Abstract
Human TopBP1 is a key mediator protein involved in DNA replication checkpoint control. In this study, we report a specific interaction between TopBP1 and Bloom syndrome helicase (BLM) that is phosphorylation and cell cycle dependent. Interestingly, TopBP1 depletion led to decreased BLM protein level and increased sister chromatid exchange (SCE). Moreover, our data indicated that BLM was ubiquitinated by E3 ligase MIB1 and degraded in G1 cells, but was stabilized by TopBP1 in S phase cells. Depletion of MIB1 restored BLM protein level and rescued the elevated SCE phenotype in TopBP1-depleted cells. In addition, cells expressing an un-degradable BLM mutant showed radiation sensitivity, probably by triggering end resection and inhibiting NHEJ pathway in G1 phase. Taken together, these data suggest that while BLM is down-regulated in G1 phase to promote NHEJ-mediated DNA repair, it is stabilized by TopBP1 in S phase cells to suppress SCE and thereby prevent genomic instability.
INTRODUCTION
Human TopBP1 plays essential roles in DNA replication and replication checkpoint control. TopBP1 possesses eight BRCA1 C-Terminus (BRCT) phosphopeptide recognition motifs and an ATR-activating domain (AAD) (Bartek and Mailand, 2006; Burrows and Elledge, 2008; Cimprich and Cortez, 2008; Kumagai et al., 2006; Manke et al., 2003). The AAD, which is located between the 6th and 7th BRCT repeats of TopBP1, is necessary and sufficient for ATR activation both in vitro and in vivo (Delacroix et al., 2007; Kumagai et al., 2006). The multiple BRCT repeats in TopBP1 mediate many protein-protein interactions, which are important for the regulation of DNA replication and DNA damage checkpoints. The N-terminal BRCT domains of TopBP1 are known to be involved in several protein-protein interactions; they interact with the phosphorylated RAD9 tail in the 9-1-1 complex and are required for ATR-mediated CHK1 activation in mammalian cells (Delacroix et al., 2007) and Xenopus egg extracts (Lee et al., 2007). The N-terminal tandem BRCT domain is also required for binding to Treslin/ticrr, which functions in DNA replication initiation (Kumagai et al., 2010; Sansam et al., 2010), and for binding to NBS1, which recruits ATM to TopBP1 in response to DSBs (Yoo et al., 2009). The 5th BRCT domain (BRCT5) of TopBP1 is required for its localization to DNA damage sites (Yamane et al., 2002). We recently demonstrated that the BRCT5 domain is responsible for the interaction of TopBP1 with phosphorylated MDC1 and required for efficient CHK1 phosphorylation following replication stress (Wang et al., 2011), while another study revealed that the recruitment of TopBP1 to sites of DNA double-strand breaks (DSBs) during the G1 phase depends on 53BP1 and ATM (Cescutti et al., 2010). As for the C-terminal tandem BRCT domains (the 7th and 8th BRCT repeats) in TopBP1, we reported that this region associates with BACH1, which is required for early replication checkpoint control (Gong et al., 2010). In addition, Zou and colleagues showed that this region of TopBP1 binds to phosphorylated ATR and enables TopBP1 to engage ATR-ATRIP and stimulate ATR kinase activity (Liu et al., 2011). Thus, TopBP1 acts as a signal integrator, which functions primarily in DNA replication and replication checkpoint control.
In this study, we found a functional connection between TopBP1 and Bloom syndrome helicase (BLM). It is well known that BLM mutations lead to Bloom Syndrome, a disease characterized by growth retardation and predisposition to cancer (Hanada and Hickson, 2007). The hallmark cellular phenotype of Bloom syndrome is elevated sister chromatid exchange (SCE) (Ray and German, 1984), suggesting that the critical function of BLM is to suppress illegitimate recombination, preventing genomic instability due to loss of heterozygosity and therefore inhibiting tumor development. Here, we identified BLM as a TopBP1-binding protein and provide evidence suggesting that TopBP1 has an unexpected role in suppressing SCE that is independent of its function in replication checkpoint control.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmids
HeLa and 293T cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified incubator with 5% CO2. Wild-type and BLM-deficient cells were cultivated with the same conditions. TopBP1, BLM, and MIB1 cDNA were cloned using Gateway Technology (Invitrogen). All internal deletion mutants were generated by site-directed mutagenesis and verified by sequencing.
Antibodies
The rabbit polyclonal anti-TopBP1 antibody was described previously (Gong et al., 2010; Kim et al., 2005; Wang et al., 2011). The anti-BLM polyclonal antibody was obtained from Abcam and Bethyl Laboratories. The anti-MIB1 antibody was purchased from Epitomics and Abcam. The monoclonal anti-FLAG M2, anti-HA, and anti–β-actin antibodies were purchased from Sigma-Aldrich. The anti-Myc (9E10) antibody was obtained from Covance. Anti-BLMpS338 polyclonal antibody was raised against phospho-peptide CKEDVLST-(phospho-S)-KDLLSKPE and affinity purified.
SCE Assay
HeLa cells were transfected with the indicated small interfering RNA (siRNA), cultured for 24 hours, and transfected again with the same siRNA to achieve optimal knockdown efficiency. Sixteen hours after the second transfection, BrdU (100 µM) was added to the medium, and cells were cultured for another 48 hr. Cells were then incubated with colcemid (200 ng/ml) for 1 hour, collected by trypsinization, and washed with phosphate buffered saline (PBS). Cells were swollen in 75 mM KCl for 15 min and then fixed with a mixture of methanol and acetic acid (3:1) and dropped onto slides. After 2 days, the slides were stained with Hoechst 33258 and ultraviolet irradiation, followed by incubation in 2×SSC at 65°C for 30 min. They were then stained with 4% Giemsa solution for 10 min and viewed under the microscope. For each condition at least 20 cells/metaphase and ~1,000 or more chromosomes were analyzed for measuring SCE frequency.
Co-precipitation and Western Blotting
Cells were lysed with NETN buffer (20 mM Tris-HCl, pH 8.0; 100 mM NaCl; 1 mM EDTA; and 0.5% Nonidet P-40) containing protease inhibitors on ice for 20 min. The soluble fractions were collected after centrifugation and incubated with protein A agarose beads coupled with anti-TopBP1 antibody, anti-BLM antibody, or S-protein beads for 4 hours at 4°C. The precipitates were then washed and boiled in 2 × sodium dodecyl sulfate (SDS) loading buffer. Samples were resolved on SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene fluoride membrane, and immunoblotting was carried out with antibodies as indicated.
In Vivo and In Vitro Ubiquitination Assays
For the in vivo ubiquitination assay, 293T cells were co-transfected with constructs encoding SFB-BLM, Myc-MIB1 (or Myc-MIB1-D3), and His-ubiquitin. Cells were lysed in 6 M urea, 300 mM NaCl, and 10 mM imidazole in 100 mM sodium phosphate buffer, pH 7.4, and then incubated with Ni-NTA agarose beads for 2–4 hours at 22°C. The purified complexes were then washed and subjected to Western blot analysis. For the in vitro ubiquitination assay, SFB-BLM or MBP-BLM proteins that were respectively isolated from 293T cell lysates and Escherichia coli were incubated with 0.5 µg E1, 1.5 µg ubiquitin, 1 µg UBCH5c, and 2.5 mM ATP in a final 30-µl reaction buffer (1.5 mM MgCl2, 5 mM KCl, 1 mM DTT, and 20 mM HEPES, pH 7.4) at 30°C for 2 hours. The ubiquitination of BLM was detected by Western blotting.
RNA Interference
Briefly, HeLa cells were transfected twice at 24-hr intervals with the indicated siRNA using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. The siRNA against human TopBP1 was previously described (Gong et al., 2010; Kim et al., 2005; Wang et al., 2011). The sequence of the second TopBP1 siRNA was ACAAAUACAUGGCUGGUUA. The sequence of the MIB1 siRNA was GACCUGAGCAUUCGAAAUAUU. The sequence of the control siRNA was UUCAAUAAAUUCUUGAGGUUU. The smart pool siRNA against ATR was purchased from Dharmacon, Inc. The sequence of the NEK11 shRNA was GTCACTGGCATGCATTTTC.
NHEJ assay
NHEJ repair was assessed by a cell-based plasmid-integration as previously described (Feng and Chen, 2012)
Tandem Affinity Purification
293T cells stably expressing SFB-TopBP1 BRCT5, SFB-BLM, or SFB-MIB1 were used for tandem affinity purification. For MIB1 purification, MG132 was added for 6 hr before the cells were collected. Cells stably expressing SFB-TopBP1 BRCT5, SFB-BLM, or SFB-MIB1 were lysed with NETN buffer on ice for 20 min. After removal of cell debris by centrifugation, crude lysates were incubated with streptavidin Sepharose beads for 1 hour at 4°C. The bead-bound proteins were washed three times with NETN buffer and eluted twice with 2 mg/ml biotin (Sigma) for 1 hour at 4°C. The eluates were combined and then incubated with S-protein agarose (Novagen) for 1 hour at 4°C. The S beads were washed three times with NETN buffer. The proteins bound to S-protein agarose beads were separated by SDS-PAGE and visualized by Coomassie Blue staining. The eluted proteins were identified by mass spectrometry analysis, performed by the Taplin Biological Mass Spectrometry Facility (Harvard Medical School).
Cell Synchronization and Fluorescence-Activated Cell Sorting
HeLa cells were treated with 2 mM thymidine for 18 hours and then released in fresh medium for 10 hours. Thymidine (2 mM) was added again, and cells were incubated for another 9 hours to arrest cells in G1 phase before releasing them again in fresh medium. Cells were collected at the indicated time points. For cell cycle analysis, synchronized cells were washed twice with PBS, resuspended in 300 µl of PBS, and then fixed with the addition of 700 µl of 100% ethanol. After storage at −20°C overnight, fixed cells were washed and incubated in sodium citrate buffer containing RNAse A for 30 min and then stained with propidium iodide for 30 min. Cells were then run on a FACScan, and cell cycle analysis was performed.
Glutathione-S-transferase (GST) Pull-Down Assay
GST fusion proteins were expressed in E. coli and purified. GST fusion proteins were immobilized on glutathione Sepharose 4B beads and incubated with lysates prepared from cells transiently transfected with plasmids encoding the indicated proteins. The samples were subjected to SDS-PAGE and analyzed by Western blotting.
In Vitro Kinase Assay
The SFB tagged CDC7, CDK2 and NEK11 were expressed in 293T cells and purified by S protein beads. GST-BLM-F2 (aa250–480) was expressed in E.Coli, purified using glutathione Sepharose 4B beads, and eluted with 10mM reduced glutathione. The 100 µl reaction mixture contained 100 ng GST-BLM-F2, 100 µM AMP, 100 µM ATP (0.5 µCi 33P-ATP per reaction), and 50 µM SAMs in a buffer (40 mM HEPES, pH 7.0, 80 mM NaCl, 0.8 mM EDTA, 5 mM MgCl2, 0.025% BSA, and 0.8 mM DTT). The reaction was initiated with addition of the enzyme. After 60-minute incubation at 30°C, the supernatant were transferred to a new tube and the reaction was stopped by addition of SDS loading buffer. The mixtures were subjected for autoradiography. Proteins bound to the beads were also eluted with addition of SDS loading buffer and subjected to Western blotting analysis using anti-Flag antibody.
RESULTS
TopBP1 Interacts with BLM
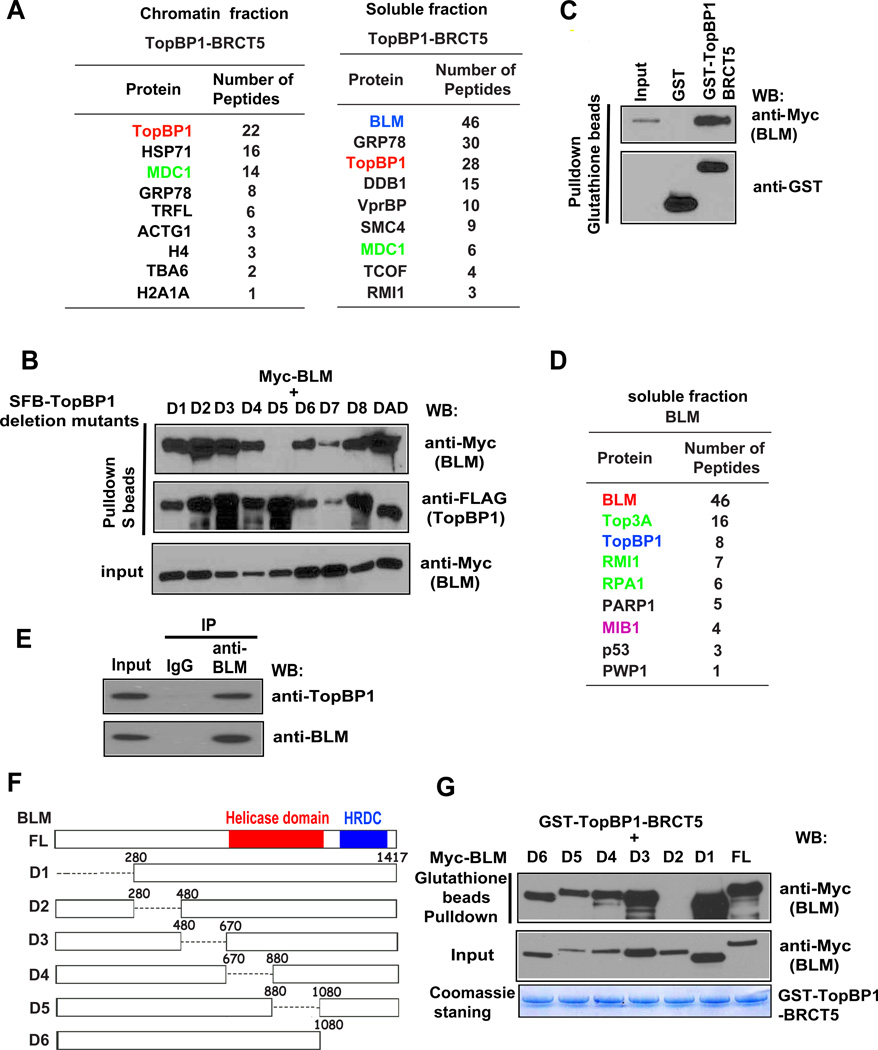
Proteins associated with TopBP1 BRCT domains appear to be important for TopBP1 functions. We found that a region containing BRCT5 of TopBP1 is sufficient for TopBP1 focus formation following replication stress (Wang et al., 2011; Yamane et al., 2002). As previously reported (Wang et al., 2011), we generated stable cell lines expressing the triple-tagged (S-protein, FLAG, and streptavidin-binding peptide) BRCT5 of TopBP1. Tandem affinity purification followed by mass spectrometry analysis identified MDC1 as a major TopBP1-associated protein in the chromatin fraction (Wang et al., 2011). Interestingly, we found that BLM is the major TopBP1-associated protein in the soluble fraction (Figure 1A). Using SFB-tagged wild-type TopBP1 and a series of TopBP1 deletion mutants, we showed that deletion of BRCT5 abolished the TopBP1/BLM interaction (Figure 1B), confirming that the BRCT5 domain is important for TopBP1 binding to BLM. Furthermore, using a bacterially expressed and purified GST-fused BRCT5 of TopBP1 protein, we showed that the TopBP1 BRCT5 binds to BLM in vitro (Figure 1C). We also showed that TopBP1 interacts directly with BLM using GST-TopBP1 and SFB-BLM proteins expressed and purified from insect cells (Figure S1A).
Figure 1. TopBP1 Interacts with BLM Through its BRCT5 Domain.
(A) Lists of TopBP1 BRCT5-associated proteins in chromatin and soluble fractions identified by mass spectrometric analysis.
(B) The BRCT5 domain of TopBP1 is required for TopBP1/BLM interaction. 293T cells were transfected with plasmids encoding Myc-tagged BLM together with plasmids encoding deletion mutants of SFB-tagged TopBP1. Precipitation reactions were conducted using S-protein beads and then subjected to Western blotting using the indicated antibodies. D1 to D8 are TopBP1 BRCT domain deletion mutants that each has a deletion of one of the TopBP1 BRCT domains. DAD is a deletion mutant that removes the ATR activation domain of TopBP1.
(C) TopBP1 BRCT5 specifically binds to BLM. Beads coated with bacterially expressed GST-fused TopBP1 BRCT5 were incubated with cell lysates containing exogenously expressed Myc-tagged BLM. Immunoblotting experiments were carried out using the indicated antibodies.
(D) A list of BLM-associated proteins in soluble fraction identified by mass spectrometry analysis.
(E) Interaction between endogenous TopBP1 and BLM. HeLa cells were collected and immunoprecipitation was carried out using control or anti-BLM antibodies. Immunoprecipitates were separated by SDS-PAGE and immunoblotted with the indicated antibodies.
(F) Schematic diagram of wild-type and deletion mutants of BLM used in this study.
(G) Beads coated with bacterially expressed GST-TopBP1 BRCT5 were incubated with cell lysates containing exogenously expressed, Myc-tagged wild-type or deletion mutants of BLM. Immunoblotting experiments were carried out using the indicated antibodies.
To further validate the interaction between TopBP1 and BLM, we performed a reverse tandem affinity purification using 293T cells stably expressing SFB-tagged BLM. Mass spectrometry analysis not only revealed several known BLM-binding partners, including TOP3A and RMI1, but also confirmed the TopBP1/BLM interaction (Figure 1D). Co-immunoprecipitation demonstrated that endogenous BLM associates with TopBP1 (Figure 1E).
Next, we sought to define the TopBP1 binding region(s) on BLM. We generated and purified recombinant GST-fused TopBP1-BRCT5 protein from bacteria. Using a series of BLM deletion mutants, we mapped the TopBP1-binding region to residues 280 to 480 of BLM (Figures 1F and 1G).
TopBP1/BLM Interaction Is Cell Cycle–Regulated and Phosphorylation Dependent
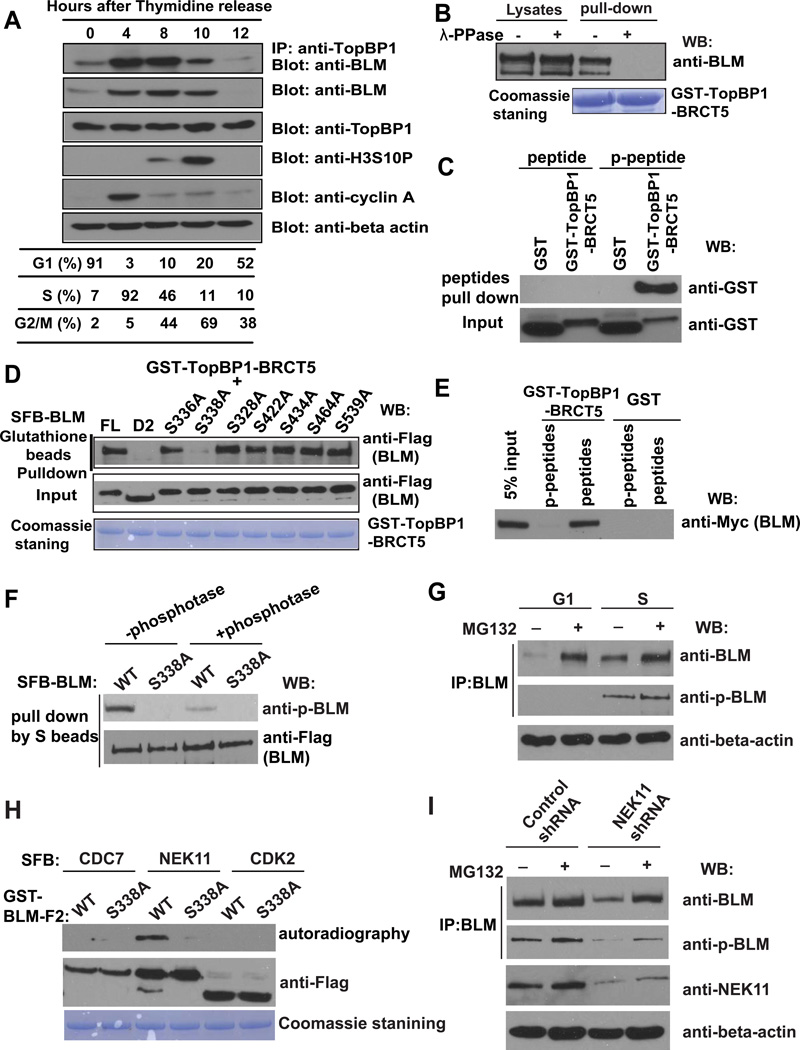
Both BLM and TopBP1 function primarily in S and G2 phases. Although the TopBP1 protein level remained constant throughout the cell cycle, BLM protein was significantly enriched in S and G2 cells, and this enrichment appeared to correlate with its interaction with TopBP1 (Figure 2A).
Figure 2. TopBP1/BLM Interaction Is Cell Cycle–Regulated.
(A) The TopBP1/BLM interaction is cell cycle–regulated. HeLa cells were synchronized by double thymidine block, and then released in fresh medium without thymidine and collected at the indicated time points. Cell lysates were prepared, immunoprecipitation and immunoblotting experiments were performed using antibodies as indicated. Cell cycle distributions were confirmed by fluorescence-activated cell sorting analysis and summarized in the table.
(B) TopBP1/BLM interaction is phosphorylation dependent. Beads coated with bacterially expressed GST-TopBP1 BRCT5 fusion protein were incubated with cell lysates containing exogenously expressed, SFB-tagged BLM that was mock treated or treated with λ lambda protein phosphatase. Immunoblotting experiments were carried out with the indicated antibodies.
(C) Mapping phosphorylation site on BLM that is important for its interaction with TopBP1. S336A, S338A, S328A, S422A, S434A, S464A, and S539A are mutants in which a known phosphorylated serine site was changed to alanine. Beads coated with bacterially expressed GST-TopBP1 BRCT5 fusion protein were incubated with cell lysates containing exogenously expressed, SFB-tagged wild-type or mutant BLM. Immunoblotting experiments were carried out using indicated antibodies.
(D) Phosphorylated or control BLM peptides were incubated with purified GST or GST-BRCT5 fusion proteins. Input (lower panel) and GST-fusion proteins associated with peptides (upper panel) were assessed by immunoblotting using anti-GST antibodies.
(E) Beads coated with bacterially expressed GST or GST-BRCT5 fusion proteins were incubated with cell lysates containing exogenously expressed, Myc-tagged BLM together with phosphorylated or control BLM peptides. Immunoblotting experiments were carried out using indicated antibodies.
(F) Cell lysates containing SFB-tagged wild-type or S338A mutant of BLM were treated with or without λ-phosphotase and subjected to immunoprecipitation using S protein beads. The immunoprecipitates were immunoblotted with indicated antibodies.
(G) HeLa cells were synchronized in G1 phase or S phase. Cell lysates were prepared, and immunoprecipitated with BLM antibody. Western blotting was performed using antibodies as indicated.
(H) SFB-CDC7, SFB-NEK11 and SFB-CDK2 were purified from 293T cells by S protein beads and incubated with eluted GST-BLM-F2 (aa 250–480) and γ-33P-ATP for 60 minutes in kinase buffer. The supernatant were subjected for autoradiography and the beads were subjected to Western blotting with anti-Flag antibody.
(I) HeLa cells were infected with NEK11 shRNA or control shRNA. Cell lysates were subjected to immunoprecipitation using anti-BLM antibody. The immunoprecipitates were blotted with indicated antibodies.
The BRCT domain is a phosphoprotein-binding domain. To determine whether the interaction between TopBP1 and BLM is phosphorylation dependent, cell lysates were treated with λ protein phosphatase in a GST-pull down assay. The interaction between TopBP1 and BLM was abolished after treatment with lambda protein phosphatase, suggesting that the interaction between TopBP1 and BLM is phosphorylation dependent (Figure 2B).
We isolated BLM protein from G1 or S phase cells and performed mass spectrometry analysis to identify all potential phosphorylation sites on BLM. A total of 11 phosphorylation sites were identified, nine in both G1 and S phases and two only in S phase sample (Figure S1B). Among these 11 residues, six were located within the D2 region, which is required for BLM/TopBP1 interaction. We mutated each of these six residues and one additional phosphorylation site adjacent to the D2 region (Ser539). We found that only mutation of residue Ser338, which is the S phase–specific phosphorylation site, greatly reduced the BLM/TopBP1 interaction (Figure 2C). Mutation of the other S phase–specific phosphorylation site Ser336 did not notably affect the BLM/TopBP1 interaction (Figure 2C), and mutation of both the Ser336 and Ser338 sites behaved like Ser338 mutation alone (Figure S1C), suggesting that the Ser338 site is the major S phase phosphorylation site involved in the BLM/TopBP1 interaction.
To further confirm that the Ser338 phosphorylation site on BLM is required for its interaction with TopBP1, we synthesized phosphorylated and un-phosphorylated peptides containing this residue to perform pull-down experiments. As a result, GST-TopBP1-BRCT5 could be pulled down by the phosphorylated BLM peptide but not by the control un-phosphorylated peptide (Figure 2D). Moreover, a competition assay showed that the phosphorylated BLM peptide could disrupt the TopBP1/BLM interaction, whereas the un-phosphorylated peptide failed to do so (Figure 2E). Thus, we concluded that the S phase–specific phosphorylation of Ser338 is required for BLM/TopBP1 interaction.
We generated a phospho-specific antibody raised against a peptide containing phospho-Serine 338 of BLM. This antibody p-BLM (Ser 338) specifically recognized wild-type BLM, but not the S338A mutant of BLM (Figure 2F). Moreover, λ-phosphatase treatment dramatically decreased the signal detected by Western blotting using this phospho-specific antibody. Furthermore, this antibody could detect phosphorylated BLM in S phase cells, but not in G1 phase (Figure 2G), indicating that Serine 338 of BLM is indeed phosphorylated in vivo and this phosphorylation is cell cycle regulated.
To further explore which kinase is responsible for BLM phosphorylation at S338 site, we searched the database and found this site fits well with the consensus motif (LXXS338) phosphorylated by NEK family. KinasePhos 2.0 program predicted NEK11 as the top candidate that can phosphorylate S338 site. Thus, we performed in vitro kinase assay using SFB tagged NEK11, CDC7 and CDK2 purified from 293T cells and incubated them with GST tagged wild-type or S338A mutant of BLM-F2 (aa 250–480). Excitingly, only NEK11, but not the other two S phase specific kinases, could efficiently phosphorylate wild-type BLM-F2 (Figure 2H). NEK11 protein is known to increase at S or G2 phase and plays a role in S phase checkpoint control (Noguchi et al., 2002, 2004; Sorensen et al., 2010). We confirmed that NEK11 protein level peaks at S and G2 phases (Figure S1D), which is similar to that of BLM. Moreover, knock-down NEK11 significantly reduced BLM protein level and BLM phosphorylation at S338 site (Figure 2I), indicating that NEK11 is responsible for BLM phosphorylation and regulation in vivo.
Together, these data indicate that the interaction between TopBP1 and BLM is phosphorylation and cell cycle dependent and that this interaction may play a role in the S and G2 phases of the cell cycle.
TopBP1 Suppresses SCE by Maintaining BLM Expression
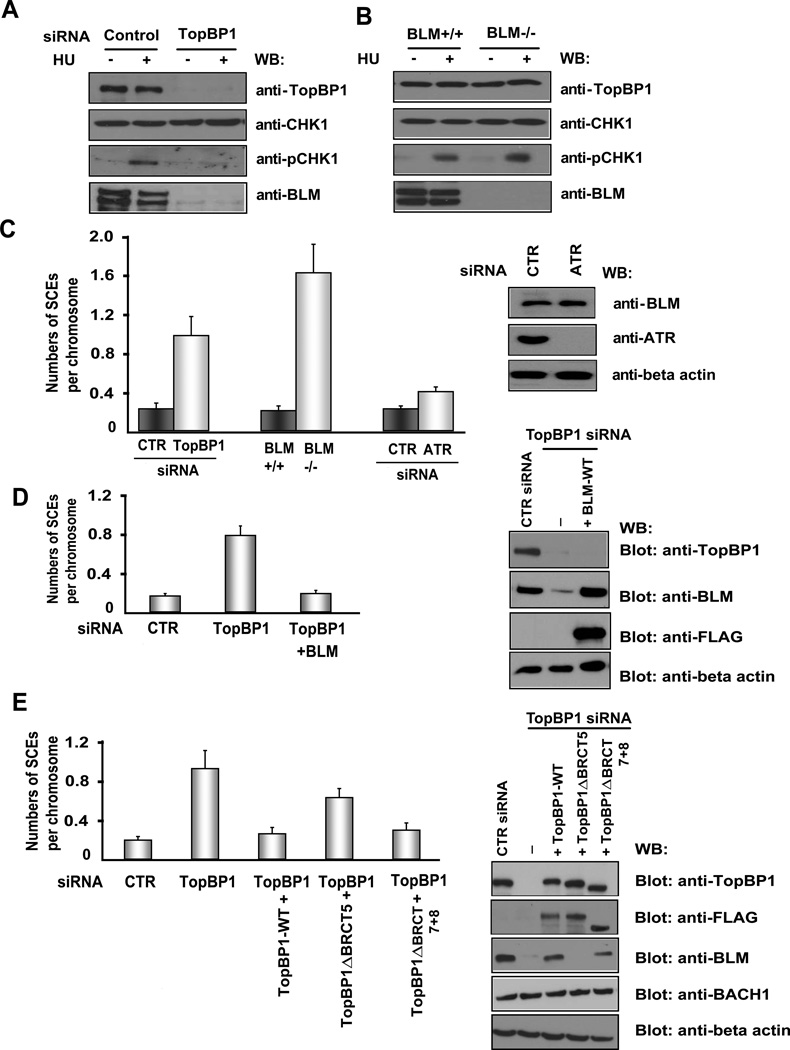
Given that TopBP1 interacts with BLM, we next wanted to know the functional significance of this interaction. TopBP1 is required for CHK1 activation following replication stress (Gong et al., 2010; Kim et al., 2005; Wang et al., 2011). Downregulation of TopBP1 inhibited hydroxyurea-induced CHK1 phosphorylation. Surprisingly, we observed that TopBP1 depletion led to a severe decrease in BLM protein level (Figure 3A). This dramatic reduction of BLM protein level was observed in TopBP1-depleted cells with or without hydroxyurea treatment (Figure 3A), implying that this decrease may be independent of TopBP1’s function in replication checkpoint control. We used another TopBP1 siRNA to further validate this result and showed that TopBP1 downregulation led to decreased BLM protein level (Figure S1D).
Figure 3. TopBP1 Suppresses SCE by Maintaining BLM Expression.
(A) Depletion of TopBP1 decreases BLM protein level. HeLa cells transfected with control (CTR) or TopBP1 siRNA were mock treated or treated with hydroxyurea (HU, 10 mM). Cells were harvested and immunoblotted with the indicated antibodies.
(B) BLM is not required for Chk1 activation in response to replication stress. Wild-type and BLM-deficient cells were mock treated or treated with HU (10 mM). Cell lysates were immunoblotted with antibodies as indicated.
(C) Depletion of TopBP1 increases SCE frequency. HeLa cells were transfected with CTR, TopBP1, or ATR siRNAs. SCE assay were performed. Numbers of SCEs per chromosome were evaluated under the microscope. The results were the average of three independent experiments and presented as mean ± SD (left panel). Western blotting was carried out to verify the knockdown of ATR and BLM protein level in ATR knockdown cells (right panel).
(D) BLM overexpression suppressed SCE in TopBP1-depleted cells. HeLa cells were transfected with CTR or TopBP1 siRNAs twice in a 24-hr interval with or without co-transfection with constructs encoding wild-type, SFB-tagged BLM. Numbers of SCEs per chromosome were evaluated under the microscope in these transfected cells. The results were the average of three independent experiments and presented as mean ± SD (left panel). Western blotting was carried out to verify the overexpression of wild-type SFB-BLM (right panel).
(E) U2OS cells stably expressing siRNA-resistant constructs encoding wild-type, ΔBRCT5 mutant, or ΔAD mutant SFB-TopBP1 were transfected with CTR siRNA or TopBP1 specific siRNA. Numbers of SCEs per chromosome were evaluated under the microscope. The results were the average of three independent experiments and presented as mean ± SD (left panel). Western blotting was carried out to confirm the expression of wild-type or mutant TopBP1 in these cells (right panel).
Since a major function of TopBP1 is to regulate replication checkpoint, we asked whether or not BLM would have a similar function in checkpoint control. However, we did not detect any significant difference in CHK1 phosphorylation between wild-type and BLM-deficient cells following hydroxyurea treatment (Figure 3B), indicating that BLM is not critical for CHK1 activation in response to replication stress.
The well-known function of BLM is to suppress SCE and thereby prevent genomic instability (Ray and German, 1984). In concordance with the phenotype induced by BLM deficiency, TopBP1 depletion led to increased SCE (Figure 3C). However, depletion of ATR, which is a key player in replication checkpoint control, did not have any drastic effects on SCE or BLM protein level (Figure 3C), implying that the role of TopBP1 in maintaining BLM expression and suppressing SCE is independent of its function in replication checkpoint control.
To determine whether TopBP1 suppresses SCE mainly through maintaining BLM expression, we over-expressed BLM in TopBP1-depleted cells; indeed, we observed a reduction of SCE in these cells (Figure 3D). Next, we explored whether the interaction of TopBP1 with BLM is required for suppressing SCE: we reconstituted TopBP1-depleted cells with wild-type TopBP1, a TopBP1 mutant defective in BLM binding (ΔBRCT5), or a TopBP1 mutant defective in ATR activation (ΔAD) and examined SCE phenotypes in these cells. Reconstitution with wild-type TopBP1 and with a TopBP1 mutant defective in ATR activation, but not with the BRCT5 deletion mutant of TopBP1, significantly reduced SCE and restored BLM expression in TopBP1-depleted cells (Figure 3E), suggesting that the BLM-binding and specifically its ability to stabilize BLM are important for TopBP1’s function in SCE suppression. Furthermore, the expression of BRCT7+8 deletion mutant of TopBP1, which fails to bind to BACH1, in TopBP1-depleted cells decreased SCE similar to that of wild-type TopBP1 but did not affect BACH1 protein level (Figure 3E). These data suggest that the effect of TopBP1 on BLM expression is not mediated by BACH1.
BLM is Ubiquitinated by E3 Ligase MIB1
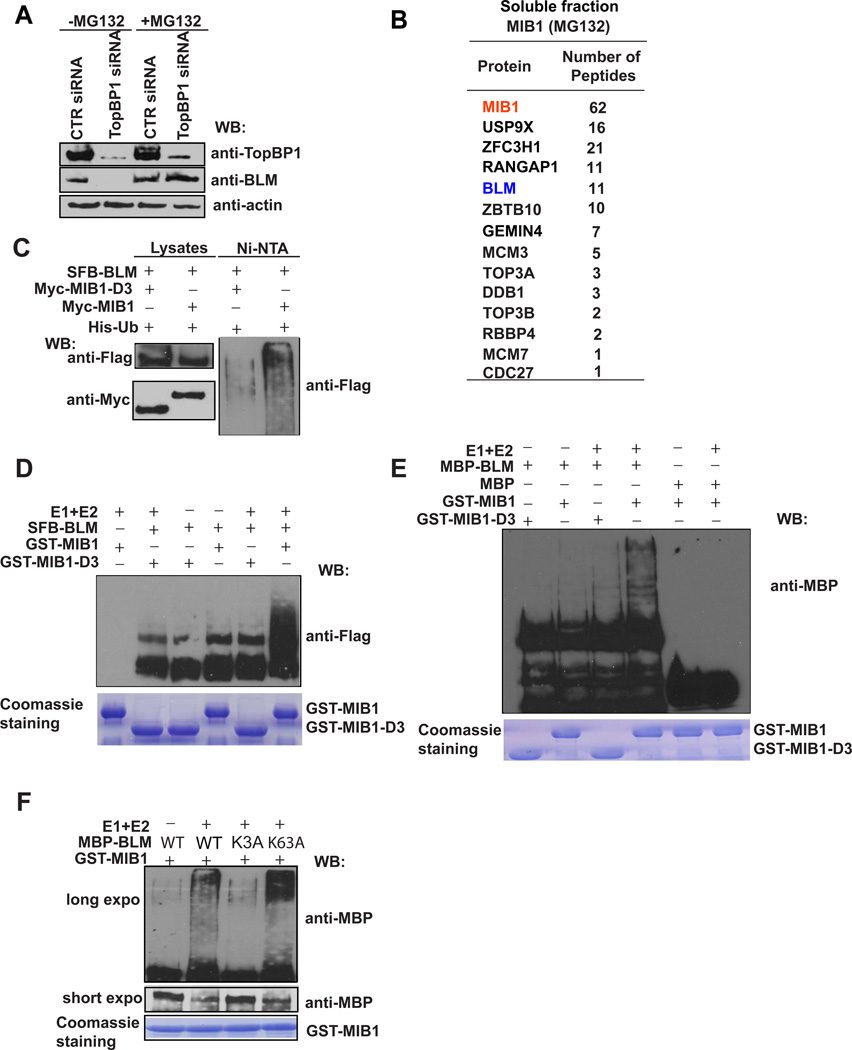
To better understand how TopBP1 maintains BLM expression in the cell, we treated TopBP1-depleted cells with the proteasome inhibitor MG132 and found that BLM expression was restored (Figure 4A), suggesting that TopBP1 may bind to BLM and protect BLM from degradation in vivo. Unfortunately, we did not know until this study how BLM is regulated in the cell. Mass spectrometry analysis of BLM-associated proteins revealed an E3 ubiquitin ligase MIB1 as a putative BLM-binding protein (Figure 1D). To confirm this observation, we performed reverse tandem affinity purification using cells expressing SFB-tagged MIB1. Mass spectrometry analysis verified the MIB1/BLM interaction (Figure 4B), indicating that BLM may be a substrate of MIB1. Moreover, we confirmed that BLM interacts with MIB1 via its helicase domain (Figure S1E) and this interaction is not phosphorylation dependent (Figure S1F).
Figure 4. BLM Is Ubiquitinated by E3 Ligase MIB1.
(A) BLM is degraded by a proteasome-mediated pathway in TopBP1-depleted cells. HeLa cells transfected with control or TopBP1 siRNA were treated with MG132 for 6 hr. Cell lysates were subjected to Western blotting with the indicated antibodies.
(B) A list of MIB1-associated proteins in soluble fraction identified by mass spectrometry analysis. 293T cells stably expressing SFB-tagged MIB1 were incubated with MG132 for 6 hr. Cells were then collected and underwent tandem affinity purification and mass spectrometry analysis.
(C) BLM is ubiquitinated by MIB1 in vivo. 293T cells were transfected with constructs encoding 6xHis-ubiquitin, and SFB-BLM together with constructs encoding Myc-MIB1 or Myc-MIB1-D3. Cells were lysed with denaturing buffer. Ubiquitinated proteins were purified by Ni-NTA agarose beads and subjected to Western blotting with the indicated antibodies.
(D, E) BLM is ubiquitinated by MIB1 in vitro. SFB-tagged BLM purified from 293T cells (D) or MBP-tagged BLM purified from E. coli (E) were incubated with E. coli expressed and purified GST-MIB1 or GST-MIB1-D3 for in vitro ubiquitination assays. BLM ubiquitination was evaluated by Western blotting using the indicated antibodies.
(F) MBP-tagged wild-type or mutant BLM was purified from E. coli and subjected to in vitro ubiquitination assays using MIB1 as the E3 ligase. BLM ubiquitination was assessed by anti-MBP immunoblotting.
MIB1 is an E3 ligase known to ubiquitinate delta receptors, which act as Notch ligands (Itoh et al., 2003). We further explored whether MIB1 could promote BLM ubiquitination. Overexpression of wild-type MIB1, but not of a RING domain deletion mutant (D3) of MIB1, increased BLM ubiquitination in vivo (Figure 4C). We also performed in vitro ubiquitination assays using SFB-tagged BLM purified from 293T cells. Indeed, only wild-type GST-MIB1, but not the D3 mutant of MIB1, could polyubiquitinate BLM in vitro (Figure 4D). To exclude any contamination from mammalian cells, we repeated these in vitro ubiquitination assays using E. coli–expressed and -purified MBP-tagged BLM and GST-tagged MIB1. Similarly, GST-MIB1, but not GST-MIB1-D3 mutant, could polyubiquitinate BLM (Figure 4E). Next, we sought to identify ubiquitination sites on BLM. A series of BLM deletion mutants was constructed. We observed that the N-terminus of BLM was ubiquitinated by MIB1 (data not shown). To further map the ubiquitination sites, several lysine residues at the N-terminus of BLM were mutated to alanine. While wild-type BLM and a K63A mutant of BLM could be efficiently ubiquitinated by MIB1 in vitro, a K3A mutant (K38A/K39A/K40A) of BLM dramatically decreased MIB1-mediated BLM ubiquitination (Figure 4F), suggesting that these residues are potential ubiquitination sites on BLM. As a control experiment, we showed that the K3A mutant did not affect BLM/MIB1 interaction (Figure S1G). Together, these data suggest that MIB1 is an E3 ligase that ubiquitinates BLM both in vitro and in vivo.
TopBP1 and E3 Ligase MIB1 Have Opposite Roles in Regulating BLM Protein Level
We further investigated how TopBP1 and MIB1 would participate in the regulation of BLM protein level. BLM protein level decreased in G1 phase and peaked in S phase (Figure 2C), suggesting that BLM protein is degraded in G1 but stabilized in S phase. Overexpression of wild-type TopBP1, but not of the BRCT5 deletion mutant of TopBP1, significantly decreased the binding of MIB1 to BLM (Figure 5A), suggesting that the binding of TopBP1 to BLM competes for its recruitment by MIB1. Indeed, when we checked the BLM/TopBP1 and BLM/MIB1 interactions in G1 and S phase cells, we only observed the BLM/MIB1 interaction in G1 phase (Figure 5B). In contrast, BLM/TopBP1 interaction peaked in S phase (Figure 5B; also see Figure 2C), indicating that TopBP1 interacts with BLM in S phase and thus prevents BLM degradation by MIB1. In support of this hypothesis, we detected BLM ubiquitination in G1 but not in S phase cells (Figure S2A). Moreover, we showed that BLM was rapidly degraded In G1 cells (Figure S2B). Since TopBP1 does not interact with BLM in G1 (Figure 2C), the presence or absence of TopBP1 did not affect BLM half-life in G1 (Figure S2B). However, in S phase, BLM was quite stable in the presence of TopBP1, but its half-life notably decreased in TopBP1-depleted cells (Figure S2B). Thus, BLM is targeted for degradation by MIB1 in G1 phase, but protected by TopBP1 in S phase of the cell cycle. In agreement with this hypothesis, we found that the K3A mutant of BLM was more stable than wild-type BLM in G1 cells (Figure S2C). On the other hand, the half-life of BLM S338A mutant, which cannot bind to TopBP1, was shorter than that of wild-type BLM in the cell (Figure S2D).
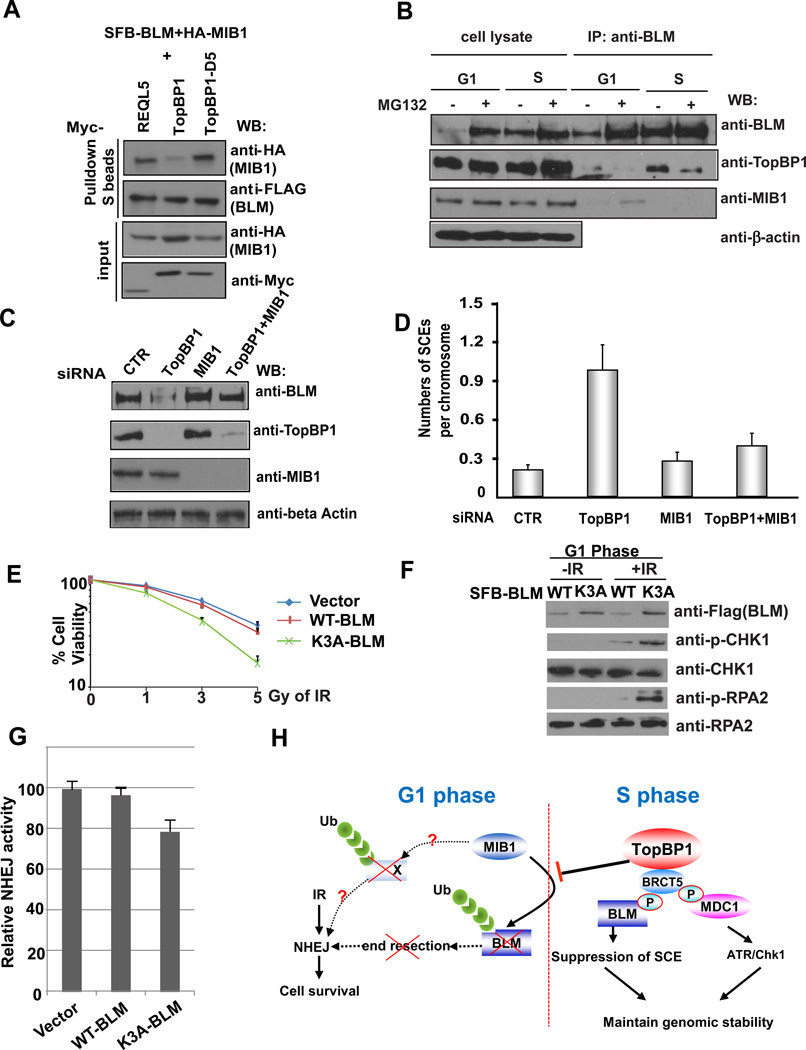
Figure 5. TopBP1 and E3 Ligase MIB1 Serve Opposite Roles in Regulating BLM Protein Level.
(A) TopBP1 competes with MIB1 for its binding to BLM. 293T cells were transfected with plasmids encoding SFB-tagged BLM, HA-tagged MIB1 together with plasmids encoding Myc-tagged RECQL5, wild-type TopBP1, or TopBP1 with deletion of the BRCT5 (TopBP1-D5). Precipitation reactions were conducted using S-protein beads and then subjected to Western blotting using the indicated antibodies.
(B) BLM/TopBP1 and BLM/MIB1 interactions are cell cycle–regulated. HeLa cells were synchronized with nocodazole and then released into fresh medium with or without MG132 treatment for 2 hr just before collecting at the G1 phase (4 hr after release) or S phase (12 hr after release). Cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies.
(C) Depletion of MIB1 restores BLM expression in TopBP1-depleted cells. HeLa cells were transfected with control (CTR) siRNA, TopBP1 siRNA, MIB1 siRNAs, or TopBP1 and MIB1 siRNAs together. Cell lysates were immunoblotted with the indicated antibodies.
(D) Depletion of MIB1 reverses the increase of SCE in TopBP1-depleted cells. HeLa cells were treated with the indicated siRNAs, and SCE assays were performed. Numbers of SCEs per chromosome were evaluated under the microscope. The results were the average of three independent experiments and presented as mean ± SD.
(E) Cells expressing the K3A mutant of BLM were more sensitive to irradiation than cells stably expressing wild-type BLM. HeLa cells stably expressing SFB-vector, wild-type BLM, or K3A mutant BLM were synchronized in G1 phase and irradiated by the indicated dose of X-ray. Cell survival following irradiation was measured by clonogenic assay.
(F) HeLa cells stably expressing wild-type or the K3A mutant of BLM were synchronized in G1 phase with or without irradiation treatment. Cell lysates were immunoblotted with the indicated antibodies.
(G) HeLa cells stably expressing wild-type or the K3A mutant of BLM were transfected with linearized plasmid pcDNA3.1/hygro. Cells were incubated in selective media containing 100 µg ml−1 hygromycin for 14 days and the numbers of colonies were determined. Results are means (±SD) of three independent experiments.
(H) A revised model of TopBP1 functions in genome maintenance via its involvement in both replication checkpoint control and SCE suppression.
Moreover, we showed that depletion of MIB1 restored BLM protein level in TopBP1-depleted cells (Figure 5C) and restored BLM foci formation following DNA damage (Figure S2E). In addition, while depletion of TopBP1 led to increased SCE, co-depletion of MIB1 reversed this phenotype (Figure 5D), suggesting that MIB1 and TopBP1 compete to regulate BLM protein stability and thus control SCE in the cell.
BLM downregulation in G1 cells is important for DNA repair via NHEJ pathway
An interesting question is why BLM needs to be degraded in G1 cells. We speculated that because BLM promotes end resection as shown in previous publications (Gravel et al., 2008), BLM may need to be degraded in G1 to limit end resection and thus promote non-homologous end joining (NHEJ) repair of DSB in G1 cells. To test this hypothesis, we used cell lines stably expressing wild-type or the K3A mutant of BLM. Since BLM K3A mutant could not be targeted for degradation by MIB1 in G1, the expression level of this mutant was higher in G1 cells (Figure S2C). Interestingly, when we synchronized cells stably expressing wild-type or the K3A mutant of BLM in G1 and irradiated them, we found that cells expressing BLM K3A mutant were more sensitive to radiation than cells stably expressing wild-type BLM (Figure 5E). Moreover, in G1, CHK1 and RPA phosphorylation levels were higher in cells expressing BLM K3A mutant than those in cells expressing wild-type BLM (Figure 5F), indicating that while end resection is normally limited in G1, but this is not the case in cells expressing the K3A mutant of BLM, which cannot be degraded. Furthermore, cells expressing BLM K3A mutant has lower NHEJ activity than that in cells expressing wild-type BLM (Figure 5G). Taken together, we propose that BLM degradation in G1 phase is needed to limit end resection and thus allow the use of NHEJ pathway for DSB repair in G1 cells (Figure 5H). In agreement with this conclusion, we showed that MIB1-depleted cells displayed normal cell cycle distribution (Figure S2F), but exhibited increased radiation sensitivity, possibly by preventing BLM degradation in G1 cells (Figure S3A and S3B), indicated that MIB1 may regulate the stability of BLM and probably other substrates and thus play an indirect role in DNA repair.
DISCUSSION
DNA damage checkpoints play important roles in maintaining genomic stability. Increasing evidence has implicated TopBP1 in DNA replication checkpoint control (Bartek and Mailand, 2006; Burrows and Elledge, 2008; Cimprich and Cortez, 2008; Gong et al., 2010; Wang et al., 2011; Yan and Michael, 2009). In this study, we demonstrated that TopBP1 interacts with BLM, maintains BLM protein level, and thereby suppresses SCE. This function of TopBP1 appears to be independent of its role in replication checkpoint control; we did not observe similar phenotypes in ATR-depleted cells. Thus, we propose that TopBP1 maintains genomic stability by suppressing SCE in addition to its well-established role in replication checkpoint control (Figure 5H). We suspect that both of these functions are important for preventing genomic instability and tumorigenesis.
We showed that TopBP1 binds to BLM via its BRCT5 domain. Given that the BRCT domains are known phosphoprotein-binding domains (Manke et al., 2003; Yu et al., 2003), we reasoned that this interaction is regulated in a phosphorylation-dependent manner. Indeed, we showed that the BLM/TopBP1 interaction is cell cycle–regulated and that the Ser338 residue of BLM is important for its interaction with TopBP1. These data, together with S phase-specific phosphorylation of BLM at Ser338 site, support the hypothesis that the Ser338 site of BLM is phosphorylated in vivo, which contributes to the cell cycle-dependent regulation of TopBP1/BLM interaction. Of course, it remains to be determined which kinase is responsible for this phosphorylation event and how it regulates TopBP1/BLM interaction in S/G2 cells.
In vertebrates TopBP1 plays important roles in both DNA replication and replication checkpoint control (Bartek and Mailand, 2006; Burrows and Elledge, 2008; Cescutti et al., 2010). Of course if the replication is stopped, there would be no sister-chromatid recombination in TopBP1-depleted cells. However, our previous study showed that siRNA-mediated depletion of TopBP1 does not dramatically affect DNA replication (Gong et al., 2010). It is possible that our siRNA-mediated depletion of TopBP1 is not complete. The residual TopBP1 in siRNA-treated cells may be sufficient to support replication initiation, but not adequate to maintain replication checkpoint control. This could explain that under the conditions we used, we did not observe any dramatic defect in DNA replication, but noticed significant defects in replication checkpoint control and SCE suppression as reported in this study.
The significant reduction of BLM protein level in TopBP1-depleted cells raises the possibility that BLM may be actively degraded in the cell. We identified the E3 ligase MIB1 as a BLM-associated protein. MIB1 contains a zinc finger domain at its N-terminus, ankyrin repeats in the middle, and a RING finger domain at its C-terminus. It was reported that MIB1 mediates ubiquitination of delta receptors, which act as Notch ligands (Itoh et al., 2003). We observed MIB1 localization in both nuclei and cytoplasm (Figure S3C), which is consistent with previous report (Itoh et al., 2003). In this study, we showed that MIB1 ubiquitinates BLM both in vivo and in vitro (Figure 4). More importantly, we demonstrated that co-depletion of MIB1 restored BLM expression and SCE suppression function in TopBP1-depleted cells (Figures 5C and 5D), suggesting that the critical function of TopBP1 in BLM regulation is to prevent MIB1-dependent degradation.
Moreover, MIB1-mediated BLM degradation in G1 phase appears to be important for DNA double-strand break repair. Cells expressing an un-degradable BLM mutant showed radiation sensitivity, probably by trigging end resection and inhibiting NHEJ pathway in G1 cells (Figure 5 E–G). MIB1 depletion also led to increased radiation sensitivity (Figure S3A), possible by increasing BLM protein level and inhibiting NHEJ in G1 cells. However, we cannot exclude the possibility that other MIB1 substrates may also contribute to the repair defect observed in MIB1 knockdown cells. Future studies will be devoted to identify additional MIB1 substrates and elucidate how MIB1 may regulate BLM and other substrate to control DNA repair. In addition, we would like to determine whether deregulation of MIB1-dependent BLM degradation contributes to genomic instability and tumorigenesis in humans.
Supplementary Material
Highlights.
TopBP1 stabilizes BLM in S phase cells to suppress sister chromatid exchange.
BLM is ubiquitinated by E3 ligase MIB1 and degraded in G1 cells.
BLM downregulation in G1 limits resection and promotes NHEJ-mediated DNA repair.
ACKNOWLEDGMENTS
We thank our colleagues in Dr. Chen’s laboratory for insightful discussions and technical assistance. We thank Dr. Lei Li at The University of Texas MD Anderson Cancer for providing wild-type and BLM-deficient cells. This work was supported in part by grants from the National Institutes of Health (CA089239 and CA092312 to J.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.C. is a recipient of an Era of Hope Scholar award from the United States Department of Defense (W81XWH-05-1-0470) and is a member of MD Anderson Cancer Center, which is supported in part by the National Institutes of Health through a Cancer Center Support Grant (CA016672).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest.
REFERENCE
- Bartek J, Mailand N. TOPping up ATR activity. Cell. 2006;124:888–890. doi: 10.1016/j.cell.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes & development. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescutti R, Negrini S, Kohzaki M, Halazonetis TD. TopBP1 functions with 53BP1 in the G1 DNA damage checkpoint. EMBO J. 2010;29:3723–3732. doi: 10.1038/emboj.2010.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes & development. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nature structural & molecular biology. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes & development. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Kim JE, McAvoy SA, Smith DI, Chen J. Human TopBP1 ensures genome integrity during normal S phase. Mol Cell Biol. 2005;25:10907–10915. doi: 10.1128/MCB.25.24.10907-10915.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. The Journal of biological chemistry. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Fukazawa H, Murakami Y, Uehara Y. Nek11, a new member of the NIMA family of kinases, involved in DNA replication and genotoxic stress responses. The Journal of biological chemistry. 2002;277:39655–39665. doi: 10.1074/jbc.M204599200. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Fukazawa H, Murakami Y, Uehara Y. Nucleolar Nek11 is a novel target of Nek2A in G1/S-arrested cells. The Journal of biological chemistry. 2004;279:32716–32727. doi: 10.1074/jbc.M404104200. [DOI] [PubMed] [Google Scholar]
- Ray JH, German J. Bloom's syndrome and EM9 cells in BrdU-containing medium exhibit similarly elevated frequencies of sister chromatid exchange but dissimilar amounts of cellular proliferation and chromosome disruption. Chromosoma. 1984;90:383–388. doi: 10.1007/BF00294165. [DOI] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes & development. 2010;24:183–194. doi: 10.1101/gad.1860310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Melixetian M, Klein DK, Helin K. NEK11: linking CHK1 and CDC25A in DNA damage checkpoint signaling. Cell Cycle. 2010;9:450–455. doi: 10.4161/cc.9.3.10513. [DOI] [PubMed] [Google Scholar]
- Wang J, Gong Z, Chen J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol. 2011;193:267–273. doi: 10.1083/jcb.201010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Dunphy WG. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell. 2009;20:2351–2360. doi: 10.1091/mbc.E08-12-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.