes molecules using a 3D discrete function that distinguishes the surface from the interior of the target molecule. Molecules are scanned in relative orientation in three dimensions, and the extent of overlap between molecules is determined using a correlation function calculated from a Fourier transform. Best overlap is determined from a list of overlap functions (Katchalskikatzir et al., 1992). Physiochemical properties may be represented on the grid by storing energy potentials on surface grid points.
C. Sampling Algorithms for Protein-Ligand Docking
Docking methods can be classified as rigid-body docking and flexible docking applications depending on the degree to which they consider ligand and protein flexibility during the docking process (Halperin et al., 2002; Dias and de Azevedo, 2008). Rigid body docking methods consider only static geometric/physiochemical complementarities between ligand and target and ignore flexibility and induced-fit (Halperin et al., 2002) binding models. More advanced algorithms consider several possible conformations of ligand or receptor or both at the same time according to the conformational selection paradigm (Changeux and Edelstein, 2011). Rigid docking simulations are generally preferred when time is critical, i.e., when a large number of compounds are to be docked during an initial vHTS. However, flexible docking methods are still needed for refinement and optimization of poses obtained from an initial rigid docking procedure. With the evolution of computational resources and efficiency, flexible docking methods are becoming more commonplace. Some of the most popular approaches include systematic enumeration of conformations, molecular dynamic simulations, Monte Carlo search algorithms with Metropolis criterion (MCM), and genetic algorithms.
1. Systematic Methods.
Systematic algorithms incorporate ligand flexibility through a comprehensive exploration of a molecule’s degrees of freedom. In systematic algorithms, the current state of the system determines the next state. Starting from the same exact state and same set of parameters, systematic methods will yield exactly the same final state. Systematic methods can be categorized into (1) exhaustive search algorithms and (2) fragmentation algorithms.
Exhaustive searches elucidate ligand conformations by systematically rotating all possible rotatable bonds at a given interval. Large conformational space often prohibits an exhaustive systematic search. Algorithms such as GLIDE (Friesner et al., 2004) use heuristics to focus on regions of conformational space that are likely to contain good scoring ligand poses. GLIDE precomputes a grid representation of target’s shape and properties. Next, an initial set of low-energy ligand conformations in ligand torsion-angle space is created. Initial favorable ligand poses are identified by approximate positioning and scoring methods (shape and geometric complementarities). This initial screen reduces the conformational space over which the high-resolution docking search is applied. High-resolution search involves the minimization of the ligand using standard molecular mechanics energy function followed by a Monte Carlo procedure for examining nearby torsional minima.
Fragmentation methods sample ligand conformation by incremental construction of ligand conformations from fragments obtained by dividing the ligand of interest. Ligand conformations are obtained by docking fragments in the binding site one at a time and incrementally growing them or by docking all fragments into the binding site and linking them covalently. Desjarlais et al. (1986) modified the DOCK algorithm to allow for ligand flexibility by separately docking fragments into the binding site and subsequently joining them. FLEXX (Rarey et al., 1996) uses the “anchor and grow method” for ligand conformational sampling. A base fragment has to be interactively selected by the user, which is followed by automatic determination of placements for the fragment that maximize favorable interactions with the target protein. The base fragment is grown incrementally by adding new fragments in all possible conformations, and the extended fragment is selected if no significant steric clashes (overlap volume ≤ 4.5 Å3) are observed between ligand and target atoms. Extended ligands are optimized (1) if new interactions are found and (2) if minor steric interactions exist (Rarey et al., 1996). Fully automated “anchor and grow” methods have been implemented in several methods such as FLOG (Miller et al., 1994), SURFLEX (Jain, 2003), and SEED (Majeux et al., 2001). In a benchmark study in which performance of eight docking algorithm was compared on 100 protein-ligand complex, GLIDE and SURFLEX were among the methods that showed best accuracy (Kellenberger et al., 2004). GLIDE and SURFLEX generated poses close to X-ray conformation for 68 protein-ligand complexes in the Directory of Useful Decoys (Cross et al., 2009).
a. Example application in computer-aided drug design.
Human Pim-1 kinase, responsible for cell survival/apoptosis, differentiation, and proliferation, is a valuable anticancer target as it is overexpressed in a variety of leukemia. Pierce et al. (2008) used GLIDE to dock approximately 700,000 commercially available compounds and identified four compounds with Ki values less than 5 µM. Chiu et al. (2009) used SURFLEX to identify novel inhibitors of anthrax toxin lethal factor responsible for anthrax-related cytotoxicity. Docking study of a compound library derived from seven data bases, including DrugBank (Wishart et al., 2006), ZINC (Irwin and Shoichet, 2005), National Cancer Institute data base (Milne et al., 1994), etc., identified lead compounds that eventually led to the development of nanomolar inhibitors upon optimization. Table 2 illustrates some examples of drug discovery campaigns in which systematic docking algorithms have been used.
TABLE 2.
Successful docking applications of some widely used docking software
The table lists some of the most widely used docking softwares along with some successful applications in drug-discovery campaigns
| Algorithm | Target |
|---|---|
| SEED | Plasmepsin (Friedman and Caflisch, 2009), target for malaria |
| Flavivirus Proteases (Ekonomiuk et al., 2009a,b), target for WNV and Dengue virus | |
| Tyrosine kinase erythropoietin-producing human hepatocellular carcinoma receptor B4 (EphB4) (Lafleur et al., 2009) | |
| FlexX | Plasmepsin II and IV inhibitors (Luksch et al., 2008), malaria |
| Anthrax edema factor (Chen et al., 2008) | |
| Pneumococcal peptidoglycan deacetylase inhibitors (Bui et al., 2011) | |
| Glide | Aurora kinases inhibitors (Warner et al., 2006) |
| Falcipain inhibitors (Shah et al., 2011) | |
| Cytochrome P450 inhibitors (Caporuscioi et al., 2011) | |
| Surflex | Topoisomerase I, anticancer (optimization) |
| DOCK | FK506 immunophilin (Zhao et al., 2006) |
| BCL6, oncogene in B-cell lymphomas (Cerchietti et al., 2010) |
2. Molecular Dynamics Simulations.
Molecular dynamics (MD) simulation calculates the trajectory of a system by the application of Newtonian mechanics. However, standard MD methods depend heavily on the starting conformation and are not readily appropriate for simulation of ligand-target interactions. Because of its nature, MD is not able to cross high-energy barriers within the simulation’s lifetime and is not efficient for traversing the rugged hyper surface of protein-ligand interactions. Strategies like simulated annealing have been applied for more efficient use of MD in docking. Mangoni et al. (1999) described a MD protocol for docking small flexible ligands to flexible targets in water. They separated the center of mass movement of ligand from its internal and rotational motions. The center of mass motion and internal motions were coupled to different temperature baths, allowing independent control to the different motions. Appropriate values of temperature and coupling constants allowed flexible or rigid ligand and/or receptor.
The McCammon group developed a “relaxed-complex” approach that explores binding conformations that may occur only rarely in the unbound target protein. A 2-ns MD simulation of ligand free target is carried out to extensively sample its conformations. Docking of ligands is then performed in target conformation snapshots taken at different time points of the MD run. This relaxed complex method was used to discover novel modes of inhibition for HIV integrase and led to the discovery of the first clinically approved HIV integrase inhibitor, raltegravir. This MD method was also used in several other campaigns to identify inhibitors of target of interest (Amaro et al., 2008; Durrant et al., 2010a,b).
Metadynamics is a MD-based technique for predicting and scoring ligand binding. The method maps the entire free energy landscape in an accelerated way as it keeps track of history of already sampled regions. During the MD simulation of a protein-ligand complex, a Gaussian repulsive potential are added on explored regions, steering the simulation toward new-free energy regions (Durrant and McCammon, 2010; Leone et al., 2010; Biarnes et al., 2011).
Millisecond timescale MD simulations are now possible with special purpose machines like Anton (Shaw et al., 2008). Such long simulations have allowed study of drug binding events to their protein target (Shan et al., 2011). Anton has been used successfully for full atomic resolution protein folding (Lindorff-Larsen et al., 2011). Advances in computer hardware capabilities mean protein flexibility can be accessed more routinely on longer timescales. This would allow better descriptions of conformational flexibility in future.
3. Monte Carlo Search with Metropolis Criterion.
Stochastic algorithms make random changes to either ligand being docked or to its target binding site. These random changes could be translational or rotational in the case of ligand or random conformational sampling of residue side-chains in the target binding site. Whether a step is accepted or rejected in such a stochastic search is decided based on the Metropolis criterion, which generally accepts steps that lower the overall energy and occasionally accepts steps that increase energy to enable departure from a local energy minimum. The probability of acceptance of an uphill step decreases with increasing energy gap and depends on the "temperature" of the MCM simulation (Sousa et al., 2006). MCM simulations have been adopted for flexible docking applications such as in MCDOCK (Liu and Wang, 1999), Internal Coordinate Mechanics (ICM) (Abagyan et al., 1994), and ROSETTALIGAND (Meiler and Baker, 2006; Davis and Baker, 2009). MCM samples conformational space faster than molecular dynamics in that it requires only energy function evaluation and not the derivative of the energy functions. Although traditional MD drives a system toward a local energy minimum, the randomness introduced with Monte Carlo allows hopping over the energy barriers, preventing the system from getting stuck in local energy minima. A disadvantage is that any information about the timescale of the motions is lost.
ROSETTALIGAND (Kaufmann et al., 2010; RosettaCommons, 2013) uses a knowledge-based scoring procedure with a Monte Carlo-based energy minimization scheme that reduces the number of conformations that must be sampled while providing a more rapid scoring system than offered through molecular mechanics force fields. ROSETTALIGAND incorporates side-chain and ligand flexibility during a high-resolution refinement step through a Monte Carlo-based sampling of torsional angles. All torsion angles of protein and ligand are optimized through gradient-based minimization, mimicking an induced fit scenario (Davis and Baker, 2009). MCDOCK uses two stages of docking and a final energy minimization step for generating target-ligand structure. In the first docking stage, the ligand and docking site are held rigid while the ligand is placed randomly into the binding site. Scoring is done completely on the basis of short contacts. This allows identification of nonclashing binding poses. In the next stage, energy-based Metropolis sampling is done to sample the binding pocket (Liu and Wang, 1999). QXP (McMartin and Bohacek, 1997) optimizes grid map energy and internal ligand energy for searching ligand-target structure. The algorithm performs a rigid body alignment of ligand-target complex followed by MCM translation and rotation of ligand. This step is followed by another rigid body alignment and scoring using energy grid map. ICM (Totrov and Abagyan, 1997) relies on a stochastic algorithm for global optimization of entire flexible ligand in receptor potential grid. The relative positions of ligand and target molecule make up the internal variables of the method. Internal variables are subject to random change followed by local energy minimization and selection by Metropolis criterion. ICM performed satisfactorily in generating protein-ligand complexes for 68 diverse, high-resolution X-ray complexes found in DUD (Cross et al., 2009).
a. Example application in computer-aided drug design.
ROSETTALIGAND was used by Kaufmann et al. (2009) to predict the binding mode of serotonin with serotonin transporters. The binding site predicted to be deep within the binding pocket was consistent with mutagenesis studies. QXP has been used to optimize inhibitors of human β-secretase (BACE1) (Malamas et al., 2009, 2010; Nowak et al., 2010), which is an important therapeutic target for treating Alzheimer’s disease by diminishing β-amyloid deposit formation. ICM was used successfully to identify inhibitors for a number of targets, including tumor necrosis factor-α (Chan et al., 2010), dysregulation of which is implicated in tumorigenesis and autoinflammatory diseases like rheumatoid arthritis and psoriatic arthritis. Computational screening of 230,000 compounds from the NCI data base against neuraminidase using ICM identified 4-{4-[(3-(2-amino-4-hydroxy-6-methyl-5-pyrimidinyl)propyl)amino]phenyl}-1-chloro-3-buten-2-one, which inhibited influenza virus replication at a level comparable to known neuraminidase inhibitor oseltamivir (An et al., 2009).
4. Genetic Algorithms.
Genetic algorithms introduce molecular flexibility through recombination of parent conformations to child conformations. In this simulated evolutionary process, the “fittest” or best scoring conformations are kept for another round of recombination. In this way, the best possible set of solutions evolves by retaining favorable features from one generation to the next. In docking, a set of values that describe the ligand pose in the protein are state variable, i.e., the genotype. State variables may include set of values describing translation, orientation, conformation, number of hydrogen bonds, etc. The state corresponds to the genotype; the resulting structural model of the ligand in the protein corresponds to the phenotype, and binding energy corresponds to the fitness of the individual. Genetic operators may swap large regions of parent’s genes or randomly change (mutate) the value of certain ligand states to give rise to new individuals.
Genetic Optimization for Ligand Docking (GOLD) (Jones et al., 1997) explores full ligand flexibility with partial target flexibility using a genetic algorithm. The GOLD algorithm optimizes rotatable dihedrals and ligand-target hydrogen bonds. The fitness of a generation is evaluated based on a maximization of intermolecular hydrogen bonds. The fitness function is the sum of a hydrogen bonding term, a term for steric energy interaction between the protein and the ligand and a Lennard-Jones potential for internal energy of ligand. AutoDock (Morris et al., 1998) uses the Lamarckian genetic algorithm, which allows favorable phenotypic characteristics to become inheritable. GOLD has demonstrated better accuracy than most docking algorithms, except GLIDE, in various benchmark studies (Kellenberger et al., 2004; Kontoyianni et al., 2004; Li et al.,2010b).
a. Example application in computer-aided drug design.
Inhibition of α-glucosidase has shown to retard glucose absorption and decrease postprandial blood glucose level, which makes it an attractive target for curing diabetes and obesity. Park et al. (2008) used AUTODOCK to identify four novel inhibitors of α-glucosidase by screening a library of 85,000 compounds obtained from INTERBIOSCREEN chemical data base (http://www.ibscreen.com). AUTODOCK was also used to identify inhibitors of RNA Editing Ligase-1 enzyme of T. brucei, the causative agent of human African trypanosomniasis (Durrant et al., 2010a).
5. Incorporating Target Flexibility in Docking.
Conformational variability is seen in unbound form and different apo structures (B-Rao et al., 2009; Sinko et al., 2013). It is widely believed that the ligand-bound state is selected from an ensemble of protein conformations by the ligand (Carlson, 2002). Accounting for receptor flexibility in the form of protein side-chain and backbone movement is essential for predicting correct binding pose. An ensemble of nonredundant low energy target structures will cover a large conformational space as against a single conformation, resulting in more realistic target-ligand bound states. Methods for inducing receptor flexibility include induced-fit docking and ensemble generated from MD simulation snapshots. Induced-fit algorithms allow small overlap between the ligand and the target along with side-chain movements, resulting in elasticity. GLIDE uses an induced fit model in which all side-chain residues are changed to alanine before initial docking. Side-chain sampling is followed by energy minimization of the binding site and ligand. ROSETTALIGAND allows for full protein backbone and side-chain flexibility in the active site. Multiple fix receptor conformations are used in docking protocols, known as ensemble-based screening, to incorporate receptor flexibility (Abagyan et al., 2006). Receptor conformations may either be experimentally determined by crystallography or NMR or computationally generated from MD simulations, normal mode analysis, and MC sampling (Cozzini et al., 2008). Schames et al., (2004) used the relaxed complex scheme (RCS) to describe a novel trench in HIV integrase, which led to the discovery of the integrase inhibitor raltegravir. In RCS, multiple conformations are determined from MD simulations to perform docking studies against. Other sampling methods include umbrella sampling, metadynamics, accelerated MD, etc. (Sinko et al., 2013).
D. Scoring Functions for Evaluation Protein-Ligand Complexes
Docking applications need to rapidly and accurately assess protein-ligand complexes, i.e., approximate the energy of the interaction. A ligand docking experiment may generate hundreds of thousands of target-ligand complex conformations, and an efficient scoring function is necessary to rank these complexes and differentiate valid binding mode predictions from invalid predictions. More complex scoring functions attempt to predict target-ligand binding affinities for hit-to-lead and lead-to-drug optimization. Scoring functions can be grouped into four types: (1) force-field or molecular mechanics-based scoring functions, (2) empirical scoring functions, (3) knowledge-based scoring functions, and (4) consensus scoring functions.
1. Force-Field or Molecular Mechanics-Based Scoring Functions.
Force-field scoring functions use classic molecular mechanics for energy calculations. These functions use parameters derived from experimental data and ab initio quantum mechanical calculations. The parameters for various force terms including prefactor variables are obtained by fitting to high-quality ab initio data on intermolecular interactions (Halgren, 1996). The binding free energy of protein-ligand complexes are estimated by the sum of van der Waals and electrostatic interactions. DOCK uses the AMBER force fields in which van der Waals energy terms are represented by the Lennard-Jones potential function while electrostatic terms are accounted for by coulomb interaction with a distance-dependent dielectric function. Standard force fields are however biased to select highly charged ligands. This can be corrected by handling ligand solvation during calculations (Shoichet et al., 1999; Kukic and Nielsen, 2010). Terms from empirical scoring functions (discussed below) are often added to force-field functions to treat solvation and electronic polarizability. A semi-empirical force field has been implemented in AUTODOCK to evaluate the contribution of water surrounding the receptor-ligand complex in the form of empirical enthalpic and entropic terms, for example (Huey et al., 2007).
2. Empirical Scoring Functions.
Empirical scoring functions fit parameters to experimental data. An example is binding energy, which is expressed as a weighted sum of explicit hydrogen bond interactions, hydrophobic contact terms, desolvation effects, and entropy. Empirical function terms are simple to evaluate and are based on approximations. The weights for different parameters are obtained from regression analysis using experimental data obtained from molecular data. Empirical functions have been used in several commercially available docking suits like LUDI (Bohm, 1992), FLEXX (Rarey et al., 1996), and SURFLEX (Jain, 2003).
3. Knowledge-Based Scoring Function.
Knowledge-based scoring functions use the information contained in experimentally determined complex structures. They are formulated under the assumption that interatomic distances occurring more often than average distances represent favorable contacts. On the other hand, interactions that are found to occur with lower frequencies are likely to decrease affinity. Several knowledge based potentials have been developed to predict binding affinity like potential of mean force (Shimada et al., 2000), DRUGSCORE (Velec et al., 2005), SMOG (DeWitte and Shakhnovich, 1997), and BLEEP (Mitchell et al., 1999).
4. Consensus-Scoring Functions.
More recently, consensus-scoring functions have been demonstrated to achieve improved accuracies through a combination of advantages of basic scoring functions. Consensus approaches rescore predicted poses several times using different scoring functions. These results can then be combined in different ways to rank solutions (Feher, 2006). Some strategies for combining scores include (1) weighted combinations of scoring functions, (2) a voting strategy in which cutoffs established for each scoring method is followed by decision based on number of passes a molecule has, (3) a rank by number strategy ranks each compound by its average normalized score values, and (4) a rank by rank method sorts compounds based on average rank determined by individual scoring functions. O'Boyle et al. (2009) evaluated consensus scoring strategies to investigate the parameters for the success of properly combined rescoring strategies. It turns out that combining scoring functions that have complementary strengths leads to better results over those that have consensus in their predictions. For example, scoring functions whose strengths are distinguishing actives from inactive compounds are complemented by scoring functions that can distinguish correct from incorrect binding poses. A disadvantage of consensus scoring methods could be a possible loss of active compound if poorly scored by one of the scoring functions.
a. Example application in computer-aided drug design.
Okamoto et al. (2009) have used consensus scoring technique for identifying inhibitors of death-associated protein kinases that are targets for ischemic diseases in the brain, kidney, and other organs. The consensus scoring function used in the study was implemented in DOCK4.0 program and included three scoring functions: (1) empirical scoring function (implemented in FLEXX), (2) a knowledge-based scoring function (Muegge and Martin, 1999), and (3) a force-field function from DOCK4.0. Approximately 400,000 compounds from a corporate compound library were docked followed by simultaneous scoring with the three functions. The consensus score was defined as the score that was highest among the three. In another successful application of consensus scoring scheme, Friedman and Caflisch (2009) discovered plasmepsin inhibitors for use as antimalarial agents using a scoring based on median ranking of four field-based scoring functions.
E. Structure-Based Virtual High-Throughput Screening
Structure-based virtual high-throughput screening (SB-vHTS), the in silico method for identifying putative hits out of hundreds of thousands of compounds to targets of known structure, relies on a comparison of the 3D structure of the small molecule with the putative binding pocket. SB-vHTS selects for ligands predicted to bind a particular binding site as opposed to traditional HTS that experimentally asserts general ability of a ligand to bind, inhibit, or allosterically alter the protein’s function. To make screening of large compounds libraries in finite time feasible, SB-vHTS often uses limited conformational sampling of protein and ligand and a simplified approximation of binding energy that can be rapidly computed. The inaccuracies introduced by these approximations lead to false-positive hits that ideally can at least in part be removed by subsequent refinement of the best ranking molecules and binding poses with more sophisticated methods involving iterative docking and clustering of ligand poses. The key steps in SB-vHTS are as follows: (1) preparation of the target protein and compound library for docking, (2) determining a favorable binding pose for each compound, and (3) ranking the docked structures. SB-vHTS has been used successfully in identifying novel and potent hits in several drug discovery campaigns (Becker et al., 2006; Lu et al., 2006; Zhao et al., 2006; Ruiz et al., 2008; Triballeau et al., 2008; Li et al., 2009; Budzik et al., 2010; Izuhara et al., 2010; Simmons et al., 2010; Roughley et al., 2012). We discuss two examples in which SB-vHTS played pivotal role in discovery of lead compounds.
1. Inhibitors of Hsp90.
Hsp90 is a molecular chaperone that modulates the activity of multiple oncogenic processes, which makes it an important therapeutic target for oncology. Roughley et al. (2012) virtually screened 0.7 million compounds from rCat (Baurin et al., 2004) with Hsp90 to identify leads that led to the development of potent inhibitors of Hsp90. Crystal structures of Hsp90 bound to previously known inhibitors were used in the docking-based virtual screen. From over 9000 nonredundant hits identified after screen, a set of 719 compounds were purchased after culling based on chemical diversity analysis. A total of 13 compounds with IC50 < 100 µM and seven with IC50 < 10 µM was identified. Determination of structure of hit-protein complex identified resorcinol-pyrazole series of compounds as lead compounds for further optimization. Compound AUY922, which was obtained after lead optimization, was evaluated for multiple myeloma, breast, lung, and gastric cancers.
2. Discovery of M1 Acetylcholine Receptor Agonists.
Selective agonism of M1 mAChR, which belong to the GPCR family A, has therapeutic potential for treating dementia, including Alzheimer’s disease and cognitive impairment associated with schizophrenia. Budzik et al. (2010) used a homology model of M1 mAChR based on crystal structure of bovine rhodopsin for virtual screening of a corporate compound collection. The docking of compounds into a previously known allosteric binding site yielded approximately 1000 putative hits. In vitro testing of these hits identified a lead compound, which is shown in Fig. 5. Optimization for improving potency and selectivity for M1 mAChR led to development of a series of novel 1-(N-substituted piperidin-4-yl) benzimidazolones, which resulted in compounds that were potent, central nervous system penetrant, and orally active M1 mAChR agonists.
Fig. 5.
Lead compound obtained through virtual screening of a library of compounds against M1 mAChR. Adapted from Budzik et al. (2010).
F. Atomic-Detail/High-Resolution Docking
The goal of SB-vHTS is to identify most probable hits that can bind to a target structure. As mentioned, scoring function and sampling algorithms are kept simple to evaluate large libraries of compounds in realistic time frames. The most promising hit compounds often are evaluated with more sophisticated scoring functions, for example, using an electrostatic solvation model for evaluating energetics of protein-ligand interaction. The implicit electrostatic solvation model is achieved by assuming the solvent as a continuum high-dielectric-constant medium through the use of numerical solutions of Poisson equation (Honig and Nicholls, 1995) or a generalized-Born approximation (Bashford and Case, 2000). Realistic conformational sampling, for example, through the inclusion of protein conformational changes is often done for lead compounds. The objective of this atomic-detail refinement of initial docking poses is threefold: (1) improved judgment if ligand will actually engage the target, (2) accurate prediction of complex conformation, and (3) accurate prediction of binding affinity. We describe some recent studies to highlight the success of high-resolution docking in identifying therapeutically important compounds.
1. Inhibitors of Casein Kinase by Hierarchical Docking.
Casein kinase 2 (CK2), a target for antineoplastic and anti-infectious drugs, is involved in a large variety of important cell functions, and many viruses exploit CK2 as phosphorylating agent of proteins essential to their life cycle. Cozza et al. (2006, 2009) used a hierarchical docking process to identify a potent inhibitor from an in-house molecular data base containing approximately 2000 compounds that included several families of polyphenolic compounds like catechins, coumarins, etc. A rigid body docking step using FRED was used to dock ligand conformations generated by OMEGA v.1.1. The top 50% of poses ranked by FRED score were selected, and one unique pose for each of the best-scored compounds was used for subsequent steps. The selected poses were optimized via a flexible ligand-docking protocol with three different programs: MOE-DOCK, GLIDE, and GOLD. A consensus scoring scheme was developed in which each docked set, i.e., FRED-DOCK, FRED-GLIDE, and FRED-GOLD, was scored by five different scoring functions: MOE-Score, GlideScore, GoldScore, ChemScore, and Xscore, leading to three docking/scoring sets. Common compounds among the top 5% of compounds ranked by consensus scores from each list were prioritized for in vitro testing. The hierarchical docking process allowed identification of nanomolar CK2 inhibitors like ellagic acid (IC50 = 40 nM) and quinalizarin (IC50 = 50 nM), which are shown in Fig. 6. Segers et al. (2007) applied the same hierarchical docking process to identify inhibitors of protein-membrane interaction. The C2 domain of coagulation factor V, a cofactor in blood coagulation, interacts with platelet membrane to cause platelet aggregation. Inhibitors of the protein-membrane interaction could be used for treating thrombotic disease, which is excessive blood coagulation due to overexpression of thrombin. A virtual screening of 300,000 compounds at the interface of C2 domain of coagulation factor V and the membrane identified four low micromolar inhibitors (IC50 < 10 µM).
Fig. 6.
Potent inhibitors of protein kinase CK2—ellagic acid and quinalizarin.
2. Discovery of Peroxisome Proliferator-Activated Receptor γ Agonists.
Lu et al. (2006) successfully combined a docking study and analog search for designing agonists of peroxisome proliferator-activated receptor (PPAR) γ, the receptor of thiazolidinedione antidiabetic drugs. A shape-based screening of the Maybridge data base using the shape of the bound conformation of a previously known agonist (PDB code 1K74) yielded 163 compounds. Screening of these compound in an in vitro binding assay identified [N-[1-(4-fluorophenyl)-3-(2-thienyl)-1H-pyrazol-5-yl]-5-[5-(tri-fluoromethyl) isoxazol-3-yl] thiophene-2-sulfonamide] (Compound 7) with an IC50 value of 175 nM, which is shown in Fig. 7. The binding mode of compound 7 was predicted by high-resolution docking using GOLD. With the exception of isoxazole group, all other four aromatic rings and the sulfonamide group of compound 7 made significant interactions with the protein. Thus, the scaffold containing the four aromatic rings along with the sulfonamide moiety was used for further analog search. The analog search in the Maybridge compound data base yielded 37 compounds that were tested in an in vitro binding assay. Compound 1 shown in Fig. 6 exhibited the strongest binding affinity, with an IC50 of 22.7 nM. Compound 1 is selective for PPARγ with no activity to PPARα or PPARδ. Administration at a daily dose of 30 mg/kg for 5 days to KKAγ mice, which exhibit obesity, insulin resistance, and type 2 diabetes-like symptoms, decreased the blood glucose level by 35.7%, demonstrating its glucose-lowering efficacy.
Fig. 7.
Outline of discovery process of novel family of PPAR-γ partial agonists. A, Conformation of compound 6 bound to active site of PPAR-γ was used as a pharmacophore. B, The bound conformation of compound 6 was used to screen the compound library. Compound 7 identified as a hit in the compound library screen. The binding mode of compound 7 obtained through docking study was used to define a core structure that was used for further similarity search which identified compound 1 as a potent agonist of PPAR-γ. From Lu et al. (2006).
3. Discovery of Novel Serotonin Receptor Agonists.
Becker et al. (2006) used virtual screening tools to guide a drug design campaign for novel serotonin receptor subtype 1A (5-HT1A) agonists. 5-HT1A agonists have been clinically demonstrated to be effective in treatment of anxiety and depression. The 3D structure of 5-HT1A was modeled using PREDICT (Becker et al., 2004; Shacham et al., 2004), a de novo modeling method for packing transmembrane helical bundles. A screening library of 150,000 compounds was selected from PREDIX corporate compound data base containing approximately 2,100,000 drug-like compounds. The selection was based on desired properties such as molecular weight range, compound diversity, and conformity to binding site characteristics. A docking study of a subset of screening library containing 40,000 compounds using DOCK4.0 was followed by ranking based on a rank-by-vote and rank-by-number consensus scoring approach (Bar-Haim et al., 2009). The top 10% of the library based on the best DOCK scores was filtered using a consensus score method using DOCK, CSCORE, and CHARMM. The hit list was clustered for diverse scaffolds and best-scored representatives of clusters yielded 78 compounds, which were further tested in vitro. The in vitro 5-HT1A identified 16 hits with binding affinities less than 5 µM, reflecting a hit rate of 21%. Nine hits had a Ki < 1 µM and the best hit, arylpiperazinylsulfonamide, which had a binding affinity of 1 nM was selected as the lead compound for further optimization. Further evaluation of arylpiperazinylsulfonamide (compound 8 in Table 3) revealed that it was suboptimal in both its DMPK/ADMET properties and selectivity profile. Thus, the goal of optimization process was to introduce selectivity for serotonin receptor (5-HT1A) over α1- and α2-adrenergic receptors. Analysis and comparison of docked poses of compound 8 with 5-HT1A and α1-adrenergic receptor models (obtained using PREDICT) shown in Fig. 8 was used to design the optimization strategy for compound 8. Prior to synthesis, each proposed compound was analyzed for (1) selectivity for 5-HT1A by docking with 5-HT1A and α1 adrenergic receptors, (2) potency of inhibition of hERG (human ether-a-go-go-related gene) K+ channel by docking to assess cardiovascular safety, (3) ADME-related properties like brain penetration predictions and physiochemical parameters such as polar surface area, cLogP, and more. The first round of optimization for 5-HT1A and ADME-related properties led to discovery of compounds shown in Table 3. Compound 20d, shown in Table 3, exhibited excellent DMPK/ADMET profile, but it was identified as a potent hERG blocker inhibiting the channel with IC50 of 300 nM compared with compound 8 with IC50 > 5000 nM. The structural analysis of the binding modes of the two compounds docked into a hERG channel model (based on the 3D structure of a related bacterial channel) was used for further optimization of compound 20d. Figure 9 illustrates the optimization strategy for reducing hERG affinity of compound 20d that led to identification of a preclinical candidate compound 20m as shown in Table 3. The greater hERG affinity of compound 20d is probably due to its interaction with the Ser660 region, which is not formed by compound 8. However, removing this interaction would annul previous optimization strategies. The authors thus decided to focus on the p-toluenesulfonyl region of compound 20d to reduce affinity of compound 20d for hERG. An optimization strategy that replaced p-toluenesulfonyl group with a nonaromatic hydrophobic group led to identification of compound 20m, which had lower affinity to hERG, while maintaining substantial binding affinity to 5-HT1A. Compound 20m was found to decrease the hyperthermic response to stress in the stress-induced hyperthermia model, which is based on the observation that stressful events cause a rise in core body temperature in mammals and anxiolytics. In silico approaches used in the study allowed the discovery of a phase three clinical trial drug candidate in fewer than 2 years.
TABLE 3.
Novel serotonin receptor agonists, which were identified during optimization stage
The chemical structure in the table represents the basic scaffolds of the receptor agonists. Derivatives of this structure were tested for affinity to 5-HT1A and hERG receptors. Some derivatives are listed in table below the structure. For example, compound 8 is derivative that has 2-OMe as the R group and 4-Me-Ph as the R2 group. Compound 8 exhibits a Ki value of 1 nM for 5-HT1A with an IC50 value of > 5000 nM for hERG receptor.
 |
||||
|---|---|---|---|---|
| Compound | R | R2 | 5-HT1A Ki | hERG: IC50 |
| nM | nM (%) | |||
| 8 | 2-OMe | 4-Me-Ph | 1 | >5000 (11%) |
| 20d | 3-MeCONH | 4-Me-Ph | 6.8 | 300 (73%) |
| 20m | 3-MeCONH | CH2-chexyl | 5.1 | 3800 (21%) |
Fig. 8.
Optimization of compound 8 for selective binding to 5-HT1A over α1-adrenergic receptor. 1a and 1b, Interactions of compound 8 with 5-HT and α1, respectively. The dotted box represents the structural differences between the two target molecules. The authors leveraged this difference between the protein molecules to design a virtual analog of compound 8, identified as 20h. 2b and 2b, docking models of 20h into 5-HT and α1. These docking modes indicate that the piperazine atom and aspartic acid interaction is maintained for 20h-5-HT complex and not for 20h-α1 complex. An optimization strategy based on this observation was used to design the novel agonist PRX-00023 for treatment of anxiety and depression. Adapted from Becker et al. (2006).
Fig. 9.
A comparison of the hERG binding modes of compounds 8 and 20d. Shown are a detailed 3D view of the binding of compounds 8 in the hERG pore, as well as two schematic views of the binding of compounds 8 and 20d next to each other. The four main interaction regions are highlighted in all views: an aromatic region formed by the four Tyr652 residues, a K+ pocket, an aromatic region formed by the four Phe656 residues, and a polar region formed by four Ser660 residues (shown only schematically).
4. Molecular Dynamics for High-Resolution Docking.
Human African trypanosomiasis caused by T. brucei affects approximately 70,000 people living in sub-Saharan Africa. Durrant et al. (2010a) used a virtual screening method that accounts for full protein flexibility to identify low micromolar inhibitors of RNA editing ligase 1 (TbREL1). A substructure search for previously known inhibitors of TbREL1 in several compound data bases such as ZINC, Hit2Lead, National Cancer Institute, and Sigma-Aldrich was used to generate a library of compounds. These compounds were docked into the ATP-binding pocket of a 1.20-Å resolution TbREL1 crystal structure. The docked poses were clustered at a tolerance of 2.0 Å, and the lowest-energy pose of the most populated cluster was judged to be the correct docked pose. A 2-ns explicit solvent MD simulation was used to generate 400 target conformations, one every 50 ps. A total of 33 conformations obtained after removing redundant conformations were used for further studies. The top 7.5% of the screening library by score was redocked into the full MD ensemble using AutoDock and rescored. This hybrid method of combining docking method with dynamic approach provided by MD simulations is called the relaxed complex scheme (RCS). The RCS rescoring scheme involved computation of simple mean of ensemble-average binding energy of each ligand predicted by AutoDock. Experimental evaluation of top 12 compounds ranked by RCS scoring scheme identified four low micromolar binders of TbREL1. RCS was also successfully used in identifying low micromolar inhibitors of T. brucei uridine diphosphate galactose 4′-epimerase (Durrant et al., 2010b).
G. Binding Site Characterization
The success of SB-CADD methods depends on the understanding of physiochemical interactions between molecules. Optimization of lead molecules into high-affinity compounds that are worth studying in vivo often requires optimization of its binding affinity along with pharmacological properties. This process of optimization requires a deep understanding of the molecular interactions between ligand and target. Structural studies to understand binding modes are commonly done using experimental methods such as X-ray and NMR. However, because of long turnover time for generating samples and structure determination, these methods are often unsuitable for repetitive cycles of lead optimization. The optimization process can be accelerated by the use of computational methods like molecular docking, molecular dynamics simulation, and quantum-mechanical simulations.
Experimentally determined protein structures in complex with ligand often serve as starting point for SB-CADD campaigns. For example, the cocrystal structure (PDB code 2BEL) of 11β-hydroxysteroid dehydrogenase (11β-HSD1) and its inhibitor, a semisynthetic derivative of 18β-glycyrrhetnic acid (carbenoxolone, shown in Fig. 10), were used to generate a model of the binding site. Increased expression of 11β-HSD1 in liver and adipose tissue has been linked to obesity, insulin resistance, diabetes, and cardiovascular diseases in humans. The crystal structure illustrates interaction of carbenoxolone with active site residues Ser170, Tyr183, and Lys187, as shown in Fig. 10. In addition, two hydrophobic pockets exist on either side of the catalytic site, which is exploited by a number of adamantine containing 11β-HSD1 inhibitors. A proprietary structure-based drug design program, Contour, was used to develop binding models of inhibitors containing an N-(2-adamantyl) amide moiety. Structural insight of binding site allowed the investigators to apply ligands containing an N-(2-adamantyl) amide moiety in a drug design program. With the help of the model and modeling studies, the authors discovered an 11β-HSD1 inhibitor that is orally bioavailable in three species and is active in a primate pharmacodynamic model (Tice et al., 2010).
Fig. 10.
Carboxynoxolone and 10j2. Overlap of carenoxolone (yellow) and urea 10j2 (green) in binding site of 11β-HSD1.
1. Helicase Inhibitor.
Hepatitis C virus (HCV) infection affects 180 million people worldwide and is implicated in serious life-threatening liver diseases, including cirrhosis, which may progress to hepatocellular carcinoma. Kandil et al. (2009) used docking and MD simulations to design selective inhibitors of HCV NS3 helicase. A helicase domain cocrystallized with a strand of DNA (PDB code 1A1V) was used as a starting point for de novo design of inhibitors that could compete with nucleic acid strand for binding. The authors identified two residues with which ligands could interact to inhibit helicase activity: (1) Arg393, which interacts with DNA strand, and (2) Cys431, which is situated 10 Å away from Arg393, whose sulfur atom could anchor ligand to the DNA binding site. A de novo designed inhibitor, structure B shown in Fig. 11, was able to interact with Arg393 but could not interact with Cys431 because of lack of any functional group. For optimization of compound B, virtual libraries were generated using MOE by varying linkers between the two aromatic rings, whereas replacing one carboxylic acid group with acceptors that have the ability to react with thiol. MD simulation was performed on these ligand/target complexes to evaluate stability of binding. The 1-ns simulation revealed slow drifting of compound 3 away from Cys431, which was attributed to steric hindrance of the aromatic ring. Smaller heterocycles in place of the aromatic ring were investigated, and linker chains were removed in subsequent molecules after MD simulations showed that they provided no particular advantage. Stable interactions with Arg481 and Cys431 during MD simulations prompted synthesis and evaluation of compound 4, shown in Fig. 11, in helicase assay, and it exhibited an IC50 of 0.26 µM.
Fig. 11.
Evolution of the design of novel HCV helicase inhibitor.
H. Pharmacophore Model
A pharmacophore model of the target binding site summarizes steric and electronic features needed for optimal interaction of a ligand with a target. Most common properties that are used to define pharmacophores are hydrogen bond acceptors, hydrogen bond donors, basic groups, acidic groups, partial charge, aliphatic hydrophobic moieties, and aromatic hydrophobic moieties. Pharmacophore features have been used extensively in drug discovery for virtual screening, de novo design, and lead optimization (Yang, 2010). A pharmacophore model of the target binding site can be used to virtually screen a compound library for putative hits. Apart from querying data base for active compounds, pharmacophore models can also be used by de novo design algorithms to guide the design of new compounds.
Structure-based pharmacophore methods are developed based on an analysis of the target binding site or based on a target-ligand complex structure. LigandScout (Wolber and Langer, 2005) uses protein-ligand complex data to map interactions between ligand and target. A knowledge based rule set obtained from the PDB is used to automatically detect and classify interactions into hydrogen bond interactions, charge transfers, and lipophilic regions (Wolber and Langer, 2005). The Pocket v.2 (Chen and Lai, 2006) algorithm is capable of automatically developing a pharmacophore model from a target-ligand complex. The algorithm creates regularly spaced grids around the ligand and the surrounding residues. Probe atoms that represent a hydrogen bond donor, a hydrogen bond acceptor, and a hydrophobic group are used to scan the grids. An empirical scoring function, SCORE, is used to describe the binding constant between probe atoms and the target. SCORE includes terms to account for van der Waals interactions, metal-ligand bonding, hydrogen bonding, and desolvation effects upon binding (Wang et al., 1998). A pharmacophore model is developed by rescoring the grids followed by clustering and sorting to extract features essential for protein-ligand interaction. During rescoring, hydrogen bond donor/acceptor scores lower than 0.2 and hydrophobic scores lower than 0.47 are reset to zero. Grids with three zero scores are filtered out, and the “neighbor number” for each grid is determined by counting the number of grids within 2 Å having non-zero score for a particular type. Grids with less than 50 donor neighbors, 30 acceptor neighbors, and 40 hydrophobic neighbors are reset to zero for their donor score, acceptor score, and hydrophobic scores, respectively. Grids are filtered by eliminating those with three zero scores, leaving only those grids that represent key interaction sites. The algorithm then superimposes the ligand on the grid, and a given grid is selected as a candidate if it is close to an atom type that can mediate the same interaction. Candidates with non-zero donor, acceptor, or hydrophobic scores are gathered into separate clusters, and the grid with highest score is defined as the center of donor, acceptor, or hydrophobic property.
1. Virtual Screening Using a Pharmacophore Model.
17β-Hydroxysteroid dehydrogenase type 1 (17β-HSD1) plays an important role in the synthesis of the most potent estrogen estradiol. Its inhibition could be important for breast cancer prevention and treatment. Schuster et al. (2008) used LigandScout2.0 to generate pharmacophore models of 17β-HSD1 from cocrystallization complexes with inhibitors (PDB codes 1EQU and 1I5R). These pharmacophore models represent the binding mode of a steroidal compound and small hybrid compounds (consisting of a steroidal part and an adenosine), respectively. The 1I5R-based pharmacophore model was used to screen the NCI and SPECS data bases for new inhibitors using CATALYST. Best scoring hit compounds were docked into the binding pocket of 1EQU using GOLD, and final selection for in vitro testing was performed according to the best fit value, visual inspection of predicted docking pose and the ChemScore (GOLD scoring function) value. Four of 14 compounds tested in vitro showed an IC50 value of less than 50 µM, with the most potent being 5.7 µM. Brvar et al. (2010) applied pharmacophore models to discover novel inhibitors of bacterial DNA gyrase B, a bacterial type II topoisomerase originating from gyrase and a target for antibacterial drugs. A pharmacophore model obtained using LigandScout was used to screen the ZINC data base, which yielded a novel class of thiazole-based inhibitors with IC50 value of 25 µM.
2. Multitarget Inhibitors Using Common Pharmacophore Models.
Wei et al. (2008) used Pocket v.2 to identify a common pharmacophore for two targets involved in inflammatory signaling, human leukotriene A4 hydrolase (LTA4H-h) and human nonpancreatic secretory phospholipase A2 (PLA2). The cocrystal structure (PDB code 1HS6) of LTA4H-h with 2-(3-amino-2-hydroxy- 4-phenylbutyrylamino)-4-methyl-pentanoic acid (bestatin) and the structure (PDB code 1DB4) of PLA2 with [3-(1-benzyl-3-carbamoylmethyl-2-methyl-1H-indol-5-yloxy)propyl]phosphonic acid (indole 8) were used to derive pharmacophores of the two targets. For LTA4H-h, six pharmacophore centers were identified that included four hydrophobic centers, one hydrogen bond acceptor, and one zinc metal coordination pharmacophore. In the binding pocket of PLA2, three hydrophobic centers, one hydrogen bond acceptor, and two calcium ion coordination centers were identified. The comparison of two sets of pharmacophore models revealed that two hydrophobic pharmacophores and a pharmacophore that coordinated with metal, shown in Fig. 12, was common to both proteins. The authors hypothesized that compounds that satisfy the common pharmacophores would inhibit both the proteins. The MDL chemical data base was screened virtually with LTA4H-h and PLA2 using Dock4.0 and binding conformation of the top 150,000 compounds (60% of data base) ranked by Dock score was extracted and checked for conformity to common pharmacophores. This identified 163 compounds whose binding conformations were reanalyzed using Autodock3.5 followed by comparison with common pharmacophores. Finally, nine compounds whose conformations matched the common pharmacophores were tested in vitro for binding with PLA2 and LTA4H-h. The best inhibitor, compound 10, shown in Fig. 13, inhibited LTA4H-h at submicromolar range and PLA2 with an IC50 value of 7.3 µM.
Fig. 12.
Extracting common pharmacophores of LTA4H-h and human-PLA2. Cyan spheres depict hydrophobic centers, red spheres represent H-bond acceptor while yellow spheres stand for feature that coordinates with a metal. Adapted from Wei et al. (2008).
Fig. 13.
(A) A reported inhibitor of LTA4H-h. (B) Compound 11.
3. Dynamic Pharmacophore Models That Account for Protein Flexibility.
The overexpression of murine double minute 2 oncoprotein (MDM2), which inhibits p53 tumor suppressor, is responsible for approximately one-half of all human cancers. Reactivation of p53-MDM2 integration has been shown to be a novel approach for enhancing cancer cell death (Bowman et al., 2007). Bowman et al. (2007) extracted snapshots at every 100 ps from a 2-ns MD simulation of MDM3 bound to p53. The resulting 21 structures for MDM2 were used to generate a six-site pharmacophore model of the active site, which included three aromatic/hydrophobic sites and three hydrogen-bond donor sites. A virtual screening of a library of 35,000 compounds identified 27 hits, 23 of which were tested in a competitive binding assay. Four of the tested compounds were identified as true hits, with the best inhibitor having a Ki value of 110 ± 30 nM. The dynamic pharmacophore model was also used successfully to identify low micromolar inhibitors of HIV-1 integrase (Deng et al., 2005).
I. Automated De Novo Design of Ligands
De novo structure-based ligand design can be accomplished by either a ligand-growing or ligand-linking approach. With the ligand-growing approach, a fragment is docked into the binding site and the ligand is extended by adding functional groups to the fragment. The linking method is similar in that multiple small fragments are docked into adjacent binding pockets of the target. Subsequently, the fragments are linked to each other to form a single compound. This approach is a computational version of the popular structure-activity relationship by NMR technique introduced by Shuker et al. (1996).
Several methods have been developed that implement both ligand-growing and ligand-linking strategies for designing ligands that can bind to a given target. LigBuilder (Wang et al., 2000b) builds ligands in a step by step fashion using a library of fragments. The design process can be carried out by various operations like ligand growing and linking and the construction process is controlled by a genetic algorithm. The target-ligand complex binding affinity is evaluated by using an empirical scoring function. The program first reads the target protein and analyzes the binding pocket. Depending on the choice of the user, it can then either use a growing or a linking strategy. In the growing strategy, a seed structure is placed in a binding pocket and then the program replaces user-defined growing sites with candidate fragments. This gives rise to a new seed structure that can then be used in further rounds of growing. For the linking strategy, several fragments placed at different locations on the target protein serve as seed structure. The growing scheme happens simultaneously on each fragment. In the process, the program seeks to link these fragments. The LUDI (Bohm, 1992) algorithm, which precedes LigBuilder, primarily uses a linking strategy for ligand design. It positions seed fragments into binding pockets of the target structure, optimizing their interactions individually. This step is followed by linking the fragments into a single molecule. The synthetic accessibility of ligands can be taken into account. For example, LigBuilder 2.0 analyzes designed using a chemical reaction data base and a retrosynthesis analyzer (Yuan et al., 2011).
The biggest challenge of de novo drug design is inseparable from its greatest advantage. By defining compounds that have never been seen before, one is invariably necessitating synthetic effort for acquisition prior to testing. This forces any de novo protocol to incorporate synthesizability metrics into its scoring. This increases the effort required in terms of cost, yield, time, and expertise necessary. Synthesizability thus becomes increasingly important when designing a large number of different compounds and scaffolds. Tools have been designed and used to approach synthesizability constraints. SYNOPSIS (SYNthesize and OPtimize System in Silico) (Vinkers et al., 2003) is a commonly used tool that enforces synthesizability throughout the design process by starting with available compounds and creating novel compounds by virtually using known chemical reactions. This tool contains a set of 70 reaction types that are selected based on the presence of different functional groups in the evolving molecule. SYNOPSIS also provides additional restraints for desired properties such as solubility. Krier et al. (2005) proposed an approach called the Scaffold-Linker-Functional Group (SLF) approach, which has been implemented in de novo strategies. This method is designed to create a de novo scaffold-focused library that maximizes diversity and minimizes size. A limited number of nonoverlapping functional groups were selected that are added or removed from the static scaffold core. The linker plays the role of varying the distances between the scaffold and functional groups. RECAP (Retrosynthetic Combinatorial Analysis Procedure) was the first fragment generation method to incorporate rules that limit the chemical reactions to ones used in typical combinatorial chemistry techniques, thereby limiting the possible fragments as well as possible recombination patterns (Lewell et al., 1998; Degen et al., 2008).
1. Example Application in Computer-Aided Drug Design.
De novo design by linking fragments has been successfully applied in the design of inhibitors of p38 MAPK (Cogan et al., 2008), which is a key regulator in signaling pathways that control the production of cytokines such as tumor necrosis factor-α and interleukin-1β. Inhibitors of MAPK can potentially be used for the treatment of various autoimmune diseases. Figure 14A shows four classes of interactions of a clinical compound BIRB 796 with MAPK: (1) interaction with residues in ATP binding site (Met109), (2) interaction with the “Phe pocket” (dotted arc), (3) hydrophobic interaction with the kinase specificity pocket (solid arc), and (4) interaction of the urea with backbone NH-bond of Asp168 and carboxylate of Glu71. A design strategy for exploring structurally distinct scaffolds by leveraging the interactions of BIRB 796 [1-(5-tert-butyl-2-p-tolyl-2H-pyrazol-3-yl)-3-[4-(2-morpholin-4-yl-ethoxy)naphthalen-1-yl] urea] was devised as follows: (1) a tert-butyl group was used as “Phe pocket” seed structure in place of pyrazole ring of BIRB 796, (2) an N-formyl group was appended to tert-butyl fragment to access the hydrogen bonds with Glu71 and Asp168, and (3) a carbonyl group was used as the second seed fragment to access the hydrogen bond with Met109 as shown in Fig. 14B. LigandBuilder software was used to link the two seed fragments, the tert-butyl linked to N-formyl group, and the carbonyl group. The program consistently introduced a 4-tolyl group in the kinase specificity pocket. However, LigandBuilder failed to predict favorable rigid linkers for connecting the tolyl group to carbonyl group, which would be essential for carbonyl display at the proper distance to interact with Met109. Modeling indicated N-linked azoles connected to tolyl group via an N-linkage as a suitable linker. Derivatives of this designed molecule were synthesized, leading to the discovery of compound 28 shown in Fig. 14D, which exhibited an IC50 value of 83 nM.
Fig. 14.
Design strategy for inhibitors of p38 MAPK. (A) Key interactions of BIRB-796 inhibitor with MAPK. (B) A fragment linking strategy to link two seed structures was applied using LigBuilder. A tert-butyl phenyl fragment was used in the first pocket, whereas a carbonyl fragment was used to access the hydrogen bond with Met109 in the second site. An N-formyl group was attached to the first seed fragment to access hydrogen bonds with Glu71 and Asp168. (C) General structure of optimized structures which showed potent activity. (D) R group for compound 28, which showed IC50 value of 83 nM. Adapted from Cogan et al. (2008).
The fragment extension approach was used by Zhang et al. (2011) in the discovery of inhibitors of vascular endothelial growth factor (VEGF) receptor 2 (VEGFR2), a therapeutic target for tumor-induced angiogenesis. The authors used quinazoline as the seed fragment, because three of the nine clinically approved kinase inhibitor drugs are 4-anilinoquinazoline derivatives (Li et al., 2010a). These inhibitors bind the active site of their respective targets such that the quinazoline ring is located at the front of ATP binding pocket. The ligand building process involved placing the quinazoline fragment in the binding pocket in the same orientation as found for known inhibitors. The design strategy sought to create a ligand that would extend to fit a specific hydrophobic pocket at the back of the ATP binding cleft. An NH2, OH, or SH group was added in the C4 position of the quinazoline ring to allow for a turn owing to orientation of quinazoline and the spatial arrangement of the hydrophobic pocket. A fragment-growth-based de novo method was applied in which various fragments (approximately 1200 fragments) were allowed to grow on the turn fragment to extend into the hydrophobic pocket. Designed molecules were then rescored and ranked using GOLD. The design process led to the development of a potent and specific VEGFR2 inhibitor, SKLB1002 [2-((6,7-dimethoxyquinazolin-4-yl)thio)-5-methyl-1,3,4-thiadiazole], shown in Fig. 15. The inhibitor was successful in inhibiting angiogenic processes in zebrafish embryo and athymic mice with human tumor xenografts.
Fig. 15.
(A) Chemical structure of SKLB1002. (B) SKLB1002 is docked into the active site of VEGFR2, showing interactions between SKLB1002 and VEGFR2 by using the in silico model. (C) A 2D interaction map of SKLB1002 and VEGFR2. Adapted from Zhang et al. (2011).
J. Strategies for Important Classes of Drug Targets
Protein kinases and GPCRs are the most frequently targeted classes in drug discovery. About a dozen drugs that target kinases have been approved for clinical use in the field of cancer and more than a hundred are undergoing clinical trials. Kinase inhibitors have been found to be useful in several other conditions like inflammatory diseases, treatment of hypertension, and Parkinson’s disease (Cohen and Alessi, 2013). Modulators of GPCRs represent 27% of all clinically approved drugs. Diseases associated with loss of function include congenital hypothyroidism, congenital bowel obstruction, abnormal breast and bone development, and loss of function mutation in type 5 chemokine receptor leading to resistance to HIV infection. In addition, there are several gain-of-function disorders such as night blindness due to constitutive active rhodopsin, hyperthyrodism, neonatal hyperparathyroidism, etc. In general, drugs targeting GPCRs come under two categories: (1) agonists stimulate GPCRs and (2) antagonists that block the activation.
Kinase 3D structures are abundantly available in the PDB. A large number of 3D structures have also been developed by means of homology modeling based on templates having highest sequence similarity with targets. Docking studies with kinases, however, indicate that similarity of binding site influences docking prediction (Tuccinardi et al., 2010). In addition, kinases’ binding sites have high plasticity, allowing adaptation to interact with ligands (Rabiller et al., 2010). This makes for a good argument to go for ligand-induced homology modeling. A straightforward approach is to use homology models created from a template cocrystallized with a ligand similar to the one of interest. Tuccinardi et al. (2010) found that this approach yielded models that could be reliable for docking studies. Ideally homology models based on multiple-templates may be used in docking and selected on the ability to generate conserved interactions. ATP-competing inhibitors typically form at least one hydrogen bond with a backbone amide or carbonyl group in the hinge region (Cheng et al., 2012). Docking results can be improved by keeping constraints to preserve conserved interactions (Ravindranathan et al., 2010). Kinases undergo receptor rearrangement upon ligand binding (Rabiller et al., 2010). The ATP binding pocket has highly conserved residues, which is an obstacle for development selective kinase inhibitors and requires the exploitation of an adjacent “allosteric pocket.” Mimicking conformational change of kinases, and not just side-chain flexibility, improves the success of kinase inhibitor docking (Cavasotto and Abagyan, 2004; Rabiller et al., 2010). Cavasotto and Abagyan (2004) reported that incorporating protein flexibility in ligand docking is essential.
GPCRs play a central role in human physiology and are prime targets for drug discovery for different indications such as cardiovascular, metabolic, neurodegenerative, and oncologic diseases. High-resolution crystal structures of GPCRs have become available only recently and are sparse. de Graaf and Rognan (2009) studied the effect of template choice on docking. The results indicate that multiple-template-based models performed slightly better than single-template models if all templates shared low sequence identity with the target. Fragment-based methods like I-TASSER (Zhang, 2008) have performed well in CASP. I-TASSER takes a hierarchical approach to homology modeling by using fragments from template structures and assembling multiple fragments based on threading alignment. Ligand-induced homology modeling using LiBERO (Katritch et al., 2010) has shown promise in terms of percentage of correctly predicted native contacts. MD refinement of homology models of GPCRs has demonstrated benefits (Yarnitzky et al., 2010). The accurate modeling of extracellular loops is essential because they are important for ligand recognition as has been demonstrated by several site-directed mutagenesis studies (Bokoch et al., 2010). Although considerable progress has been made in de novo loop modeling, loopless models provide practical alternatives in cases where de novo modeling fails. de Graaf et al. (2008) recommended loopless models of GPCRs for virtual screening unless high homology targets or receptor specific data were available. Finally, receptor ensemble docking studies have shown promising results compared with one binding site conformation in terms of significant improvement in virtual screening yields (Vilar et al., 2011).
III. Ligand-Based Computer-Aided Drug Design
The ligand-based computer-aided drug discovery (LB-CADD) approach involves the analysis of ligands known to interact with a target of interest. These methods use a set of reference structures collected from compounds known to interact with the target of interest and analyze their 2D or 3D structures. The overall goal is to represent these compounds in such a way that the physicochemical properties most important for their desired interactions are retained, whereas extraneous information not relevant to the interactions is discarded. It is considered an indirect approach to drug discovery in that it does not necessitate knowledge of the structure of the target of interest. The two fundamental approaches of LB-CADD are (1) selection of compounds based on chemical similarity to known actives using some similarity measure or (2) the construction of a QSAR model that predicts biologic activity from chemical structure. The difference between the two approaches is that the latter weights the features of the chemical structure according to their influence on the biologic activity of interest, whereas the former does not. The methods are applied for in silico screening for novel compounds possessing the biologic activity of interest, hit-to-lead and lead-to drug optimization, and also for the optimization of DMPK/ADMET properties. LB-CADD is based on the Similar Property Principle, published by Johnson et al. (1990), which states that molecules that are structurally similar are likely to have similar properties. LB-CADD approaches in contrast to SB-CADD approaches can also be applied when the structure of the biologic target is unknown. Additionally, active compounds identified by ligand-based virtual high-throughput screening (LB-vHTS) methods are often more potent than those identified in SB-vHTS (Stumpfe et al., 2012).
A. Molecular Descriptors/Features
LB-CADD techniques use different methods for describing features of small molecules using computational algorithms that balance efficiency and information content. The optimal descriptor set depends on the biologic function predicted as well as on the LB-CADD technique used, and therefore many different algorithms for deriving chemical information have been developed and used. Molecular descriptors can be structural as well as physicochemical and can be described on multiple levels of increasing complexity. Information described can include properties such as molecular weight, geometry, volume, surface areas, ring content, rotatable bonds, interatomic distances, bond distances, atom types, planar and nonplanar systems, molecular walk counts, electronegativities, polarizabilities, symmetry, atom distribution, topological charge indices, functional group composition, aromaticity indices, solvation properties, and many others (Cramer et al., 1988; Randic, 1995; Schuur et al., 1996; Bravi et al., 1997; Hemmer et al., 1999; Pearlman and Smith, 1999; Hong et al., 2008; Roberto Todeschini, 2010). These descriptors are generated through knowledge-based, graph-theoretical methods, molecular-mechanical, or quantum-mechanical tools (Acharya et al., 2011; Marrero-Ponce et al., 2012) and are classified according to the “dimensionality” of the chemical representation from which they are computed (Ekins et al., 2007): 1D, scalar physicochemical properties such as molecular weight; 2D, molecular constitution-derived descriptors; 2.5D, molecular configuration-derived descriptors; 3D, molecular conformation-derived descriptors. These different levels of complexity, however, are overlapping with the more complex descriptors, often incorporating information from the simpler ones. For example, many 2D and 3D descriptors use physicochemical properties to weight their functions and to describe the overall distribution of these properties.
1. Functional Groups.
Functional groups are defined by the International Union of Pure and Applied Chemistry as atoms or groups of atoms that have similar chemical properties across different compounds. These groups are attached to a central backbone of the molecule, also called scaffold or chemotype. The spatial positioning of the functional groups achieved by the backbone defines the physical and chemical properties of compounds. Therefore, the location and nature of functional groups for a given compound contain key information for most ligand-based CADD methods. There are many different kinds of functional groups including those that contain hydrocarbons, halogens, oxygens, nitrogens, sulfur, phosphorous, etc. Functional groups include alcohols, esters, amides, carboxylates, ethers, nitro group, thiols, etc. (March, 1977)
Functional groups can either be described explicitly by their atomic composition and bonding or may be implicitly encoded by their general properties. For example, under physiologic conditions carboxyl groups are often negatively charged, whereas amine groups are positively charged. This property is accurately reflected in the structure of the functional group but also in the charge computed from that structure. Because it is the properties conferred by the functional groups that are most important to the biochemical activity of a given compound, many CADD applications treat functional groups containing different atoms but conferring the same properties as similar or even identical. For example, the capacity for hydrogen bonding can heavily influence a molecule’s properties. These interactions frequently occur between a hydrogen atom and an electron donor such as oxygen or nitrogen. Hydrogen bonding interactions influence the electron distribution of neighboring atoms and the site’s reactivity, making it an important functional property for therapeutic design. Commonly, hydrogen bonding groups are separated as hydrogen bond donors with strong electron-withdrawing substituents (OH, NH, SH, and CH) and hydrogen bond acceptor groups (PO, SO, CO, N, O, and S) (Pimentel and McClellan, 1960; Vinogradov and Linnell, 1971). The applications Phase, Catalyst, DISCO, and GASP (Genetic Algorithm Superposition Program) as well as Pharmacophore mapping algorithms discussed in greater detail below focus primarily on hydrogen-bond donors, hydrogen-bond acceptors, hydrophobic regions, ionizable groups, and aromatic rings.
2. Prediction of Psychochemical Properties.
Descriptors within the same dimensionality can show a range of complexity. The simplest ones, such as molecular weight and number of hydrogen bond donors, are relatively simple to compute. These can be rapidly and accurately computed. More complex descriptors such as solubility and partial charge are more difficult to compute. However, the higher information content provided by these descriptors makes them extremely useful for model development. (Zhou et al., 2010). Therefore, prediction of physicochemical properties is a critical step in developing effective molecular descriptors. The trade-off in computing such descriptors is between the high speed needed to encode thousands of molecules and sufficient accuracy.
a. Electronegativity and partial charge.
Electron distribution plays an important role in a molecule’s properties and activities. Therefore, it was important to develop a descriptor that is capable of modeling the charge distribution over an entire molecule. A useful form of this descriptor was to assign a partial charge to all atoms in a molecule. Initially, electron distribution could be assigned to individual atoms through quantum mechanical calculations. However, when screening thousands or millions of compounds, a much faster and more efficient method is necessary. Gasteiger and Marsili (1980) developed a method for assigning partial charges to individual atoms called the Partial Equalization of Orbital Electronegativity (PEOE). This method is based on a definition of electronegativity introduced by Mulliken (1934) that relates electronegativity of an atom to its ionization potential I and electron affinity E with the equation electronegativity = 1/2(I + E). The values for E and I depend on the valence state of the atom, and Hinze and Jaffe (1962) and Hinze et al. (1963) introduced the concept of orbital electronegativity, which was capable of defining electronegativity of a specific orbital in a given valence state. Orbital electronegativities depend on hybridization and occupation number of the orbital.
Electronegativity equalization was proposed by Sanderson (1951, 1960) and stated that bonded atoms changed electron density until total equalization of electronegativity was reached. However, this simple model led to chemically unacceptable calculations. The PEOE method is an improvement to this electronegativity equalization model that produces more appropriate results by adding some complexity to the equalization of electronegativities. Gasteiger and Marsili (1980) first introduced an approximation function that joins the electronegativity values of an atom in its anionic, neutral, and cationic state with appropriate ionization potentials and electron affinities and relates orbital occupation with orbital electronegativity. They also added a damping function to account for the fact that when charge transfer is occurring an electrostatic field is generated, inhibiting further electron transfer and preventing a complete equalization. Finally, they introduced an iterative procedure to account for the fact that modified electronegativities after charge transfer give rise to new charge separations. Progressive iterations included wider spheres of neighboring atoms until the total transfer dropped below a cutoff. The total charge of an atom is then calculated as the sum of the individual charge transfers after the iteration.
For small-member rings, special bonds based on the valence bond model (Coulson and Moffitt, 1947) were used as additional parameters in the PEOE method (Guillen and Gasteiger, 1983). The valence bond model states that the bonds of three- and four-membered ring systems arise from orbitals with varying amounts of s and p character depending on the type and number of rings involved and whether exo- or endocyclic bonds are considered. The extra coefficients provided charge dependence for the different hybridization states interpolated from the values of electronegativities for sp3, sp2, sp, and p states (Hinze and Jaffe, 1962).
Gasteiger and Saller (1985) introduced a method for applying the PEOE method to molecules with multiple resonance structures. Charge distribution in π systems could be calculated on the basis of resonance structure weights. These weights were calculated by including a topological weight and electronic weight. The topological weight was based on whether resonance structures involved the loss of covalent bonds, decrease in aromatic systems, or charge separation. The electronic weight was based on the idea that resonance structures are more important when a negative charge is localized on the more strongly electronegative atom. Therefore, it was a measure of how well the donor atom can donate its lone pair of electrons and how stable is a negative charge on the acceptor atom. To calculate this weight, the electronegativity concept is applied. Finally, by adding the changes in charge of the individual resonance structures to the scaling factor the charge distribution could be calculated. Orbital electronegatives are often implemented into σ and π bond systems. Standard connection tables describe localized connections between two atoms that contain twice the number of electrons per bond order (single bonds contain two electrons, double bonds contain four, etc.). This valence bond structure, however, is insufficient to describe some compounds and to distinguish between the different excited states of a molecule. Separating σ and π electrons has been shown to be advantageous to this representation scheme (Gasteiger, 1979). Bauershmidt and Gasteiger (1997) describe computational representation of chemical species using three electron systems: σ-electron systems, π-electron systems, and coordinative bonds.
σ-Electron systems contain electrons localized in the σ part of a bond and single bond electrons. These systems may consist of more than two atoms when multicenter bonds are described, including overlapping orbitals that point into a central region between bonded atoms and open bridging α-electron systems where one atom is located between the other atoms part of the same system. π-Electron systems encode free electrons. One π-electron system is generated for each electron pair. For example, the electrons of a triple bond are distributed into one σ-electron system and two π-electron systems, each with two electrons. Properties such as orbital electronegativity and partial charges are more accurately described using the σ- and π-electron systems. Therefore, it is common to implement descriptors separated as σ charges, π charges, σ electronegativity, and π electronegativity.
These methods provide a means to quantitatively calculate electronegativity and partial charge on a per-atom basis without the need for quantum mechanics. PEOE charges have been shown to be useful information for predicting chemical properties such as taste (Belitz et al., 1979). Additionally, these properties are often used to weight three-dimensional descriptors that would, on their own, only capture purely structural information. By weighting these descriptors with these properties, information regarding the three-dimensional distribution of electrons is available.
b. Polarizability.
Effective polarizability or mean molecular polarizability is another widely used molecular descriptor. It quantifies the response of electron density to an external field to give an induced dipole moment (Le Fèvre, 1965). Polarizability contributes to dispersion forces and influences intermolecular interactions (a fast empirical method for the calculation of molecular polarizability). Brauman and Blair (1968) described stabilization effects of substituent polarizability. For example, induced dipole moments in unsubstituted alkyl groups are believed to stabilize charges in gaseous ions formed by protonation or deprotonation (Gasteiger and Hutchings, 1984). The magnitude of the induced dipole is calculated as the product of the electric field operator and the polarizability tensor of the molecule. The average polarizability of a molecule is calculated as the average of the three principal components of this tensor (Glen, 1994).
Miller and Savchik (1979) introduced a formula for calculation of mean molecular polarizabilities using a polarizability contribution for each atom based on its atom type and hybridization state and the total number of electrons in the molecule. Gasteiger and Hutchings (1983) improved this formula to account for the attenuation of substituent influence. This was accomplished through the introduction of a damping factor dependent on the distance in bonds between the atom and the charged reaction center.
Glen (1994) defined a method for calculating static molecular polarizability using a modified calculation of atomic nuclear screening constants based on effective nuclear charge described by Slater (1930). This calculation divides electrons into different groups with different shielding constants. These shielding constants reflect the fact that inner-shell electrons modify the view of the nucleus for outer-shell electrons and adjust the field of nuclear charge for each group of electrons.
c. Octanol/water partition coefficient.
LogP (logarithm of partition coefficient between n-octanol and water) is an important molecular descriptor that has been widely used in QSAR since the work of Leo et al. (1971). Lipinksi’s rule of five, a class set of rules describing the “druggability” of a compound, includes measurement of the compound’s logP. Traditionally, logP can be calculated experimentally by measuring its partitioning behavior in the insoluble mixture of n-octanol and water and reflects the molecule’s hydrophobicity. This molecular property has been shown to be important in solubility, oral availability, transport, penetration of the blood-brain-barrier, receptor binding, and toxicity (Hansch et al., 1962, 1987). For virtual screening applications, several methods for calculating logP based on molecular constitution have been established.
LogP calculations largely rely on an additive method introduced by Rekker and Mannhold (1992) in which the contributions to logP by basic fragments of a molecule (atoms and functional groups) are summed. Additivity methods improved with the incorporation of molecular properties have also been used to calculate logP (Kellogg et al., 1991; Meng et al., 1994).
Wang et al. (1997) developed the very popular additivity method called XLOGP. This method originally defined 80 basic atom types for carbon, nitrogen, oxygen, sulfur, phosphorous, and halogen atoms. Hydrogen atoms are implicitly included in the different atom types. Additionally, correction factors were applied to account for specific intramolecular interactions that can affect a molecule’s logP beyond each of the fragments on their own. This method was later improved to include 90 atom types and 10 correction factors (Wang et al., 2000a).
Correction factors were necessary and determined empirically due to the fact that many logP calculations based on simple summations were incorrect. For example, compounds with long hydrocarbon chains had underestimated logP because of their flexibility and aggregation behavior, atoms bonded to two or more halogen atoms had altered properties due to dipole shielding, internal hydrogen bonding, the unusually strong internal hydrogen bonding with salicylic acids, and the existence of α-amino acids as zwitterions. Additionally, correction factors are included for aromatic nitrogen pairs, ortho sp3 oxygen pairs, para donor pairs, sp2 oxygen pairs, and amino sulfonic acids.
Xing and Glen (2002) introduced an alternative logP calculation that was based on the evidence that molecular size and hydrogen-bonding ability account for a major part of logP. They created a statistical model by combining molecular size and dispersion interactions using molecular polarizability and the sum of squared partial atomic charges on oxygen and nitrogen atoms. The final model showed that molecular polarizability is more significant than atomic charges and that an increase in polarizability is correlated with an increase in logP, whereas a decrease in charge densities on nitrogen and oxygen correlated with a decrease in logP. They theorized that the importance of molecular polarizability on logP was due in part to the relative energy required for a larger molecule to create a cavity in water or octanol.
3. Converting Properties into Descriptors.
Molecule properties are converted into numerical vectors of descriptors for analysis. This conversion is needed to ensure that descriptions of molecules have a constant length independent of size. Each position in the vector of descriptors encodes a well-defined property or feature that facilitates comparison by mathematical algorithms.
a. Binary molecular fingerprints.
Fingerprints are bit string representations of molecular structure and/or properties (Bajorath 2001, 2002). They encode various molecular descriptors as predefined bit settings (Auer and Bajorath, 2008), i.e., representation as 1 or 0, where 1 means descriptor is present or 0 if not. This allows chemical identity to be unambiguously assigned by the presence or absence of features (Hutter, 2011). The features described in a molecular fingerprint can vary in number and complexity (from hundreds of bits for structural fragments to thousands for connectivity fingerprints and millions for the complex pharmacophore-like fingerprints) (Auer and Bajorath, 2008), depending on the computational resources available and the intended application. Fingerprints that rely solely on interatomic connectivity, i.e., molecular constitution, are known as 2D fingerprints (Hutter, 2011). In the prototypic 2D keyed fingerprint design, each bit position is associated with the presence or absence of a specific substructure pattern, for example carbonyl group attached to sp3 carbon, hydroxyl group attached to sp3 carbon, etc. (Barnard and Downs, 1997).
Molecular structure itself comprises several levels of organization between the atoms within a molecule, and, therefore, fingerprints may differ in their levels of organization too. For example, the simplest fingerprint may contain the information that a given compound contains six carbon atoms and six hydrogen atoms. However, up to 217 different isomers can contain this fingerprint. Adding connectivity increases the specificity of the fingerprints but does not necessarily provide discrimination between stereoisomers. These molecules are not identical despite having equal constitutions and 2D fingerprints that are insufficient to describe their structures. Therefore, considerable effort is taken to ensure the efficient application of fingerprints without sacrificing important molecular characteristics. One extension to fingerprints is the use of hash codes. These are bit strings of fixed length that contain information about connectivity, stereo centers, isotope labeling, and further properties. This information is then compressed to avoid redundancies (Ihlenfeldt and Gasteiger, 1994). Unfortunately, it is not always obvious which of these characteristics are important in a given context and which are not (Hutter, 2011).
Commonly used fingerprints include the ISIS (Integrated Scientific Information System) keys with 166 bits and the MDL (Molecular Design Limited) MACCS (Molecular ACCess System) keys (Durant et al., 2002) with 960 bits. The ISIS keys are small topological substructure fragments, whereas the MACCS keys consist of the ISIS keys plus algorithmically generated more abstract atom-pair descriptors. MDL keys are commonly used when optimizing diversity (McGregor and Pallai, 1997; Roberto Todeschini, 2010). For example, the PubChem data base uses a fingerprint that is 881 bits long to rank substances against a query compound. This fingerprint is comprised of the number and type of elements, ring systems (saturated and unsaturated up to a size of 10), pairwise atom combinations, sequences, and substructures (Hutter, 2011).
b. 2D description of molecular constitution.
2D descriptors can be computed solely from the constitution or topology of a molecule, whereas 3D descriptors are obtained from the 3D structure of the molecule (Ekins et al., 2007). Many 2D molecular descriptors are based on molecular topology derived from graph-theoretical methods. Topological indices treat all atoms in a molecule as vertices and index-specific information for all pairs of vertices. A simple topological index, for example, will contain only constitutional information such as which atoms are directly bound to each other. This is known as an adjacency matrix and an entry of 1 for vertices vi and vj if their corresponding atoms are bonded, and an entry of 0 for vi and vi indicates that the corresponding atoms are not directly bonded (Trinajstić, 1992). For an adjacency matrix, the sum of all entries is equal to twice the total number of bonds in the molecule.
Complex topological indices are created by performing specific operations to an adjacency matrix that allow for the encoding of more complex constitutional information. These indices are based on local graph invariants that can represent atoms independent of their initial vertex numbering (Devillers and Balaban, 1999). For example, topological indices may contain entries for the number of bonds linking the vertices. Information gathered from such an index can include the number of bonds linking all pairs of atoms and the number of distinct ways a path can be superimposed on the molecular graph. A topological index that includes information such as heteroatoms and multiple bonds through the weighting of vertices and edges was introduced by Bertz (1983). Randic and Basak (2001) introduced an augmented adjacency matrix by replacing the zero diagonal entries (where vi = vj) with empirically obtained atomic properties. This adjacency matrix includes atom type information as well as connectivity (Randic and Basak, 2001). Topological indices that describe the molecular charge distribution as evaluated by charge transfers between pairs of atoms and global charge transfers have also been developed (Galvez et al., 1994, 1995). Additionally, topological indices known as geometrical indices have been derived to describe molecular shape. For example, the shape index E measures how elongated is the molecular graph (Galvez et al., 1995, 1998). Statistical methods such as linear discriminant analysis are often applied to topological indices and biologic properties to create predictive descriptors relating indices to molecular activity (Galvez et al., 1994, 1995).
Topological autocorrelation (2D autocorrelation) is designed to represent the structural information of a molecular diagram as a fixed-length vector that can be applied to molecules of any shape or size. It encodes the constitutional information as well as atom property distribution by analyzing the distances between all pairs of atoms. Topological autocorrelations are independent of conformational flexibility because all distances are measured as the shortest path of bonds between the two atoms. The autocorrelation vector is created by summing all products for atom pairs within increasing distance intervals in terms of number of bonds. In other words, it creates a frequency plot for a specific range of atom pair distances. By including atom property coefficients for all atom pairs, autocorrelations are capable of plotting the arrangement of specific atom properties. For example, information such as the frequency at which two negatively charged atoms are three bonds apart versus four bonds apart is stored in an autocorrelation plot that has been weighted by partial atomic charge (Moreau and Broto, 1980).
c. 3D Description of molecular configuration and conformation.
The physicochemical meaning of topological indices and autocorrelations is unclear and incapable of representing some qualities that are inherently three-dimensional (stereochemistry). 3D molecular descriptors were developed to address some of these issues (Kubinyi., 1998).
The 3D autocorrelation is similar to the 2D autocorrelation but measures distances between atoms as Euclidian distances between their 3D coordinates in space. This allows a continuous measure of distances and encodes the spatial distribution of physicochemical properties. Instead of summing all pairs within discrete shortest path differences, the pairs are summed into interval steps (Broto et al., 1984).
Radial distribution functions (RDFs) is another very popular 3D descriptor. It maps the probability distribution to find an atom in a spherical volume of radius r. In its simplest form, the RDF maps the interatomic distances within the entire molecule. Often it is combined with characteristic atom properties to fit the requirements of the information to be represented (Hemmer et al., 1999). RDFs not only provide information regarding interatomic distances between atoms and properties, they reflect other information such as bond distances, ring types, and planar versus nonplanar molecules. These functions allow estimation of molecular flexibility through the use of a “fuzziness” coefficient that extends the width of all peaks to allow for small changes in interatomic distances.
GRIND (Grid-Independent Descriptor) is another 3D descriptor that does not require prior alignment (Pastor et al., 2000). This set of descriptors was designed to retain characteristics that could be directly traced to the molecules themselves, rather than producing purely mathematical descriptors that are not obviously related to the molecular structures they describe. GRIND is comprised of three steps. The first step is to calculate a molecular-interaction field (MIF). The MIF is calculated using probes with different chemical properties to scan the molecule and identify regions showing favorable interaction energy (Goodford, 1985).
The initial MIFs generated may contain up to 100,000 nodes. Therefore, the second step of GRIND reduces this set of nodes to focused regions of greatest favorable interaction energies. Initial implementation of GRIND used a Fedorov-like optimization algorithm (Fedorov, 1972) to reduce the number of nodes to several hundred by considering both the intensity of a field and the mutual node-node distances between the selected nodes. In the second iteration of GRIND (GRIND-2), this method was replaced with a new algorithm called AMANDA (Duran et al., 2008). Although the original GRIND requires users to define the number of nodes to extract per molecule, AMANDA allows GRIND-2 to automatically adjust the number of nodes per compound. After a prefiltering step in which all nodes failing an energy cutoff are removed, every atom in the molecule is assigned a set of nodes and the number of nodes to extract per atom is calculated using a weighting factor and function that automatically assigns more nodes to larger regions. The node selection uses a recursive technique that is designed to initially assign selection weight to energy values. As the iterations continue through lower energy nodes, however, the internode distances become more important than the individual energy score of each node.
The final step of GRIND-2 (and GRIND) encodes this set of nodes into descriptors using auto- and cross-correlation methods. Pairs of interaction energies are multiplied and only the greatest product is retained for each internode distance. This is called maximum auto- and cross-correlation and allows for GRIND-2 (and GRIND) to contain information that directly correlates with the initial molecular structure.
GRIND-PP (Duran et al., 2009) improves GRIND-2 by removing much of the inherent repetition in the calculated descriptors. Structural features are repeated across many GRIND-2 variables and this can artificially assign importance to some structural features while reducing computational efficiency (Pastor, 2006). Principle properties replace the original variables in GRIND and are calculated using principle component analysis. These variables are linear combinations of the original variables selected to explain as much of the variance in the original set of variables as possible.
Comparative field molecular analysis (CoMFA) (Cramer et al., 1988) is a 3D-QSAR technique that aligns molecules and extracts aligned features that can be related to biologic activity. This method focuses on the alignment of molecular interaction fields rather than the features of each individual atom. CoMFA was established over 20 years ago as a standard technique for constructing 3D models in the absence of direct structural data of the target. In this method, molecules are aligned based on their 3D structures on a grid and the values of steric (Van der Waals interactions) and electrostatic potential energies (Coulombic interactions) are calculated at each grid point. Comparative molecular similarity indices (CoMSIA) is an important extension to CoMFA. In CoMSIA, the molecular field includes hydrophobic and hydrogen-bonding terms in addition to the steric and Coulombic contributions. Similarity indices are calculated instead of interaction energies by comparing each ligand with a common probe and Gaussian-type functions are used to avoid extreme values (Klebe et al., 1994). One important limitation to these methods, however, is that their applicability is limited to static structures of similar scaffolds while neglecting the dynamical nature of the ligands (Acharya et al., 2011).
B. Molecular Fingerprint and Similarity Searches
Molecular fingerprint-based techniques attempt to represent molecules in such a way as to allow rapid structural comparison in an effort to identify structurally similar molecules or to cluster collections based on structural similarity. These methods are less hypothesis driven and less computationally expensive than pharmacophore mapping or QSAR models (see sections III.C and III.E). They rely entirely on chemical structure and omit compound known biologic activity, making the approach more qualitative in nature than other LB-CADD approaches (Auer and Bajorath, 2008). Additionally, fingerprint-based methods consider all parts of the molecule equally and avoid focusing only on parts of a molecule that are thought to be most important for activity. This is less error prone to overfitting and requires smaller datasets to begin with. However, model performance suffers from the influence of unnecessary features and the often narrow chemical space evaluated (Auer and Bajorath, 2008). Despite this drawback, 2D fingerprints continue to be the representation of choice for similarity-based virtual screening (Willett, 2006). Not only are these methods the computationally least expensive way to compare molecular structures (Hutter, 2011), but their effectiveness has been demonstrated in many comparative studies (Willett, 2006).
1. Similarity Searches in LB-CADD.
Fingerprint methods may be used to search data bases for compounds similar in structure to a lead query, providing an extended collection of compounds that can be tested for improved activity over the lead. In many situations, 2D similarity searches of data bases are performed using chemotype information from first generation hits, leading to modifications that can be evaluated computationally or ordered for in vitro testing (Talele et al., 2010). Bologa et al. (2006) used 2D fingerprint and 3D shape-similarity searches to identify novel agonists of the estradiol receptor family receptor GPR30. Estrogen is an important hormone responsible for many aspects of development of physiology of tissues (Hall et al., 2001; Osborne and Schiff, 2005). The GPCR GPR30 has recently been shown to bind estrogen with high affinity and its specific role in estrogen-regulated signaling is being studied (Revankar et al., 2005). This group used virtual screening to identify compounds selective for GPR30 that could be used to study this target. 10,000 molecules provided by Chemical Diversity Laboratories were enriched with GPCR binding ligands and screened for fingerprint-based similarity to the reference molecule 17β-estradiol. Fingerprints used were Daylight and MDL and similarities were scored using Tanimoto and Tversky scores. The top 100 ranked hits were selected for biologic testing and a first-in-class selective agonist with a Ki of 11 nM for GPR30 was discovered (Bologa et al., 2006).
Stumpfe et al. (2010) used SecinH3 and analogs as reference compounds for a combined fingerprint and fingerprint-based support vector machine modeling screen aimed at inhibitors targeting the multifunctional cytohesins. Cytohesins are small guanine nucleotide exchange factors that stimulate Ras-like GTPases, which control various regulatory networks implicated in a variety of diseases (Klarlund et al., 1997; Ogasawara et al., 2000; Fuss et al., 2006). For example, cytohesin-1 has been shown to be involved in MAPK signaling in tumor cell proliferation and T-helper cell activation (Kliche et al., 2001; Perez et al., 2003), and cytohesin-3 was identified as an essential component of the phosphatidylinositol 3-kinase-based insulin signaling in liver cells (Fuss et al., 2006; Hafner et al., 2006). The group screened approximately 2.6 million compounds in the ZINC data base (Irwin and Shoichet, 2005), and the top 145 candidates were selected for biologic testing. Of those tested, 40 compounds showed measurable activity, and 26 were more potent than SecinH3 (Stumpfe et al., 2010).
Ijjaali et al. (2007) created 2D pharmacophoric fingerprints using a query dataset of 19 published T-type calcium channel blockers. T-type calcium channels underlie the generation of rhythmical firing patters in the central nervous system and have been implicated in the pathologies of epilepsy and neuropathic pain (Huguenard and Prince, 1992; Perez-Reyes, 2003; Bourinet and Zamponi, 2005). Specifically, T-type calcium channel 3.2 has been identified as a promising target for novel analgesic drugs for pathologic pain syndromes (Bourinet and Zamponi, 2005). A data base of two million compounds was collected from various commercial catalogs and filtered for drug-like qualities, uniqueness, and standardization. The group used ChemAxon’s PF and CGC GpiDAPH3 fingerprints and tested a subset of 38 unique hits biologically. Sixteen hits showed more than 50% blockade of CaV3.2-mediated T-type current. These compounds proved to be an interesting collection of T-type calcium channel blockers. Some showed reversible inhibition, whereas others resulted in irreversible inhibition, and one of the compounds caused alterations in depolarization/repolarization kinetics (Ijjaali et al., 2007).
In addition to the enrichment of lead compound population, fingerprints are also used to increase molecular diversity of test compounds. Fingerprints can be used to cluster large libraries of hits to allow the sampling of a wide range of compounds without the need to sample the entire library. In this case, fingerprints are being used to optimize the sampling of diversity space. The Jarvis-Patrick method that calculates a list of nearest neighbors for each molecule has been shown to perform well for chemical clustering. Two structures cluster together if they are in each other's list of nearest neighbors, and they have at least K of their J nearest neighbors in common. The MDL keys also provide a way to eliminate compounds that are least likely to satisfy the drug-likeness criterion (McGregor and Pallai, 1997).
2. Polypharmacology: Similarity Networks and Off-Target Predictions.
Recently, chemical similarity measures such as Tanimoto coefficients are being used to generate networks capable of clustering drugs that bind to multiple targets in an effort to predict novel off-target effects. Keiser et al. (2009) used a similarity ensemble approach (SEA) (Keiser et al., 2007) to compare drug targets based on the similarity of their ligands. SEA predicts whether a ligand and target will interact using a statistical model to control for chemical similarity due to chance. Sets of ligands that interact with each target are compared by calculating Tanimoto coefficients based on standard 2D Daylight fingerprints (Daylight Chemical Information Systems, 2013) for each pair of molecules between two sets. Raw similarity scores between all pairs of ligand sets are calculated as the sum of all Tanimoto coefficients between the sets greater than 0.57. Because the probability of achieving Tanimoto coefficients greater than 0.57 increases with set size, this is normalized by expected similarity due entirely to chance. This model for random chemical similarity is achieved by randomly generating 300,000 pairs of molecule sets spanning logarithmic size intervals from 10 to 1000 molecules. Expectation scores are calculated based on raw scores and the probability of achieving the raw score by random chance and used to sequentially link ligand sets into a clustered map. Keiser et al. (2007, 2009) collected over 900,000 drug-target comparisons from 65,241 ligands and 246 targets in the MDL Drug Data Report data base (Schuffenhauer et al., 2002) to generate a target similarity network. Another drug data base, WOMBAT (Olah et al., 2005), included interactions not listed in the MDDR data base, and the authors tested the predictability of their networks by searching their networks for interactions found in WOMBAT but not MDDR. They found that 19% of the off-target effects listed in WOMBAT but not in MDDR were captured in their network. In addition to those found in MDDR and WOMBAT, 257 additional drug-target predictions were captured in their network, 184 of which had not been documented. The authors tested 30 of these undocumented predictions using radioligand competition assays and verified 23 interactions with binding constants less than 15 μM. Some of these interactions may help to explain well-known side effects. For example, the authors discovered an interaction between β-adrenergic receptors and selective serotonin reuptake inhibitors Prozac (fluoxetine) and Paxil (paroxetine). This may explain the selective serotonin reuptake inhibitors discontinuation syndrome seen with these drugs that are analogous to discontinuation syndrome seen with β-blockers.
Lounkine et al. (2012) used the SEA approach combined with adverse drug reaction (ADR) information to generate a drug-target-ADR network. This network was then used to predict off-target interactions that may explain specific ADRs. The authors experimentally tested 694 predictions and verified that 151 interactions showed IC50 values less than 30 μM. The clinical relevance of these off-target interactions was explored through the enrichment of target-ADR pairs within their network. For example, abdominal pain has been reported for 45 drugs that interact with COX-1, and based on their network, the ADR-target pair abdominal pain-COX-1 was enriched (represented in a greater degree within the network than average) 2.3-fold, reflecting a predicted correlation between abdominal pain and COX-1 interaction. Another target-ADR correlation is predicted for sedation and H1 interaction with an enrichment of 4.9.
3. Fingerprint Extensions.
Current research focuses improving fingerprint-based LB-CADD methods. As mentioned, one drawback is that all features of a query molecule are equally important for ranking candidate molecules, regardless of any effect of these features on the biologic activity at a target. Hessler et al. (2005) proposed a method that combines the advantages of similarity and pharmacophore searching on the basis of 2D structural representations only. In their proposed method, a set of query molecules is converted into a topological model (MTree) based on chemically reasonable matching of corresponding functional groups. This creates a topological map of the most similar fragments from a set of structurally diverse but active molecules, and conserved features are characterized by high similarity scores of the corresponding nodes in the MTree model (Hessler et al., 2005). Because of the low dependence on chemical substructures, they argue that the MTree model is especially useful for identification of alternative novel molecular scaffolds or chemotypes. Methods for forming multiple feature tree models and multiple feature tree scoring schemes are also presented.
C. Quantitative Structure-Activity Relationship Models
Quantitative structure-activity relationship (QSAR) models describe the mathematical relation between structural attributes and target response of a set of chemicals (Zhang, 2011). Classic QSAR is known as the Hansch-Fujita approach and involves the correlation of various electronic, hydrophobic, and steric features with biologic activity. In the 1960s, Hansch (1964) and others began to establish QSAR models using various molecular descriptors to physical, chemical, and biologic properties focused on providing computational estimates for the bioactivity of molecules. In 1964, Free and Wilson (1964) developed a mathematical model relating the presence of various chemical substituents to biologic activity (each type of chemical group was assigned an activity contribution), and the two methods were later combined to create the Hansch/Free-Wilson method (Free and Wilson, 1964; Tmej et al., 1998).
The general workflow of a QSAR-based drug discovery project is to first collect a group of active and inactive ligands and then create a set of mathematical descriptors that describe the physicochemical and structural properties of those compounds. A model is then generated to identify the relationship between those descriptors and their experimental activity, maximizing the predictive power. Finally, the model is applied to predict activity for a library of test compounds that were encoded with the same descriptors. Success of QSAR, therefore, depends not only on the quality of the initial set of active/inactive compounds but also on the choice of descriptors and the ability to generate the appropriate mathematical relationship. One of the most important considerations regarding this method is the fact that all models generated will be dependent on the sampling space of the initial set of compounds with known activity, the chemical diversity. In other words, divergent scaffolds or functional groups not represented within this “training” set of compounds will not be represented in the final model, and any potential hits within the library to be screened that contain these groups will likely be missed. Therefore, it is advantageous to cover a wide chemical space within the training set. For a comprehensive guide on performing a QSAR-based virtual screen, please see the review by Zhang (2011).
1. Multidimensional QSAR: 4D and 5D Descriptors.
Multidimensional QSAR (mQSAR) seeks to quantify all energy contributions of ligand binding including removal of solvent molecules, loss of conformational entropy, and binding pocket adaptation.
4D-QSAR is an extension of 3D-QSAR that treats each molecule as an ensemble of different conformations, orientations, tautomers, stereoisomers, and protonation states. The fourth dimension in 4D-QSAR refers to the ensemble sampling of spatial features of each molecule. A receptor-independent (RI) 4D-QSAR method was proposed by Hopfinger et al. (1997). This method begins by placing all molecules into a grid and assigning interaction pharmacophore elements to each atom in the molecule (polar, nonpolar, hydrogen bond donor, etc.). Molecular dynamic simulations are used to generate a Boltzmann weighted conformational ensemble of each molecule within the grid. Trial alignments are performed within the grid across the different molecules, and descriptors are defined based on occupancy frequencies within each of these alignments. These descriptors are called grid cell occupancy descriptors. A conformational ensemble of each compound is used to generate the grid cell occupancy descriptors rather than a single conformation.
5D-QSAR has been developed to account for local changes in the binding site that contribute to an induced fit model of ligand binding. In a method developed by Vedani and Dobler (2002), induced fit is simulated by mapping a “mean envelope” for all ligands in a training set on to an “inner envelope” for each individual molecule. Their method involves several protocols for evaluating induced-fit models including a linear scale based on the adaptation of topology, adaptations based on property fields, energy minimization, and lipophilicity potential. By using this information, the energetic cost for adaptation of the ligand to the binding site geometry is calculated.
2. Receptor-Dependent 3D/4D-QSAR.
Although QSAR methods are especially useful when structural information regarding target binding site is not available, QSAR methods that specifically include such information have been developed. One method, known as free energy force field 3D-QSAR trains a ligand-receptor force field QSAR model that describes all thermodynamic contributions for binding (Pan et al., 2003). A 4D-QSAR version of free energy force-field has also been developed to apply this method to the RI-4D-QSAR methods described above (Pan et al., 2003). Structurally, the analysis is focused solely on the site of interaction between the ligand and target, and all atoms of interest are assigned partial charges. Molecular dynamic simulations are applied to these structures to generate a conformational ensemble following energy minimization. This approach avoids any alignment issues present in the RI-4D-QSAR method, because the binding site constrains the three-dimensional orientations of the ligands. The conformation ensembles of receptor-ligand complexes generated are placed in a similar grid-cell lattice as used in RI-4D-QSAR, and occupancy profiles are calculated to generate receptor-dependent 4D-QSAR models. When tested alongside RI-4D-QSAR against a set of glucose analog inhibitors of glycogen phosphorylase, predictability of receptor-dependent 4D-QSAR models outperformed those of RI-4D-QSAR (Pan et al., 2003).
3. Linear Regression and Related Methods.
Linear models used include multivariable linear regression analysis (MLR), principal component analysis (PCA), or partial least square analysis (PLS) (Acharya et al., 2011). MLR computes biologic activity as a weighted sum of descriptors or features. The method requires typically 4 or 5 data points for every descriptor used. PCA increases the efficiency of MLR by extracting information from multiple variables into a smaller number of uncorrelated variables. Analysis of results is, however, not always straightforward (Wold et al., 1987; Kubinyi, 1997). It can be applied with smaller sets of compounds than MLR. PLS combines MLR and PCA and extracts the dependent variable (biologic activity) into new components to optimize correlations (Zheng and Tropsha, 2000). PCA or PLS are commonly used for developing models for the molecular interaction field algorithm CoMFA and CoMSIA (Acharya et al., 2011). Advantage of these models is that they can be trained rapidly using the tools of linear algebra. The major drawback is that chemical structure often relates with biologic activity in a non-linear fashion.
4. Nonlinear Models Using Machine Learning Algorithms.
Artificial neural networks (ANNs) are one of the most popular nonlinear regression models applied to QSAR-based drug discovery (Livingstone, 2008). These models belong to the class of self-organizing algorithms in which the neural network learns the relationship between descriptors and biologic activity through iterative prediction and improvement cycles (Acharya et al., 2011). A major drawback of neural networks is the fact that they are sensitive to overtraining, resulting in excellent performance within the training set but reduced ability to assess novel compounds. Therefore, care is taken to always measure ANN performance on “independent” datasets not used for model generation.
SVM is a kernel-based supervised learning method that was introduced by Vapnik and Lerner (Vapnik and Lerner, 1963; Boser et al., 1992). It is based on statistical learning theory and the Vapnik-Chervonenkis dimension (Blumer et al., 1989; Vapnik, 1999) and seeks to divide sets of patterns (molecules described with descriptors) based on their classification (biologic function). Once this separation is performed on a training dataset, novel patterns can be classified based on which side of the boundary they fall. The simplest form of separation can be imagined as a straight line down the center of a graph with the two classes clustered in opposite corners of the graph. Because there are many different lines that can be defined to separate these classes, SVM is described as a maximal margin classifier as it seeks to define the hyperplane with the widest margin between these two classes. The patterns (compounds) that line the closest border of each class define the two hyperplanes separated by that margin. These patterns (molecules) are known as support vectors and represent the maximal margin solution and are used to predict classes for novel unclassified patterns. All patterns that lie further from these boundaries are not support vectors and have no influence on the classification of novel patterns. Hyperplanes defined by the lowest number of support vectors are preferred. The solution is a parallel decision boundary that lies equidistant from the two hyperplanes defined by their respective support vectors (Ivanciuc, 2007; Boyle, 2011; Liang, 2011).
Ideally, the margin between hyperplanes contains no patterns (molecules). However, to account for noise within datasets and other issues that prevent a linear solution from being reached, a soft-margin classifier is used that allows for misclassification of some data and the existence of patterns within the margin between hyperplanes. In this approach, a penalization constant can be adjusted, with higher values stressing classification accuracy and lower values providing more flexibility.
SVM was initially designed for datasets that could be separated linearly. However, especially in CADD application, this is not always possible. Therefore, SVM incorporated a high-dimensional space in which linear classification was once again possible. This involves the preprocessing of input data using feature functions where the input variables are mapped into a Hilbert space of finite or infinite dimension (Ivanciuc, 2007). Although it cannot be predicted which feature functions will allow for linear classification because the input vector is mapped into higher space, this becomes more possible. This strategy, however, must be offset by the fact that higher dimensional space creates more computational burden and contributes to overfitting (Cristianini and Shawe-Taylor, 2000).
SVM utilizes kernel functions to ease the computational demand imposed by the existence of higher dimensional data. These special nonlinear functions combine the feature functions in a way that avoids explicit transformation and preprocessing using feature functions (Ivanciuc, 2007). In other words, the higher dimensional space that allows for linear separation does not need to be dealt with directly.
A kernel is essentially a function in which the solution for two inputs is equal to the dot product of their mapping from input space to Hilbert space. Based on this fact, any novel kernels a researcher seeks to develop must be a dot product in a mapped feature space. This can be tested mathematically applying Mercer’s condition (Cristianini and Shawe-Taylor, 2000). The definition of new kernels, however, is not usually necessary because multiple useful kernels have already been well established for different problem types. Which kernel is necessary for any given problem cannot be predicted but is generally best selected a priori by researching which kernels have been successfully used in similar applications. It is not recommended to select the best kernel based on performance with the dataset being researched, because this can often lead to overfitting and poor generalizability. Some of the most commonly used kernels include the linear (dot) kernel, used mainly as a test of nonlinearity and reference for classification improvement after the application of nonlinear kernels; the polynomial kernel, which can be adjusted based on its degree to allow for larger feature space; radial basis function kernel; analysis of variance kernel; Fourier series kernel; spline kernel; additive kernel; and tensor product kernel. Addition, multiplication, and composition of these kernels all result in valid kernels (Ivanciuc, 2007). When implementing a novel kernel function, however, the researcher must ensure that it is the dot product in a feature space for some mapping. This condition can be tested by applying Mercer’s condition (Cristianini and Shawe-Taylor, 2000). It should be considered, however, that overfitting can be induced with more complex kernel functions.
Several methods of SVM optimization have been considered. SVM parameter optimization is accomplished by solving the quadratic programming problem with a termination condition called the Klarush-Kuhn-Tucker condition that defines when parameters are at their minima. This can be computationally demanding and difficult to implement. Therefore, decompositional methods have been used to discard all zero parameters (Vapnik, 2006). The sequential minimization optimization algorithm is a commonly used alternative introduced by Platt (1999). This method breaks the overall quadratic programming problem into subproblems and solves the smallest possible optimization problem at every stop involving only two parameters. One problem with the sequential minimization optimization, however, is that it can result in selection of support vectors that include more than those necessary for the optimal model. Researchers have found that identical solutions can be achieved even after several of these support vectors have been removed (Zhan and Shen, 2005). Because the time needed to predict a pattern classification with an SVM model is dependent on the number of support vectors, it is beneficial to eliminate unnecessary or redundant support vectors. Zhan and Shen (2005) describe a four-step method for removing unnecessary support vectors. Once the SVM has completed training, the support vectors that contribute to the most curvature along the hypersurface are removed. The SVM model is then retrained and the hypersurface is further approximated with a subset of support vectors.
Decision tree learning is a supervised learning algorithm that works by iteratively grouping the training dataset into small and more specific groups. The resulting classification resembles a tree in which each feature is broken into different values and each of these values is subsequently divided based on values of a different feature. The order in which features are divided is usually based on an information gain (difference between information before and after the branching) parameter with the highest valued features appearing first (Mitchell, 1997; Han and Kamber, 2006). Various methods are used to sort the features, with the overall goal of the smallest possible decision tree providing the best performance. C4.5 is a widely used decision tree algorithm that calculates information gain based on information entropy (Quinlan, 1993; Fukunishi, 2009). The information entropy of a given classification that can divide the dataset into two classes is calculated based on the number of compounds in either class. The information entropy of the system when dividing the dataset into two subsets using a specific feature is calculated based on the number of compounds from each class in either of the feature subsets. Finally, the information gain for that specific feature is calculated as the difference between the information entropy of the classification and the information entropy of the system.
Once the decision tree has been optimized for the training set, new compounds can be classified by applying their descriptors to the decision tree and activities can be predicted based on which subset they fall into and the activities of the training compounds that are contained in that subset.
5. Quantitative Structure-Activity Relationship Application in Ligand-Based Computer-Aided Drug Design.
QSAR has been used to screen for novel therapeutics in the same way both pharmacophore models and fingerprint similarity methods have been applied to virtual libraries. Casañola-Martin et al. (2007) used Dragon (Talete S.R.L., Italy) software to define descriptors for tyrosinase inhibitors. Tyrosinase is a copper-containing enzyme that catalyzes two reactions in the melanin biosynthesis pathway (Sanchezferrer et al., 1995; Briganti et al., 2003). Altered melanin synthesis is found in multiple disease states including hyperpigmentation, melisma, and age spots. Additionally, this protein has been implicated in dopamine neurotoxicity in diseases such as Parkinson’s disease (Xu et al., 1997). Descriptors were generated using a highly variable training set of 245 active tyrosinase inhibitors and 408 inactive molecules. These descriptors include constitutional, topological, BCUT, Galvez, topological charge, 2D autocorrelations, and empirical properties and descriptors. Seven models were created using linear discriminant analysis. In vitro testing revealed their most potent inhibitor with an IC50 of 1.72 µM. This presents a more potent inhibition of tyrosinase than the current reference drug l-mimosine (IC50 = 3.68 μM) (Casañola-Martin et al., 2007).
Mueller et al. (2012) used ANN QSAR models to identify novel positive and negative allosteric modulators of mGlu5. This receptor has been implicated in neurologic disorders including anxiety, Parkinson’s disease, and schizophrenia (Gasparini et al., 2008; Conn et al., 2009). For the identification of positive allosteric modulators, they first performed a traditional high-throughput screen of approximately 144,000 compounds. This screen yielded a total of 1356 hits, a hit rate of 0.94%. The dataset from this HTS was then used to develop a QSAR model that could be used in a virtual screen. To generate the QSAR model, a set of 1252 different descriptors across 35 categories was calculated using the ADRIANA (Molecular Networks GmbH, Erlangen, Germany) software package. The descriptors included scalar, 2D, and 3D descriptor categories. The authors iteratively removed the least-sensitive descriptors to create the optimal set. This final set included 276 different descriptors, including scalar descriptors such as molecular weight up to 3D descriptors, including the radial distribution function weighted by lone-pair electronegativity and π electronegativity. A virtual screen was performed against approximately 450,000 commercially available compounds in the ChemBridge data base. Eight hundred twenty-four compounds were tested experimentally for the potentiation of mGlu5 signaling. Of these compounds, 232 were confirmed as potentiators or partial agonists. This hit rate of 28.2% was approximately 30 times greater than that of the original HTS, and the virtual screen took approximately 1 hour to complete once the model had been optimized (Fig. 16) (Mueller et al., 2012).
Fig. 16.
QSAR-based virtual screening of mGlu5 negative allosteric modulators yields lead compounds that contain substructure combinations taken across several known actives used for model generation. Adapted from Mueller et al. (2012).
In a separate study, Mueller et al. (2010) used a similar approach to identify negative allosteric modulators for mGlu5. Rodriguez et al. (2010) previously performed a traditional HTS screen of 160,000 compounds for allosteric modulators of mGlu5 and found 624 antagonists. The QSAR model was used to virtually screen over 700,000 commercially available compounds in the ChemDiv Discovery data base. Hits were filtered for drug-like properties, and fingerprint techniques were used to remove hits that were highly similar to known actives to identify new chemotypes. Seven hundred forty-nine compounds were tested in vitro, and 27 compounds were found to modulate mGlu5 signaling. This hit rate of 3.6% was a significant increase over the 0.2% hit rate of the traditional HTS screen. The most potent of the compounds showed in vitro IC50 values of 75 and 124 nM, respectively, and contained a previously unidentified scaffold. After analog synthesis and stability optimization, the experimenters tested the effect of their best lead in vivo against two behaviors known to involve mGlu5: operant sensation seeking behavior (Olsen et al., 2010) and the burying of foreign objects in deep bedding (Deacon, 2006). Both behaviors were found to be inhibited given intraperitoneal administration of their lead analog.
In addition to predicting the behavior of novel compounds within a virtual library, QSAR has been used to improve compound libraries used in traditional HTS. Although many chemical libraries are constructed in a combinatorial manner, it was reported that the chemical space covered by combinatorially synthesized libraries is different from the chemical space of known drugs and natural products. Because of this, along with the overall chemical space estimated to be more than 1060, it is critical to design HTS compound collections to cover the widest possible space of drug-like chemicals (Bohacek et al., 1996). QSAR can be used to direct combinatorial library synthesis for constructing libraries that will later be screened against targets of a particular class or classes. This allows the researcher to cover a wide range of chemical space that has been enriched with compounds more likely to be hits for their target of interest. This strategy has been used to create several libraries directed at particular target types. For example, Erickson et al. (2010) generated seven libraries meant to be screened for kinase inhibitors. The group initially generated a fragment library from over 1400 known kinase inhibitors. Potential scaffold fragments were identified using substructure and similarity searching, and break points for fragment generation were guided with structure-based pharmacophores. These data were also used to train SVM-based QSAR models. Compounds were generated from this fragment library, and their activity was predicted using the QSAR model. The final library included compounds predicted to have some activity against kinase targets and showed good hit rates against six different kinases. These compounds, however, did not exhibit the desirable specificity, and the authors suggested that more specific pharmacophores may be necessary (Erickson et al., 2010). Rolland et al. (2005) used a similar strategy to design a library that could be screened with GPCR targets. They collected binding profiles for 1939 compounds against 40 GPCR targets and used this information to train a global QSAR model. The model was used to screen for putative GPCR active compounds within a library of 16,000 compounds. Additionally, 50 focused libraries of 200 compounds each were generated using medicinal-chemistry-based scaffolds guided by the QSAR model. The researchers found significant hit rates within these libraries not only among the original panel of GPCRs but against previously untested GPCR targets (Rolland et al., 2005).
QSAR has also been applied to de novo drug design techniques when structural information regarding the target is unknown. Descriptor and model generation is performed and is used to score the de novo-generated molecules in place of other structure-based scoring techniques such as docking. Most commonly, compound generation involves iterative algorithms in which structures are repeatedly modified and their biologic activities are estimated using QSAR models. In the simplest case, modifications can be achieved by randomly swapping parts of the structure such as functional groups. Ligand-based de novo drug design, however, is less practiced than structure-based de novo design because of the inherent challenges of accurately evaluating a new molecule in the absence of the receptor structure. To address the challenge of scoring the newly generated molecules, similarity based methods have been applied in addition to QSAR models (Brown et al., 2004).
Feher et al. (2008) used five selective norepinephrine reuptake inhibitors as a training set to generate 2200 molecules using a combination of structural similarity, 2D pharmacophore similarity, and properties to drive the evolution. One of the top scoring compounds was found to be highly active and has been selected as a lead compound in a project at Neurocrine (Feher et al., 2008).
Golla et al. (2012) applied QSAR-based methods to the design of novel chemical penetration enhancers (CPEs) to be used in transdermal drug delivery. This group used a genetic algorithm to design novel CPEs. In this paradigm, new molecules are generated based on crossover and mutation operations randomly applied to candidates. All generated molecules are scored based on the QSAR model, and predicted property values and the highest scoring molecules are retained for new rounds of evolution. Two hundred seventy-two CPEs were used to both generate the QSAR model and provide seed molecules for the genetic algorithm. The QSAR model was created using sequential regression analysis and heuristic analysis using CODESSA and contained a final set of 40 descriptors that optimally predicted properties, including skin penetration coefficient, logP, melting point, skin sensitization, and irritation. The top scoring molecules were validated experimentally for permeation and toxicity using Franz Cell with porcine skin and HPLC analysis as well as toxicity effects on human foreskin fibroblasts and porcine abdominal skin. The study resulted in the identification of 18 novel CPEs, four of which showed minimal or no toxic effects (Golla et al., 2012).
Hoeglund et al. (2010) used QSAR modeling combined with synthetic optimization in a follow up to their most potent hit from a 2008 in silico screen for inhibitors of autotaxin. Autotaxin is an autocrine motility factor and has been linked to cancer progression, multiple sclerosis, obesity, diabetes, Alzheimer’s disease, and chronic pain through the production of lysophosphatidic acid (LPA) (Kawagoe et al., 1997; Euer et al., 2002; Baumforth et al., 2005; Boucher et al., 2005; Umemura et al., 2006; Inoue et al., 2008). Analogs of the lead compound were tested, and 4 of the 30 exhibited IC50 less than or equal to the lead. The most potent compound showed 3-fold higher affinity for autotaxin than the lead, whereas another compound showed twofold higher affinity (Hoeglund et al., 2010).
CoMFA and CoMSIA 3D-QSAR methods have also been used to predict novel therapeutic compounds for a variety of disease targets. Ke et al. (2013) generated CoMFA and CoMSIA models using 66 previously discovered pyrazole- and furanopyrimidine-based aurora kinase inhibitors (Coumar et al., 2009, 2010a,b). Aurora kinase A is a serine/threonine kinase involved in mitosis (Li et al., 2010) that has been shown to be involved in various different forms of cancer (Agnese et al., 2007; Fu et al., 2007). By using the model that showed the best predictive performance, the group synthesized a novel compound (compound 67). This compound was tested in vitro and displayed an IC50 of 25 nM against aurora kinase A. Additionally, compound 67 displayed antiproliferative activity with an IC50 of 23 nM against the HCT-116 colon cancer cell line.
Chai et al. (2011) used 26 previously identified anti-hepatitis B (HBV) compounds (Chai et al., 2006; Zhao et al., 2006a,b) to generate CoMFA models based on steric and electrostatic fields and CoMSIA models based on steric, electrostatic, hydrophobic, and H-bond acceptor fields. Three compounds were designed using these models and subsequently tested against replication of HBV DNA in HBV-infected 2.2.15 cells. The most potent compound displayed an IC50 of 3.1 μM, whereas the other two showed IC50 values of 5.1 and 3.3 μM. These compounds were comparatively more potent than the control lamivudine, which displays an IC50 of 994 μM.
Jiao et al. (2010) generated CoMFA models using 38 styrylquinoline derivatives in an effort to understand and design potential HIV integrase inhibitors. Their model suggested that a bulky group near the carboxyl group at C-7 in the quinolone ring may confer increased inhibition. Additionally, the presence of an H-bonding donor is favorable near the C-7 atom. On the basis of these predictions, they designed several compounds that were tested against purified HIV Integrase to determine inhibitory activity on the strand transfer reaction of integrase. Four of these compounds showed higher inhibitory activity than their positive control baicalein (Sigma-Aldrich, St. Louis, MO).
Over the past several decades, over 18,000 QSAR models have been reported for a variety of targets with a variety of descriptors. C-QSAR was used to generate a comprehensive database of QSAR models (Kurup, 2003). This collection has provided not only access to models for novel applications, but allows the analysis of QSAR models to identify challenges for the field. Kim (2007) examined the C-QSAR data base for outlier patterns, i.e., compounds that showed poor prediction when the average prediction for the model was good. They found that of over 47 QSAR models examined, the number of compounds scoring as outliers ranged from 3 to 36%. Twenty-six of the 47 datasets showed 20% or more compound outliers (Kim, 2007). They presented several theories as to why QSAR models are so sensitive to the generation of outliers. One possibility came from analysis of the RCSB protein databank (RCSB 2013) where they discovered examples where related analogs were shown to bind in very different poses. Another explanation offered was protein flexibility, leading to multiple binding modes and or binding sites on the same protein. These different binding modes/sites may reflect different structure-activity relationships for molecules within a given dataset. In other words, analogous compounds that do not share the same binding mode can present difficulties in the classifications of ligands (Kim, 2007).
D. Selection of Optimal Descriptors/Features
Hristozov et al. (2007) analyzed the performance of different descriptors across a range of benchmarking datasets and found that the performance of a particular descriptor was often dependent on the activity class. It was found that topological autocorrelation usually offers the best dimensionality/performance ratio. The fusion of the ranked lists obtained with RDF codes and 2D descriptor improved results because RDF codes, while giving similar results, covered different parts of the activity spaces under investigation (Hristozov et al., 2007). As a result, it is not possible to choose a small optimal set of descriptors independent of the problem; a custom-optimized descriptor set is needed for optimal performance of LB-CADD.
Excessive numbers of descriptors or features can add noise to a model, reducing its predictive power. Feature selection techniques remove unnecessary features to minimize the number of degrees of freedom of the model. Thus, the ratio of data points versus degrees of freedom increases, leading to models of increased predictive power. Techniques that have proven successful in QSAR modeling include selecting features by measures such as information gain (Kent, 1983) and F-score (Chen and Lin, 2006), sequential feature forward selection or feature backward elimination (Mao, 2004), genetic algorithm (Davis, 1991; Goodarzi et al., 2009), swarm optimization (Goodarzi et al., 2009), and input sensitivity analysis (Mueller et al., 2010).
Information gain measures the change of information entropy from the data distribution of two classes (active and inactive compounds) of one feature compared with the entropy of the feature overall. Thus, discriminatory power of the individual feature increases with information gain. An F-score is calculated that considers the mean and standard deviation of each feature across data classes. The higher the f-score value, the greater discriminatory power of that feature. Selecting features by individual benchmarks has the disadvantage that correlation between features is ignored. For example, let us assume a feature has a high information gain. However, if a second feature highly correlated is already part of the model, no improved model will result from adding the feature. More complex feature selection schemes address this limitation:
Sequential feature forward selection is a deterministic, greedy search algorithm. In each round, the best feature set from the previous round N appends a single feature from the pool of M remaining features and trains the M models using the N + 1 features. The best performing feature set from this round then advances to the next round. This continues until all features are used in a final feature set. The best performing model over all iterations is then chosen as the best feature set. This process is time consuming and not guaranteed to yield the optimal feature set; the single best performing feature will always be part of the model. However, there is no guarantee that it is needed. Feature backward elimination inverts the process starting from a model trained from all features, eliminating one after the other. Although the process is more robust in terms of identifying the optimal model, it also requires substantial computer time. Therefore, alternative approaches have been explored to optimize feature sets.
Genetic algorithms mimic the process of evolution to create an efficient search heuristic. This method uses a population of individuals (distinct feature sets) to encode candidate solutions. The initial individuals can be generated randomly. In each iteration, or generation, the fitness of each individual is evaluated, i.e., the predictive power of the derived LB-CADD model. This fitness function is the performance metric of a model trained using that individual as the feature set. Individuals are then selected based on the fitness and undergo recombination and/or mutation to form the next generation. The algorithm continues until a desired fitness score is achieved or a set number of generations has been completed.
Swarm optimization algorithms, such as ant colony optimization (Zhou et al., 2012), particle swarm optimization, and artificial bee colony optimization (Lv et al., 2012), are optimization techniques based on the organized behavior of social animals such as birds. The algorithm iteratively searches for a best solution by moving individuals around the search space guided by both the local best solution as well as the best solutions found so far in the entire population. The best overall solution is constantly updated, letting the swarm converge toward the optimal solutions.
Input sensitivity analysis seeks to combine speed of individual benchmark values with accuracy of methods that take correlation into account. First, a model is constructed using all features. Next, the influence of each feature on the model output is determined: Each feature xi is perturbed, and the change in output y is computed. This procedure numerically estimates the partial derivative of the output with respect to each input, a measure that is effective in selecting optimal descriptor sets (Mueller et al., 2010).
E. Pharmacophore Mapping
In 1998, the International Union of Pure and Applied Chemistry formally defined a pharmacophore as “the ensemble of steric and electronic features that is necessary to ensure the optimal supramolecular interactions with a specific biological target structure and to trigger (or to block) its biological response” (Wermuth, 2006). In terms of drug activity, it is the spatial arrangement of functional groups that a compound or drug must contain to evoke a desired biologic response. Therefore, an effective pharmacophore will contain information about functional groups that interact with the target, as well as information regarding the type of noncovalent interactions and interatomic distances between these functional groups/interactions. This arrangement can be derived either in a structure-based manner by mapping the sites of contact between a ligand and binding site or in a ligand-based approach. The former can be achieved by analyzing one or several cocrystal structures with lead or drug compounds bound and will not be discussed in more detail here. We focus on the latter, more challenging problem.
To generate a ligand-based pharmacophore, multiple active compounds are overlaid in such a way that a maximum number of chemical features overlap geometrically (Wolber et al., 2008). This can involve rigid 2D or 3D structural representations or, in more precise applications, incorporate molecular flexibility to determine overlapping sites. This conformational flexibility can be incorporated by precomputing the conformational space of each ligand and creating a general-purpose conformational model or conformations can be explored by changing molecule coordinates as needed by the alignment algorithm (Wolber et al., 2008). For example, one popular pharmacophore-generating software package, Catalyst (Accelrys, Inc., San Diego, CA), uses the “polling” algorithm (Smellie et al., 1995) to generate approximately 250 conformers that it uses in its pharmacophore generation algorithm (Acharya et al., 2011). In a study targeting HSP90α, Al-Sha’er and Taha (2010) used 83 known reference molecules to generate pharmacophore queries and identified 25 diverse inhibitors including three with IC50 values below 10 nM.
1. Superimposing Active Compounds to Create a Pharmacophore.
Molecules are commonly aligned through either a point-based or property-based technique. The point-based technique involves superposing pairs of points (atoms or chemical features) by minimizing Euclidean distances. These alignment methods typically use a root-mean-square distance to maximize overlap (Poptodorov et al., 2006). Property-based alignment techniques, on the other hand, use molecular field descriptors to generate alignments (Wolber et al., 2008). These fields define 3D grids around compounds and calculate the interaction energy for a specific probe at each point. The distribution of interaction energies is represented by Gaussian functions, and the degree of overlap between Gaussian functions of two aligned compounds is used as the objective scoring function to maximize alignment (Poptodorov et al., 2006). One popular field generation method for property-based alignments is GRID (Goodford, 1985).
Molecular flexibility is always an important consideration when aligning compounds of interest, and several approaches are used to most efficiently sample conformational space. These approaches include rigid, flexible, and semiflexible methods. Rigid methods require knowledge of the active conformation of known ligands and align only the active conformations. This is only applicable, however, when the active conformation is known with confidence. Semiflexible methods begin with pregenerated ensembles of static conformations to overlay, and flexible methods, being the most computationally expensive, perform conformational search during the alignment process, often using molecular dynamics or randomly sampling of rotatable bonds. Because the conformational space can increase substantially with an increase in the number of rotatable bonds, strategies are often used to limit the exploration of conformational space through the use of reference geometry (often an active ligand with low flexibility). This method is known as the active analog approach (Marshall et al., 1979).
2. Pharmacophore Feature Extraction.
A pharmacophore feature map is carefully constructed so as to balance generalizability with specificity. A general definition might categorize all functional groups having similar physiochemical properties (i.e., similar hydrogen-bonding behavior, ionizability) into one group, whereas specific feature definitions may include specific atom types at specific locations. More general feature definitions increase the population of compounds that match the pharmacophore. They allow the identification of novel scaffolds but also increase the ratio of false-positives. The level of feature definition generalizability is usually determined by the algorithm used to extract feature maps and through user-specified parameters. The most common features used to define pharmacophore maps are hydrogen bond acceptors and donors, acidic and basic groups, aliphatic hydrophobic moieties, and aromatic hydrophobic moieties (Acharya et al., 2011). Features are commonly implemented as spheres with a certain tolerance radius for pharmacophore matching (Wolber et al., 2008).
3. Pharmacophore Algorithms and Software Packages.
The most common software packages used for ligand-based pharmacophore generation include Phase (Dixon, Smondyrev et al., 2006), MOE (Chemical Computing Group, Quebec, Canada), Catalyst (Kurogi and Güner, 2001), LigandScout (Inte:Ligand, Vienna, Austria), DISCO (Martin et al., 1993), and GASP (Jones et al., 1995). These packages use different approaches to molecular alignment, flexibility, and feature extraction. Catalyst approaches alignment and feature extraction by identifying common chemical features arranged in certain positions in three-dimensional space. These chemical features focus on those expected to be important for interaction between ligand and protein and include hydrophobic regions, hydrogen-bond donors, hydrogen-bond acceptors, positive ionizable, and negative ionizable regions. Chemical groups that participate in the same type of interaction are treated as identical. Catalyst contains two algorithms that can be used for pharmacophore construction. HipHop is the simpler of the two algorithms and looks for common 3D arrangements of features only for compounds with a threshold activity against the target. It begins with best alignment of only two features (scored by RMS deviations) and continues expanding the model to include more features until no further improvements are possible. This method is only capable of producing a qualitative distinction between active and inactive predictions. HypoGen, on the other hand, uses biologic assay data such as IC50 values for active compounds as well as a set of inactive compounds. Initial pharmacophore construction in HypoGen is identical to HipHop but includes additional algorithms that incorporate inactive compounds and experimental values. These algorithms compare the best pharmacophore from the "HipHop" stage with the inactive compounds and features common to the inactive set are removed. Finally, HypoGen performs an optimization routine that attempts to improve the predictive power of the pharmacophore by making adjustments and scoring the accuracy in predicting the specific experimental activities (Güner, 2000; Kurogi and Guner, 2001). This results in models that are capable of quantitative predictions that can predict specific levels of activity. Ten different models are created following a simulated annealing optimization (Chang and Swaan, 2006). Both Catalyst methods incorporate molecular flexibility by storing compounds as multiple conformations per molecule. The Poling algorithm published by Smellie et al. (1995) is used to increase the conformational variation within the set of conformations per molecule. This allows Catalyst to cover the greatest extent of conformational space while keeping the number of conformations at a minimum.
Phase approaches alignment and feature extraction using a tree-based partitioning algorithm and an RMS deviation-based scoring function that considers the volume of heavy atom overlap. It incorporates molecular flexibility through a preparation step where conformational space is sampled using a Monte Carlo or torsional search (Poptodorov et al., 2006).
DISCO regards compounds as sets of interpoint distances between heavy atoms containing features of interest. Alignments are based on the spatial orientation of common point among all active compounds. DISCO considers multiple conformations that have been prespecified by the user during the alignments and uses a clique-detection algorithm for scoring alignments (Güner, 2000).
GASP uses a genetic algorithm with iterative generations of the best models for pharmacophore construction (Jones et al., 1995). Flexibility is handled during the alignment process through random rotations and translations. Conformations are optimized by fitting them to similarity constraints and weighing the conformations that fit these constraints more than conformations that do not (Chang and Swaan, 2006).
Different software packages can produce different results for the same datasets, and their strengths and weaknesses should be considered prior to any application. For example, Catalyst only permits a single bonding feature per heavy atom, whereas LigandScout allows a hydrogen-bond donor or acceptor to be involved in more than one hydrogen-bonding interaction (Wolber et al., 2008). MOE, on the other hand, allows a more customizable approach to hydrogen-bonding features. Lipophilic areas are generally represented as spheres located on hydrophobic atom chains, branches, or groups in a similar manner across software packages but with slight nuances. Although subtle, these differences have important consequences on prediction models. Additionally, software packages that do not attach a hydrophobic feature to an aromatic ring are unable to predict that an aromatic group may be positioned in a lipophilic binding pocket (Wolber et al., 2008). The level of customizability also differs across pharmacophore software packages and can influence predictions. Catalyst allows the specification of one or more chemical groups that satisfy a particular feature, whereas Phase allows not only matching chemical groups but also a list of exclusions for a given feature. MOE offers a level of customization that allows the user to implement entirely novel pharmacophore schemes as well as modification of existing schemes. However, this requires additional levels of expertise to program (Wolber et al., 2008). For a comprehensive analysis of the differences between commercial pharmacophore software packages, please see the 2008 review by Wolber et al. (2008) and a 2002 comparison of Catalyst, DISCO, and GASP by Patel et al. (2002).
Ligand-based pharmacophore methods have been used for the discovery of novel compounds across a variety of targets. New compounds can have activity in the micromolar and nanomolar range and reflect proof of concept with in vivo disease models. Al-Sha’er and Taha (2010) used a diverse set of 83 known Hsp90-α inhibitors and the HypoGen module of Catalyst to generate a pharmacophore model. Hsp90-α is a molecular chaperone that is involved in protein folding, stability, and function (Prodromou and Pearl, 2003). By interacting with many oncogenic proteins, it has been shown to be a valid anticancer drug target (Chiosis et al., 2006; Solit and Rosen, 2006). The pharmacophore model was used to screen the NCI list of compounds (238,000) using the “best flexible” search option. The top 100 hits were evaluated in vitro, and their most potent compound had an IC50 of 25 nM (Al-Sha'er and Taha, 2010).
Schuster et al. (2011) used three steroidal inhibitors and two nonsteroidal inhibitors of 17β-HSD3 and Catalyst to create a pharmacophore model that was used to screen for novel 17β-HSD3 inhibitors. Hydroxysteroid dehydrogenases (HSD3) catalyze the oxidoreduction of alcohols or carbonyls and the final step in male and female sex hormone biosynthesis. Therefore, these enzymes are suggested therapeutic targets for control of estrogen- and androgen-dependent diseases such as breast and prostate cancer, acne, and hair loss (Poirier, 2009). Eight commercial data bases were screened, and the 15 top scoring hits were tested in vitro at 2 µM; five were verified to be inhibitors of 17β-HSD3. The most potent compound was able to inhibit 17β-HSD3 by 67.1% at 2 µM (Schuster et al., 2011).
Noha et al. (2011) developed 5-point pharmacophore models using the HipHop algorithm of Catalyst based on a training set of compounds with IC50 < 100 nM against IKK-β as potential anti-inflammatory and chemosensitizing agents. The authors used 128 active and 44 inactive compounds to develop a pharmacophore model (Noha et al., 2011). Their model was further refined with exclusion volume spheres and shape constraints to improve the scoring of compounds in their virtual high-throughput screen against the National Cancer Institute molecular data base. Ten compounds were selected, and the most potent compound (NSC719177, C26H31NO4) showed inhibitory activity against IKK-β in a cell-free in vitro assay with IC50 of 6.95 µM. Additionally, this compound inhibited NF-κB activation induced by tumor necrosis factor-α in HEK293 cells with an IC50 of 5.85 µM (Noha et al., 2011).
Chiang et al. (2009) used the HypoGen module of Catalyst to generate four-feature pharmacophore models based on an indole series of 21 compounds that showed antiproliferative activity through the inhibition of tubulin polymerization/microtubule depolymerization. Disruption of microtubules during the mitotic phase of the cell cycle can induce cell-cycle arrest and apoptosis (Valiron et al., 2001). Therefore, inhibitors of tubulin polymerization are useful cancer treatments. One hundred thirty thousand compounds of the ChemDiv data base and in-house compound collection were screened, and the top 142 hits were tested in vitro. Four novel compounds were discovered with antiproliferative activity. The most potent compound displayed antiproliferative activity in human cancer KB cells with an IC50 of 187 nM. This compound also inhibited the proliferation of other cancer cell types, including MCF-7, NCI-H460, and SF-268, and demonstrated anticancer effects in a histoculture system. In vitro assays revealed that this compound inhibited tubulin polymerization with an IC50 of 4.4 µM (Chiang et al., 2009).
Doddareddy et al. (2007) generated a pharmacophore model containing three hydrophobic regions, one positive ionizable center, and two hydrogen bond acceptor groups for the identification of novel selective T-type calcium channel blockers. The most potent hit showed an IC50 of 100 nM (Annoura et al., 2002; Doddareddy et al., 2007). T-type calcium channels are involved in rhythmical firing patterns in the central nervous system and present therapeutic targets for the treatment of epilepsy and neuropathic pain (Ijjaali et al., 2007).
Lanier et al. (2007) generated pharmacophores containing five feature points using Catalyst and CombiCode (Deltagen Research Laboratories, San Diego CA) software and an exclusion sphere generated in MOE based on a training set of 100 active and 1000 inactive compounds. This model was used to guide and evaluate variations of a core molecule, leading them to a gonadotropin releasing hormone GnRH receptor antagonist with receptor affinity below 10 nM (Lanier et al., 2007). GnRH is involved in the regulatory pathways of follicle stimulating hormone and luteinizing hormone. It is a target for disease therapeutics, including endometriosis, uterine fibroids, and prostate cancer (Cheng and Leung, 2000; Huirne and Lambalk, 2001).
Roche and Rodriguez Sarmiento (2007) used known H3 antagonists to generate a pharmacophore model with four features including a distal positive charge, an electron-rich position, a central aromatic ring, and either a second basic amine or another aromatic. Histamine is a central modulator in the central and peripheral nervous systems through four receptors (H1–H4) (Hough, 2001). H3 is a presynaptic autoreceptor that modulates production and release of histamine and other neurotransmitters (Alguacil and Perez-Garcia, 2003). H3 antagonists have been studied in Alzheimer’s disease, attention deficit disorder, and schizophrenia (Witkin and Nelson, 2004). Additionally, it has been suggested to be involved in appetite and obesity (Hancock and Brune, 2005). This model was used in a de novo approach with the Skelgen software (Stahl et al., 2002) to generate novel compounds from fragment libraries that match the pharmacophoric restraints. They found a series of four compounds with high potency and selectivity for H3. Their most potent compound showed inverse agonist activity with an EC50 of 200 pM in a guanosine 5[prime]-O-(3-thio)triphosphate functional assay and a binding affinity Ki toward H3 of 9.8 nM (Roche and Rodriguez Sarmiento, 2007).
Chao et al. used (2007) pharmacophore-based design to take advantage of the therapeutic benefits of indole-3-carbinol (I3C) in the treatment of cancer. I3C is known to suppress proliferation and induce apoptosis of various cancer cells through the inhibition of Akt activation (Howells et al., 2002; Li et al., 2005). I3C, however, has a poor metabolic profile and low potency, likely due to the fact that its therapeutic behavior comes from only four of its metabolites. By overlaying these low-energy conformers of these four metabolites, Chao et al. (2007) was able to identify similar N-N′ distances and overlapping indole rings. This led them to design SR13650, which showed an IC50 of 80 nM. Tumor xenograft studies using MCF-7 cells revealed antitumor effects at 10 mg/kg for 30 days. Computational analysis was also applied to increase the bioavailability, and three compounds showed 45–60% tumor growth inhibition in vivo compared with the 26% growth inhibition of SR13650. SR13668 was the most potent compound and also displayed antitumor effects in other xenograft models. In vitro, SR13668 was shown to inhibit Akt activation by blocking growth-factor stimulated phosphorylation and showed favorable toxicological profiles (Chao et al., 2007). This drug is currently in phase 0 trials for the treatment of cancer (Reid et al., 2011) (Fig. 17).
Fig. 17.
SR13668, an anticancer therapeutic was discovered using ligand-based pharmacophore screening based on active components of indole-3-carbinol. Adapted from Chao et al. (2007).
Dayam et al. (2008) used pharmacophore modeling in an effort to identify novel HIV-1 integrase (IN) inhibitors. IN is the third viral enzyme in HIV and is responsible for integration of viral DNA into host cell chromosomes through 3′-processing and strand transfer (Gordon et al., 2007; Palmisano, 2007). This model was created with the HipHop algorithm within Catalyst and was based on the quinolone 3-carboxylic acid class of IN inhibitors that show IC50 values ranging from 43.5 to 7.2 nM and EC50 against HIV-1 replication of 805 to 0.9 nM (Sato et al., 2006). The final pharmacophore hypothesis consisted of four features including a negatively ionizable feature, hydrogen-bond acceptor, and two hydrophobic aromatic features (Fig. 18). Three hundred sixty-two thousand two hundred sixty commercially available compounds were screened and 56 selected for in vitro evaluation. Eleven of those tested inhibited the IN catalytic activity with an IC50 value < 100 µM. Five compounds had an IC50 less than 20 µM, and the most potent compound inhibited both the 3′-processing (IC50 14 µM) as well as strand transfer activities (IC50 5 µM) of IN (Dayam et al., 2008). Mugnaini et al. (2007) created a pharmacophore model using 30 known inhibitors of the 3′-processing step of HIV-1 IN and screened the ASINEX gold data base of over 200,000 compounds for inhibitors of IN. Twelve hits were tested in vitro and one compound was discovered with a novel scaffold and anti-integrase activity with IC50 of 164 µM. Further improvement of this compound yielded an analog with IC50 of 12 µM (Mugnaini et al., 2007).
Fig. 18.
(I, A) Novel HIV-1 Integrase inhibitor using ligand-based virtual screening with a pharmacophore model of quinolone 3-carboxylic acid IN inhibitors [from Dayam et al. (2008)]. (B) Pharmacophore query generated from the quinolone 3-carboxylic acid IN inhibitors accompanied with an overlay onto a known HIV-1 integrase inhibitor. Features are color-coded, and their 3D arrangement/distances are shown in angstroms. Green sphere represent H-bond acceptor regions, blue spheres represent negatively ionizable regions, and cyan spheres represent hydrophobic aromatic regions. (II) Pharmacophore query overlayed with 3 potent hits from the ligand-based virtual screen: compounds 8 (A), 9 (B), and 17 (C).
Noeske et al. (2007) used 2D-pharmacophore-based virtual screening to identify novel mGlu1 antagonists. Antagonism of this receptor has been studied in regards to therapeutic potential in neurodegenerative diseases, anxiety, pain, and schizophrenia (Bordi and Ugolini, 1999; Spooren et al., 2003). Six reference mGlu1 antagonists were used to construct 2D-pharmacophores with the CATS software package (Schneider et al., 1999). This software assigns all atoms in a compound as either a hydrogen-bond donor, hydrogen-bond acceptor, positively charged, negatively charged, lipophilic, or noninterest atom type. Then, all compounds of a library are compared with the distances between these different atom types in the reference molecule, and similarity scores are calculated to rank molecules that most closely fit this 2D-pharmacophore. Screening the Gold Collection of Asinex Ltd. yielded six different hit lists (one for each reference molecule). The top hits were collected from all lists as well as hits that appeared in three or more different lists, and 23 compounds were tested experimentally for mGlu1 antagonism. Their most potent compound yielded an IC50 of 360 nM and was further optimized to a compound with an IC50 of 123 nM.
IV. Prediction and Optimization of Drug Metabolism and Pharmacokinetics Properties Including Absorption, Distribution, Metabolism, Excretion, and the Potential for Toxicity Properties
In addition to high biologic activity and selectivity for the target of interest, drug metabolism and pharmacokinetics (DMPK) properties including absorption, distribution, metabolism, excretion, and the potential for toxicity (ADMET) in humans are critical to the success of any candidate therapeutic. After lead discovery or design, there is considerable attention given to improving the compounds' in vivo DMPK/ADMET properties without losing its biologic activity. It is common to apply some DMPK/ADMET-based restrictions early on in the discovery process to reduce the number of compounds necessary to evaluate and save time and resources. Therefore, computational techniques extend to predicting this very important aspect of drug design and discovery. Methods used are structure-based to study the interaction of candidate compounds with key proteins involved in DMPK/ADMET and ligand-based to predict of key properties using quantitative structure property relation (QSPR) models.
A. Compound Library Filters
Computational tools are routinely used to filter large data bases so that compounds predicted to have poor DMPK/ADMET profiles may be avoided. One of the earliest and still the most popular filters to apply to any compound data base when performing a vHTS is Lipinski’s rule of five. These rules are: (1) molecular weight of 500 or less, (2) logP coefficient less than 5, (3) 5 or fewer hydrogen-bond donor sites, (4) 2×5 or fewer hydrogen-bond accepting sites (Lipinski et al., 1997). The rule set is based on an analysis of 2245 compounds from the World Drug Index that had reached phase II trials or higher. The rules were based on distributions for molecular weight, logP, hydrogen bond donors, and hydrogen bond acceptors for the top percentile of these compounds (Lipinski et al., 1997). This set of rules suggests the necessary properties for good oral bioavailability (Lajiness et al., 2004) and reflects the notion that pharmacokinetics, toxicity, and other adverse effects are directly linked to the chemical structure of a drug. Although this criteria is well established and offers a relatively fast and simple way to apply DMPK/ADMET filters before any sort of screening is performed, it is incapable of predicting with any certainty whether a compound will make an appropriate therapeutic. It has been estimated that almost 69% of available compounds in the Available Chemical Directory (ACD) Screening Database (2.4 million compounds) and 55% of the compounds in the ACD (240,000) do not violate this rule of 5 (Hou et al., 2006). Accordingly, this rule set has always been intended to be a guide and not necessarily a hard-set filter. It is expected that such a simple rule of thumb will remove lead compounds; for example, many peptidomimetics, transporter substrates, and natural products will violate Lipinski’s rule. Approximately 16% of oral drugs violate at least one criterion and 6% fail two or more criteria, and multiple examples exist of highly successful drugs that fail one or more of Lipinksi’s criteria including Lipitor and Singulair (Bickerton et al., 2012). At the same time the Lipinski’s rule will not, for example, recognize and remove compounds with structural features that give rise to toxicity. It is limited to evaluating oral bioavailability through passive transport only. When used to train models with machine learning, Lipinski’s rule failed to provide better than random classification of drugs and nondrugs (Frimurer et al., 2000). Additionally, it is not designed to provide any discrimination beyond a binary pass or fail. Any compound that violates two or more criteria is treated as an equal fail, whereas any compound that does not is treated as an equal pass.
On the basis of its shortcomings, several improvements and replacements have been proposed for the rule of 5. For example, two additional criteria have been suggested that include the number of rotatable bonds being less than or equal to ten and the polar surface area being less than 140 Å2 (Veber et al., 2002). Bickerton et al. (2012) introduced the quantitative estimate of drug-likeness that is a score ranging from 0 (all properties unfavorable) to 1 (all properties favorable). This score is taken as a geometric mean of individual desirability functions, each of which corresponds to a different molecular descriptor. These descriptors include molecular weight, logP, hydrogen bond donors and acceptors, rotatable bonds, aromatic rings, and the number of structural alerts (Brenk et al., 2008).
However, the simple application of filters such as these during a lead compound search can be problematic by nature of the limitation of these descriptors and the evolution of lead compound to drug. For example, Hann et al. (2001) found that, on average over a set of 470 lead-drug pairs, lead compounds had lower molecular weight, lower logP, fewer aromatic rings, and fewer hydrogen-bond acceptors compared with their eventual drugs. Therefore, it can be problematic to apply filters designed around the average properties of drugs to libraries that are intended for the discovery of lead compounds.
Additionally, some of the properties used in these filters can depend on conformation and environment. Kulkarni et al. (2002) state that permeability and hydrophobicity can change depending on the free energy of solvation, interaction of the drug with a phospholipid monolayer, and the drug’s flexibility. Vistoli et al. (2008) state that hydrophobicity and hydrogen bonding are both dependent on the dynamic nature of molecules and that chemical information is limited without the use of dynamic descriptors. For a comprehensive review on the concept of drug likeness, please see the 2011 review by Ursu et al. (2011).
The same computational tools used to predict activity can be applied to predict a more detailed DMPK/ADMET profile, including solubility, membrane permeability, metabolism, interaction with influx/efflux transporter proteins, interaction with transcription proteins, and different aspects of toxicity. For example, QSAR-based techniques have been especially important in predicting the toxicology profiles for drugs very early on in their development. These tools collect information regarding known toxins such as carcinogens, neurotoxins, and skin irritating agents, and create statistical models that can predict the likelihood that a particular compound will reflect these undesirable properties (Schnecke and Bostrom, 2006).
B. Lead Improvement: Metabolism and Distribution
Aside from general filters applied to compound libraries preceding a screen, computational tools can be used to guide hit-to-lead optimization where a compound’s metabolic profile is fine tuned. This requires a precise balancing act as the changes necessary to improve a compound’s metabolic profile may also significantly reduce its target affinity. During this stage of drug development, efforts are made in changing the compound’s structure not only to improve affinity but also to improve its metabolism. Therefore, although computational tools are useful in predicting the effects on target affinity from any proposed changes to the lead structure, they can be used in parallel to predict the affinity and interactions the compound may have with metabolizing enzymes and their regulators (Sun and Scott, 2010). The metabolism of a drug can have significant impacts not only on its bioavailability but also on its half-life and generation of harmful metabolites. When metabolic stability is lowered, a drug can lose its efficacy. Increasing stability can amplify harmful side effects attributed to a long half-life. Physiologically, there are two important phases in drug metabolism that have been studied extensively. The phase I reactions include hydrolysis, reduction, and oxidation and are primarily performed by cytrochrome P450 enzymes. Phase II reactions are more diverse and include glucuronidation, sulfation, acetylation, methylation, and glutathione conjugation (Goldstein, 1974). These reactions accelerate the drug’s elimination from the body but can result in toxic products like highly reactive electrophiles or free radicals (Sun and Scott, 2010).
Computational tools have been developed to address the phase I metabolism reactions performed by cytrochrome P450 enzymes, mainly through docking and QSAR procedures to predict the likelihood that a particular compound will bind to a cytochrome P450. At least 57 P450 isoforms exist in the human body, but phase I metabolism is dominated by the isoforms 1A2, 2C9, 2C19, 2D6, and 3A4 (Ortiz de Montellano, 2005) and computational methods are routinely directed against these particular P450 isoforms. In addition to the elimination of the drug and generation of metabolites, P450s can also be the source of drug-drug interactions in that one drug can reduce the elimination of another drug by blocking access to metabolizing enzymes or can increase elimination by upregulating expression of those enzymes. For example, in the early development of CCR5 antagonists, experimenters discovered hits that contained functional groups that are common among CYP2D6 inhibitors. By modeling the binding of these ligands to CYP2D6, imidazopyridines were replaced with benzimidazoles so that possible drug-drug interactions arising from inhibition of CYP2D6 were avoided early on (Armour et al., 2006).
Structure-based methods are the most popular computational tools for predicting the interaction between a compound and P450 enzymes. Binding poses predicted through docking studies may provide further insight into the specific sites of metabolism within the compound. For example, structure-based methods successfully predicted the metabolism of celecoxib and its 13 analogs through CYP2C9 (Ahlstrom et al., 2007a,b). In addition to some P450 isoforms, X-ray structures of the ligand-binding domain of prenane X receptor (Xue et al., 2007), the transcription regulator of CYP3A4 (Yano et al., 2004), glutathione S-transferases (Udomsinprasert et al., 2005), and drug transporters such as P-glycoprotein (Aller et al., 2009) have been determined. Structural information about prenane X receptor and drug transporters can be used to predict drug-drug interactions through the induction of CYP3A4 or transport channels.
One of the major challenges in modeling P450 binding is the dynamic nature of the binding site that accommodates a wide variety of ligands. Another challenge with docking studies involving P450 enzymes is the fact that the goal is often fundamentally opposite to that of most docking studies in that weaker binding is usually preferred over stronger binding. Monte Carlo and stochastic simulations of a wide variety of cocrystal structures have allowed development of several dynamic models of P450 binding sites exploring the different orientations amino acid side chains (Sun and Scott, 2010). GOLD, FlexX, DOCK, AutoDock, and the scoring function C-Score are most commonly used for structure-based methods with P450 predictions (de Graaf et al., 2005). For modeling the catalytic reaction encountered when the ligand binds to the P450 enzyme, ab initio calculations using Hartree-Fock or density functional theory have been used (Sun and Scott, 2010).
For example, the formation of the hydroquinone metabolite and electrophilic quinonone from remoxipride was calculated using hybrid density functional theory. This information was then used to redesign remoxipride (Erve et al., 2004). Density functional theory calculations were used to eliminate the formation of reactive metabolites from a series of tyrosine kinase-2 inhibitors. These calculations correctly predicted the necessary changes that avoided the formation of these harmful metabolites (Sun et al., 2009). Park and Harris (2003) used DFT on CYP2E1 homology models along with docking and MD to predict the metabolism profiles for seven compounds. Li et al. (2008) used homology modeling and MD to dock ligands into CYP2J2 in an effort to describe the binding characteristics of this enzyme. CYP2J2 is involved in the creation of eicosatrienoic acids from arachidonic acid. They were able to identify key residues that were important for the substrate specificity of CYP2J2. Additionally, they discovered that different ligands, although sharing the same scaffold, show different binding modes (Li et al., 2008). Bazeley et al. (2006) used structural information of CYP2D6 to identify invariant segments and performed conformational sampling with MD. Combining this data with neural-network based feature selection they found that only three out of 20 conformations are relevant for CYP2D6 binding. They also analyzed the docking of 82 compounds and showed that the most important attributes that conferred a compound’s affinity for CYP2D6 was the number of hydrogen-bonding sites, molecular weight, the number of rotatable bonds, AlogP, formal charge, number of aromatic rings, and the number of positive atoms. With these findings, they were able to achieve a prediction accuracy of 85% (Bazeley et al., 2006).
In addition to these structural methods, reactivity rules are also used to predict the metabolism of small molecules. Data bases such as Accelrys Metabolite (Accelrys, 2013) contain curated metabolic transformations from the literature. This information can be used to predict the various metabolic transformations that will be produced from an input structure. META (Talafous et al., 1994) is a model of mammalian xenobiotic metabolism that incorporates metabolic data from literature, textbooks, and monographs to define chemical transformation rules called transforms, which can identify and substitute functional groups. These focus on both phase 1 and phase 2 metabolism.
Another method uses electronics and intramolecular sterics to predict sits of CYP3A4 metabolism. This approach focuses on the rate-limiting step of the hydroxylation by CYP3A4, namely the removal of the hydrogen-atom (Shaik et al., 2002). The model assumes that the susceptibility for removal depends mainly on the electronic environment surrounding the hydrogen. Therefore, the method calculates a hydrogen abstraction energy for each hydrogen atom and this information is used to predict sites of metabolism (Singh et al., 2003b).
SMARTCyp (Rydberg et al., 2010) is another rule-based method that determines the reactivity of molecular fragments based on activation energies calculated by quantum mechanical methods. It combines a reactivity descriptor and accessibility descriptor. The reactivity descriptor estimates energy required for P450 metabolism at a given site by looking up fragments in an energy table for each atom. The accessibility descriptor is a calculation that determines the 2D distance from the center of the molecule a given atom is and always ranges between 0.5 and 1.
The activation energy table used for the reactivity descriptor combines 11 previously defined rules for aliphatic, aromatic, and alkene carbon atoms for 50 carbon sites (Rydberg et al., 2009) with new data generated by the authors. This produced a collection of 139 transition states that can represent different types of P450 reactions.
Other aspects of a drug’s DMPK/ADMET profile that are predicted with computational tools include membrane permeability, which is a large part of bioavailability as well as volume of distribution and penetration of the blood-brain barrier and blood plasma protein binding, involved in a drug’s volume of distribution and effective plasma concentrations. The evolution of predictive models for blood-brain barrier penetration is reviewed in detail by Norinder and Haeberlein (2002). Additionally, the structure of human serum albumin is used to predict plasma protein binding and volume of distribution changes (Davis and Riley, 2004).
C. Prediction of Human Ether-a-go-go Rrelated Gene Binding
The human ether-a-go-go related gene (hERG) protein is a voltage-gated potassium channel expressed in the heart and nervous system. The tetramer has six transmembrane spanning regions per protamer and is important for repolarization during the cardiac action potential (Mitcheson and Perry, 2003; Recanatini et al., 2005; Sanguinetti and Tristani-Firouzi, 2006). The delayed rectifier repolarizing current, an outward potassium current comprised of a rapid and slow component, is involved in plateau repolarization and the configuration of the action potential. Alterations in this channel’s conductance, especially blockade of the channel, can lead to an altered refractory period and action potential duration (Recanatini et al., 2005), often resulting in what is known as drug-induced QT syndrome and a severe cardiac side effect called torsades de points (Hancox and Mitcheson, 2006). The QT interval is the period of a cardiac cycle where ventricular repolarization occurs (Sanguinetti and Tristani-Firouzi, 2006), and drug-induced QT syndrome can lead to sudden death (Keating and Sanguinetti, 1996). Because of its importance in the proper regulation of cardiac action potential, off-target interactions with hERG have caused several drugs to be removed from the market and/or linked to arrhythmias and sudden death (Mitcheson and Perry, 2003). hERG has been termed an “antitarget” in the pharmaceutical industry (Aronov, 2005). It has been estimated that 2–3% of prescribed medications include some unintended QT elongation (Recanatini et al., 2005). Although most drugs have been shown to inhibit the rapid component of the outward potassium current (Garg et al., 2008), interaction between drugs and hERG is not completely understood, and high-affinity ligands tend to interact with the inactivated channel with low voltage-dependency, whereas low-affinity ligands tend to interact with the activated state with high voltage-dependent kinetics (Ficker et al., 2002). However, key residues involved in the interaction between hERG and at least some ligands have been identified. For example, Phe656 and Tyr652 in the channel pore may engage in π-π and cation-π interactions with the ligand. Thr623 and Ser624 are thought to interact with the polar tails of some ligands and some evidence exists of a second binding site (Aronov, 2005; Recanatini et al., 2005; Choe et al., 2006; Sanguinetti and Tristani-Firouzi, 2006). In vitro and in vivo methods are commonly used to evaluate drug candidates for potential hERG blockade activity, especially patch clamp techniques and radioligand binding assays (Wood et al., 2004; Polak et al., 2009). However, these methods are difficult to scale to high-throughput candidate evaluation, making the computational approach attractive for this aspect of drug discovery.
SB-CADD and LB-CADD have both been used to develop models to discriminate hERG blockers and nonblockers (Bridgland-Taylor et al., 2006; Thai and Ecker, 2007). Currently, LB-CADD is more popular for hERG predictions because of the fact that there is currently no crystal structure for the hERG potassium channel (Wang et al., 2012). Therefore, SB-CADD techniques have mainly relied on docking with homology models, and this method has not been validated with large, highly diverse datasets (Wang et al., 2012). LB-CADD-based hERG models have been created using tools including ligand-based pharmacophore (Ekins et al., 2002a; Cianchetta et al., 2005), CoMFA (Cavalli et al., 2002), Bayesian classification with QSAR (Sun, 2006), and 2D fragment-based descriptors (Song and Clark, 2006).
Wang et al. (2012) developed discrimination models based on molecular property descriptors and fingerprints. Descriptors were calculated using Discovery Studio molecular simulation package (Accelrys) and included several variations on logP, molecular weight, hydrogen-bonding, the number of rotatable bonds, rings, and aromatic rings, the sum of oxygen and nitrogen atoms, and fractional polar surface area. The fingerprints included SciTegic extended-connectivity fingerprints and Daylight-style path-based fingerprints using the Morgan algorithm (Rogers and Hahn, 2010). Bayesian classifiers and decision tree methods were used to create models based on these descriptors.
Wang et al. (2012) analyzed the results of their models and found that increased hydrophobicity was correlated with increased hERG binding. Additionally, molecular weight showed a significant, although lesser impact on hERG binding, with molecules having a molecular weight under 250 being less likely to be a hERG blocker. Additionally, analysis of their fingerprints revealed that most hERG-binding fragments contained nitrogen atoms, with four of the top five containing positively charged nitrogen atoms. These top five fragments also contained at least one oxygen atom or a carboxylic acid. Despite these correlations, the authors stressed that no single molecular property can be used to discriminate between hERG blockers and nonblockers.
Obrezanova and Segall (2010) used the Gaussian process to build models for hERG inhibition as well as other ADMET properties. The Gaussian process method (Gibbs and MacKay, 2000; Rasmussen and Williams, 2006) is a nonlinear regression technique that is resistant to overtraining. It uses Bayesian inference to link the descriptors of a molecule with the probability of the molecule falling into a specific class. Eventually, a posterior probability distribution is created over functions that identify those that best describe the observed data. The mean value over all functions can provide the prediction, whereas the full distribution can provide a measure of uncertainty for each prediction. The hERG inhibitor model was trained on 117 active and 51 inactive compounds evaluated through patch clamp in mammalian cells with descriptors generated in StarDrop’s Auto-Modeler (Obrezanova et al., 2008). These 2D descriptors were based on SMARTS and included atom type counts, functionality, and molecular properties such as logP, molecular weight, and polar surface areas. Datasets were also clustered using 2D fingerprints and tanimoto similarity.
Nisius and Goller (2009) used the Tripos Topomer Search technology (Cramer et al., 2002) to design a modeling approach termed topoHERG. This method screens reference datasets for molecules similar to a query compound and returns pharmacophore and shape-based distances between a query molecule and its neighbors. The dataset contained 115 inactive compounds, 90 moderately active hERG blockers, and 70 highly active hERG blockers. The topomer is defined as a 3D representation of a molecular fragment that is based on 2D topology and a rule set that generates an absolute conformation (Jilek and Cramer, 2004) so that distances between topomers of different molecules in large data bases can be calculated. To differentiate between hERG active and inactive neighbors, the inverse of the topomer search distance was multiplied by one if the topomor search neighbor was active and negative one if it was inactive. A molecule was predicted to be an active hERG blocker if its overall sum was greater than zero. A two-stage approach using two optimized models yielded a prediction accuracy of 76–81% (Nisius and Goller, 2009).
Garg et al. (2008) used a genetic function approximation to generate quantitative structure-toxicity relationship (QSTR) models using 2D descriptors generated using the QSAR+ module of Cerius (Accelrys). These models were trained with 56 hERG blockers and descriptors included electrotopological descriptors that contained information regarding the topological environments for all atoms in the molecule as well as electronic interactions with other atoms in the molecule. To perform genetic function approximation, the authors generated a number of random equations that were randomly selected as pairs. These parent pairs underwent random crossover operations to generate new equations, and those that showed improved fitness scores were kept (Rogers and Hopfinger, 1994). In parallel, the authors generated a toxicophore (pharmacophore-based toxicity model) using Catalyst’s HypoGen that included hydrogen-bonding, hydrophobic, aromatic, and positive ionizable features. Upon analysis of their models, the authors noted that both basic and neutral hERG blockers had highly flexible linkers and various molecular fragments.
D. Drug Metabolism and Pharmacokinetics/Absorption, Distribution, Metabolism, and Excretion and the Potential for Toxicity Prediction Software Packages and Algorithms
There is currently a great deal of models available for predicting absorption, bioavailability, transporter binding, metabolism, volume of distribution, and P450 interactions (Yoshida and Topliss, 2000; de Groot and Ekins, 2002; Ekins et al., 2002a,b; Lewis, 2003; Pintore et al., 2003; Turner et al., 2003; Lombardo et al., 2004). Comprehensive software packages have been developed such as QikProp, which can be used to predict an array of ADMET-related properties such as solubility, membrane permeability, partition coefficients, blood-brain barrier penetration, plasma protein binding, and the formation of metabolites (Jorgensen and Duffy, 2002). These predictions mainly come from statistical models such as regression and neural networks that are trained on known ADMET properties for many compounds. The OSIRIS Property Explorer allows scientists to draw chemical structures and predict ADMET profile (Mandal et al., 2009). The software package MetaSite (Molecular Discovery Ltd, Middlesex UK) is used to predict the site of metabolism using structural information from both the ligand and the enzyme. A probability function is created for the site(s) of metabolism using the free energy of P450-ligand binding and reactivity. This software uses structure-based techniques to identify the relevant amino acids and proposes compound modifications that can optimize its metabolism profile (Cruciani et al., 2005). Ahlstrom et al. (2007) proposed a three-step procedure using MetaSite to identify metabolic sites, in silico modification of these sites, and docking of new compounds. These software packages aim at predicting overall ADMET properties with convenient and accessible tools and have shown great benefit in drug development. For example, computational modeling of ADMET properties prevented a potential blood pressure-lowering drug from being lost early in the development process. The proposed compound showed low EC50 values, indicating that it was less potent than another compound of consideration. However, pharmacokinetic modeling showed that this compound would actually have greater efficacy than the one that showed higher potency. This compound did indeed show superior efficacy in the clinic (Rajman, 2008).
E. Drug Metabolism and Pharmacokinetics/Absorption, Distribution, Metabolism, and Excretion and the Potential for Toxicity: Clinical Trial Prediction and Dosing
Computational tools are also being developed to address the possibility of simulating early clinical trials to avoid the waste resources inherent in testing drugs with poor ADMET profiles. This is a prevalent problem in drug development because up to 90% of drugs fail during clinical development, and the time between reaching clinical trials and approval is up to 8 years (Holford et al., 2010). These simulations aim at modeling the pathophysiology of biologic systems and the pharmacology of treatments and can often incorporate things such as disease progression, placebo response, and dropout rates.
For example, clinical trial simulation was used by Laer et al. (2005) to propose appropriate doses for Sotalol [CAS 959-24-0; N-[4-[1-hydroxy-2-[(1-methylethyl)amino]ethyl]phenyl]methanesulfonamide hydrochloride] in children and the Food and Drug Administration approved dosing changes for Etanercept (Immunex Corporation, Thousand Oaks CA) in juvenile rheumatoid arthritis due to clinical trial simulations performed by Yim et al. (2005). SimCYP (Simcyp Ltd, Sheffield UK) is a software package that creates virtual populations of participants with specifiable genetic and physiologic characteristics using literature data. In vitro metabolism data can be applied to the in vitro-in vivo extrapolation process to simulate whole-live and hepatic clearances for these virtual populations (Jamei et al., 2009). Kowalski et al. (2008) used the NONMEM software package (ICON plc, Dublin, Ireland) and PK/PD modeling to suggest a dosing regimen for SC-75416, a selective COX-2 inhibitor that would be comparable to the pain relief afforded from 50 mg of rofecoxib. This simulation saved an estimated 9 months of development.
V. Conclusions
The extensive variety of computational tools used in drug discovery campaigns suggests that there are no fundamentally superior techniques. The performance of methods varies greatly with target protein, available data, and available resources. For example, Kruger and Evers (2010) completed a performance benchmark between structure- and ligand-based vHTS tools across four different targets, including angiotensin-converting enzyme, cyclooxygenase-2, thrombin, and HIV-1 protease. Docking methods including Glide, GOLD, Surflex, and FlexX were used to dock ligands into rigid target crystal structures obtained from PDB. A single ligand was used as a reference for ligand-based similarity search strategies such as 2D (fingerprints and feature trees) and 3D [rapid overlay of chemical structures (ROCS; OpenEye Scientific Software, Santa Fe, NM)], a similarity algorithm that calculates maximum volume overlap of two 3D structures (Rush et al., 2005). In general the authors found that docking methods performed poorly for HIV-1 protease and thrombin attributable to the flexible nature of the targets and the fact that the known ligands for these proteins have large molecular weight and peptidomimetic character.
Enrichments based on 3D similarity searches were poor for HIV-1 protease and thrombin datasets compared with ACE, which is likely due to the higher level of diversity in the HIV-1 protease and thrombin ligand datasets. Similarity scoring algorithms like ShapeTanimoto, ColorScore, and ComboScore were compared with the performance of ROCS (Kruger and Evers, 2010). It was found that even within the scoring, algorithm performance varied across targets. For example, ColorScore performed best for ACE and HIV-1 protease, whereas ShapeTanimoto for COX-2 and ComboScore was the method of choice for thrombin. All vHTS tools performed comparatively well for ACE, but ligand-based 2D fingerprint approach generally outperformed docking methods. The authors also note an important observation in that, especially for HIV-1 protease, the structure-based and ligand-based approaches yielded complimentary hit lists. Therefore, performance metrics are not the only benchmark to consider when comparing CADD techniques. In some cases, discovery of novel chemotypes is more important than high hit rates or high activity. In the current study, Kruger and Evers (2010) found that ROCS and feature trees were more successful in retrieving compounds with novel scaffolds compared with other fingerprints.
Warren et al. (2006) published an in-depth assessment of the capabilities and shortcomings for docking programs and their scoring techniques against eight proteins of seven evolutionarily diverse target types. They found that docking programs were well adept at generating poses that included ones similar to those found in complex crystal structures. In general, although the molecular conformation was less precise across docking programs, they were fairly accurate in terms of the ligand’s overall positioning. With regards to scoring, their findings agree with others that docking programs lack reliable scoring algorithms. So while the tools were able to predict a set of poses that included those that were seen in the crystal structure, the preference for the crystal structure pose was not necessarily reflected in the scoring. For five of the seven targets that were evaluated, the success rate, however, was greater than 40%. It was found that the enrichment of hits could be increased by applying previous knowledge regarding the target. However, there was little statistically significant correlation between docking scores and ligand affinity across the targets. The study concluded that a docking program’s ability to reproduce accurate binding poses did not necessarily mean that the program could accurately predict binding affinities. This analysis underscores the necessity not only to re-rank the top hits from a docking-based vHTS using computationally expensive tools but also to continue evaluating novel scoring functions that can efficiently and accurately predict binding affinities (Warren et al., 2006).
Improvements in scoring functions involve the use of consensus scoring methods and free energy scoring with docking techniques. Consensus scoring methods have been shown to improve enrichments and prediction of bound conformations and poses by balancing out errors of individual scoring functions. In 2008, Enyedy and Egan (2008) compared docking scores of ligands with known IC50 and found that docking scores were incapable of correctly ranking compounds and were sometimes unable to differentiate active from inactive compounds. They concluded that individual scoring methods can be used successfully to enrich a dataset with increased population of actives but are insufficient to identify actives against inactives. Page and Bates (2006) concluded that although binding energy calculations such as MM-PBSA are one of the more successful methods of estimating free energy of complexes, these techniques are more applicable to providing insights into the nature of interactions rather than prediction or screening. Consensus scoring functions where free energy scores of different algorithms have been combined or averaged have been shown to substantially improve performance (Fukunishi et al., 2008; Teramoto and Fukunishi, 2008; Bar-Haim et al., 2009; Plewczynski et al., 2011).
In their literature survey, Ripphausen et al. (2010) reported that structure-based virtual screening was used much more frequently than ligand-based virtual screening (322 to 107 studies). Despite a preference for structure-based methods, ligand-based methods on average yield hits with higher potency than structure-based methods. Most ligand-based hits had activities better than 1 µM, whereas structure-based hits fall frequently in the range of 1–100 µM. Scoring algorithms in docking functions have been found to be biased toward known protein ligand complexes; for example, more potent hits against protein kinase targets are discovered when compared with other target classes (Stumpfe et al., 2012) (Fig. 19).
Fig. 19.
Ripphausen et al. (2010) report that ligand-based computationally approaches yield compounds with higher affinity than structure-based computationally approaches. Adapted from Ripphausen et al. (2010).
One CADD approach that has been gaining considerable momentum is the combination of structure-based and ligand-based computation techniques (Nicolotti et al., 2008). For example, the GRID-GOLPE method docks a set of ligands at a common binding site using GRID and then calculates descriptors for the binding interactions by probing these docking poses with GOLPE (Baroni et al., 1993). Multivariate regression is then used to create a statistical model that can explain the biologic activity of these ligands. Structure-based interactions between a ligand and target can also be used in similarity-based searches to find compounds that are similar only in the regions that participate in binding rather than cross the entire ligand. LigandScout uses such a technique to define a pharmacophore based on hydrogen bonding and charge-transfer interactions between a ligand and its target. Another technique known as the pseudoreceptor technique (Tanrikulu and Schneider, 2008) uses pharmacophore mapping-like overlaying techniques for a collection of ligands that bind to the same binding site to establish a virtual representation of the binding site’s structure, which is then used as a template for docking and other structure-based vHTS. This approach has been used by VirtualToxLab (Vedani et al., 2007) for the creation of nuclear receptors and cytochrome P450 binding site models in ADMET prediction tools and by Tanrikulu et al. (2009). in the modeling of the H4 receptor binding site subsequently used to identify novel active scaffolds (Tanrikulu et al., 2009). In a recent review by Wilson and Lill (2011), these methods are grouped into a major class of combined techniques called interaction based methods. A second major class involves the use of QSAR and similarity methods to enrich a library of virtual compounds prior to a molecular docking project. This can increase the efficiency of the project by reducing the number of compounds to be docked. This is similar to the application of CADD to enrich libraries prior to traditional HTS projects. This review also presents comprehensive descriptions of software packages using a combination of ligand- and structure-based techniques as well as several case studies testing the performance of these tools.
As discussed earlier, these methods are often used in serial where ligand-based methods are first used to enrich libraries that will subsequently be used in structure-based vHTS. The most common application is at the ligand library creation stage through the use of QSAR techniques to filter out compounds with low similarity to a query compound or no predicted activity based on a statistical model. QSAR has also been used as a means to refine the docking scores of a structure-based virtual screen. 2D and 3D QSAR can also be used to track docking errors. This method has been used by Novartis where a QSAR model is built from docking scores rather than observed activities, and this model is applied to that set to provide additional score weights for each compound (Klon et al., 2004).
Although CADD has been applied quite extensively in drug discovery campaigns, certain lucrative therapeutic targets like protein-protein interaction and protein-DNA interactions are still formidable problems, mainly because of the relatively massive size of interaction sites (in excess of 1500 Å2) (Van Drie, 2007). Lastly, accessibility has also been a problem with CADD as many tools are not designed with a friendly user interface in mind. In many cases, there can be an overwhelming number of variables that must be configured on a case-by-case basis and the interfaces are not always straightforward. A great deal of expertise is often required to use these tools to get desired measure of success. Increasingly, efforts are being made to develop user friendly interfaces, especially in commercially available tools. For example, ICM-Pro (MolSoft L.L.C., San Diego, CA) is a software package that is designed to be a user friendly docking tool and replaces the front-end of current docking algorithms with an interface that is manageable to a wider audience (Abagyan et al., 2006). More recently, gamification of the ROSETTA folding program, known as Foldit (Khatib et al., 2011), has allowed individuals from nonscientific community to help solve the structure of M-PMV retroviral protease (Khatib et al., 2012) and for predicting backbone remodeling of computationally designed biomolecular Diels-Alderase that increased its activity (Eiben et al., 2012). The successful application of crowd-sourced biomolecule design and prediction suggests further potential of CADD methods in drug discovery.
Abbreviations
- 3D
three dimensional
- 11β-HSD1
11β-hydroxysteroid dehydrogenase
- ACD
Available Chemical Directory
- ACE
angiotensin-converting enzyme
- ADMET
absorption, distribution, metabolism, and excretion and the potential for toxicity
- ADR
adverse drug reaction
- ANN
artificial neural networks
- CADD
computer-aided drug discovery/design
- CK2
casein kinase 2
- CPE
chemical penetration enhancers
- DMPK
drug metabolism and pharmacokinetics
- GPCR
G-protein–coupled receptor
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- HTS
high-throughput screening
- ICM
internal coordinate mechanics
- IN
integrase
- KIC
kinematic closure
- LB-CADD
ligand-based CADD
- LTA4H-h
human leukotriene A4 hydrolase
- mAChR
M1 acetylcholine receptor
- MAPK
mitogen-activated protein kinase
- MD
molecular dynamics
- MDM
murine double minute
- MIF
molecular-interaction field
- MLR
multivariate linear regression analysis
- p38 MAPK
p38 mitogen-activated protein kinase
- P450
cytochrome P450
- PCA
principal component analysis
- PEOE
partial equalization of orbital electronegativity
- PLA2
phospholipase A2
- PLS
partial least squares analysis
- PPAR
peroxisome proliferator-activated receptor
- QSAR
quantitative structure activity relationship
- RCS
relaxed complex scheme
- RDF
radical distribution functions
- RI
receptor independent
- SB-CADD
structure-based CADD
- SEA
similarity ensemble approach
- SVM
support vector machine
- VEGF
vascular endothelial growth factor
- vHTS
virtual-HTS
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Sliwoski, Kothiwale, Meiler, Lowe.
Footnotes
This work was supported by the National Science Foundation through the Office for Cyber Infrastructure Transformative Computational Sciences Fellowship [OCI-1122919] (E.W.L.); the National Institutes of Health National Institute of Mental Health [Grant R01 MH090192] (to the Meiler laboratory); the National Institutes of Health National Institute of General Medical Sciences [Grant R01 GM099842]; and the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01 DK097376].
References
- Abagyan R, Lee WH, Raush E, Budagyan L, Totrov M, Sundstrom M, Marsden BD. (2006) Disseminating structural genomics data to the public: from a data dump to an animated story. Trends Biochem Sci 31:76–78 [DOI] [PubMed] [Google Scholar]
- Abagyan R, Totrov M,, Kuznetsov D. (1994) ICM - a new method for protein modeling and design–applications to docking and structure prediction from the distorted native conformation. J Comput Chem 15:488–506 [Google Scholar]
- Abrams CF, Vanden-Eijnden E. (2010) Large-scale conformational sampling of proteins using temperature-accelerated molecular dynamics. Proc Natl Acad Sci USA 107:4961–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accelrys (2013) Accelrys metabolite. Available from http://accelrys.com/products/databases/bioactivity/metabolite.html
- Acharya C, Coop A, Polli JE, Mackerell AD., Jr (2011) Recent advances in ligand-based drug design: relevance and utility of the conformationally sampled pharmacophore approach. Curr Comput Aided Drug Des 7:10–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal AK, Fishwick CW. (2010) Structure-based design of anti-infectives. Ann N Y Acad Sci 1213:20–45 [DOI] [PubMed] [Google Scholar]
- Agnese V, Bazan V, Fiorentino FP, Fanale D, Badalamenti G, Colucci G, Adamo V, Santini D, Russo A. (2007) The role of Aurora-A inhibitors in cancer therapy. Ann Oncol 18 (Suppl 6):vi47–vi52 [DOI] [PubMed] [Google Scholar]
- Ahlström MM, Ridderström M, Zamora I. (2007. a) CYP2C9 structure-metabolism relationships: substrates, inhibitors, and metabolites. J Med Chem 50:5382–5391 [DOI] [PubMed] [Google Scholar]
- Ahlström MM, Ridderström M, Zamora I, Luthman K. (2007. b) CYP2C9 structure-metabolism relationships: optimizing the metabolic stability of COX-2 inhibitors. J Med Chem 50:4444–4452 [DOI] [PubMed] [Google Scholar]
- Al-Sha’er MA, Taha MO. (2010) Elaborate ligand-based modeling reveals new nanomolar heat shock protein 90α inhibitors. J Chem Inf Model 50:1706–1723 [DOI] [PubMed] [Google Scholar]
- Alguacil LF, Pérez-García C. (2003) Histamine H3 receptor: a potential drug target for the treatment of central nervous system disorders. Curr Drug Targets CNS Neurol Disord 2:303–313 [DOI] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, et al. (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323:1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. (2008) Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci USA 105:17278–17283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Lee DCW, Law AH, Yang CL, Poon LL, Lau AS, Jones SJ. (2009) A novel small-molecule inhibitor of the avian influenza H5N1 virus determined through computational screening against the neuraminidase. J Med Chem 52:2667–2672 [DOI] [PubMed] [Google Scholar]
- Anderson AC. (2012) Structure-based functional design of drugs: from target to lead compound. Methods Mol Biol 823:359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annoura H, Nakanishi K, Uesugi M, Fukunaga A, Imajo S, Miyajima A, Tamura-Horikawa Y, Tamura S. (2002) Synthesis and biological evaluation of new 4-arylpiperidines and 4-aryl-4-piperidinols: dual Na(+) and Ca(2+) channel blockers with reduced affinity for dopamine D(2) receptors. Bioorg Med Chem 10:371–383 [DOI] [PubMed] [Google Scholar]
- Arakaki AK, Zhang Y, Skolnick J. (2004) Large-scale assessment of the utility of low-resolution protein structures for biochemical function assignment. Bioinformatics 20:1087–1096 [DOI] [PubMed] [Google Scholar]
- Armour D, de Groot MJ, Edwards M, Perros M, Price DA, Stammen BL, Wood A. (2006) The discovery of CCR5 receptor antagonists for the treatment of HIV infection: hit-to-lead studies. ChemMedChem 1:706–709 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- Aronov AM. (2005) Predictive in silico modeling for hERG channel blockers. Drug Discov Today 10:149–155 [DOI] [PubMed] [Google Scholar]
- Auer J, Bajorath J. (2008) Molecular similarity concepts and search calculations. Methods Mol Biol 453:327–347 [DOI] [PubMed] [Google Scholar]
- B-Rao C, Subramanian J, Sharma SD. (2009) Managing protein flexibility in docking and its applications. Drug Discov Today 14:394–400 [DOI] [PubMed] [Google Scholar]
- Bajorath J. (2001) Selected concepts and investigations in compound classification, molecular descriptor analysis, and virtual screening. J Chem Inf Comput Sci 41:233–245 [DOI] [PubMed] [Google Scholar]
- Bajorath J. (2002) Integration of virtual and high-throughput screening. Nat Rev Drug Discov 1:882–894 [DOI] [PubMed] [Google Scholar]
- Ball P. (2008) Water as an active constituent in cell biology. Chem Rev 108:74–108 [DOI] [PubMed] [Google Scholar]
- Bambini S, Rappuoli R. (2009) The use of genomics in microbial vaccine development. Drug Discov Today 14:252–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Haim S, Aharon A, Ben-Moshe T, Marantz Y, Senderowitz H. (2009) SeleX-CS: a new consensus scoring algorithm for hit discovery and lead optimization. J Chem Inf Model 49:623–633 [DOI] [PubMed] [Google Scholar]
- Barnard JM, Downs GM. (1997) Chemical fragment generation and clustering software. J Chem Inf Comput Sci 37:141–142 [Google Scholar]
- Baroni M, Costantino G, Cruciani G, Riganelli D, Valigi R, Clementi S. (1993) Generating optimal linear PLS estimations (GOLPE)–an advanced chemometric tool for handling 3d-QSAR problems. Quantitative Struct-Activity Relation 12:9–20 [Google Scholar]
- Basak SC. (2012) Chemobioinformatics: the advancing frontier of computer-aided drug design in the post-genomic era. Curr Comput Aided Drug Des 8:1–2 [DOI] [PubMed] [Google Scholar]
- Bashford D, Case DA. (2000) Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem 51:129–152 [DOI] [PubMed] [Google Scholar]
- Bauerschmidt S, Gasteiger J. (1997) Overcoming the limitations of a connection table description: a universal representation of chemical species. J Chem Inf Comput Sci 37:705–714 [Google Scholar]
- Baumforth KR, Flavell JR, Reynolds GM, Davies G, Pettit TR, Wei W, Morgan S, Stankovic T, Kishi Y, Arai H, et al. (2005) Induction of autotaxin by the Epstein-Barr virus promotes the growth and survival of Hodgkin lymphoma cells. Blood 106:2138–2146 [DOI] [PubMed] [Google Scholar]
- Baurin N, Baker R, Richardson C, Chen I, Foloppe N, Potter A, Jordan A, Roughley S, Parratt M, Greaney P, et al. (2004) Drug-like annotation and duplicate analysis of a 23-supplier chemical database totalling 2.7 million compounds. J Chem Inf Comput Sci 44:643–651 [DOI] [PubMed] [Google Scholar]
- Bazeley PS, Prithivi S, Struble CA, Povinelli RJ, Sem DS. (2006) Synergistic use of compound properties and docking scores in neural network modeling of CYP2D6 binding: predicting affinity and conformational sampling. J Chem Inf Model 46:2698–2708 [DOI] [PubMed] [Google Scholar]
- Becker OM, Dhanoa DS, Marantz Y, Chen D, Shacham S, Cheruku S, Heifetz A, Mohanty P, Fichman M, Sharadendu A, et al. (2006) An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression. J Med Chem 49:3116–3135 [DOI] [PubMed] [Google Scholar]
- Becker OM, Marantz Y, Shacham S, Inbal B, Heifetz A, Kalid O, Bar-Haim S, Warshaviak D, Fichman M, Noiman S. (2004) G protein-coupled receptors: in silico drug discovery in 3D. Proc Natl Acad Sci USA 101:11304–11309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitz HD, Chen W, Jugel H, Treleano R, Wieser H, Gasteiger J, Marsili M. (1979) Sweet and bitter compounds: structure and taste relationship. Food Taste Chemistry 115:93–131 [Google Scholar]
- Bertz SH. (1983) On the complexity of graphs and molecules. Bull Math Biol 45:849–855 [Google Scholar]
- Biarnés X, Bongarzone S, Vargiu AV, Carloni P, Ruggerone P. (2011) Molecular motions in drug design: the coming age of the metadynamics method. J Comput Aided Mol Des 25:395–402 [DOI] [PubMed] [Google Scholar]
- Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. (2012) Quantifying the chemical beauty of drugs. Nat Chem 4:90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum LC, Reymond JL. (2009) 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. J Am Chem Soc 131:8732–8733 [DOI] [PubMed] [Google Scholar]
- Blumer A, Ehrenfeucht A, Haussler D, Warmutii MD. (1989) Learnability and the Vapnik-Chervonenkis dimension. J ACM 36:929–965 [Google Scholar]
- Bohacek RS, McMartin C, Guida WC. (1996) The art and practice of structure-based drug design: a molecular modeling perspective. Med Res Rev 16:3–50 [DOI] [PubMed] [Google Scholar]
- Böhm HJ. (1992) The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J Comput Aided Mol Des 6:61–78 [DOI] [PubMed] [Google Scholar]
- Bokoch MP, Zou YZ, Rasmussen SG, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi HJ, Thian FS, Kobilka TS, et al. (2010) Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologa CG, Revankar CM, Young SM, Edwards BS, Arterburn JB, Kiselyov AS, Parker MA, Tkachenko SE, Savchuck NP, Sklar LA, et al. (2006) Virtual and biomolecular screening converge on a selective agonist for GPR30. Nat Chem Biol 2:207–212 [DOI] [PubMed] [Google Scholar]
- Bordi F, Ugolini A. (1999) Group I metabotropic glutamate receptors: implications for brain diseases. Prog Neurobiol 59:55–79 [DOI] [PubMed] [Google Scholar]
- Boser BE, Guyon IM, and Vapnik VN (1992) A training algorithm for optimal margin classifiers, in Proceedings of the Fifth Annual Workshop on Computational Learning Theory; Association for Computing Machinery; 27--29 July 1992; Pittsburgh, PA, pp. 144–152. [Google Scholar]
- Boucher J, Quilliot D, Pradères JP, Simon MF, Grès S, Guigné C, Prévot D, Ferry G, Boutin JA, Carpéné C, et al. (2005) Potential involvement of adipocyte insulin resistance in obesity-associated up-regulation of adipocyte lysophospholipase D/autotaxin expression. Diabetologia 48:569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourinet E, Zamponi GW. (2005) Voltage gated calcium channels as targets for analgesics. Curr Top Med Chem 5:539–546 [DOI] [PubMed] [Google Scholar]
- Bower MJ, Cohen FE, Dunbrack RL., Jr (1997) Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol 267:1268–1282 [DOI] [PubMed] [Google Scholar]
- Bowman AL, Nikolovska-Coleska Z, Zhong H, Wang S, Carlson HA. (2007) Small molecule inhibitors of the MDM2-p53 interaction discovered by ensemble-based receptor models. J Am Chem Soc 129:12809–12814 [DOI] [PubMed] [Google Scholar]
- Boyle BH. (2011) Support Vector Machines: Data Analysis, Machine Learning, and Applications, Nova Science Publishers, New York [Google Scholar]
- Brauman JI, Blair LK. (1968) Gas-phase acidities of alcohols. Effects of alkyl groups. J Am Chem Soc 90:6561–6562 [Google Scholar]
- Bravi G, Gancia E, Mascagni P, Pegna M, Todeschini R, Zaliani A. (1997) MS-WHIM, new 3D theoretical descriptors derived from molecular surface properties: a comparative 3D QSAR study in a series of steroids. J Comput Aided Mol Des 11:79–92 [DOI] [PubMed] [Google Scholar]
- Brenk R, Schipani A, James D, Krasowski A, Gilbert IH, Frearson J, Wyatt PG. (2008) Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem 3:435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgland-Taylor MH, Hargreaves AC, Easter A, Orme A, Henthorn DC, Ding M, Davis AM, Small BG, Heapy CG, Abi-Gerges N, et al. (2006) Optimisation and validation of a medium-throughput electrophysiology-based hERG assay using IonWorks HT. J Pharmacol Toxicol Methods 54:189–199 [DOI] [PubMed] [Google Scholar]
- Briganti S, Camera E, Picardo M. (2003) Chemical and instrumental approaches to treat hyperpigmentation. Pigment Cell Res 16:101–110 [DOI] [PubMed] [Google Scholar]
- Broto P, Moreau G, Vandycke C. (1984) Molecular-structures—perception, auto-correlation descriptor, and SAR studies—perception of molecule-topological structure and 3-dimensional structure. Eur J Med Chem 19:71–78 [Google Scholar]
- Brown N, McKay B, Gilardoni F, Gasteiger J. (2004) A graph-based genetic algorithm and its application to the multiobjective evolution of median molecules. J Chem Inf Comput Sci 44:1079–1087 [DOI] [PubMed] [Google Scholar]
- Brvar M, Perdih A, Oblak M, Masic LP, Solmajer T. (2010) In silico discovery of 2-amino-4-(2,4-dihydroxyphenyl)thiazoles as novel inhibitors of DNA gyrase B. Bioorg Med Chem Lett 20:958–962 [DOI] [PubMed] [Google Scholar]
- Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. (2010) Protein annotation and modelling servers at University College London. Nucleic Acids Res 38: W563–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik B, Garzya V, Walker G, Woolley-Roberts M, Pardoe J, Lucas A, Tehan B, Rivero RA, Langmead CJ, et al. (2010) Novel N-substituted benzimidazolones as potent, selective, CNS-penetrant, and orally active M(1) mAChR agonists. Acs Medicinal Chemistry Letters 1:244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui NK, Turk S, Buckenmaier S, Stevenson-Jones F, Zeuch B, Gobec S, Vollmer W. (2011) Development of screening assays and discovery of initial inhibitors of pneumococcal peptidoglycan deacetylase PgdA. Biochem Pharmacol 82:43–52 [DOI] [PubMed] [Google Scholar]
- Canutescu AA, Dunbrack RL., Jr (2003) Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci 12:963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporuscio F, Rastelli G, Imbriano C, Del Rio A. (2011) Structure-based design of potent aromatase inhibitors by high-throughput docking. J Med Chem 54:4006–4017 [DOI] [PubMed] [Google Scholar]
- Carlson HA. (2002) Protein flexibility and drug design: how to hit a moving target. Curr Opin Chem Biol 6:447–452 [DOI] [PubMed] [Google Scholar]
- Casañola-Martín GM, Marrero-Ponce Y, Khan MT, Ather A, Khan KM, Torrens F, Rotondo R. (2007) Dragon method for finding novel tyrosinase inhibitors: Biosilico identification and experimental in vitro assays. Eur J Med Chem 42:1370–1381 [DOI] [PubMed] [Google Scholar]
- Cavalli A, Poluzzi E, De Ponti F, Recanatini M. (2002) Toward a pharmacophore for drugs inducing the long QT syndrome: insights from a CoMFA study of HERG K(+) channel blockers. J Med Chem 45:3844–3853 [DOI] [PubMed] [Google Scholar]
- Cavasotto CN, Abagyan RA. (2004) Protein flexibility in ligand docking and virtual screening to protein kinases. J Mol Biol 337:209–225 [DOI] [PubMed] [Google Scholar]
- Cavasotto CN, Phatak SS. (2011) Docking methods for structure-based library design. Methods Mol Biol 685:155–174 [DOI] [PubMed] [Google Scholar]
- Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, Bunting KL, Polo JM, Farès C, Arrowsmith CH, et al. (2010) A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 17:400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai HF, Liang XX, Li L, Zhao CS, Gong P, Liang ZJ, Zhu WL, Jiang HL, Luo C. (2011) Identification of novel 5-hydroxy-1H-indole-3-carboxylates with anti-HBV activities based on 3D QSAR studies. J Mol Model 17:1831–1840 [DOI] [PubMed] [Google Scholar]
- Chai HF, Zhao YF, Zhao C, Gong P. (2006) Synthesis and in vitro anti-hepatitis B virus activities of some ethyl 6-bromo-5-hydroxy-1H-indole-3-carboxylates. Bioorg Med Chem 14:911–917 [DOI] [PubMed] [Google Scholar]
- Chan DSH, Lee HM, Yang F, Che CM, Wong CC, Abagyan R, Leung CH, Ma DL. (2010) Structure-based discovery of natural-product-like TNF-α inhibitors. Angew Chem Int Ed Engl 49:2860–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Swaan PW. (2006) Computational approaches to modeling drug transporters. Eur J Pharm Sci 27:411–424 [DOI] [PubMed] [Google Scholar]
- Changeux JP, Edelstein S. (2011). Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol Rep 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WR, Yean D, Amin K, Green C, Jong L. (2007) Computer-aided rational drug design: a novel agent (SR13668) designed to mimic the unique anticancer mechanisms of dietary indole-3-carbinol to block Akt signaling. J Med Chem 50:3412–3415 [DOI] [PubMed] [Google Scholar]
- Chen D, Misra M, Sower L, Peterson JW, Kellogg GE, Schein CH. (2008) Novel inhibitors of anthrax edema factor. Bioorg Med Chem 16:7225–7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lai LH. (2006) Pocket v.2: further developments on receptor-based pharmacophore modeling. J Chem Inf Model 46:2684–2691 [DOI] [PubMed] [Google Scholar]
- Chen J, Swamidass SJ, Dou Y, Bruand J, Baldi P. (2005) ChemDB: a public database of small molecules and related chemoinformatics resources. Bioinformatics 21:4133–4139 [DOI] [PubMed] [Google Scholar]
- Chen JH, Linstead E, Swamidass SJ, Wang D, Baldi P. (2007) ChemDB update—full-text search and virtual chemical space. Bioinformatics 23:2348–2351 [DOI] [PubMed] [Google Scholar]
- Chen Y-W and Lin C-J (2006) Combining SVMs with various feature selection strategies. Studies in Fuzziness and Soft Computing 207:315–324 [Google Scholar]
- Cheng KW, Leung PC. (2000) The expression, regulation and signal transduction pathways of the mammalian gonadotropin-releasing hormone receptor. Can J Physiol Pharmacol 78:1029–1052 [PubMed] [Google Scholar]
- Cheng TJ, Li QL, Zhou Z, Wang Y, Bryant SH. (2012) Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J 14:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YK, Kuo CC, Wu YS, Chen CT, Coumar MS, Wu JS, Hsieh HP, Chang CY, Jseng HY, Wu MH, et al. (2009) Generation of ligand-based pharmacophore model and virtual screening for identification of novel tubulin inhibitors with potent anticancer activity. J Med Chem 52:4221–4233 [DOI] [PubMed] [Google Scholar]
- Chikhi R, Sael L, Kihara D. (2010) Real-time ligand binding pocket database search using local surface descriptors. Proteins 78:2007–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosis G, Rodina A, Moulick K. (2006) Emerging Hsp90 inhibitors: from discovery to clinic. Anticancer Agents Med Chem 6:1–8 [DOI] [PubMed] [Google Scholar]
- Chiu TL, Solberg J, Patil S, Geders TW, Zhang X, Rangarajan S, Francis R, Finzel BC, Walters MA, Hook DJ, et al. (2009) Identification of novel non-hydroxamate anthrax toxin lethal factor inhibitors by topomeric searching, docking and scoring, and in vitro screening. J Chem Inf Model 49:2726–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivian D, Baker D. (2006) Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res 34:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H, Nah KH, Lee SN, Lee HS, Lee HS, Jo SH, Leem CH, Jang YJ. (2006) A novel hypothesis for the binding mode of HERG channel blockers. Biochem Biophys Res Commun 344:72–78 [DOI] [PubMed] [Google Scholar]
- Cianchetta G, Li Y, Kang J, Rampe D, Fravolini A, Cruciani G, Vaz RJ. (2005) Predictive models for hERG potassium channel blockers. Bioorg Med Chem Lett 15:3637–3642 [DOI] [PubMed] [Google Scholar]
- Cleves AE, Jain AN. (2006) Robust ligand-based modeling of the biological targets of known drugs. J Med Chem 49:2921–2938 [DOI] [PubMed] [Google Scholar]
- Cogan DA, Aungst R, Breinlinger EC, Fadra T, Goldberg DR, Hao MH, Kroe R, Moss N, Pargellis C, Qian KC, et al. (2008) Structure-based design and subsequent optimization of 2-tolyl-(1,2,3-triazol-1-yl-4-carboxamide) inhibitors of p38 MAP kinase. Bioorg Med Chem Lett 18:3251–3255 [DOI] [PubMed] [Google Scholar]
- Cohen P, Alessi DR. (2013) Kinase drug discovery—what’s next in the field? ACS Chem Biol 8:96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn PJ, Christopoulos A, Lindsley CW. (2009) Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov 8:41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly ML. (1983) Analytical molecular-surface calculation. J Appl Cryst 16:548–558 [Google Scholar]
- Coulson CA, Moffitt WE. (1947) Strain in Non-Tetrahedral Carbon Atoms. J Chem Phys 15:151 [Google Scholar]
- Coumar MS, Chu CY, Lin CW, Shiao HY, Ho YL, Reddy R, Lin WH, Chen CH, Peng YH, Leou JS, et al. (2010a) Fast-forwarding hit to lead: aurora and epidermal growth factor receptor kinase inhibitor lead identification. J Med Chem 53:4980–4988 [DOI] [PubMed] [Google Scholar]
- Coumar MS, Leou JS, Shukla P, Wu JS, Dixit AK, Lin WH, Chang CY, Lien TW, Tan UK, Chen CH, et al. (2009) Structure-based drug design of novel Aurora kinase A inhibitors: structural basis for potency and specificity. J Med Chem 52:1050–1062 [DOI] [PubMed] [Google Scholar]
- Coumar MS, Tsai MT, Chu CY, Uang BJ, Lin WH, Chang CY, Chang TY, Leou JS, Teng CH, Wu JS, et al. (2010b) Identification, SAR studies, and X-ray co-crystallographic analysis of a novel furanopyrimidine aurora kinase A inhibitor. ChemMedChem 5:255–267 [DOI] [PubMed] [Google Scholar]
- Coutsias EA, Seok C, Jacobson MP, Dill KA. (2004) A kinematic view of loop closure. J Comput Chem 25:510–528 [DOI] [PubMed] [Google Scholar]
- Cozza G, Bonvini P, Zorzi E, Poletto G, Pagano MA, Sarno S, Donella-Deana A, Zagotto G, Rosolen A, Pinna LA, et al. (2006) Identification of ellagic acid as potent inhibitor of protein kinase CK2: a successful example of a virtual screening application. J Med Chem 49:2363–2366 [DOI] [PubMed] [Google Scholar]
- Cozza G, Mazzorana M, Papinutto E, Bain J, Elliott M, di Maira G, Gianoncelli A, Pagano MA, Sarno S, Ruzzene M, et al. (2009) Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2. Biochem J 421:387–395 [DOI] [PubMed] [Google Scholar]
- Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. (2009) Evaluation of template-based models in CASP8 with standard measures. Proteins 77 (Suppl 9):18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzini P, Kellogg GE, Spyrakis F, Abraham DJ, Costantino G, Emerson A, Fanelli F, Gohlke H, Kuhn LA, Morris GM, et al. (2008) Target flexibility: an emerging consideration in drug discovery and design. J Med Chem 51:6237–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer RD, Jilek RJ, Andrews KM. (2002) Dbtop: topomer similarity searching of conventional structure databases. J Mol Graph Model 20:447–462 [DOI] [PubMed] [Google Scholar]
- Cramer RD, Patterson DE, Bunce JD. (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967 [DOI] [PubMed] [Google Scholar]
- Cristianini N, Shawe-Taylor J. (2000) An Introduction to Support Vector Machines: and Other Kernel-Based Learning Methods, Cambridge University Press, Cambridge, New York [Google Scholar]
- Cross JB, Thompson DC, Rai BK, Baber JC, Fan KY, Hu Y, Humblet C. (2009) Comparison of several molecular docking programs: pose prediction and virtual screening accuracy. J Chem Inf Model 49:1455–1474 [DOI] [PubMed] [Google Scholar]
- Cruciani G, Carosati E, De Boeck B, Ethirajulu K, Mackie C, Howe T, Vianello R. (2005) MetaSite: understanding metabolism in human cytochromes from the perspective of the chemist. J Med Chem 48:6970–6979 [DOI] [PubMed] [Google Scholar]
- Davis AM, Riley RJ. (2004) Predictive ADMET studies, the challenges and the opportunities. Curr Opin Chem Biol 8:378–386 [DOI] [PubMed] [Google Scholar]
- Davis IW, Baker D. (2009) RosettaLigand docking with full ligand and receptor flexibility. J Mol Biol 385:381–392 [DOI] [PubMed] [Google Scholar]
- Davis L. (1991) Handbook of Genetic Algorithms, Van Nostrand Reinhold, New York [Google Scholar]
- Dayam R, Al-Mawsawi LQ, Zawahir Z, Witvrouw M, Debyser Z, Neamati N. (2008) Quinolone 3-carboxylic acid pharmacophore: design of second generation HIV-1 integrase inhibitors. J Med Chem 51:1136–1144 [DOI] [PubMed] [Google Scholar]
- Daylight Chemical Information Systems (2008) Daylight Theory: SMARTS—A language for describing molecular patterns. Available from http://www.daylight.com/dayhtml/doc/theory/theory.smarts.html#RTFToC35
- Daylight Chemical Information Systems (2013) Daylight theory manual. Available from http://www.daylight.com/dayhtml/doc/theory/
- de Beer SB, Vermeulen NP, Oostenbrink C. (2010) The role of water molecules in computational drug design. Curr Top Med Chem 10:55–66 [DOI] [PubMed] [Google Scholar]
- de Graaf C, Foata N, Engkvist O, Rognan D. (2008) Molecular modeling of the second extracellular loop of G-protein coupled receptors and its implication on structure-based virtual screening. Proteins 71:599–620 [DOI] [PubMed] [Google Scholar]
- de Graaf C, Pospisil P, Pos W, Folkers G, Vermeulen NP. (2005) Binding mode prediction of cytochrome P450 and thymidine kinase protein-ligand complexes by consideration of water and rescoring in automated docking. J Med Chem 48:2308–2318 [DOI] [PubMed] [Google Scholar]
- de Graaf C, Rognan D. (2009) Customizing G protein-coupled receptor models for structure-based virtual screening. Curr Pharm Des 15:4026–4048 [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Ekins S. (2002) Pharmacophore modeling of cytochromes P450. Adv Drug Deliv Rev 54:367–383 [DOI] [PubMed] [Google Scholar]
- Deacon RM. (2006) Digging and marble burying in mice: simple methods for in vivo identification of biological impacts. Nat Protoc 1:122–124 [DOI] [PubMed] [Google Scholar]
- Degen J, Wegscheid-Gerlach C, Zaliani A, Rarey M. (2008) On the art of compiling and using ‘drug-like’ chemical fragment spaces. ChemMedChem 3:1503–1507 [DOI] [PubMed] [Google Scholar]
- Deng JX, Lee KW, Sanchez T, Cui M, Neamati N, Briggs JM. (2005) Dynamic receptor-based pharmacophore model development and its application in designing novel HIV-1 integrase inhibitors. J Med Chem 48:1496–1505 [DOI] [PubMed] [Google Scholar]
- DesJarlais RL, Sheridan RP, Dixon JS, Kuntz ID, Venkataraghavan R. (1986) Docking flexible ligands to macromolecular receptors by molecular shape. J Med Chem 29:2149–2153 [DOI] [PubMed] [Google Scholar]
- DesJarlais RL, Sheridan RP, Seibel GL, Dixon JS, Kuntz ID, Venkataraghavan R. (1988) Using shape complementarity as an initial screen in designing ligands for a receptor binding site of known three-dimensional structure. J Med Chem 31:722–729 [DOI] [PubMed] [Google Scholar]
- Desmet J, De Maeyer M, Hazes B, Lasters I. (1992) The dead-end elimination theorem and its use in protein side-chain positioning. Nature 356:539–542 [DOI] [PubMed] [Google Scholar]
- Devillers J, Balaban AT. (1999) Topological Indices and Related Descriptors in QSAR and QSPR, Gordon and Breach, Amsterdam [Google Scholar]
- DeWitte RS, Shakhnovich E. (1997) SMoG: De novo design method based on simple, fast and accurate free energy estimates. J Am Chem Soc 119:4608–4617 [Google Scholar]
- Dias R, de Azevedo WF., Jr (2008) Molecular docking algorithms. Curr Drug Targets 9:1040–1047 [DOI] [PubMed] [Google Scholar]
- Dimitropoulos D, Ionides J, and Henrick K (2006) Using PDBeChem to Search the PDB Ligand Dictionary, in Current Protocols in Bioinformatics; John Wiley & Sons, 14.13.11-14.13.13. [DOI] [PubMed]
- Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Friesner RA. (2006) PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 20:647–671 [DOI] [PubMed] [Google Scholar]
- Doddareddy MR, Choo H, Cho YS, Rhim H, Koh HY, Lee JH, Jeong SW, Pae AN. (2007) 3D pharmacophore based virtual screening of T-type calcium channel blockers. Bioorg Med Chem 15:1091–1105 [DOI] [PubMed] [Google Scholar]
- Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet BK. (2002) Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem 45:2213–2221 [DOI] [PubMed] [Google Scholar]
- Dunbrack RL, Karplus M., Jr (1993) Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J Mol Biol 230:543–574 [DOI] [PubMed] [Google Scholar]
- Dunbrack RL, Karplus M., Jr (1994) Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nat Struct Biol 1:334–340 [DOI] [PubMed] [Google Scholar]
- Durán A, Martínez GC, Pastor M. (2008) Development and validation of AMANDA, a new algorithm for selecting highly relevant regions in Molecular Interaction Fields. J Chem Inf Model 48:1813–1823 [DOI] [PubMed] [Google Scholar]
- Durán A, Zamora I, Pastor M. (2009) Suitability of GRIND-based principal properties for the description of molecular similarity and ligand-based virtual screening. J Chem Inf Model 49:2129–2138 [DOI] [PubMed] [Google Scholar]
- Durant JL, Leland BA, Henry DR, Nourse JG. (2002) Reoptimization of MDL keys for use in drug discovery. J Chem Inf Comput Sci 42:1273–1280 [DOI] [PubMed] [Google Scholar]
- Durrant JD, Hall L, Swift RV, Landon M, Schnaufer A, Amaro RE. (2010a) Novel naphthalene-based inhibitors of Trypanosoma brucei RNA editing ligase 1. PLoS Negl Trop Dis 4:e803;1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant JD, McCammon JA. (2010) Computer-aided drug-discovery techniques that account for receptor flexibility. Curr Opin Pharmacol 10:770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant JD, Urbaniak MD, Ferguson MA, McCammon JA. (2010b) Computer-aided identification of Trypanosoma brucei uridine diphosphate galactose 4′-epimerase inhibitors: toward the development of novel therapies for African sleeping sickness. J Med Chem 53:5025–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiben CB, Siegel JB, Bale JB, Cooper S, Khatib F, Shen BW, Players F, Stoddard BL, Popovic Z, Baker D. (2012) Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. Nat Biotechnol 30:190–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Crumb WJ, Sarazan RD, Wikel JH, Wrighton SA. (2002a) Three-dimensional quantitative structure-activity relationship for inhibition of human ether-a-go-go-related gene potassium channel. J Pharmacol Exp Ther 301:427–434 [DOI] [PubMed] [Google Scholar]
- Ekins S, Kim RB, Leake BF, Dantzig AH, Schuetz EG, Lan LB, Yasuda K, Shepard RL, Winter MA, Schuetz JD, et al. (2002b) Application of three-dimensional quantitative structure-activity relationships of P-glycoprotein inhibitors and substrates. Mol Pharmacol 61:974–981 [DOI] [PubMed] [Google Scholar]
- Ekins S, Mestres J, Testa B. (2007) In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br J Pharmacol 152:9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekonomiuk D, Su XC, Ozawa K, Bodenreider C, Lim SP, Otting G, Huang D, Caflisch A. (2009a) Flaviviral protease inhibitors identified by fragment-based library docking into a structure generated by molecular dynamics. J Med Chem 52:4860–4868 [DOI] [PubMed] [Google Scholar]
- Ekonomiuk D, Su XC, Ozawa K, Bodenreider C, Lim SP, Yin Z, Keller TH, Beer D, Patel V, Otting G, et al. (2009b) Discovery of a non-peptidic inhibitor of West Nile virus NS3 protease by high-throughput docking. PLoS Negl Trop Dis 3:e356:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedy IJ, Egan WJ. (2008) Can we use docking and scoring for hit-to-lead optimization? J Comput Aided Mol Des 22:161–168 [DOI] [PubMed] [Google Scholar]
- Erickson JA, Mader MM, Watson IA, Webster YW, Higgs RE, Bell MA, Vieth M. (2010) Structure-guided expansion of kinase fragment libraries driven by support vector machine models. Biochim Biophys Acta 1804:642–652 [DOI] [PubMed] [Google Scholar]
- Erve JC, Svensson MA, von Euler-Chelpin H, Klasson-Wehler E. (2004) Characterization of glutathione conjugates of the remoxipride hydroquinone metabolite NCQ-344 formed in vitro and detection following oxidation by human neutrophils. Chem Res Toxicol 17:564–571 [DOI] [PubMed] [Google Scholar]
- Euer N, Schwirzke M, Evtimova V, Burtscher H, Jarsch M, Tarin D, Weidle UH. (2002) Identification of genes associated with metastasis of mammary carcinoma in metastatic versus non-metastatic cell lines. Anticancer Res 22 (Suppl. 2A):733–740 [PubMed] [Google Scholar]
- Evers A, Gohlke H, Klebe G. (2003) Ligand-supported homology modelling of protein binding-sites using knowledge-based potentials. J Mol Biol 334:327–345 [DOI] [PubMed] [Google Scholar]
- Evers A, Klebe G. (2004) Successful virtual screening for a submicromolar antagonist of the neurokinin-1 receptor based on a ligand-supported homology model. J Med Chem 47:5381–5392 [DOI] [PubMed] [Google Scholar]
- Fauman EB, Rai BK, Huang ES. (2011) Structure-based druggability assessment—identifying suitable targets for small molecule therapeutics. Curr Opin Chem Biol 15:463–468 [DOI] [PubMed] [Google Scholar]
- Fedorov VV. (1972) Theory of Optimal Experiments, Academic Press, New York [Google Scholar]
- Feher M. (2006) Consensus scoring for protein-ligand interactions. Drug Discov Today 11:421–428 [DOI] [PubMed] [Google Scholar]
- Feher M, Gao Y, Baber JC, Shirley WA, Saunders J. (2008) The use of ligand-based de novo design for scaffold hopping and sidechain optimization: two case studies. Bioorg Med Chem 16:422–427 [DOI] [PubMed] [Google Scholar]
- Ferrè F, Ausiello G, Zanzoni A, Helmer-Citterich M. (2004) SURFACE: a database of protein surface regions for functional annotation. Nucleic Acids Res 32 (Database issue):D240–D244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficker E, Obejero-Paz CA, Zhao S, Brown AM. (2002) The binding site for channel blockers that rescue misprocessed human long QT syndrome type 2 ether-a-gogo-related gene (HERG) mutations. J Biol Chem 277:4989–4998 [DOI] [PubMed] [Google Scholar]
- Fink T, Bruggesser H, Reymond JL. (2005) Virtual exploration of the small-molecule chemical universe below 160 Daltons. Angew Chem Int Ed Engl 44:1504–1508 [DOI] [PubMed] [Google Scholar]
- Fink T, Reymond JL. (2007) Virtual exploration of the chemical universe up to 11 atoms of C, N, O, F: assembly of 26.4 million structures (110.9 million stereoisomers) and analysis for new ring systems, stereochemistry, physicochemical properties, compound classes, and drug discovery. J Chem Inf Model 47:342–353 [DOI] [PubMed] [Google Scholar]
- Foloppe N, Chen IJ. (2009) Conformational sampling and energetics of drug-like molecules. Curr Med Chem 16:3381–3413 [DOI] [PubMed] [Google Scholar]
- Free SM, Wilson JW., Jr (1964) A mathematical contribution to structure-activity studies. J Med Chem 7:395–399 [DOI] [PubMed] [Google Scholar]
- Frembgen-Kesner T, Elcock AH. (2006) Computational sampling of a cryptic drug binding site in a protein receptor: explicit solvent molecular dynamics and inhibitor docking to p38 MAP kinase. J Mol Biol 359:202–214 [DOI] [PubMed] [Google Scholar]
- Friedman R, Caflisch A. (2009) Discovery of plasmepsin inhibitors by fragment-based docking and consensus scoring. ChemMedChem 4:1317–1326 [DOI] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, et al. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749 [DOI] [PubMed] [Google Scholar]
- Frimurer TM, Bywater R, Naerum L, Lauritsen LN, Brunak S. (2000) Improving the odds in discriminating “drug-like” from “non drug-like” compounds. J Chem Inf Comput Sci 40:1315–1324 [DOI] [PubMed] [Google Scholar]
- Fu J, Bian M, Jiang Q, Zhang C. (2007) Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res 5:1–10 [DOI] [PubMed] [Google Scholar]
- Fukunishi H, Teramoto R, Takada T, Shimada J. (2008) Bootstrap-based consensus scoring method for protein-ligand docking. J Chem Inf Model 48:988–996 [DOI] [PubMed] [Google Scholar]
- Fukunishi Y. (2009) Structure-based drug screening and ligand-based drug screening with machine learning. Comb Chem High Throughput Screen 12:397–408 [DOI] [PubMed] [Google Scholar]
- Fuss B, Becker T, Zinke I, Hoch M. (2006) The cytohesin Steppke is essential for insulin signalling in Drosophila. Nature 444:945–948 [DOI] [PubMed] [Google Scholar]
- Galvez J, Garcia-Domenech RY, de Julian-Oritz JV. (1998) Design of new antineoplastic lead drugs by molecular topology. Expert Opin Ther Targets 2:265–268 [Google Scholar]
- Galvez J, Garcia R, Salabert MT, Soler R. (1994) Charge indexes. New topological descriptors. J Chem Inf Comput Sci 34:520–525 [Google Scholar]
- Gálvez J, García-Domenech R, de Julián-Ortiz JV, Soler R. (1995) Topological approach to drug design. J Chem Inf Comput Sci 35:272–284 [DOI] [PubMed] [Google Scholar]
- Gálvez J, Garcia-Domenech R, De Julián-Ortiz V, Soler R. (1994) Topological approach to analgesia. J Chem Inf Comput Sci 34:1198–1203 [DOI] [PubMed] [Google Scholar]
- Garg D, Gandhi T, Gopi Mohan C. (2008) Exploring QSTR and toxicophore of hERG K+ channel blockers using GFA and HypoGen techniques. J Mol Graph Model 26:966–976 [DOI] [PubMed] [Google Scholar]
- Gasparini F, Bilbe G, Gomez-Mancilla B, Spooren W. (2008) mGluR5 antagonists: discovery, characterization and drug development. Curr Opin Drug Discov Devel 11:655–665 [PubMed] [Google Scholar]
- Gasteiger J. (1979) A representation of π systems for efficient computer manipulation. J Chem Inf Comput Sci 19:111–115 [Google Scholar]
- Gasteiger J, Hutchings MG. (1983) New empirical-models of substituent polarizability and their application to stabilization effects in positively charged species. Tetrahedron Lett 24:2537–2540 [Google Scholar]
- Gasteiger J, Hutchings MG. (1984) Quantitative models of gas-phase proton-transfer reactions involving alcohols, ethers, and their thio analogs - correlation analyses based on residual electronegativity and effective polarizability. J Am Chem Soc 106:6489–6495 [Google Scholar]
- Gasteiger J, Marsili M. (1980) Iterative partial equalization of orbital electronegativity - a rapid access to atomic charges. Tetrahedron 36:3219–3228 [Google Scholar]
- Gasteiger J, Saller H. (1985) Calculation of the charge distribution in conjugated systems by a quantification of the resonance concept. Angew Chem Int Ed Engl 24:687–689 [Google Scholar]
- Gibbs MN, MacKay DC. (2000) Variational Gaussian process classifiers. IEEE Trans Neural Netw 11:1458–1464 [DOI] [PubMed] [Google Scholar]
- Glen RC. (1994) A fast empirical method for the calculation of molecular polarizability. J Comput Aided Mol Des 8:457–466 [DOI] [PubMed] [Google Scholar]
- Goldstein A. (1974) Principles of Drug Action; the Basis of Pharmacology, Wiley, New York [Google Scholar]
- Golla S, Neely BJ, Whitebay E, Madihally S, Robinson RL, Jr, Gasem KA. (2012) Virtual design of chemical penetration enhancers for transdermal drug delivery. Chem Biol Drug Des 79:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AC, Oprea TI. (2008) Optimization of CAMD techniques 3. Virtual screening enrichment studies: a help or hindrance in tool selection? J Comput Aided Mol Des 22:169–178 [DOI] [PubMed] [Google Scholar]
- Goodarzi M, Freitas MP, Jensen R. (2009) Feature selection and linear/nonlinear regression methods for the accurate prediction of glycogen synthase kinase-3beta inhibitory activities. J Chem Inf Model 49:824–832 [DOI] [PubMed] [Google Scholar]
- Goodford PJ. (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem 28:849–857 [DOI] [PubMed] [Google Scholar]
- Gordon CP, Griffith R, Keller PA. (2007) Control of HIV through the inhibition of HIV-1 integrase: a medicinal chemistry perspective. Med Chem 3:199–220 [DOI] [PubMed] [Google Scholar]
- Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. (2002) LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res 30:402–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubmüller H. (1995) Predicting slow structural transitions in macromolecular systems: Conformational flooding. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics 52:2893–2906 [DOI] [PubMed] [Google Scholar]
- Guillen MD, Gasteiger J. (1983) Extension of the method of iterative partial equalization of orbital electronegativity to small ring-systems. Tetrahedron 39:1331–1335 [Google Scholar]
- Güner OF. (2000) Pharmacophore Perception, Development, and Use in Drug Ddesign, International University Line, LaJolla, CA [Google Scholar]
- Hafner M, Schmitz A, Grüne I, Srivatsan SG, Paul B, Kolanus W, Quast T, Kremmer E, Bauer I, Famulok M. (2006) Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature 444:941–944 [DOI] [PubMed] [Google Scholar]
- Hajduk PJ, Huth JR, Tse C. (2005) Predicting protein druggability. Drug Discov Today 10:1675–1682 [DOI] [PubMed] [Google Scholar]
- Halgren TA. (1996) Merck molecular force field. 1. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519 [Google Scholar]
- Hall JM, Couse JF, Korach KS. (2001) The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem 276:36869–36872 [DOI] [PubMed] [Google Scholar]
- Halperin I, Ma B, Wolfson H, Nussinov R. (2002) Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins 47:409–443 [DOI] [PubMed] [Google Scholar]
- Han J and Kamber M (2006) Data mining: concepts and techniques. Amsterdam; Boston [Google Scholar]
- Hancock AA, Brune ME. (2005) Assessment of pharmacology and potential anti-obesity properties of H3 receptor antagonists/inverse agonists. Expert Opin Investig Drugs 14:223–241 [DOI] [PubMed] [Google Scholar]
- Hancox JC, Mitcheson JS. (2006) Combined hERG channel inhibition and disruption of trafficking in drug-induced long QT syndrome by fluoxetine: a case-study in cardiac safety pharmacology. Br J Pharmacol 149:457–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann MM, Leach AR, Harper G. (2001) Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci 41:856–864 [DOI] [PubMed] [Google Scholar]
- Hansch C. (1964) Rho-Sigma-Pi-analysis—a method for the correlation of biological-activity and chemical-structure. J Amer Chem Soc 86:1616–1626 [Google Scholar]
- Hansch C, Björkroth JP, Leo A. (1987) Hydrophobicity and central nervous system agents: on the principle of minimal hydrophobicity in drug design. J Pharm Sci 76:663–687 [DOI] [PubMed] [Google Scholar]
- Hansch C, Maloney PP, Fujita T, Muir RM. (1962) Correlation of biological activity of phenoxyacetic acids with Hammett substituent constants and partition coefficients. Nature 194:178–180 [Google Scholar]
- Harris CJ, Hill RD, Sheppard DW, Slater MJ, Stouten PF. (2011) The design and application of target-focused compound libraries. Comb Chem High Throughput Screen 14:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman GD, Egbertson MS, Halczenko W, Laswell WL, Duggan ME, Smith RL, Naylor AM, Manno PD, Lynch RJ, Zhang G, et al. (1992) Non-peptide fibrinogen receptor antagonists. 1. Discovery and design of exosite inhibitors. J Med Chem 35:4640–4642 [DOI] [PubMed] [Google Scholar]
- Hemmer MC, Steinhauer V, Gasteiger J. (1999) Deriving the 3D structure of organic molecules from their infrared spectra. Vib Spectrosc 19:151–164 [Google Scholar]
- Hendlich M, Rippmann F, Barnickel G. (1997) LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model 15:359–363, 389 [DOI] [PubMed] [Google Scholar]
- Henrich S, Salo-Ahen OMH, Huang B, Rippmann FF, Cruciani G, Wade RC. (2010) Computational approaches to identifying and characterizing protein binding sites for ligand design. J Mol Recognit 23:209–219 [DOI] [PubMed] [Google Scholar]
- Hessler G, Zimmermann M, Matter H, Evers A, Naumann T, Lengauer T, Rarey M. (2005) Multiple-ligand-based virtual screening: methods and applications of the MTree approach. J Med Chem 48:6575–6584 [DOI] [PubMed] [Google Scholar]
- Hillisch A, Pineda LF, Hilgenfeld R. (2004) Utility of homology models in the drug discovery process. Drug Discov Today 9:659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinze J, Jaffe HH. (1962) Electronegativity. I. Orbital electronegativity of neutral atoms. J Am Chem Soc 84:540–546 [Google Scholar]
- Hinze J, Whitehead MA, Jaffe HH. (1963) Electronegativity. II. Bond and orbital electronegativities. J Am Chem Soc 85:148–154 [Google Scholar]
- Hoeglund AB, Bostic HE, Howard AL, Wanjala IW, Best MD, Baker DL, Parrill AL. (2010) Optimization of a pipemidic acid autotaxin inhibitor. J Med Chem 53:1056–1066 [DOI] [PubMed] [Google Scholar]
- Holford N, Ma SC, Ploeger BA. (2010) Clinical trial simulation: a review. Clin Pharmacol Ther 88:166–182 [DOI] [PubMed] [Google Scholar]
- Hong H, Xie Q, Ge W, Qian F, Fang H, Shi L, Su Z, Perkins R, Tong W. (2008) Mold(2), molecular descriptors from 2D structures for chemoinformatics and toxicoinformatics. J Chem Inf Model 48:1337–1344 [DOI] [PubMed] [Google Scholar]
- Honig B, Nicholls A. (1995) Classical electrostatics in biology and chemistry. Science 268:1144–1149 [DOI] [PubMed] [Google Scholar]
- Hopfinger AJ, Wang S, Tokarski JS, Jin B, Albuquerque M, Madhav PR, Duraiswami C. (1997) Construction of 3D-QSAR models using the 4D-QSAR analysis formalism. J Am Chem Soc 119:10509–10524 [Google Scholar]
- Horvath D. (1997) A virtual screening approach applied to the search for trypanothione reductase inhibitors. J Med Chem 40:2412–2423 [DOI] [PubMed] [Google Scholar]
- Hou T, Wang J, Zhang W, Wang W, Xu X. (2006) Recent advances in computational prediction of drug absorption and permeability in drug discovery. Curr Med Chem 13:2653–2667 [DOI] [PubMed] [Google Scholar]
- Hough LB. (2001) Genomics meets histamine receptors: new subtypes, new receptors. Mol Pharmacol 59:415–419 [PubMed] [Google Scholar]
- Howells LM, Gallacher-Horley B, Houghton CE, Manson MM, Hudson EA. (2002) Indole-3-carbinol inhibits protein kinase B/Akt and induces apoptosis in the human breast tumor cell line MDA MB468 but not in the nontumorigenic HBL100 line. Mol Cancer Ther 1:1161–1172 [PubMed] [Google Scholar]
- Hristozov DP, Oprea TI, Gasteiger J. (2007) Virtual screening applications: a study of ligand-based methods and different structure representations in four different scenarios. J Comput Aided Mol Des 21:617–640 [DOI] [PubMed] [Google Scholar]
- Huang N, Shoichet BK, Irwin JJ. (2006) Benchmarking sets for molecular docking. J Med Chem 49:6789–6801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T, van Gunsteren WF. (1998) SWARM-MD: Searching conformational space by cooperative molecular dynamics. J Phys Chem A 102:5937–5943 [Google Scholar]
- Huey R, Morris GM, Olson AJ, Goodsell DS. (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152 [DOI] [PubMed] [Google Scholar]
- Huguenard JR, Prince DA. (1992) A novel T-type current underlies prolonged Ca(2+)-dependent burst firing in GABAergic neurons of rat thalamic reticular nucleus. J Neurosci 12:3804–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huirne JA, Lambalk CB. (2001) Gonadotropin-releasing-hormone-receptor antagonists. Lancet 358:1793–1803 [DOI] [PubMed] [Google Scholar]
- Hutter MC. (2011) Graph-based similarity concepts in virtual screening. Future Med Chem 3: 485–501 [DOI] [PubMed] [Google Scholar]
- Ihlenfeldt WD, Gasteiger J. (1994) Hash codes for the identification and classification of molecular structure elements. J Comput Chem 15:793–813 [Google Scholar]
- Ijjaali I, Barrere C, Nargeot J, Petitet F, Bourinet E. (2007) Ligand-based virtual screening to identify new T-type calcium channel blockers. Channels (Austin) 1:300–304 [DOI] [PubMed] [Google Scholar]
- InChITRUST (2013) InChI FAQ. Available from http://www.inchi-trust.org/fileadmin/user_upload/html/inchifaq/inchi-faq.html
- Inoue M, Ma L, Aoki J, Ueda H. (2008) Simultaneous stimulation of spinal NK1 and NMDA receptors produces LPC which undergoes ATX-mediated conversion to LPA, an initiator of neuropathic pain. J Neurochem 107:1556–1565 [DOI] [PubMed] [Google Scholar]
- Irwin JJ. (2008) Community benchmarks for virtual screening. J Comput Aided Mol Des 22:193–199 [DOI] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanciuc O (2007) Applications of support vector machines in chemistry, in Reviews in Computational Chemistry pp 291–400, John Wiley & Sons, Inc., Hoboken, NJ.
- Izuhara Y, Yamaoka N, Kodama H, Dan T, Takizawa S, Hirayama N, Meguro K, van Ypersele de Strihou C, Miyata T. (2010) A novel inhibitor of plasminogen activator inhibitor-1 provides antithrombotic benefits devoid of bleeding effect in nonhuman primates. J Cereb Blood Flow Metab 30:904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AN. (2003) Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 46:499–511 [DOI] [PubMed] [Google Scholar]
- Jamei M, Marciniak S, Feng K, Barnett A, Tucker G, Rostami-Hodjegan A. (2009) The SimCYP population-based ADME simulator. Expert Opin Drug Metab Toxicol 5:211–223 [DOI] [PubMed] [Google Scholar]
- Jiao ZG, He HQ, Zeng CC, Tan JJ, Hu LM, Wang CX. (2010) Design, synthesis and anti-HIV integrase evaluation of N-(5-chloro-8-hydroxy-2-styrylquinolin-7-yl)benzenesulfonamide derivatives. Molecules 15:1903–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilek RJ, Cramer RD. (2004) Topomers: a validated protocol for their self-consistent generation. J Chem Inf Comput Sci 44:1221–1227 [DOI] [PubMed] [Google Scholar]
- Joffe E. (1991) Complication during root canal therapy following accidental extrusion of sodium hypochlorite through the apical foramen. Gen Dent 39:460–461 [PubMed] [Google Scholar]
- Johnson MA, Maggiora GM. (1990) Concepts and Applications of Molecular Similarity, Wiley, New York [Google Scholar]
- Jones G, Willett P, Glen RC. (1995) A genetic algorithm for flexible molecular overlay and pharmacophore elucidation. J Comput Aided Mol Des 9:532–549 [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL. (2004) The many roles of computation in drug discovery. Science 303:1813–1818 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL. (2010) Drug discovery: Pulled from a protein’s embrace. Nature 466:42–43 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Duffy EM. (2002) Prediction of drug solubility from structure. Adv Drug Deliv Rev 54:355–366 [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Chen YP. (2011) Structure-based drug design to augment hit discovery. Drug Discov Today 16:831–839 [DOI] [PubMed] [Google Scholar]
- Kandil S, Biondaro S, Vlachakis D, Cummins AC, Coluccia A, Berry C, Leyssen P, Neyts J, Brancale A. (2009) Discovery of a novel HCV helicase inhibitor by a de novo drug design approach. Bioorg Med Chem Lett 19:2935–2937 [DOI] [PubMed] [Google Scholar]
- Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. (1992) Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA 89:2195–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Rueda M, Lam PC, Yeager M, Abagyan R. (2010) GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex. Proteins 78:197–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann KW, Dawson ES, Henry LK, Field JR, Blakely RD, Meiler J. (2009) Structural determinants of species-selective substrate recognition in human and Drosophila serotonin transporters revealed through computational docking studies. Proteins 74:630–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. (2010) Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry 49:2987–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe H, Stracke ML, Nakamura H, Sano K. (1997) Expression and transcriptional regulation of the PD-Ialpha/autotaxin gene in neuroblastoma. Cancer Res 57:2516–2521 [PubMed] [Google Scholar]
- Ke YY, Shiao HY, Hsu YC, Chu CY, Wang WC, Lee YC, Lin WH, Chen CH, Hsu JT, Chang CW, et al. (2013) 3D-QSAR-assisted drug design: identification of a potent quinazoline-based Aurora kinase inhibitor. ChemMedChem 8:136–148 [DOI] [PubMed] [Google Scholar]
- Keating MT, Sanguinetti MC. (1996) Molecular genetic insights into cardiovascular disease. Science 272:681–685 [DOI] [PubMed] [Google Scholar]
- Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. (2007) Relating protein pharmacology by ligand chemistry. Nat Biotechnol 25:197–206 [DOI] [PubMed] [Google Scholar]
- Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. (2009) Predicting new molecular targets for known drugs. Nature 462:175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelder J, Grootenhuis PDJ, Bayada DM, Delbressine LP, Ploemen JP. (1999) Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm Res 16:1514–1519 [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Rodrigo J, Muller P, Rognan D. (2004) Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins 57:225–242 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371 [DOI] [PubMed] [Google Scholar]
- Kellogg GE, Semus SF, Abraham DJ. (1991) HINT: a new method of empirical hydrophobic field calculation for CoMFA. J Comput Aided Mol Des 5:545–552 [DOI] [PubMed] [Google Scholar]
- Kent JT. (1983) Information gain and a general measure of correlation. Biometrika 70:163–173 [Google Scholar]
- Khatib F, Cooper S, Tyka MD, Xu K, Makedon I, Popovic Z, Baker D, Players F. (2011) Algorithm discovery by protein folding game players. Proc Natl Acad Sci USA 108:18949–18953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib F, DiMaio F, Cooper S, Kazmierczyk M, Gilski M, Krzywda S, Zabranska H, Pichova I, et al. (2012) Crystal structure of a monomeric retroviral protease solved by protein folding game players. Nat Struct Mol Biol 18:1175–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37 (Database issue):D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Cho CH, Cho Y, Ryu J, Bhak J, Kim DS. (2008) Pocket extraction on proteins via the Voronoi diagram of spheres. J Mol Graph Model 26:1104–1112 [DOI] [PubMed] [Google Scholar]
- Kim KH. (2007) Outliers in SAR and QSAR: 2. Is a flexible binding site a possible source of outliers? J Comput Aided Mol Des 21:421–435 [DOI] [PubMed] [Google Scholar]
- Kitchen DB, Decornez H, Furr JR, Bajorath J. (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3:935–949 [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. (1997) Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 275:1927–1930 [DOI] [PubMed] [Google Scholar]
- Klebe G, Abraham U, Mietzner T. (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37:4130–4146 [DOI] [PubMed] [Google Scholar]
- Kliche S, Nagel W, Kremmer E, Atzler C, Ege A, Knorr T, Koszinowski U, Kolanus W, Haas J. (2001) Signaling by human herpesvirus 8 kaposin A through direct membrane recruitment of cytohesin-1. Mol Cell 7:833–843 [DOI] [PubMed] [Google Scholar]
- Klon AE, Glick M, Thoma M, Acklin P, Davies JW. (2004) Finding more needles in the haystack: A simple and efficient method for improving high-throughput docking results. J Med Chem 47:2743–2749 [DOI] [PubMed] [Google Scholar]
- Kontoyianni M, McClellan LM, Sokol GS. (2004) Evaluation of docking performance: comparative data on docking algorithms. J Med Chem 47:558–565 [DOI] [PubMed] [Google Scholar]
- Kortvelyesi T, Dennis S, Silberstein M, Brown L, 3rd, Vajda S. (2003) Algorithms for computational solvent mapping of proteins. Proteins 51:340–351 [DOI] [PubMed] [Google Scholar]
- Kowalski KG, Olson S, Remmers AE, Hutmacher MM. (2008) Modeling and simulation to support dose selection and clinical development of SC-75416, a selective COX-2 inhibitor for the treatment of acute and chronic pain. Clin Pharmacol Ther 83:857–866 [DOI] [PubMed] [Google Scholar]
- Krier M, Araújo-Júnior JX, Schmitt M, Duranton J, Justiano-Basaran H, Lugnier C, Bourguignon JJ, Rognan D. (2005) Design of small-sized libraries by combinatorial assembly of linkers and functional groups to a given scaffold: application to the structure-based optimization of a phosphodiesterase 4 inhibitor. J Med Chem 48:3816–3822 [DOI] [PubMed] [Google Scholar]
- Krivov GG, Shapovalov MV, Dunbrack RL., Jr (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77:778–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger DM, Evers A. (2010) Comparison of structure- and ligand-based virtual screening protocols considering hit list complementarity and enrichment factors. ChemMedChem 5:148–158 [DOI] [PubMed] [Google Scholar]
- Kubinyi H. (1997) QSAR and 3D QSAR in drug design. 1. methodology. Drug Discov Today 2:457–467 [Google Scholar]
- Kubinyi H. (1998) 3D QSAR in Drug Design, Kluwer Academic, Dordrecht [Google Scholar]
- Kukić P, Nielsen JE. (2010) Electrostatics in proteins and protein-ligand complexes. Future Med Chem 2:647–666 [DOI] [PubMed] [Google Scholar]
- Kulkarni A, Han Y, Hopfinger AJ. (2002) Predicting Caco-2 cell permeation coefficients of organic molecules using membrane-interaction QSAR analysis. J Chem Inf Comput Sci 42:331–342 [DOI] [PubMed] [Google Scholar]
- Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. (1982) A geometric approach to macromolecule-ligand interactions. J Mol Biol 161:269–288 [DOI] [PubMed] [Google Scholar]
- Kurogi Y, Güner OF. (2001) Pharmacophore modeling and three-dimensional database searching for drug design using catalyst. Curr Med Chem 8:1035–1055 [DOI] [PubMed] [Google Scholar]
- Kurup A. (2003) C-QSAR: a database of 18,000 QSARs and associated biological and physical data. J Comput Aided Mol Des 17:187–196 [DOI] [PubMed] [Google Scholar]
- Ladbury JE. (1996) Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Chem Biol 3:973–980 [DOI] [PubMed] [Google Scholar]
- Läer S, Elshoff JP, Meibohm B, Weil J, Mir TS, Zhang W, Hulpke-Wette M. (2005) Development of a safe and effective pediatric dosing regimen for sotalol based on population pharmacokinetics and pharmacodynamics in children with supraventricular tachycardia. J Am Coll Cardiol 46:1322–1330 [DOI] [PubMed] [Google Scholar]
- Lafleur K, Huang DZ, Zhou T, Caflisch A, Nevado C. (2009) Structure-based optimization of potent and selective inhibitors of the tyrosine kinase erythropoietin producing human hepatocellular carcinoma receptor B4 (EphB4). J Med Chem 52:6433–6446 [DOI] [PubMed] [Google Scholar]
- Lajiness MS, Vieth M, Erickson J. (2004) Molecular properties that influence oral drug-like behavior. Curr Opin Drug Discov Devel 7:470–477 [PubMed] [Google Scholar]
- Landon MR, Amaro RE, Baron R, Ngan CH, Ozonoff D, McCammon JA, Vajda S. (2008) Novel druggable hot spots in avian influenza neuraminidase H5N1 revealed by computational solvent mapping of a reduced and representative receptor ensemble. Chem Biol Drug Des 71:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier MC, Feher M, Ashweek NJ, Loweth CJ, Rueter JK, Slee DH, Williams JP, Zhu YF, Sullivan SK, Brown MS. (2007) Selection, synthesis, and structure-activity relationship of tetrahydropyrido[4,3-d]pyrimidine-2,4-diones as human GnRH receptor antagonists. Bioorg Med Chem 15:5590–5603 [DOI] [PubMed] [Google Scholar]
- Laskowski RA. (1995) SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph 13:323–330, 307–308 [DOI] [PubMed] [Google Scholar]
- Laurie ATR, Jackson RM. (2005) Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics 21:1908–1916 [DOI] [PubMed] [Google Scholar]
- Laurie ATR, Jackson RM. (2006) Methods for the prediction of protein-ligand binding sites for structure-based drug design and virtual ligand screening. Curr Protein Pept Sci 7:395–406 [DOI] [PubMed] [Google Scholar]
- Le Fèvre RJW. (1965) Molecular refractivity and polarizability, in Advances in Physical Organic Chemistry (Gold V. ed.), Academic Press, New York [Google Scholar]
- Leo A, Hansch C. (1971) Partition coefficients and their uses. Chem Rev 71:525–616 [Google Scholar]
- Leone V, Marinelli F, Carloni P, Parrinello M. (2010) Targeting biomolecular flexibility with metadynamics. Curr Opin Struct Biol 20:148–154 [DOI] [PubMed] [Google Scholar]
- Levitt DG, Banaszak LJ. (1992) POCKET: a computer graphics method for identifying and displaying protein cavities and their surrounding amino acids. J Mol Graph 10:229–234 [DOI] [PubMed] [Google Scholar]
- Levitt M, Park BH. (1993) Water: now you see it, now you don’t. Structure 1:223–226 [DOI] [PubMed] [Google Scholar]
- Lewell XQ, Judd DB, Watson SP, Hann MM. (1998) RECAP—retrosynthetic combinatorial analysis procedure: a powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J Chem Inf Comput Sci 38:511–522 [DOI] [PubMed] [Google Scholar]
- Lewis DF. (2003) Quantitative structure-activity relationships (QSARs) within the cytochrome P450 system: QSARs describing substrate binding, inhibition and induction of P450s. Inflammopharmacology 11:43–73 [DOI] [PubMed] [Google Scholar]
- Li M, Jung A, Ganswindt U, Marini P, Friedl A, Daniel PT, Lauber K, Jendrossek V, Belka C. (2010) Aurora kinase inhibitor ZM447439 induces apoptosis via mitochondrial pathways. Biochem Pharmacol 79:122–129 [DOI] [PubMed] [Google Scholar]
- Li N, Wang F, Niu S, Cao J, Wu K, Li Y, Yin N, Zhang X, Zhu W, Yin Y. (2009) Discovery of novel inhibitors of Streptococcus pneumoniae based on the virtual screening with the homology-modeled structure of histidine kinase (VicK). BMC Microbiol 9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tang Y, Liu H, Cheng J, Zhu W, Jiang H. (2008) Probing ligand binding modes of human cytochrome P450 2J2 by homology modeling, molecular dynamics simulation, and flexible molecular docking. Proteins 71:938–949 [DOI] [PubMed] [Google Scholar]
- Li WW, Chen JJ, Zheng RL, Zhang WQ, Cao ZX, Yang LL, Qing XY, Zhou LX, Yang L, Yu LD, et al. (2010a) Taking quinazoline as a general support-Nog to design potent and selective kinase inhibitors: application to FMS-like tyrosine kinase 3. ChemMedChem 5:513–516 [DOI] [PubMed] [Google Scholar]
- Li X, Li Y, Cheng T, Liu Z, Wang R. (2010b) Evaluation of the performance of four molecular docking programs on a diverse set of protein-ligand complexes. J Comput Chem 31:2109–2125 [DOI] [PubMed] [Google Scholar]
- Li Y, Chinni SR, Sarkar FH. (2005) Selective growth regulatory and pro-apoptotic effects of DIM is mediated by AKT and NF-kappaB pathways in prostate cancer cells. Front Biosci 10:236–243 [DOI] [PubMed] [Google Scholar]
- Li Z, Lazaridis T. (2007) Water at biomolecular binding interfaces. Phys Chem Chem Phys 9:573–581 [DOI] [PubMed] [Google Scholar]
- Liang Y. (2011) Support Vector Machines and Their Application in Chemistry and Biotechnology, CRC Press, Boca Raton [Google Scholar]
- Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. (2011) How fast-folding proteins fold. Science 334:517–520 [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26 [DOI] [PubMed] [Google Scholar]
- Liu M, Wang SM. (1999) MCDOCK: a Monte Carlo simulation approach to the molecular docking problem. J Comput Aided Mol Des 13:435–451 [DOI] [PubMed] [Google Scholar]
- Livingstone D. (2008) Artificial Neural Networks: Methods and Applications, Humana Press, Totowa, NJ: [PubMed] [Google Scholar]
- Liwo A, Czaplewski C, Ołdziej S, Scheraga HA. (2008) Computational techniques for efficient conformational sampling of proteins. Curr Opin Struct Biol 18:134–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo F, Obach RS, Shalaeva MY, Gao F. (2004) Prediction of human volume of distribution values for neutral and basic drugs. 2. Extended data set and leave-class-out statistics. J Med Chem 47:1242–1250 [DOI] [PubMed] [Google Scholar]
- Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Lavan P, Weber E, Doak AK, Côté S, et al. (2012) Large-scale prediction and testing of drug activity on side-effect targets. Nature 486:361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu IL, Huang CF, Peng YH, Lin YT, Hsieh HP, Chen CT, Lien TW, Lee HJ, Mahindroo N, Prakash E, et al. (2006) Structure-based drug design of a novel family of PPARgamma partial agonists: virtual screening, X-ray crystallography, and in vitro/in vivo biological activities. J Med Chem 49:2703–2712 [DOI] [PubMed] [Google Scholar]
- Luksch T, Chan NS, Brass S, Sotriffer CA, Klebe G, Diederich WE. (2008) Computer-aided design and synthesis of nonpeptidic plasmepsin II and IV inhibitors. ChemMedChem 3:1323–1336 [DOI] [PubMed] [Google Scholar]
- Lundstrom K. (2011) Genomics and drug discovery. Future Med Chem 3:1855–1858 [DOI] [PubMed] [Google Scholar]
- Lv J, Wang Y, Zhu L, Ma Y. (2012) Particle-swarm structure prediction on clusters. J Chem Phys 137:084104. [DOI] [PubMed] [Google Scholar]
- Majeux N, Scarsi M, Caflisch A. (2001) Efficient electrostatic solvation model for protein-fragment docking. Proteins 42:256–268 [DOI] [PubMed] [Google Scholar]
- Malamas MS, Barnes K, Hui Y, Johnson M, Lovering F, Condon J, Fobare W, Solvibile W, Turner J, Hu Y, et al. (2010) Novel pyrrolyl 2-aminopyridines as potent and selective human beta-secretase (BACE1) inhibitors. Bioorg Med Chem Lett 20:2068–2073 [DOI] [PubMed] [Google Scholar]
- Malamas MS, Erdei J, Gunawan I, Barnes K, Johnson M, Hui Y, Turner J, Hu Y, Wagner E, Fan K, et al. (2009) Aminoimidazoles as potent and selective human beta-secretase (BACE1) inhibitors. J Med Chem 52:6314–6323 [DOI] [PubMed] [Google Scholar]
- Mandal S, Moudgil M, Mandal SK. (2009) Rational drug design. Eur J Pharmacol 625:90–100 [DOI] [PubMed] [Google Scholar]
- Mandell DJ, Coutsias EA, Kortemme T. (2009) Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods 6:551–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangoni M, Roccatano D, Di Nola A. (1999) Docking of flexible ligands to flexible receptors in solution by molecular dynamics simulation. Proteins 35:153–162 [DOI] [PubMed] [Google Scholar]
- Mao KZ. (2004) Orthogonal forward selection and backward elimination algorithms for feature subset selection. IEEE Trans Syst Man Cybern B Cybern 34:629–634 [DOI] [PubMed] [Google Scholar]
- March J. (1977) Advanced Organic Chemistry: Reactions, Mechanisms, and Structure, McGraw-Hill, New York [Google Scholar]
- Marrero-Ponce Y, Santiago OM, López YM, Barigye SJ, Torrens F. (2012) Derivatives in discrete mathematics: a novel graph-theoretical invariant for generating new 2/3D molecular descriptors. I. Theory and QSPR application. J Comput Aided Mol Des 26:1229–1246 [DOI] [PubMed] [Google Scholar]
- Marshall GR, Barry CD, Bosshard H, Dammkoehler R, and Dunn D (1979) Conformational parameter in drug design - active analog approach. in Computer Assisted Drug Design pp. 205–226, ACS Publications, Washington DC.
- Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29:291–325 [DOI] [PubMed] [Google Scholar]
- Martin YC, Bures MG, Danaher EA, DeLazzer J, Lico I, Pavlik PA. (1993) A fast new approach to pharmacophore mapping and its application to dopaminergic and benzodiazepine agonists. J Comput Aided Mol Des 7:83–102 [DOI] [PubMed] [Google Scholar]
- McGregor MJ, Pallai PV. (1997) Clustering of large databases of compounds: Using the MDL ''keys'' as structural descriptors. J Chem Inf Comput Sci 37:443–448 [Google Scholar]
- McMartin C, Bohacek RS. (1997) QXP: powerful, rapid computer algorithms for structure-based drug design. J Comput Aided Mol Des 11:333–344 [DOI] [PubMed] [Google Scholar]
- Meiler J, Baker D. (2006) ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins 65:538–548 [DOI] [PubMed] [Google Scholar]
- Melo F, Sali A. (2007) Fold assessment for comparative protein structure modeling. Protein Sci 16:2412–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng EC, Kuntz ID, Abraham DJ, Kellogg GE. (1994) Evaluating docked complexes with the HINT exponential function and empirical atomic hydrophobicities. J Comput Aided Mol Des 8:299–306 [DOI] [PubMed] [Google Scholar]
- Miller KJ, Savchik J. (1979) A new empirical method to calculate average molecular polarizabilities. J Am Chem Soc 101:7206–7213 [Google Scholar]
- Miller MD, Kearsley SK, Underwood DJ, Sheridan RP. (1994) FLOG: a system to select ‘quasi-flexible’ ligands complementary to a receptor of known three-dimensional structure. J Comput Aided Mol Des 8:153–174 [DOI] [PubMed] [Google Scholar]
- Milne GWA, Nicklaus MC, Driscoll JS, Wang S, Zaharevitz D. (1994) National Cancer Institute Drug Information System 3D database. J Chem Inf Comput Sci 34:1219–1224 [DOI] [PubMed] [Google Scholar]
- Misura KMS, Baker D. (2005) Progress and challenges in high-resolution refinement of protein structure models. Proteins 59:15–29 [DOI] [PubMed] [Google Scholar]
- Misura KMS, Chivian D, Rohl CA, Kim DE, Baker D. (2006) Physically realistic homology models built with ROSETTA can be more accurate than their templates. Proc Natl Acad Sci USA 103:5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JBO, Laskowski RA, Alex A, Forster MJ, Thornton JM. (1999) BLEEP - Potential of mean force describing protein-ligand interactions: II. Calculation of binding energies and comparison with experimental data. J Comput Chem 20:1177–1185 [Google Scholar]
- Mitchell TM. (1997) Machine Learning, McGraw-Hill, New York [Google Scholar]
- Mitcheson JS, Perry MD. (2003) Molecular determinants of high-affinity drug binding to HERG channels. Curr Opin Drug Discov Devel 6:667–674 [PubMed] [Google Scholar]
- Moreau G, Broto P. (1980) The auto-correlation of a topological-structure - a new Molecular Descriptor. Nouveau Journal De Chimie-New Journal of Chemistry 4:359–360 [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olsen AJ. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comp Chem 19: 1639–1662 [Google Scholar]
- Muegge I, Martin YC. (1999) A general and fast scoring function for protein-ligand interactions: a simplified potential approach. J Med Chem 42:791–804 [DOI] [PubMed] [Google Scholar]
- Mueller R, Dawson ES, Meiler J, Rodriguez AL, Chauder BA, Bates BS, Felts AS, Lamb JP, Menon UN, Jadhav SB, et al. (2012) Discovery of 2-(2-benzoxazoyl amino)-4-aryl-5-cyanopyrimidine as negative allosteric modulators (NAMs) of metabotropic glutamate receptor 5 (mGlu₅): from an artificial neural network virtual screen to an in vivo tool compound. ChemMedChem 7:406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller R, Rodriguez AL, Dawson ES, Butkiewicz M, Nguyen TT, Oleszkiewicz S, Bleckmann A, Weaver CD, Lindsley CW, Conn PJ, et al. (2010) Identification of metabotropic glutamate receptor subtype 5 potentiators using virtual high-throughput screening. ACS Chem Neurosci 1:288–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugnaini C, Rajamaki S, Tintori C, Corelli F, Massa S, Witvrouw M, Debyser Z, Veljkovic V, Botta M. (2007) Toward novel HIV-1 integrase binding inhibitors: molecular modeling, synthesis, and biological studies. Bioorg Med Chem Lett 17:5370–5373 [DOI] [PubMed] [Google Scholar]
- Mulliken RS. (1934) A new electroaffinity scale; together with data on valence states and on valence ionization potentials and electron affinities. J Chem Phys 2:782–793 [Google Scholar]
- Nicolotti O, Miscioscia TF, Carotti A, Leonetti F, Carotti A. (2008) An integrated approach to ligand- and structure-based drug design: development and application to a series of serine protease inhibitors. J Chem Inf Model 48:1211–1226 [DOI] [PubMed] [Google Scholar]
- Nisius B, Göller AH. (2009) Similarity-based classifier using topomers to provide a knowledge base for hERG channel inhibition. J Chem Inf Model 49:247–256 [DOI] [PubMed] [Google Scholar]
- Noeske T, Jirgensons A, Starchenkovs I, Renner S, Jaunzeme I, Trifanova D, Hechenberger M, Bauer T, Kauss V, Parsons CG, et al. (2007) Virtual screening for selective allosteric mGluR1 antagonists and structure-activity relationship investigations for coumarine derivatives. ChemMedChem 2:1763–1773 [DOI] [PubMed] [Google Scholar]
- Noha SM, Atanasov AG, Schuster D, Markt P, Fakhrudin N, Heiss EH, Schrammel O, Rollinger JM, Stuppner H, Dirsch VM, et al. (2011) Discovery of a novel IKK-β inhibitor by ligand-based virtual screening techniques. Bioorg Med Chem Lett 21:577–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norinder U, Haeberlein M. (2002) Computational approaches to the prediction of the blood-brain distribution. Adv Drug Deliv Rev 54:291–313 [DOI] [PubMed] [Google Scholar]
- Nowak P, Cole DC, Aulabaugh A, Bard J, Chopra R, Cowling R, Fan KY, Hu B, Jacobsen S, Jani M, et al. (2010) Discovery and initial optimization of 5,5′-disubstituted aminohydantoins as potent beta-secretase (BACE1) inhibitors. Bioorg Med Chem Lett 20:632–635 [DOI] [PubMed] [Google Scholar]
- O’Boyle NM, Liebeschuetz JW, Cole JC. (2009) Testing assumptions and hypotheses for rescoring success in protein-ligand docking. J Chem Inf Model 49:1871–1878 [DOI] [PubMed] [Google Scholar]
- Obrezanova O, Gola JM, Champness EJ, Segall MD. (2008) Automatic QSAR modeling of ADME properties: blood-brain barrier penetration and aqueous solubility. J Comput Aided Mol Des 22:431–440 [DOI] [PubMed] [Google Scholar]
- Obrezanova O, Segall MD. (2010) Gaussian processes for classification: QSAR modeling of ADMET and target activity. J Chem Inf Model 50:1053–1061 [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Kim SC, Adamik R, Togawa A, Ferrans VJ, Takeda K, Kirby M, Moss J, Vaughan M. (2000) Similarities in function and gene structure of cytohesin-4 and cytohesin-1, guanine nucleotide-exchange proteins for ADP-ribosylation factors. J Biol Chem 275:3221–3230 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Takayama K, Shimizu T, Ishida K, Takahashi O, Furuya T. (2009) Identification of death-associated protein kinases inhibitors using structure-based virtual screening. J Med Chem 52:7323–7327 [DOI] [PubMed] [Google Scholar]
- Olah M, Mracec M, Ostopouici L, Rad R, Bora A, Hadaruga N, Olah I, Banda M, Simon Z, Mracec M, et al. (2005) WOMBAT: World of Molecular Bioactivity, in Chemoinformatics in Drug Discovery, pp 221–239, Wiley-VCH Verlag GmbH, Weinheim, Germany. [Google Scholar]
- Olsen CM, Childs DS, Stanwood GD, Winder DG. (2010) Operant sensation seeking requires metabotropic glutamate receptor 5 (mGluR5). PLoS ONE 5:e15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orry AJW, Abagyan RA, Cavasotto CN. (2006) Structure-based development of target-specific compound libraries. Drug Discov Today 11:261–266 [DOI] [PubMed] [Google Scholar]
- Ortholand JY, Ganesan A. (2004) Natural products and combinatorial chemistry: back to the future. Curr Opin Chem Biol 8:271–280 [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano PR. (2005) Cytochrome P450: Structure, Mechanism, and Biochemistry. Springer Publishing, New York [Google Scholar]
- Osborne CK, Schiff R. (2005) Estrogen-receptor biology: continuing progress and therapeutic implications. J Clin Oncol 23:1616–1622 [DOI] [PubMed] [Google Scholar]
- Page CS, Bates PA. (2006) Can MM-PBSA calculations predict the specificities of protein kinase inhibitors? J Comput Chem 27:1990–2007 [DOI] [PubMed] [Google Scholar]
- Palmisano L. (2007) Role of integrase inhibitors in the treatment of HIV disease. Expert Rev Anti Infect Ther 5:67–75 [DOI] [PubMed] [Google Scholar]
- Pan D, Tseng Y, Hopfinger AJ. (2003) Quantitative structure-based design: formalism and application of receptor-dependent RD-4D-QSAR analysis to a set of glucose analogue inhibitors of glycogen phosphorylase. J Chem Inf Comput Sci 43:1591–1607 [DOI] [PubMed] [Google Scholar]
- Park H, Bahn YJ, Ryu SE. (2009) Structure-based de novo design and biochemical evaluation of novel Cdc25 phosphatase inhibitors. Bioorg Med Chem Lett 19:4330–4334 [DOI] [PubMed] [Google Scholar]
- Park H, Hwang KY, Kim YH, Oh KH, Lee JY, Kim K. (2008) Discovery and biological evaluation of novel alpha-glucosidase inhibitors with in vivo antidiabetic effect. Bioorg Med Chem Lett 18:3711–3715 [DOI] [PubMed] [Google Scholar]
- Park JY, Harris D. (2003) Construction and assessment of models of CYP2E1: predictions of metabolism from docking, molecular dynamics, and density functional theoretical calculations. J Med Chem 46:1645–1660 [DOI] [PubMed] [Google Scholar]
- Pastor M. (2006) Alignment-independent descriptors from molecular interaction fields, in Molecular Interaction Fields, pp 117–143, Wiley-VCH Verlag GmbH, Weinheim, Germany [Google Scholar]
- Pastor M, Cruciani G, McLay I, Pickett S, Clementi S. (2000) GRid-INdependent descriptors (GRIND): a novel class of alignment-independent three-dimensional molecular descriptors. J Med Chem 43:3233–3243 [DOI] [PubMed] [Google Scholar]
- Patel Y, Gillet VJ, Bravi G, Leach AR. (2002) A comparison of the pharmacophore identification programs: Catalyst, DISCO and GASP. J Comput Aided Mol Des 16:653–681 [DOI] [PubMed] [Google Scholar]
- Pearlman RS, Smith KM. (1999) Metric validation and the receptor-relevant subspace concept. J Chem Inf Comput Sci 39:28–35 [Google Scholar]
- Perez-Reyes E. (2003) Molecular physiology of low-voltage-activated t-type calcium channels. Physiol Rev 83:117–161 [DOI] [PubMed] [Google Scholar]
- Perez OD, Mitchell D, Jager GC, South S, Murriel C, McBride J, Herzenberg LA, Kinoshita S, Nolan GP. (2003) Leukocyte functional antigen 1 lowers T cell activation thresholds and signaling through cytohesin-1 and Jun-activating binding protein 1. Nat Immunol 4:1083–1092 [DOI] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, et al. (2009) MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 37 (Database issue):D347–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AC, Jacobs M, Stuver-Moody C. (2008) Docking study yields four novel inhibitors of the protooncogene Pim-1 kinase. J Med Chem 51:1972–1975 [DOI] [PubMed] [Google Scholar]
- Pimentel GC, McClellan AL. (1960) The Hydrogen Bond, W.H. Freeman, San Francisco, CA [Google Scholar]
- Pintore M, van de Waterbeemd H, Piclin N, Chrétien JR. (2003) Prediction of oral bioavailability by adaptive fuzzy partitioning. Eur J Med Chem 38:427–431 [DOI] [PubMed] [Google Scholar]
- Platt JC. (1999) Fast training of support vector machines using sequential minimal optimization, in Advances in Kernel Methods, pp 185–208, MIT Press, Cambridge, MA [Google Scholar]
- Plewczynski D, Łaźniewski M, von Grotthuss M, Rychlewski L, Ginalski K. (2011) VoteDock: consensus docking method for prediction of protein-ligand interactions. J Comput Chem 32:568–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier D. (2009) Advances in development of inhibitors of 17beta hydroxysteroid dehydrogenases. Anticancer Agents Med Chem 9:642–660 [DOI] [PubMed] [Google Scholar]
- Polak S, Wiśniowska B, Brandys J. (2009) Collation, assessment and analysis of literature in vitro data on hERG receptor blocking potency for subsequent modeling of drugs’ cardiotoxic properties. J Appl Toxicol 29:183–206 [DOI] [PubMed] [Google Scholar]
- Poptodorov K, Luu T, Hoffmann R. (2006) Pharmacophore model generation software tools, in Pharmacophores and Pharmacophore Searches, pp 15–47, Wiley-VCH Verlag GmbH & Co, Weinheim, Germany [Google Scholar]
- Porter CT, Bartlett GJ, Thornton JM. (2004) The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res 32 (Database issue):D129–D133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Pearl LH. (2003) Structure and functional relationships of Hsp90. Curr Cancer Drug Targets 3:301–323 [DOI] [PubMed] [Google Scholar]
- Quinlan JR. (1993) C4.5: Programs for Machine Learning, Morgan Kaufmann Publishers, San Mateo, CA. [Google Scholar]
- Rabiller M, Getlik M, Klüter S, Richters A, Tückmantel S, Simard JR, Rauh D. (2010) Proteus in the world of proteins: conformational changes in protein kinases. Arch Pharm (Weinheim) 343:193–206 [DOI] [PubMed] [Google Scholar]
- Rai BK, Fiser A. (2006) Multiple mapping method: a novel approach to the sequence-to-structure alignment problem in comparative protein structure modeling. Proteins 63:644–661 [DOI] [PubMed] [Google Scholar]
- Rajman I. (2008) PK/PD modelling and simulations: utility in drug development. Drug Discov Today 13:341–346 [DOI] [PubMed] [Google Scholar]
- Randic M. (1995) Molecular shape profiles. J Chem Inf Comput Sci 35:373–382 [Google Scholar]
- Randić M, Basak SC. (2001) Characterization of DNA primary sequences based on the average distances between bases. J Chem Inf Comput Sci 41:561–568 [DOI] [PubMed] [Google Scholar]
- Rarey M, Kramer B, Lengauer T, Klebe G. (1996) A fast flexible docking method using an incremental construction algorithm. J Mol Biol 261:470–489 [DOI] [PubMed] [Google Scholar]
- Rasmussen CE, Williams CKI. (2006) Gaussian Processes for Machine Learning, MIT Press, Cambridge, MA. [Google Scholar]
- Raval A, Piana S, Eastwood MP, Dror RO, Shaw DE. (2012) Refinement of protein structure homology models via long, all-atom molecular dynamics simulations. Proteins-Structure Function and Genetics 80:2071–2079 [DOI] [PubMed] [Google Scholar]
- Ravindranathan KP, Mandiyan V, Ekkati AR, Bae JH, Schlessinger J, Jorgensen WL. (2010) Discovery of novel fibroblast growth factor receptor 1 kinase inhibitors by structure-based virtual screening. J Med Chem 53:1662–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RCSB (2013) RCSB protein data bank. Available from http://www.rcsb.org/pdb/home/home.do
- Recanatini M, Poluzzi E, Masetti M, Cavalli A, De Ponti F. (2005) QT prolongation through hERG K(+) channel blockade: current knowledge and strategies for the early prediction during drug development. Med Res Rev 25:133–166 [DOI] [PubMed] [Google Scholar]
- Reid JM, Walden CA, Qin R, Ziegler KL, Haslam JL, Rajewski RA, Warndahl R, Fitting CL, Boring D, Szabo E, et al. Cancer Prevention Network (2011) Phase 0 clinical chemoprevention trial of the Akt inhibitor SR13668. Cancer Prev Res (Phila) 4:347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekker RF, Mannhold R. (1992) Calculation of Drug Lipophilicity: The Hydrophobic Fragmental Constant Approach, VCH, Weinheim, New York [Google Scholar]
- Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. (2005) A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science 307:1625–1630 [DOI] [PubMed] [Google Scholar]
- Reynolds CA, Wade RC, Goodford PJ. (1989) Identifying targets for bioreductive agents: using GRID to predict selective binding regions of proteins. J Mol Graph 7:103–108, 100 [DOI] [PubMed] [Google Scholar]
- Ripphausen P, Nisius B, Peltason L, Bajorath J. (2010) Quo vadis, virtual screening? A comprehensive survey of prospective applications. J Med Chem 53:8461–8467 [DOI] [PubMed] [Google Scholar]
- Roberto Todeschini VC. (2010) Molecular Descriptors for Chemoinformatics Wiley-VCH Verlag GmbH & Co, Weinheim, Germany
- Roche O, Rodríguez Sarmiento RM. (2007) A new class of histamine H3 receptor antagonists derived from ligand based design. Bioorg Med Chem Lett 17:3670–3675 [DOI] [PubMed] [Google Scholar]
- Rodriguez AL, Grier MD, Jones CK, Herman EJ, Kane AS, Smith RL, Williams R, Zhou Y, Marlo JE, Days EL, et al. (2010) Discovery of novel allosteric modulators of metabotropic glutamate receptor subtype 5 reveals chemical and functional diversity and in vivo activity in rat behavioral models of anxiolytic and antipsychotic activity. Mol Pharmacol 78:1105–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D, Hahn M. (2010) Extended-connectivity fingerprints. J Chem Inf Model 50:742–754 [DOI] [PubMed] [Google Scholar]
- Rogers D, Hopfinger AJ. (1994) Application of genetic function approximation to quantitative structure-activity-relationships and quantitative structure-property relationships. J Chem Inf Comput Sci 34:854–866 [Google Scholar]
- Rohl CA, Strauss CEM, Misura KM, Baker D. (2004) Protein structure prediction using Rosetta. Methods Enzymol 383 (Pt D):66–93 [DOI] [PubMed] [Google Scholar]
- Rolland C, Gozalbes R, Nicolaï E, Paugam MF, Coussy L, Barbosa F, Horvath D, Revah F. (2005) G-protein-coupled receptor affinity prediction based on the use of a profiling dataset: QSAR design, synthesis, and experimental validation. J Med Chem 48:6563–6574 [DOI] [PubMed] [Google Scholar]
- RosettaCommons (2013) Rosetta–The premier software suite for macromolecular modeling. Available from http://www.rosettacommons.org/
- Roughley S, Wright L, Brough P, Massey A, Hubbard RE. (2012) Hsp90 inhibitors and drugs from fragment and virtual screening. Top Curr Chem 317:61–82 [DOI] [PubMed] [Google Scholar]
- Ruiz FM, Gil-Redondo R, Morreale A, Ortiz AR, Fábrega C, Bravo J. (2008) Structure-based discovery of novel non-nucleosidic DNA alkyltransferase inhibitors: virtual screening and in vitro and in vivo activities. J Chem Inf Model 48:844–854 [DOI] [PubMed] [Google Scholar]
- Rush TS, 3rd, Grant JA, Mosyak L, Nicholls A. (2005) A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction. J Med Chem 48:1489–1495 [DOI] [PubMed] [Google Scholar]
- Rydberg P, Gloriam DE, Zaretzki J, Breneman C, Olsen L. (2010) SMARTCyp: a 2D method for prediction of cytochrome P450-mediated drug metabolism. Acs Medicinal Chemistry Letters 1:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg P, Vasanthanathan P, Oostenbrink C, Olsen L. (2009) Fast prediction of cytochrome P450 mediated drug metabolism. ChemMedChem 4:2070–2079 [DOI] [PubMed] [Google Scholar]
- Sael L, Kihara D. (2012) Detecting local ligand-binding site similarity in nonhomologous proteins by surface patch comparison. Proteins 80:1177–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Ferrer A, Rodríguez-López JN, García-Cánovas F, García-Carmona F. (1995) Tyrosinase: a comprehensive review of its mechanism. Biochim Biophys Acta 1247:1–11 [DOI] [PubMed] [Google Scholar]
- Sanderson RT. (1951) An interpretation of bond lengths and a classification of bonds. Science 114:670–672 [DOI] [PubMed] [Google Scholar]
- Sanderson RT. (1960) Chemical Periodicity, Reinhold Pub. Corp., New York [Google Scholar]
- Sanguinetti MC, Tristani-Firouzi M. (2006) hERG potassium channels and cardiac arrhythmia. Nature 440:463–469 [DOI] [PubMed] [Google Scholar]
- Sato M, Motomura T, Aramaki H, Matsuda T, Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka K, et al. (2006) Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J Med Chem 49:1506–1508 [DOI] [PubMed] [Google Scholar]
- Sawyer JS, Anderson BD, Beight DW, Campbell RM, Jones ML, Herron DK, Lampe JW, McCowan JR, McMillen WT, Mort N, et al. (2003) Synthesis and activity of new aryl- and heteroaryl-substituted pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain. J Med Chem 46:3953–3956 [DOI] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ. (1995) RASMOL: biomolecular graphics for all. Trends Biochem Sci 20:374. [DOI] [PubMed] [Google Scholar]
- Schames JR, Henchman RH, Siegel JS, Sotriffer CA, Ni H, McCammon JA. (2004) Discovery of a novel binding trench in HIV integrase. J Med Chem 47:1879–1881 [DOI] [PubMed] [Google Scholar]
- Schlitter J, Engels M, Krüger P. (1994) Targeted molecular dynamics: a new approach for searching pathways of conformational transitions. J Mol Graph 12:84–89 [DOI] [PubMed] [Google Scholar]
- Schnecke V, Boström J. (2006) Computational chemistry-driven decision making in lead generation. Drug Discov Today 11:43–50 [DOI] [PubMed] [Google Scholar]
- Schneider G, Hartenfeller M, Reutlinger M, Tanrikulu Y, Proschak E, Schneider P. (2009) Voyages to the (un)known: adaptive design of bioactive compounds. Trends Biotechnol 27:18–26 [DOI] [PubMed] [Google Scholar]
- Schneider G, Neidhart W, Giller T, Schmid G. (1999) “Scaffold-hopping” by topological pharmacophore search: a contribution to virtual screening. Angew Chem Int Ed Engl 38:2894–2896 [PubMed] [Google Scholar]
- Schuffenhauer A, Zimmermann J, Stoop R, van der Vyver JJ, Lecchini S, Jacoby E. (2002) An ontology for pharmaceutical ligands and its application for in silico screening and library design. J Chem Inf Comput Sci 42:947–955 [DOI] [PubMed] [Google Scholar]
- Schuster D, Kowalik D, Kirchmair J, Laggner C, Markt P, Aebischer-Gumy C, Ströhle F, Möller G, Wolber G, Wilckens T, et al. (2011) Identification of chemically diverse, novel inhibitors of 17β-hydroxysteroid dehydrogenase type 3 and 5 by pharmacophore-based virtual screening. J Steroid Biochem Mol Biol 125:148–161 [DOI] [PubMed] [Google Scholar]
- Schuster D, Nashev LG, Kirchmair J, Laggner C, Wolber G, Langer T, Odermatt A. (2008) Discovery of nonsteroidal 17beta-hydroxysteroid dehydrogenase 1 inhibitors by pharmacophore-based screening of virtual compound libraries. J Med Chem 51:4188–4199 [DOI] [PubMed] [Google Scholar]
- Schuur JH, Selzer P, Gasteiger J. (1996) The coding of the three-dimensional structure of molecules by molecular transforms and its application to structure-spectra correlations and studies of biological activity. J Chem Inf Comput Sci 36:334–344 [Google Scholar]
- Segers K, Sperandio O, Sack M, Fischer R, Miteva MA, Rosing J, Nicolaes GA, Villoutreix BO. (2007) Design of protein membrane interaction inhibitors by virtual ligand screening, proof of concept with the C2 domain of factor V. Proc Natl Acad Sci USA 104:12697–12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano ML, Pérez HA, Medina JD. (2006) Structure of C-terminal fragment of merozoite surface protein-1 from Plasmodium vivax determined by homology modeling and molecular dynamics refinement. Bioorg Med Chem 14:8359–8365 [DOI] [PubMed] [Google Scholar]
- Shacham S, Marantz Y, Bar-Haim S, Kalid O, Warshaviak D, Avisar N, Inbal B, Heifetz A, Fichman M, Topf M, et al. (2004) PREDICT modeling and in-silico screening for G-protein coupled receptors. Proteins 57:51–86 [DOI] [PubMed] [Google Scholar]
- Shah F, Wu YS, Gut J,Pedduri Y, Legac J,Rosenthal PJ, Avery MA. (2011) Design, synthesis and biological evaluation of novel benzothiazole and triazole analogs as falcipain inhibitors. Medchemcomm 2:1201–1207 [Google Scholar]
- Shaik S, de Visser SP, Ogliaro F, Schwarz H, Schröder D. (2002) Two-state reactivity mechanisms of hydroxylation and epoxidation by cytochrome P-450 revealed by theory. Curr Opin Chem Biol 6:556–567 [DOI] [PubMed] [Google Scholar]
- Shan YB, Kim ET, Eastwood MP, Dror RO, Seeliger MA, Shaw DE. (2011) How does a drug molecule find its target binding site? J Am Chem Soc 133:9181–9183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, et al. (2008) Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM 51:91–97 [Google Scholar]
- Shekhar C. (2008) In silico pharmacology: computer-aided methods could transform drug development. Chem Biol 15:413–414 [DOI] [PubMed] [Google Scholar]
- Shimada J, Ishchenko AV, Shakhnovich EI. (2000) Analysis of knowledge-based protein-ligand potentials using a self-consistent method. Protein Sci 9:765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet BK, Leach AR, Kuntz ID. (1999) Ligand solvation in molecular docking. Proteins 34:4–16 [DOI] [PubMed] [Google Scholar]
- Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. (1996) Discovering high-affinity ligands for proteins: SAR by NMR. Science 274:1531–1534 [DOI] [PubMed] [Google Scholar]
- Simmons KJ, Chopra I, Fishwick CW. (2010) Structure-based discovery of antibacterial drugs. Nat Rev Microbiol 8:501–510 [DOI] [PubMed] [Google Scholar]
- Singh J, Chuaqui CE, Boriack-Sjodin PA, Lee WC, Pontz T, Corbley MJ, Cheung HK, Arduini RM, Mead JN, Newman MN, et al. (2003a) Successful shape-based virtual screening: the discovery of a potent inhibitor of the type I TGFbeta receptor kinase (TbetaRI). Bioorg Med Chem Lett 13:4355–4359 [DOI] [PubMed] [Google Scholar]
- Singh SB, Shen LQ, Walker MJ, Sheridan RP. (2003b) A model for predicting likely sites of CYP3A4-mediated metabolism on drug-like molecules. J Med Chem 46:1330–1336 [DOI] [PubMed] [Google Scholar]
- Sinko W, Lindert S, McCammon JA. (2013) Accounting for receptor flexibility and enhanced sampling methods in computer-aided drug design. Chem Biol Drug Des 81:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater JC. (1930) Atomic shielding constants. Physiol Rev 36:57–64 [Google Scholar]
- Smellie A, Teig SL, Towbin P. (1995) Poling - promoting conformational variation. J Comput Chem 16:171–187 [Google Scholar]
- Smithson DC, Lee J, Shelat AA, Phillips MA, Guy RK. (2010) Discovery of potent and selective inhibitors of Trypanosoma brucei ornithine decarboxylase. J Biol Chem 285:16771–16781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, Remmert M. (2011) Protein sequence comparison and fold recognition: progress and good-practice benchmarking. Curr Opin Struct Biol 21:404–411 [DOI] [PubMed] [Google Scholar]
- Solit DB, Rosen N. (2006) Hsp90: a novel target for cancer therapy. Curr Top Med Chem 6:1205–1214 [DOI] [PubMed] [Google Scholar]
- Song CM, Lim SJ, Tong JC. (2009) Recent advances in computer-aided drug design. Brief Bioinform 10:579–591 [DOI] [PubMed] [Google Scholar]
- Song MH, Clark M. (2006) Development and evaluation of an in silico model for hERG binding. J Chem Inf Model 46:392–400 [DOI] [PubMed] [Google Scholar]
- Sousa SF, Fernandes PA, Ramos MJ. (2006) Protein-ligand docking: current status and future challenges. Proteins 65:15–26 [DOI] [PubMed] [Google Scholar]
- Southan C. (2013) InChI in the wild: an assessment of InChIKey searching in Google. J Cheminform 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooren W, Ballard T, Gasparini F, Amalric M, Mutel V, Schreiber R. (2003) Insight into the function of Group I and Group II metabotropic glutamate (mGlu) receptors: behavioural characterization and implications for the treatment of CNS disorders. Behav Pharmacol 14:257–277 [DOI] [PubMed] [Google Scholar]
- Stahl M, Todorov NP, James T, Mauser H, Boehm HJ, Dean PM. (2002) A validation study on the practical use of automated de novo design. J Comput Aided Mol Des 16:459–478 [DOI] [PubMed] [Google Scholar]
- Stumpfe D, Bill A, Novak N, Loch G, Blockus H, Geppert H, Becker T, Schmitz A, Hoch M, Kolanus W, et al. (2010) Targeting multifunctional proteins by virtual screening: structurally diverse cytohesin inhibitors with differentiated biological functions. ACS Chem Biol 5:839–849 [DOI] [PubMed] [Google Scholar]
- Stumpfe D, Ripphausen P, Bajorath J. (2012) Virtual compound screening in drug discovery. Future Med Chem 4:593–602 [DOI] [PubMed] [Google Scholar]
- Sugita Y, Okamoto Y. (1999) Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett 314:141–151 [Google Scholar]
- Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, et al. (2008) Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem 51:5843–5855 [DOI] [PubMed] [Google Scholar]
- Sun HM. (2006) An accurate and interpretable bayesian classification model for prediction of HERG liability. ChemMedChem 1:315–322 [DOI] [PubMed] [Google Scholar]
- Sun H, Scott DO. (2010) Structure-based drug metabolism predictions for drug design. Chem Biol Drug Des 75:3–17 [DOI] [PubMed] [Google Scholar]
- Sun H, Sharma R, Bauman J, Walker DP, Aspnes GE, Zawistoski MP, Kalgutkar AS. (2009) Differences in CYP3A4 catalyzed bioactivation of 5-aminooxindole and 5-aminobenzsultam scaffolds in proline-rich tyrosine kinase 2 (PYK2) inhibitors: retrospective analysis by CYP3A4 molecular docking, quantum chemical calculations and glutathione adduct detection using linear ion trap/orbitrap mass spectrometry. Bioorg Med Chem Lett 19:3177–3182 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Zhou SY, Nakamura K, Tanino R, Furuichi A, Kido M, Kawasaki Y, Noguchi K, Seto H, Kurachi M, et al. (2011) A follow-up MRI study of the fusiform gyrus and middle and inferior temporal gyri in schizophrenia spectrum. Prog Neuropsychopharmacol Biol Psychiatry 35:1957–1964 [DOI] [PubMed] [Google Scholar]
- Talafous J, Sayre LM, Mieyal JJ, Klopman G. (1994) META. 2. A dictionary model of mammalian xenobiotic metabolism. J Chem Inf Comput Sci 34:1326–1333 [DOI] [PubMed] [Google Scholar]
- Talele TT, Khedkar SA, Rigby AC. (2010) Successful applications of computer aided drug discovery: moving drugs from concept to the clinic. Curr Top Med Chem 10:127–141 [DOI] [PubMed] [Google Scholar]
- Tanrikulu Y, Proschak E, Werner T, Geppert T, Todoroff N, Klenner A, Kottke T, Sander K, Schneider E, Seifert R, et al. (2009) Homology model adjustment and ligand screening with a pseudoreceptor of the human histamine H4 receptor. ChemMedChem 4:820–827 [DOI] [PubMed] [Google Scholar]
- Tanrikulu Y, Schneider G. (2008) Pseudoreceptor models in drug design: bridging ligand- and receptor-based virtual screening. Nat Rev Drug Discov 7:667–677 [DOI] [PubMed] [Google Scholar]
- Taylor JS, Burnett RM. (2000) DARWIN: a program for docking flexible molecules. Proteins 41:173–191 [PubMed] [Google Scholar]
- Teramoto R, Fukunishi H. (2008) Consensus scoring with feature selection for structure-based virtual screening. J Chem Inf Model 48:288–295 [DOI] [PubMed] [Google Scholar]
- Thai KM, Ecker GF. (2007) Predictive models for HERG channel blockers: ligand-based and structure-based approaches. Curr Med Chem 14:3003–3026 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice CM, Zhao W, Xu Z, Cacatian ST, Simpson RD, Ye YJ, Singh SB, McKeever BM, Lindblom P, Guo J, et al. (2010) Spirocyclic ureas: orally bioavailable 11 beta-HSD1 inhibitors identified by computer-aided drug design. Bioorg Med Chem Lett 20:881–886 [DOI] [PubMed] [Google Scholar]
- Tmej C, Chiba P, Huber M, Richter E, Hitzler M, Schaper KJ, Ecker G. (1998) A combined Hansch/Free-Wilson approach as predictive tool in QSAR studies on propafenone-type modulators of multidrug resistance. Arch Pharm (Weinheim) 331:233–240 [DOI] [PubMed] [Google Scholar]
- Totrov M, Abagyan R. (1997) Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins (Suppl 1):215–220 [DOI] [PubMed] [Google Scholar]
- Triballeau N, Van Name E, Laslier G, Cai D, Paillard G, Sorensen PW, Hoffmann R, Bertrand HO, Ngai J, Acher FC. (2008) High-potency olfactory receptor agonists discovered by virtual high-throughput screening: molecular probes for receptor structure and olfactory function. Neuron 60:767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinajstić N. (1992) Chemical Graph Theory, CRC Press, Boca Raton [Google Scholar]
- Tuccinardi T, Botta M, Giordano A, Martinelli A. (2010) Protein kinases: docking and homology modeling reliability. J Chem Inf Model 50:1432–1441 [DOI] [PubMed] [Google Scholar]
- Turner JV, Glass BD, Agatonovic-Kustrinc S. (2003) Prediction of drug bioavailability based on molecular structure. Anal Chim Acta 485:89–102 [Google Scholar]
- Udomsinprasert R, Pongjaroenkit S, Wongsantichon J, Oakley AJ, Prapanthadara LA, Wilce MC, Ketterman AJ. (2005) Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme. Biochem J 388:763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura K, Yamashita N, Yu X, Arima K, Asada T, Makifuchi T, Murayama S, Saito Y, Kanamaru K, Goto Y, et al. (2006) Autotaxin expression is enhanced in frontal cortex of Alzheimer-type dementia patients. Neurosci Lett 400:97–100 [DOI] [PubMed] [Google Scholar]
- Ursu O, Rayan A, Goldblum A, Oprea TI. (2011) Understanding drug-likeness. WIREs Comput Mol Sci 1:760–781 [Google Scholar]
- Valiron O, Caudron N, Job D. (2001) Microtubule dynamics. Cell Mol Life Sci 58:2069–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Drie JH. (2007) Computer-aided drug design: the next 20 years. J Comput Aided Mol Des 21:591–601 [DOI] [PubMed] [Google Scholar]
- Vangunsteren WF, Berendsen HJC. (1990) Computer-simulation of molecular-dynamics - methodology, applications, and perspectives in chemistry. Angew Chem Int Ed Engl 29:992–1023 [Google Scholar]
- Vapnik VN. (1999) An overview of statistical learning theory. IEEE Trans Neural Netw 10:988–999 [DOI] [PubMed] [Google Scholar]
- Vapnik VN. (2006) Estimation of Dependences Based on Empirical Data; Empirical Inference Science: Afterword of 2006, Springer, New York. [Google Scholar]
- Vapnik V, Lerner A. (1963) Pattern recognition using generalized portrait method. Autom Remote Control 24:774–780 [Google Scholar]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623 [DOI] [PubMed] [Google Scholar]
- Vedani A, Dobler M. (2002) 5D-QSAR: the key for simulating induced fit? J Med Chem 45:2139–2149 [DOI] [PubMed] [Google Scholar]
- Vedani A, Dobler M, Spreafico M, Peristera O, Smiesko M. (2007) VirtualToxLab - in silico prediction of the toxic potential of drugs and environmental chemicals: evaluation status and internet access protocol. ALTEX 24:153–161 [DOI] [PubMed] [Google Scholar]
- Velec HFG, Gohlke H, Klebe G. (2005) DrugScore(CSD)-knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J Med Chem 48:6296–6303 [DOI] [PubMed] [Google Scholar]
- Vijayakrishnan R. (2009) Structure-based drug design and modern medicine. J Postgrad Med 55:301–304 [DOI] [PubMed] [Google Scholar]
- Vilar S, Ferino G, Phatak SS, Berk B, Cavasotto CN, Costanzi S. (2011) Docking-based virtual screening for ligands of G protein-coupled receptors: not only crystal structures but also in silico models. J Mol Graph Model 29:614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkers HM, de Jonge MR, Daeyaert FF, Heeres J, Koymans LM, van Lenthe JH, Lewi PJ, Timmerman H, Van Aken K, Janssen PA. (2003) SYNOPSIS: SYNthesize and OPtimize System in Silico. J Med Chem 46:2765–2773 [DOI] [PubMed] [Google Scholar]
- Vinogradov SN, Linnell RH. (1971) Hydrogen Bonding, Van Nostrand Reinhold, New York [Google Scholar]
- Vistoli G, Pedretti A, Testa B. (2008) Assessing drug-likeness—what are we missing? Drug Discov Today 13:285–294 [DOI] [PubMed] [Google Scholar]
- Wade RC, Goodford PJ. (1993) Further development of hydrogen bond functions for use in determining energetically favorable binding sites on molecules of known structure. 2. Ligand probe groups with the ability to form more than two hydrogen bonds. J Med Chem 36:148–156 [DOI] [PubMed] [Google Scholar]
- Wang RX, Fang XL, Lu Y, Wang S. (2004) The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem 47:2977–2980 [DOI] [PubMed] [Google Scholar]
- Wang RX, Fu Y, Lai L. (1997) A new atom-additive method for calculating partition coefficients. J Chem Inf Comput Sci 37:615–621 [Google Scholar]
- Wang RX, Gao Y, Lai L. (2000a) Calculating partition coefficient by atom-additive method. Perspect Drug Discov Des 19:47–66 [Google Scholar]
- Wang RX, Gao Y, Lai L. (2000b) LigBuilder: A multi-purpose program for structure-based drug design. J Mol Model 6:498–516 [Google Scholar]
- Wang RX, Liu L, Lai L, Tang Y. (1998) SCORE: A new empirical method for estimating the binding affinity of a protein-ligand complex. J Mol Model 4:379–394 [Google Scholar]
- Wang S, Li Y, Wang J, Chen L, Zhang L, Yu H, Hou T. (2012) ADMET evaluation in drug discovery. 12. Development of binary classification models for prediction of hERG potassium channel blockage. Mol Pharm 9:996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner SL, Bashyam S, Vankayalapati H, Bearss DJ, Han H, Mahadevan D, Von Hoff DD, Hurley LH. (2006) Identification of a lead small-molecule inhibitor of the Aurora kinases using a structure-assisted, fragment-based approach. Mol Cancer Ther 5:1764–1773 [DOI] [PubMed] [Google Scholar]
- Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, et al. (2006) A critical assessment of docking programs and scoring functions. J Med Chem 49:5912–5931 [DOI] [PubMed] [Google Scholar]
- Wei DG, Jiang XL, Zhou L, Chen J, Chen Z, He C, Yang K, Liu Y, Pei J, Lai L. (2008) Discovery of multitarget inhibitors by combining molecular docking with common pharmacophore matching. J Med Chem 51:7882–7888 [DOI] [PubMed] [Google Scholar]
- Weininger D. (1988) Smiles, a chemical language and information-system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci 28:31–36 [Google Scholar]
- Weininger SJ, Stermitz FR. (1984) Organic Chemistry, Academic Press, Orlando [Google Scholar]
- Weisel M, Proschak E, Schneider G. (2007) PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem Cent J 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermuth CG. (2006) Pharmacophores: historical perspective and viewpoint from a medicinal chemist, in Pharmacophores and Pharmacophore Searches pp 1–13, Wiley-VCH Verlag GmbH & Co, Weinheim, Germany [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. (2006) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 34 (Database issue):D173–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett P. (2006) Similarity-based virtual screening using 2D fingerprints. Drug Discov Today 11:1046–1053 [DOI] [PubMed] [Google Scholar]
- Wilson GL, Lill MA. (2011) Integrating structure-based and ligand-based approaches for computational drug design. Future Med Chem 3:735–750 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34 (Database issue):D668–D672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiswesser WJ. (1954) A Line-Formula Chemical Notation, Crowell, New York [Google Scholar]
- Wiswesser WJ. (1985) Historic development of chemical notations. J Chem Inf Comput Sci 25:258–263 [Google Scholar]
- Witkin JM, Nelson DL. (2004) Selective histamine H3 receptor antagonists for treatment of cognitive deficiencies and other disorders of the central nervous system. Pharmacol Ther 103:1–20 [DOI] [PubMed] [Google Scholar]
- Wolber G, Langer T. (2005) LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model 45:160–169 [DOI] [PubMed] [Google Scholar]
- Wolber G, Seidel T, Bendix F, Langer T. (2008) Molecule-pharmacophore superpositioning and pattern matching in computational drug design. Drug Discov Today 13:23–29 [DOI] [PubMed] [Google Scholar]
- Wold S, Esbensen K, Geladi P. (1987) Principal component analysis. Chemom Intell Lab Syst 2:37–52 [Google Scholar]
- Wood C, Williams C, Waldron GJ. (2004) Patch clamping by numbers. Drug Discov Today 9:434–441 [DOI] [PubMed] [Google Scholar]
- Xiang Z. (2006) Advances in homology protein structure modeling. Curr Protein Pept Sci 7:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Glen RC. (2002) Novel methods for the prediction of logP, pK(a), and logD. J Chem Inf Comput Sci 42:796–805 [DOI] [PubMed] [Google Scholar]
- Xu Y, Stokes AH, Freeman WM, Kumer SC, Vogt BA, Vrana KE. (1997) Tyrosinase mRNA is expressed in human substantia nigra. Brain Res Mol Brain Res 45:159–162 [DOI] [PubMed] [Google Scholar]
- Xue Y, Chao E, Zuercher WJ, Willson TM, Collins JL, Redinbo MR. (2007) Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg Med Chem 15:2156–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY. (2010) Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today 15:444–450 [DOI] [PubMed] [Google Scholar]
- Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem 279:38091–38094 [DOI] [PubMed] [Google Scholar]
- Yarnitzky T, Levit A, Niv MY. (2010) Homology modeling of G-protein-coupled receptors with X-ray structures on the rise. Curr Opin Drug Discov Devel 13:317–325 [PubMed] [Google Scholar]
- Yim DS, Zhou H, Buckwalter M, Nestorov I, Peck CC, Lee H. (2005) Population pharmacokinetic analysis and simulation of the time-concentration profile of etanercept in pediatric patients with juvenile rheumatoid arthritis. J Clin Pharmacol 45:246–256 [DOI] [PubMed] [Google Scholar]
- Yoshida F, Topliss JG. (2000) QSAR model for drug human oral bioavailability. J Med Chem 43:2575–2585 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Pei JF, Lai L. (2011) LigBuilder 2: A Practical de Novo Drug Design Approach. J Chem Inf Model 51:1083–1091 [DOI] [PubMed] [Google Scholar]
- Zhan Y, Shen D. (2005) Design efficient support vector machine for fast classification. Pattern Recognit 38:157–161 [Google Scholar]
- Zhang S. (2011) Computer-aided drug discovery and development. Methods Mol Biol 716:23–38 [DOI] [PubMed] [Google Scholar]
- Zhang S, Cao ZX, Tian H, Shen G, Ma Y, Xie H, Liu Y, Zhao C, Deng S, Yang Y, et al. (2011) SKLB1002, a novel potent inhibitor of VEGF receptor 2 signaling, inhibits angiogenesis and tumor growth in vivo. Clin Cancer Res 17:4439–4450 [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CS, Zhao YF, Chai H, Gong P. (2006a) Synthesis and in vitro anti-hepatitis B virus activities of some ethyl 5-hydroxy-1H-indole-3-carboxylates. Bioorg Med Chem 14:2552–2558 [DOI] [PubMed] [Google Scholar]
- Zhao L, Huang W, Liu H, Wang L, Zhong W, Xiao J, Hu Y, Li S. (2006b) FK506-binding protein ligands: structure-based design, synthesis, and neurotrophic/neuroprotective properties of substituted 5,5-dimethyl-2-(4-thiazolidine)carboxylates. J Med Chem 49:4059–4071 [DOI] [PubMed] [Google Scholar]
- Zheng W, Tropsha A. (2000) Novel variable selection quantitative structure—property relationship approach based on the k-nearest-neighbor principle. J Chem Inf Comput Sci 40:185–194 [DOI] [PubMed] [Google Scholar]
- Zhou T, Huang D, Caflisch A. (2010) Quantum mechanical methods for drug design. Curr Top Med Chem 10:33–45 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lai X, Li Y, Dong W. (2012) Ant colony optimization with combining Gaussian eliminations for matrix multiplication. IEEE Trans Syst Man Cybern B Cybern 43:347–357 [DOI] [PubMed] [Google Scholar]