Abstract
All known active forms of diheme bacterial cytochrome c peroxidase (bCcP) enzymes are described by a homodimeric state. Further the majority of bCcPs reported only display activity when the high-potential electron transfer heme of the protein (FeH) is reduced by to the ferrous oxidation state. Reduction of FeH results in a set of conformational changes allowing for the low-potential peroxidatic heme (FeL) to adopt a high-spin, five-coordinate state that is capable of binding substrate. Here we examine the impact of dimerization upon the activity of the Shewanella oneidensis bCcP by the preparation of single charge-reversal mutants at the dimer interface, and use the resulting construct to illustrate why dimerization is likely a requirement for activity in bCcPs. The E258K mutant is found to form a monomeric state in solution as characterized by size exclusion chromatography and analytical ultracentrifugation analyses. The resulting E258K monomer has a folding stability comparable to wild-type So bCcP, and an activity that is only slightly diminished (kcat/Km of 23 × 10 6 M−1 s−1). Spectroscopic and potentiometric analyses reveal that while the thermodynamic stability of the activated form of the enzyme is unchanged (characterized by the Em value of the FeHII/III couple) , the kinetic stability of the activated form of the enzyme has been greatly diminished upon generation of the monomer. Together, these data suggest a model where dimerization of bCcP enzymes is required in order to stabilize the lifetime of the activated form of the enzyme against re-oxidation of FeH and deactivation of FeL.
Bacterial diheme cytochrome c peroxidase (bCcP) enzymes, which detoxify hydrogen peroxide in the periplasmic space of many gram-negative bacteria by utilizing reducing equivalents from the cytochrome c redox pool (1–3), have always been described as homodimeric enzymes. Each bCcP protomer contains, two c-type heme cofactors, plus an additional Ca2+ ion, and currently, eight different bCcP enzymes have been studied structurally through X-ray crystallographic analyses (4–12). In all cases, bCcPs crystallize as homodimers, and the dimeric state has been further substantiated by hydrodynamic studies of several different bCcP enzymes (8, 13–15). In the case of the Paracoccus denitrificans (Pd) enzyme, conditions that stabilize homodimerization have been studied in some detail: it has been illustrated by Pettigrew and co-workers that removal of the Ca2+ or artificially low ionic strengths result in an inactive, monomeric form of the enzyme (14). Ca2+ binding in bCcPs is thought to have two roles: a tight-binding Ca2+ affinity found in all bCcP enzymes that impacts electronic communication between the heme groups (5, 8, 16–17), as well as a non-specific ionic-strength effect that shifts the equilibrium between inactive monomers and active dimers toward the dimerized state, as described for the Pd and Pseudomonas nautica (Pn) enzymes (5, 8, 14). However, the basis for why the dimeric state is required to achieve catalysis is poorly understood, and here we examine this issue in further detail using the enzyme from Shewanella oneidensis (11, 18) as a model system to generate a monomerized version of the protein that retains catalytic activity. Our results suggest an unforeseen role in dimerization as a requirement to stabilize a conformation of the active site that is required for catalysis.
In contrast to the apparent required quaternary structure, the requirement for the cofactors in bCcPs is well understood: the high-potential (200 – 450 mV vs NHE) Met-His ligated heme (H-heme) serves as an electron transfer mediator to the other, low- potential heme (~ −300 mV vs NHE) that acts as the peroxidatic active site (3, 19). The Ca2+ ion is located at the interface between the H-heme and L-heme domains. The high-potential heme, which is not present in monoheme peroxidases (e.g. yeast CcP) is likely responsible for storing a second oxidizing equivalent during the catalytic cycle for many bCcP enzymes (19) and serving as an electron transfer conduit or mediator (20, 21). Despite these broadly conserved similarities, within bCcPs a functional difference arises in whether the isolated, FeLIIIFeHIII oxidation state of the protein displays activity, or not. Most characterized bCcPs are isolated in a catalytically inactive, fully oxidized (FeLIIIFeHIII) state, where the L-heme is bis-His coordinated such that substrate cannot bind to FeL. The active form of the enzyme can be achieved by the addition of one-electron to the H-heme. This reductive activation not only yields a FeHIIFeLIII oxidation state, but results in local conformational changes that cause the reorientation of ligands around the active site heme, producing an ‘Open’ conformer that allows for peroxide to bind at FeL. Collectively, these requirements have been illustrated for the canonical bCcP enzymes from Pseudomonas aeruginosa (Psa) (4), Paracoccus denitrificans (Pd) (5), Pseudomonas nautica (Pn) (6), Rhodobacter capsulatus (Rc) (7), Pseudomonas stutzeri (Ps) (8), two isoenzymes from Geobacter sulfurreducens (Gs) (9, 10), and, recently, Shewanella oneidensis (So) (11), all of which require activation through the production of the FeHIIFeLIII oxidation state, to generate the ‘Open’ conformer. In contrast to this canonical family of diheme peroxidases the enzymes from Nitrosomonas europaea (Ne) (22) and Methylococcus capsulatus Bath (Mc) (15) do not need to undergo an activation step: the as-isolated enzyme, still in the FeHIIIFeLIII oxidation state, displays a penta-coordinate active site iron as is active without further reduction (12). Regardless of the functional manifold displayed by bCcP enzymes, every known bCcP is a homodimer, suggesting that the dimeric state has an important, yet poorly understood, functional role. Here, we generate a charge reversal mutant of the Shewanella enzyme to disrupt the protein dimer interface and address the question: Why are all bCcP enzymes dimers?
MATERIALS AND METHODS
Protein expression and purification
Recombinant Shewanella oneidensis cytochrome c peroxidase was expressed and purified in the same manner as in the recent paper from Pulcu et al (18). Similarly, the recombinant So c5 (gene SO0264/plasmid pSOc5) was expressed and purified as previously by Pulcu et al. (18). So CcP mutants, E258K and E321K, were constructed using the QuikChange Mutagenesis Kit and expressed and purified as an MBP fusion as described for wild-type enzyme. The glutamic acid residue at position 258 was mutated to lysine using the following primers;
Forward:(5’-CCGACACTGCGCAATATTAAACTAACCTATC CCTAC-3’), and
Reverse:(5’-GTAGGGATAGGTTAGTTTAATATTGCGCAGTGTCGG-3’).
The presence of the correct mutations at position 258 was verified by gene sequencing (GeneWiz). This mutant required induction with 500 µg/L IPTG for 4 hours for optimal expression. Purification followed the same steps as the wild-type CcP. The glutamic acid residue at position 321 mutated to lysine using the following primers;
Forward:(5’-CCACCATCAAATAAAAAGACTCCGCGCCCAG TT-3’) and
Reverse:(5’-AACTGGGCGCGGAGTCTTTTTATTTGATGGT GG-3’).
The presence of the correct mutations at position 321 was verified by gene sequencing. Expression and purification followed the same steps as the wild-type CcP.
Chemical monomerization
Wild-type CcP was diluted to 50 µM and dialyzed in 20 mM HEPES, 150 mM NaCl, 10 mM EGTA, pH 8 for 4 hours at room temperature. Buffer was then replaced and the dialysis continued overnight at 4°C. Samples were concentrated to 25 µM and applied to an S-100 column equilibrated with 20 mM HEPES, 150 mM NaCl, 1 mM EGTA, pH 8. Fractions containing CcP were concentrated to approximately 100 µM and excess EGTA was removed by dialysis.
Analytical Ultracentfugation
Sedimentation velocity AUC experiments were carried out using a Beckman XL-I analytical ultracentrifuge (Beckman Coulter) with an absorbance optical system at the MIT Biophysical Instrumentation Facility. Fresh samples of 10 µM So CcP were extensively dialyzed against 20 mM HEPES, 50 mM NaCl, pH 7.5. The protein solution and dialysate were loaded into two-sector cells and equilibrated thermally to reach a constant temperature of 20.0 °C prior to centrifugation for ≥ 10 hours at 42,000 rpm. The protein concentration across the cell was measured by monitoring the heme absorbance at 420 nm at 2 min intervals. Data was analyzed using Sedfit, and diffusion-deconvoluted sedimentation coefficient distributions, c(s), were generated from 200 scans with the program SedFit.34. The sedimentation coefficients were converted to standard values (s20,w) using a solvent viscosity and density of 1.0221 and 1.00185 g/mL (calculated with the program Sednterp35), respectively.
Thermal denaturation monitored by CD
CD spectra of Shewanella oneidensis wild-type CcP and charge reversal mutant samples were recorded with an Applied Photophysics Circular Dichroism Spectrophotometer with a Quantum Northwest temperature controller. Data were collected using a 0.1 mM path length anaerobic quartz cuvette. CcP enzyme samples were kept in 25 mM phosphate buffer pH 7.0 with a final protein concentration of 20 µM. At each given temperature (increased stepwise by 1 °C from 20 °C – 90 °C) the protein sample was allowed to equilibrate for 2 min before the intensity at 222 nm was recorded with an averaging time of 5 sec/°C.
Activity assays
The electronic absorption spectra of the CcP enzyme were collected on a Cary 50 spectrophotometer (Varian). All enzymatic assays were performed at 23 °C in standard assay buffer of 5 mM MES, 5mM HEPES, 10 mM NaCl, 1 mM CaCl2 and pH 6. Horse heart cytochrome c (Sigma) and So cytochrome c5 were reduced by treatment with 20 mM L-sodium ascorbate (Sigma), and excess reductant was removed using a PD-10 desalting column (NEB) before the assay. CcP stock solutions were reductively activated with 1 mM L-sodium ascorbate and 10 µM diaminodurol (DAD), incubated on ice in assay buffer for at least 30 min, which prepared the active, FeHIIFeLIII, form of the enzyme. The extent of reduction of the sample was monitored by optical absorption. The oxidation of the reduced horse heart cytochrome c and So c5 are monitored at 550 nm and 553 nm, respectively, in the presence of H2O2 using an extinction coefficient difference between the oxidized and reduced proteins (∆ε 550nm = 21.5 mM−1cm−1 for horse heart and ∆ε 553nm = 12.5 mM−1cm−1 for c5) (18).
Potentiometric Redox Titrations
Anaerobic redox titrations of the H-heme of Shewanella oneidensis wild-type CcP and the E258K charge reversal mutant were carried out and followed optically in the wavelength region of 300–800 nm. Titrations were performed at 23 °C by measuring absorption changes at 553 nm of 6 µM protein solutions in 10 mM MES/HEPES pH 6.0 in the presence of 2 mM CaCl2. Redox potentials were measured using a combination Ag/AgCl Micro Redox electrode (MI-411 Microelectrodes, Inc). The solution potential was adjusted using the stepwise µL additions of reductant (25 mM sodium dithionite) or oxidant (25 mM potassium ferricyanide). Diaminodurol and phenazine methosulfate were included as mediators at final concentrations of 15 µM (inclusion of additional mediators did not improve nor change the experimental results, so triple replicates with just the two mediators were generated).
Oxidation and Reduction kinetic analysis of wild-type So CcP and E258K
The rate of semireduction of the oxidized forms of the wild-type So enzyme and E258K charge reversal mutant with L-sodium ascorbate and 1 µM DAD was monitored at 408 and 553 nm at 23 °C using a Cary 50 spectrophotometer (Varian). Various concentrations of L-sodium ascorbate (1 mM, 2.5 mM, and 5 mM) were used to ensure pseudo-first order kinetics applied to the reduction of 3 µM wild-type So CcP and the E258K mutant. kobs was then calculated according to pseudo-first order kinetic parameters. Re-oxidation of the activated enzyme was achieved by first desalting the semi-reduced enzymes anaerobically, and then exposing the samples to an ambient atmosphere. The same wavelengths were monitored for the re-oxidation as for the reduction.
RESULTS
Cytochrome c peroxidase dimer interface analysis
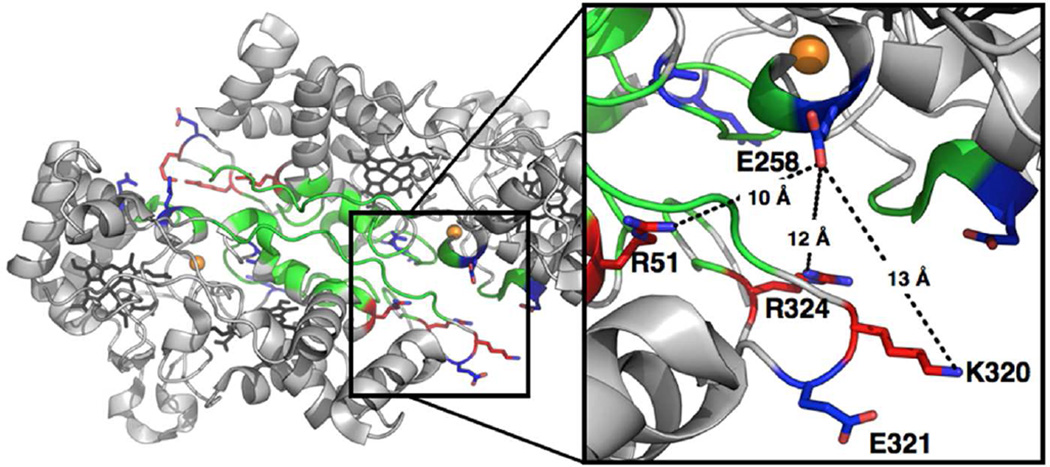
The dimer interface of the S. oneidensis cytochrome c peroxidase crystal structure was analyzed using the Protein Data Bank Europe Protein Interfaces, Surfaces, and Assemblies tool (PDBePISA) (23), a web-based server for examining potential macromolecular interfaces, and predicting probable quaternary structures as well as searching databases for structurally similar interfaces and assemblies. PDBePISA was used to predict the residues that form charged interactions across the dimer interface of the So enzyme (PDB code: 3O5C), as well as, the other crystallized dimeric bCcP enzymes. The structure of the So enzyme (24) (in the fully reduced oxidation state) and the eleven other bCcP structures were analyzed, as collected in Table S1. Eight enzymes are crystalized as dimers and between two and eight charged pairs are found across the dimer interfaces of the various enzymes. So CcP has six charged pairs in the semi-reduced enzyme. The interfacial location of E258 was confirmed by the crystal structure, shown in Figure 1, which illustrates that the charge reversal mutation is in a position that would likely disrupt three long-range charge interactions (< 10 Å) across the dimer interface. In comparison E321 is only located in close proximity to the interface but lacks discernable charge interactions with the neighboring monomer (Figure 1), providing a negative control for comparisons between characteristics of E258K that might not be linked to potential disruption of the dimer interface.
Figure 1.
Crystal structure of S. oneidensis bCcP in the fully reduced oxidation state (PDB: 3O5C). The dimer interface is highlighted by the colored residues predicted by PDBePISA and analysis of the crystal structure. Green residues represent uncharged interfacial amino acids, red are positively charged, and blue are negatively charged residues. (Inset) Local environment around residues E258 and E321. Potential long-range, charge pairing interactions with positively charged residues (R51, R320, and R324) are shown in dotted lines.
Hydrodynamic properties of the dimer and monomer enzymes
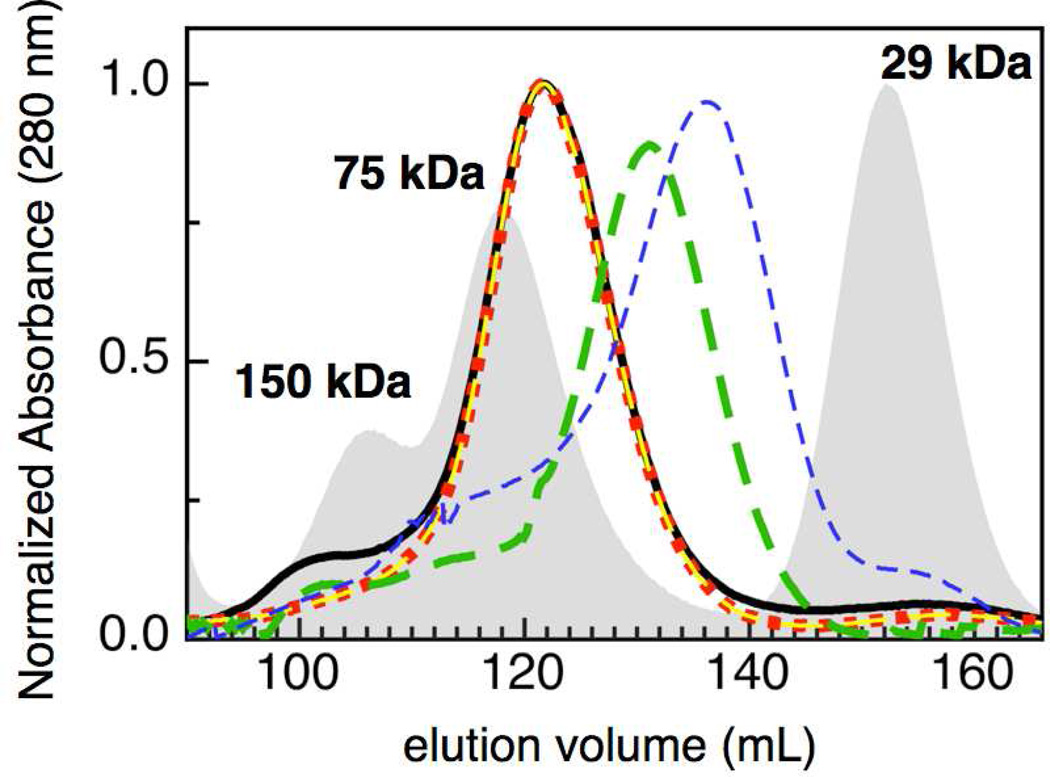
In order to verify that installation of the E258K mutation resulted in the generation of a monomeric version of So CcP, size exclusion chromatography was used to observe the changes in quaternary structure of the various forms of So CcP (i.e., monomer vs dimer). The chromatograms in Figure 2 show the elution profiles for the various So enzymes under different treatment conditions: as-isolated wild-type enzyme (solid black), EGTA incubated (green) and as-isolated E258K (blue). The as-isolated sample of wild-type protein eluted with an apparent molecular weight (Mr) of 67 kDa, consistent with the dimeric state of the enzyme. In the presence of excess calcium (1 mM CaCl2, red dots) the elution profile of the wild-type protein does not change: it is the same as that of the as-isolated sample. In contrast, evidence for higher order oligomers has been previously shown for the Ps enzyme in the presence of excess Ca2+ (8), and in the presence of 1 mM EGTA, all bCcPs examined here display a Mr of 43 kDa (green hash in Figure 2), correlating to a monomeric form of the protein. The apparent discrepancy between the observed Mr and the theoretical size of the monomer can be explained by a difference in shape of the chemically monomerized protein sample and a theoretical monomeric enzyme, as discussed in further detail below.
Figure 2.
Molecular size exclusion chromatography of S. oneidensis CcP, with respect to standards (grey). As-isolated So CcP (solid black) eluted with an apparent molecular mass of 67 kDa. Enzyme incubated with 10 mM CaCl2 (red dots) eluting with an apparent molecular mass equal to the untreated, whereas enzyme treated with EGTA (green hash) elutes with an apparent molecular mass of 43 kDa. The as-isolated E258K (blue hash) elutes with a significantly lower apparent mass (32 kDa). The as-isolated E321K (yellow hash) displays an apparent molecular mass of 67 kDa.
Size exclusion chromatography was used to observe the changes in quaternary structure of the E258K and E321K charge reversal mutants to determine under what conditions the charge reversal mutants are monomers or dimers. The chromatograms in Figure 2 show the elution profiles for the E258K (blue) and E321K (yellow) charge reversal mutants under the same treatment conditions as the wild-type enzyme. The as-isolated E258K sample elutes with Mr of 32 kDa. This is consistent with the monomeric state of the protein sample. In the presence of either excess calcium or EGTA the Mr of E258K is equal to that of the as-isolated sample with an apparent Mr of 32 kDa, which correlates to a monomeric form of the protein (data not shown). The as-isolated E321K sample elutes with an Mr of 67 kDa. This is consistent with the dimeric state of the protein sample.
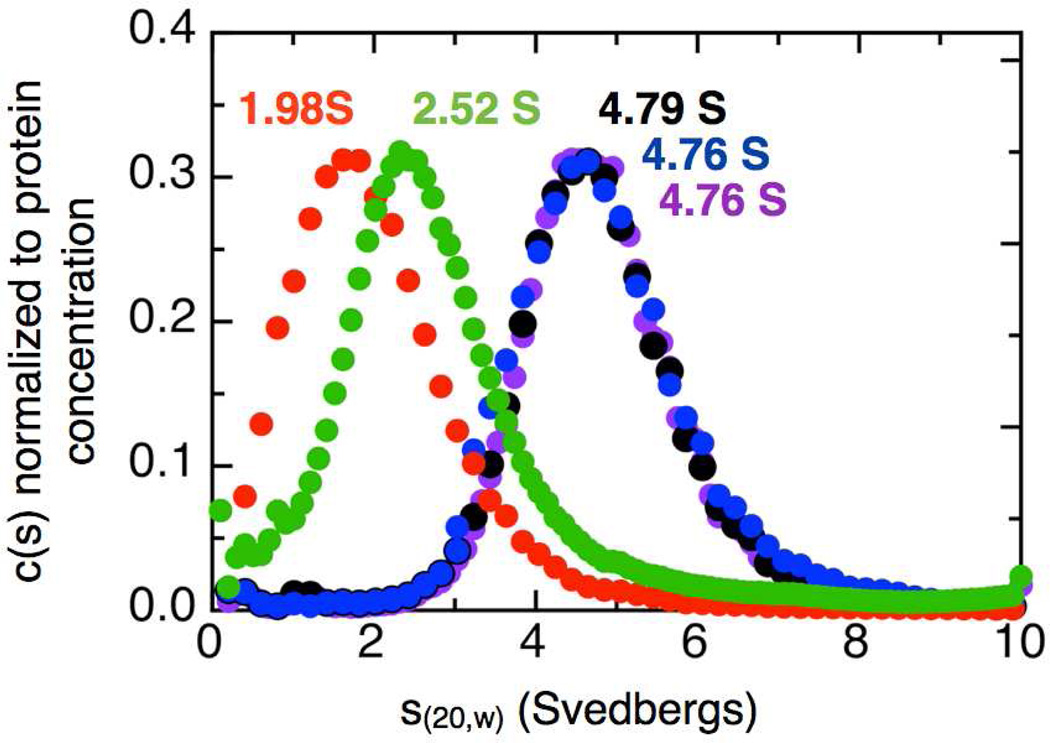
The sedimentation velocity profiles for as-isolated So bCcP in the presence of excess Ca2+ or EGTA were analyzed by the DCDT+ method (25). The sedimentation coefficient distributions are shown in Figure 3, where the maximum s20,w value of the as-isolated, wild-type So CcP is 4.79 S, nearly identical to values reported for other dimeric bCcP enzymes (Table 1) (8, 13–15). Preparation of the Ca2+-depleted enzyme, and sedimentation in the presence of EGTA yields an s20,w value 2.52 S, consistent with other chemically monomerized, EGTA-treated enzymes (8, 21). Treating the apo-protein with excess Ca2+ leads to the return of the native hydrodynamic traits: the presence of excess Ca2+ results in a maximal s20,w of 4.76, essentially identical to the value of the wild-type protein.
Figure 3.
Sedimentation velocity analytical ultracentrifugation. AUC of Soneidensis CcP shows the effect of a charge reversal mutation at the dimer interface: as-isolated bCcP (black), EGTA treated (green), E258K (red), E321K (blue) and bCcP + Ca2+ (purple). The distribution in the top panel is calculated from the 420 nm absorbance. No minor species are observed indicating that the charge reversal mutation and EGTA treatment completely monomerized the enzyme samples.
Table 1.
Sedimentation coefficients of both dimeric and monomeric bCcP enzymes.
| Enzyme | Sedimentation coefficients |
Reference | |
|---|---|---|---|
| Dimer | Monomer | ||
| P. stutzeri | 4.67 | 2.83 | (8) |
| P. denitrificans | 4.78 | 2.69/2.54 | (14) |
| P. pantotrophus | 4.8 | N.D.a | (13) |
| M. capsulatus | 4.82 | N.D.a | (15) |
| S. oneidensis | 4.79 | 2.52 /1.98 | This work |
N.D., not determined
The sedimentation velocity profiles for as-isolated E258K charge reversal mutant, (as-isolated enzyme, or in the presence of excess Ca2+, or EGTA-treated) were analyzed in the same way at the wild-type So bCcP (Figure 3). The s20,w value of as-isolated E258K is 1.98 S. The presence of either an excess of Ca2+ or EGTA does not change the s20,w value (data not shown). All of the s20,w values obtained for the E258K variant are consistent with a protein of molecular weight of the monomer, and suggest a further compacted structure for the E258K as compared to the EGTA-treated wild-type enzyme. The sedimentation velocity profiles for as-isolated E321K charge reversal mutant was analyzed in the same way at the wild-type So bCcP (Figure 3). The s20,w value of as-isolated E321K is 4.76 S virtually identical to the wild-type as-isolated enzyme sample. Notably, the data are in agreement with the calculated values of s20,w as determined by SOMO (26): where the PDB file for the So bCcP crystal structure can be used to calculate values of 4.81 S and 3.08 S, respectively.
Thermal denaturation monitored by CD
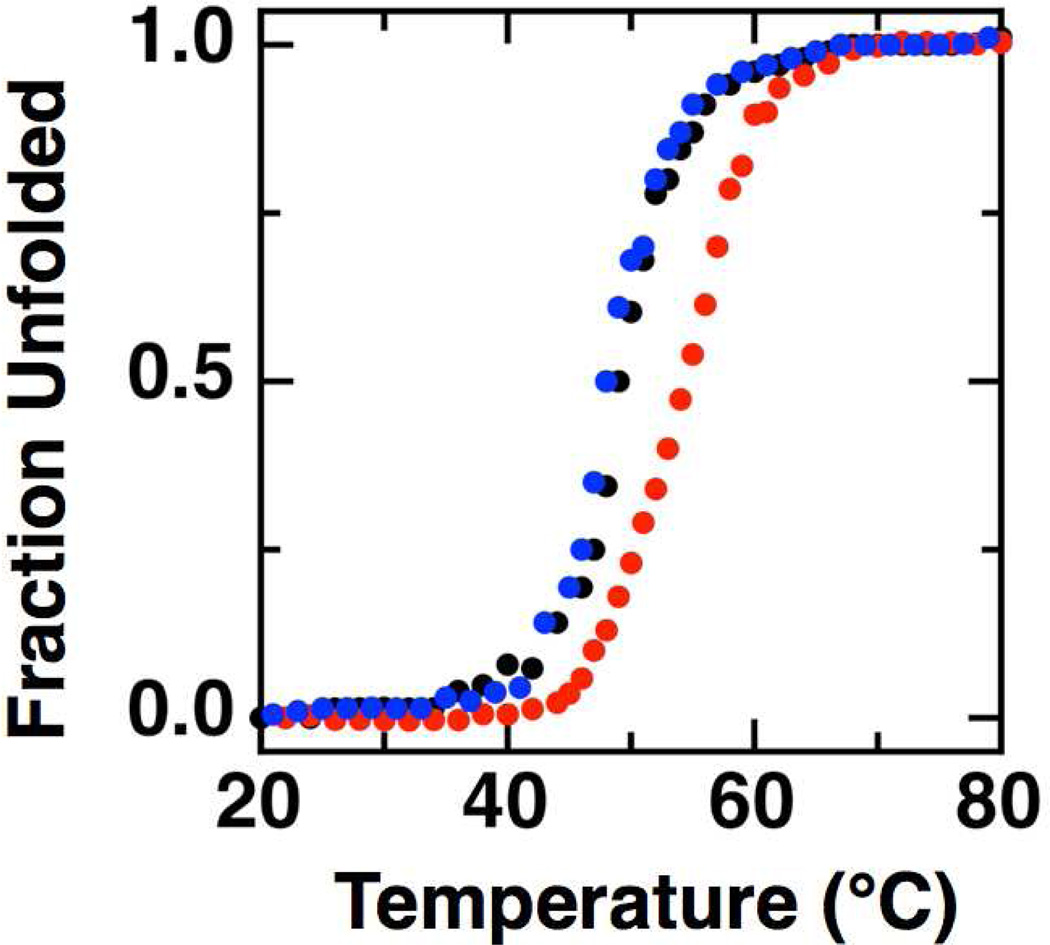
To further verify that the changes in observed quaternary structure did not represent global unfolding of the protein, CD measurements were conducted to assess the stability of the wild-type and mutant enzymes. Thermal denaturation of the wild-type So bCcP and E258K and E321K mutants was monitored at 222 nm, and the loss of the far-UV CD signal was used to monitor the temperature dependence of the thermal denaturation (Figure 4). In all cases, it was found that thermal unfolding was not freely reversible: the CD-detected protein melts let to partial, irreversible aggregation following unfolding, due to extended periods of time at elevated temperatures (> 70 °C). Yet, comparisons of the unfolding experiments allow for the direct comparison of the stability of the wild-type and mutant proteins. At pH 7.0 a single unfolding transition was observed at melting temperature (Tm) 49.2 ± 0.6 °C for the wild-type enzyme (Figure 4A), compared to the single unfolding transitions seen for the E258K and E321K charge reversal mutants showing Tm values of 53.9 ± 0.3 °C and 48.3 ± 0.4 °C, respectively. While it is surprising that disruption of the dimeric interface leads to a slight stabilization in the temperature of melting, the unforeseen enhancement of thermal stability upon monomerization suggests that the native protein may be less stable to account for its native processing and turnover in vivo. But clearly, the single amino acid changes have not resulted in significant perturbations to the thermal stability of the tertiary structure of either the E258K or E321K mutants.
Figure 4.
Thermal denaturation of So cytochrome c peroxidases monitored by CD. Typical curves of the fraction of enzyme unfolded measured at 222 nm as a function of temperature if plotted for wild-type So CcP (black), E258K (red) and E321K charge reversal mutant (blue). (Samples were 10 µM peroxidase at pH 7.0 in phosphate buffer.)
Catalytic properties of the charge reversal monomer mutant
The activity of So CcP was determined previously in the presence of micromolar concentrations of various redox partner proteins (18), where So cyt c5 is the native electron donor in the periplasm of Shewanella (27), and horse heart cyt c can been used as an artificial electron donor that enables facile comparisons with other bCcP enzymes reported in the literature. Here, we determined kinetic parameters for the E258K and E321K charge reversal mutants (Table 2). The linear initial rates at various concentrations of peroxide were used to calculate kinetic parameters using the Michaelis-Menten formalism. As previously described for the wild-type enzyme, the electron donor proteins bind extremely weakly to So bCcP (Km is much greater than 10 µM) and we could not achieve saturating concentrations in vitro using either of electron donors studied: as a result, the values reported below are not maximal turnover rates (18). As found previously with wild-type enzyme, the kinetic parameters for both the E258K and E321K mutants depend on the electron donor used. The E321K mutant behaves overall like wild-type enzyme displaying similar values of both kcat and Km. Overall, the E258K mutant shows a decreased kcat under all conditions (threefold less), with respect to the wild-type protein, but still shows the enhanced kinetic parameters when So cyt c5 is used as the electron donor, versus horse heart cyt c. With wild-type So bCcP, when horse heart cyt c is used as the electron donor turnover is slowed by a factor of 30 when compared to the native electron donor. For semi-reduced (activated) E258K, turnover with horse heart cyt c is approximately 20 times slower than reactions with the native electron donor c5. Further, fully oxidized E258K displays no reactivity with horse heart cyt c. Km values increase for E258K in all circumstances, increasing by two- to ten-fold. Taken together, under the most efficient catalytic conditions for the wild-type enzyme (pre-reduced with ascorbate, using native So cyt c5 as the electron donor), E258K shows an order of magnitude loss in kcat/Km (23 × 106 M−1s−1), indicating that regardless of spectroscopic changes detailed above, the monomeric enzyme is kinetically competent, albeit with a somewhat diminished catalytic efficiency.
Table 2.
Steady state kinetic characterization of E258K and E321K So bCcP mutants
| Wild-type | E258K | E321K | |||||
|---|---|---|---|---|---|---|---|
| Electron Donor |
Parameter | Semired | Oxidized | Semired | Oxidized | Semired | Oxidized |
| Horse heart cyt c |
Km (µM) | 0.03 ± 0.01 | 0.07± 0.01 | 0.4 ± 0.1 | no activity |
0.1 ± 0.02 | 0.9 ± 0.2 |
| kcat (s−1) | 1.9 ± 0.2 | 1.3 ± 0.1 | 0.7 ± 0.1 | 5.1 ± 0.2 | 3.5 ± 1.0 | ||
| kcat/Km (M−1s−1 × 106) | 74 | 7.5 | 2 | 51 | 4 | ||
| So cyt c5 | Km (µM) | 0.3 ± 0.1 | 0.6 ± 0.5 | 0.8 ± 0.1 | 0.7 ± 0.2 | 0.4 ± 0.1 | 0.8 ± 0.3 |
| kcat (s−1) | 73 ± 5 | 7 ± 5 | 18 ± 1 | 2.0 ± 0.4 | 84 ± 4 | 6 ± 3 | |
| kcat/Km (M−1s−1 × 106) | 240 | 12 | 23 | 3 | 210 | 8 | |
We have previously observed that assay progress curves initiated with the fully oxidized enzyme revealed a lag-phase, which has been interpreted in terms of a kinetic model where in situ conversion of the oxidized (inactive) state of the enzyme to the active state can occur, allowing for the extraction of a rate constant associated with the activation process (18). The fully oxidized forms of both charge reversal mutants also show such lag-phase kinetics, and fitting with our kinetic model confirms the results from Michealis–Menten analysis. Using this model, we have previously shown that So bCcP has an activation rate of 0.07 ± 0.02 s−1 when cytochrome c5 is used as the electron donor with the fully oxidized wild-type enzyme (18). For the charge reversal mutants, the apparent activation rates are essentially the same for E258K (0.05 ± 0.01 s−1) and within a factor of two for E321K (0.03 ± 0.01 s−1). In contrast, there are greater differences in the apparent turn-over numbers: for fully-oxidized wild-type So bCcP, the peroxide turnover rate calculated with the lag phase model is 7 ± 1 s−1 for the dimeric E321K mutant turnover of similar (2 ± 1 s−1), while for the monomerized E258K mutant the turnover number is diminished by 50-fold (0.33 ± 0.02 s−1). Together, these data suggests that E258K is less active, though possibly due to cytochrome c5 binding to the E258K monomer mutant protein more weakly, or in a conformation that gives less efficient electron transfer than is seen with the wild-type enzyme.
Spectroscopic properties of the E258K charge reversal mutant: optical absorption
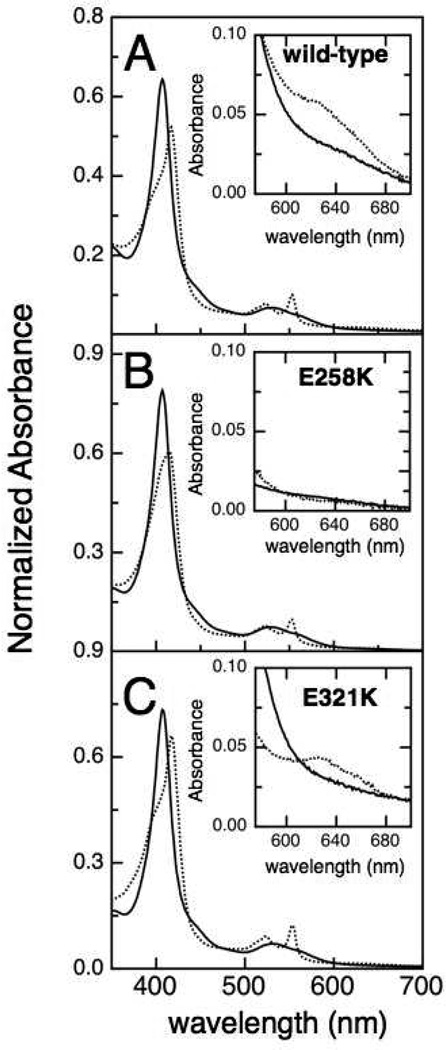
In order to assess the possible basis for the changes in catalytic properties, we first examined the optical characteristics of the charge-reversal mutants. The optical absorbance spectra of fully oxidized and ascorbate-reduced So bCcP, E258K and E321K enzymes are shown in Figure 5. For the oxidized (FeHIIIFeLIII) form of each of the enzymes, the spectral features are virtually identical, including the EGTA-treated enzyme (data not shown). All fully oxidized samples have the characteristic Soret maximum at 407 nm and broad α and β bands at 553 nm and 540 nm that are typical for the So CcP enzyme (18). As reported previously, upon reduction with L-sodium ascorbate the Soret band of the wild-type enzyme shifts to 417 nm and a shoulder appears at 407 nm while the α and β bands at 553 nm and 540 nm sharpen, consistent with the reduction of the H-heme (5). The appearance of a charge transfer band at 640 nm (Figure 5, insets) suggests a high-spin ferric heme state (28). In contrast, similar treatment of E258K with ascorbate causes the Soret band to shift to 417 nm, but with a poorly resolved shoulder at 407 nm; the α and β bands at 553 nm and 540 nm appear as with the wild-type enzyme, but the 640 nm band is absent (Figure 5B). The shoulder at 640 nm indicative of the high-spin Fe(III) is known to be a feature of weak intensity, such that quantitation of the HS state has not been achieved in bCcP enzymes (16, 18, 29–31). Thus, one impact of monomerization displayed by E258K is the diminishment of the concentration of the FeLIII HS state, which is presumably required for binding H2O2.
Figure 5.
Electronic absorption spectra of the oxidized (solid) and ascorbate-reduced (dot) (A) wild-type So bCcP, (B) the E258K charge reversal mutant, and (C) the E321K mutant. The insets show the absorption spectra of oxidized and semi-reduced bCcP at a 5-fold higher concentration.
Potentiometric Redox Titrations
While the optical measurements indicated that the semi-reduced form of E258K did not possess a high-spin, 5 coordinate heme that could bind H2O2, the kinetic data indicate that the enzyme engages in catalysis though impeded, and therefore E258K must bind peroxide. To better rationalize the kinetic differences between the constructs, we examined the electron-transfer properties of the H-heme in the charge-reversal mutants. Oxidative and reductive titrations of FeH were carried out for both the wild-type and the E258K charge reversal monomerization mutant using potentiometry (Figure S1). Because of the large separation in potential between the high potential heme and low potential heme found in bacterial cytochrome c peroxidases the contribution of the higher potential heme can be easily isolated (15, 17, 19, 29). Reductive and oxidative titrations of the FeHII/III couple for the wild-type So enzyme yielded a Em value of +246 ± 1 mV (vs SHE, at pH 6.0). E258K displayed lower potentials with slight hysteresis in the oxidative and reductive directions: reductive and oxidative titrations yielded redox potentials of + 218 mV and +201 mV vs SHE, respectively. In all cases, Nernst plots showed slopes of ~ 59 mV/decade, indicating that simple one-electron, one-proton processes are at work.
Kinetics of ascorbate reduction, and air re-oxidation
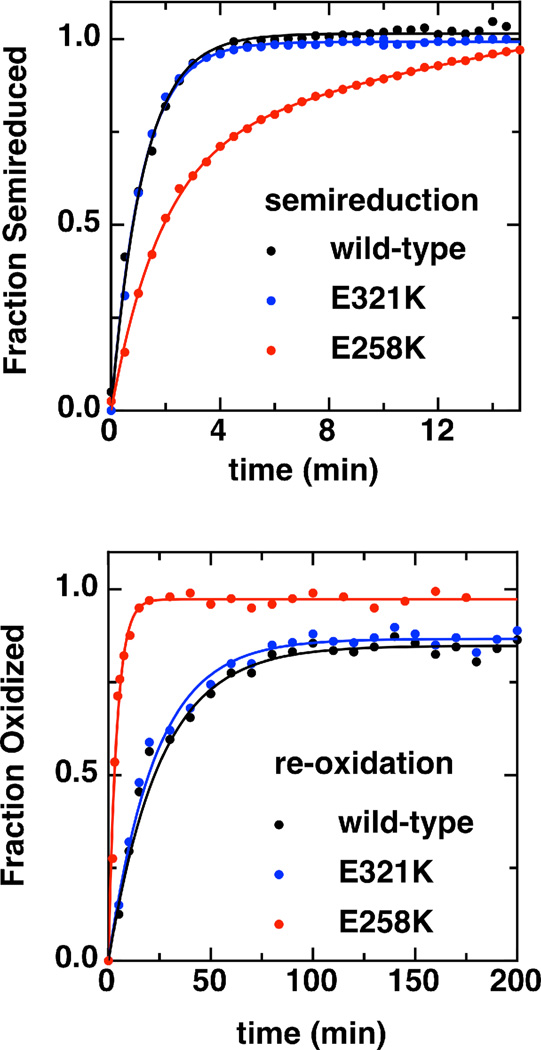
Though the E258K mutant possesses an H-heme with slightly depressed reduction potential, the possible impact upon catalytic parameters was not evident, as a decrease of roughly 15 mV in the H-heme FeII/III redox potential should not impact its ability to reduce intermediates that might be generated in the course of peroxidatic chemistry, nor receive reducing equivalents from cytochromes c. In order to address this issue, we examined the kinetic stability of the so-called “semi-reduced” state. In addition to the distinct Soret and 640 nm band features reported for E258K, the kinetics associated with ascorbate reduction of the fully oxidized enzyme to produce the FeHIIFeLIII state, and the subsequent re-oxidation with air were found to differ between wild-type and the charge-reversal mutant of So bCcP. Rates of ascorbate reduction and re-oxidation of the enzymes were examined using the absorption features described above (408 nm and 553 nm), as shown in Figure 6.
Figure 6.
Kinetics of semireduction (top) and re-oxidation (bottom) of wild-type bCcP (black) and the charge reversal mutants, E321K (blue) and E258K (red) monitored at 553 nm. Assays were conducted using 3 µM enzyme in 5 mM HEPES, 5 mM MES, 10 mM NaCl and 1 mM CaCl2, pH 6 at 23°C. Data are fit to a pseudo first order model assuming oxidant concentration is constant.
For wild-type enzyme reduction of 3 µM bCcP with 5 mM L-sodium ascorbate (pseudo-first order conditions) was monitored at either 553 or 408 nm, with reduction being achieved in minutes (kobs = 0.90 ± 0.03 min−1 at 553 nm and kobs = 0.97 ± 0.02 min−1 at 408 nm). Similar time-scales were required for reduction of E258K, though the final spectral features (i.e., Figure 4B) never exactly mimic wild-type enzyme (kobs = 0.36 ± 0.02 min−1 at 553 nm and kobs = 0.63 ± 0.04 min−1 at 408 nm). In contrast, re-oxidation of the ascorbate-reduced enzyme proceeded on distinctly different time-scales. As illustrated in Figure 6 (bottom), by first removing all exogenous reductant and then exposing the enzyme to atmospheric oxygen, the fully-oxidized spectrum was regenerated on the order of hours (after the removal of excess ascorbate and DAD, kobs = 0.04 min−1), requiring up to days for full transformation of the 553 nm bands. E321K closely mimicked the wild-type enzyme. However, similar re-oxidations of E258K were an order of magnitude faster than wild-type, occurring on the order of minutes (kobs = 0.25 min−1). Thus, upon monomerization, the FeHIIFeLIII state of the E258K mutant appears to be much less kinetically stable, unlike the stable, static “activated” conformation of bCcPs that has been amenable to crystallographic analyses (4, 16, 32).
DISCUSSION
The formation of homo-multimeric structures is found throughout biology, where alternate quaternary structures can play key roles in assisting with overall folding stability (34–36), producing allosteric affects for small molecule binding (36–38), developing high localized concentrations, and allowing for alternate (‘moonlighting’) activities (36, 39, 40). In the case of bCcP enzymes, homomultimerization is correlated to activity (8, 29), with all structurally characterized bCcPs being demonstrated to be (at least) homodimers. However, the apparent requirement for homodimerization has not been examined in detail previously. While canonical bCcP enzymes must undergo changes in secondary and tertiary structure that are tied to changes in the oxidation state of the H-heme, and result in an ‘Open’ conformation at FeL, the apparent correlation between quaternary structure and activity has not been examined. To address this open question, we have produced the first, active monomeric version of a bCcP enzyme to allow us to ask the question, “Why are all bCcP dimers?” Through the analysis of the structurally conserved dimer interface between the eight different bCcP enzymes that have been crystallized to date, we have found that a single charge reversal mutation, E258K, results in an active, yet monomeric form of the traditionally dimeric enzyme. Notably, not all charge-reversal mutations generate a monomerized protein, as the E321K mutant still retains a homodimeric structure as determined by size exclusion chromatography and analytical ultracentrifugation.
In several respects, it is easier to report on what homodimerization does not impact. For example, wild-type So bCcP does not display allostery, half-site reactivity, or ‘moonlighting’ activities, and the corresponding charge reversal constructs reported here do not evince such properties. Many traits of E258K mimic wild-type protein: the global folding stability of E258K is equal or better than the wild-type protein, by some spectroscopic measures (e.g., fully oxidized optical spectrum) E258K is identical to wild-type, the reduction potential of the H-heme is shifted modestly more negative (by 35 mV), and kcat and Km are affected, but not dramatically so. In terms of activity, the catalytic parameters given in Table 2 reveal that loss in activity displayed by E258K may be due to fundamental changes in catalytic events at the active site, or may be due to changes in the ability of electron-donors to interact with the enzyme. Thus, we conclude that E258K still acts as a reasonable cytochrome c peroxidase with the native electron donor, where kcat/Km values are similar to those found in other bacterial peroxidases (18).
Given these observations, what might homodimerization achieve? The critical differences emerge in the optical spectrum of the semi-reduced enzyme (FeHIIFeLIII) where FeL has undergone a conformation change from being six-coordinate with His71 bound, to an ‘Open’ conformation that displays activity. In the native enzyme the semi-reduced form displays absorption features at ~550 nm indicative of the reduction of FeH, but also a weak feature at 650 nm that has been interpreted in terms of a high-spin ferric FeLIII. When the same state is prepared with E258K, the former features are present, but the latter is absent, indicating that while FeH is reduced readily by ascorbate, FeL has most likely not undergone the requisite conformational change to an appreciable extent. The other key difference is found in the relative kinetics associated with the preparation and subsequent re-oxidation (by air) of the FeHIIFeLIII state: while wild-type and E258K are similar in the time-scales associated with the reduction of the totally oxidized enzyme (by monitoring either the soret region, or the α and β bands), they are hugely different when considering the re-oxidation. The wild-type FeHIIFeLIII state is stable when exposed to air for hours to days, yet the monomerized E258K protein is re-oxidized in seconds (i.e., less than a minute). This phenomenon does not seem to be entirely controlled by the actual reduction potential of the H-heme: E258K has a potential only 35 mV less than wild-type, and clearly both H-heme centers should be readily oxidized by oxygen. The possible kinetic instability of the FeHIIFeLIII state in a monomerized bCcP is further corroborated by steady-state kinetic analysis of the fully-oxidized protein, where the decrease peroxide turnover rate can be interpreted in terms of a real-time decrease of the concentration of the activated form of the protein, due to its inactivation on the timescale of seconds (i.e., kinetic instability of the FeHIIFeLIII state in E258K leads to deactivation that competes with catalytic processes).
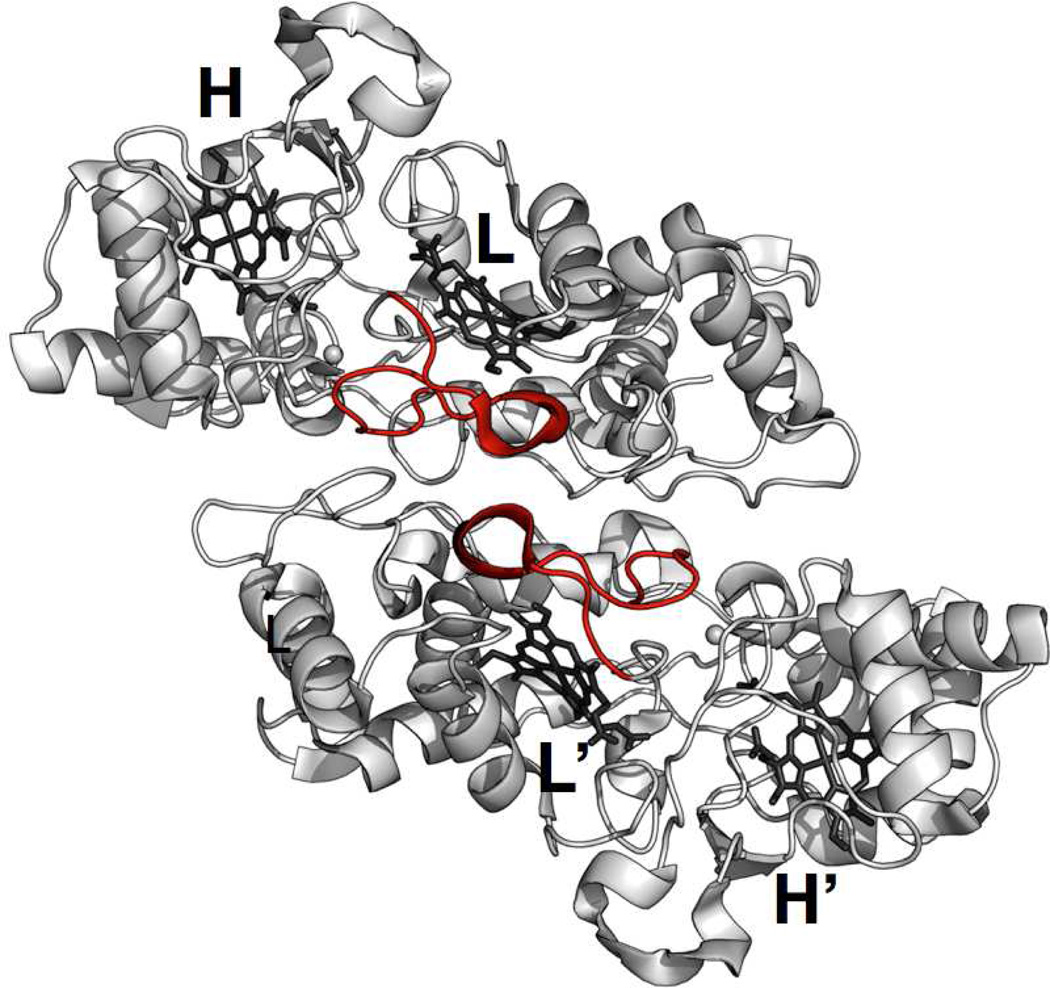
A full structural perspective of the relationship between quaternary structure and the kinetic behavior described here requires a structure of E258K. Yet, without that data, inspection of the structures of activated dimeric forms of bCcP enzymes suggest that quaternary structure may play a role in achieving kinetic stabilization of the FeHIIFeLIII state as proposed here. One of the elements of the dimer interface is comprised by a single loop that contains the His71 residue responsible for ligating the peroxidatic heme iron in the ‘Closed’ form of the active site (4, 11, 12, 16). This loop structure (termed Loop 1 by Einsle and co-workers (9)) packs against the second protomer in the dimeric structure, as shown in Figure 7. Without the presence of the second protomer molecule, Loop 1 may engage in conformational dynamics, generating either ‘Open’ or ‘Closed’ forms of the active site, and contributing to the overall kinetic instability of the activated FeHIIFeLIII state.
Figure 7.
Depiction of Loop 1 (red) of each bCcP protomer in the So wild-type enzyme (3O5C.pdb (11)). Loop 1 contains His71 that acts as a ligand at FeL in the ‘Closed’ form. However, in all structures of active enzymes Loop 1 adopts an ‘Open’ conformer that packs against the adjacent Loop1 of the second protomer in the dimer.
CONCLUSIONS
Here we have demonstrated that without removal of the obligate Ca2+ ion found in the bCcP family, a charge reversal monomeric construct can be prepared through destabilization of the interface between homodimers. Collectively, our data indicate that homodimerization of bCcPs is significant not to maintain global folding energies, but instead to stabilize the kinetic lifetime of the ‘Open’/FeHIIFeLIII state which is the entry-point for H2O2 binding and peroxidase chemistry. Thus, it appears that the bCcP family (including the constitutively active Ne enzyme) has maintained homodimeric quaternary structures in order to stabilize not the global protein fold, but the specific ‘Open’ conformation at FeL that is needed for catalysis. One may infer that if bCcP protomers exist in vivo, they may be more prone to ‘deactivate’ on the time-scale of minutes.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by National Institutes of Health grant R01-GM072663 to S.J.E. and NSF grant CHE 0840418.
ABBREVIATIONS
- bCcP
bacterial cytochrome c peroxidase
- FeH
high-potential heme
- FeL
low-potential heme
- Pd
Paracoccus denitrificans
- NHE
Normal hydrogen electrode
- Pa
Pseudomonas aeruginosa
- Pn
Pseudomonas nautica
- Rc
Rhodobacter capsulatus
- Ps
Pseudomonas stutzeri
- Gs
Geobacter sulfurreducens
- So
Shewanella oneidensis
- Ne
Nitrosomonas europaea
- Mc
Methylococcus capsulatus (Bath)
- IPTG
isopropyl-β-D-1-thiogalactopyranoside
- HEPES
4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid
- NaCl
sodium chloride
- EGTA
ethylene glycol tetraacetic acid
- AUC
Analytical ultracentrifugation
- CD
circular dichroism
- MES
2-(N-morpholino-ethanesulfonic acid
- EPR
electron paramagnetic resonance
- DAD
diaminodurol
- PMS
phenazinemethosulfate
Footnotes
ASSOCIATED CONTENT
Supporting Information. PDBePisa analysis and potentiometric titrations are given as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Van Spanning RJ, De Boer AP, Reijnders WN, Westerhoff HV, Stouthamer AH, Van Der Oost J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Molecular Microbiology. 1997;23:893–907. doi: 10.1046/j.1365-2958.1997.2801638.x. [DOI] [PubMed] [Google Scholar]
- 2.Atack JM, Kelly DJ. Structure, mechanism and physiological roles of bacterial cytochrome c peroxidases. Advanced Microbial Physiology. 2007;52:73–106. doi: 10.1016/S0065-2911(06)52002-8. [DOI] [PubMed] [Google Scholar]
- 3.Pettigrew GW, Echalier A, Pauleta SR. Structure and mechanism in the bacterial dihaem cytochrome c peroxidases. Journal of Inorganic Biochemistry. 2006;100:551–567. doi: 10.1016/j.jinorgbio.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Echalier A, Goodhew CF, Pettigrew GW, Fulop V. Activation and catalysis of the di-heme cytochrome c peroxidase from Paracoccus pantotrophus . Structure. 2006;14:107–117. doi: 10.1016/j.str.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Gilmour R, Goodhew CF, Pettigrew GW, Prazeres S, Moura JJ, Moura I. The kinetics of the oxidation of cytochrome c by Paracoccus cytochrome c peroxidase. Biochemistry Journal. 1994;300:907–914. doi: 10.1042/bj3000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alves T, Besson S, Duarte LC, Pettigrew GW, Girio FM, Devreese B, Vandenberghe I, Van Beeumen J, Fauque G, Moura I. A cytochrome c peroxidase from Pseudomonas nautica 617 active at high ionic strength: expression, purification and characterization. Biochim Biophys Acta. 1999;1434:248–259. doi: 10.1016/s0167-4838(99)00188-0. [DOI] [PubMed] [Google Scholar]
- 7.De Smet L, Savvides SN, Van Horen E, Pettigrew G, Van Beeumen JJ. Structural and mutagenesis studies on the cytochrome c peroxidase from Rhodobacter capsulatus provide new insights into structure-function relationships of bacterial di-heme peroxidases. Journal of Biological Chemistry. 2006;281:4371–4379. doi: 10.1074/jbc.M509582200. [DOI] [PubMed] [Google Scholar]
- 8.Timoteo CG, Tavares P, Goodhew CF, Duarte LC, Jumel K, Girio FM, Harding S, Pettigrew GW, Moura I. Ca2+ and the bacterial peroxidases: the cytochrome c peroxidase from Pseudomonas stutzeri . Journal of Biological Inorganic Chemistry. 2003;8:29–37. doi: 10.1007/s00775-002-0382-y. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann M, Seidel J, Einsle O. CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase. Journal of Molecular Biology. 2009;393:951–965. doi: 10.1016/j.jmb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Seidel J, Hoffmann M, Ellis KE, Seidel A, Elliott SJ, Einsle O. MacA is a Second Cytochrome c Peroxidase of Geobacter sulfurreducens . Biochemistry. 2012;13:2747–2756. doi: 10.1021/bi300249u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schutz B, Seidel J, Sturm G, Einsle O, Gescher J. Investigation of the electron transport chain to and the catalytic activity of the diheme cytochrome c peroxidase CcpA of Shewanella oneidensis . Applied Environmental Microbiology. 2011;77:6172–6180. doi: 10.1128/AEM.00606-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu H, Schuller DJ, Lanzilotta WN, Sundaramoorthy M, Arciero DM, Hooper AB, Poulos TL. Crystal structure of Nitrosomonas europaea cytochrome c peroxidase and the structural basis for ligand switching in bacterial di-heme peroxidases. Biochemistry. 2001;40:13483–13490. doi: 10.1021/bi011481h. [DOI] [PubMed] [Google Scholar]
- 13.Pauleta SR, Cooper A, Nutley M, Errington N, Harding S, Guerlesquin F, Goodhew CF, Moura I, Moura JJ, Pettigrew GW. A copper protein and a cytochrome bind at the same site on bacterial cytochrome c peroxidase. Biochemistry. 2004;43:14566–14576. doi: 10.1021/bi0485833. [DOI] [PubMed] [Google Scholar]
- 14.Pettigrew GW, Goodhew CF, Cooper A, Nutley M, Jumel K, Harding SE. The electron transfer complexes of cytochrome c peroxidase from Paracoccus denitrificans . Biochemistry. 2003;42:2046–2055. doi: 10.1021/bi027125w. [DOI] [PubMed] [Google Scholar]
- 15.Zahn JA, Arciero DM, Hooper AB, Coats JR, DiSpirito AA. Cytochrome c peroxidase from Methylococcus capsulatus Bath. Archives of Microbiology. 1997;168:362–372. doi: 10.1007/s002030050510. [DOI] [PubMed] [Google Scholar]
- 16.Echalier A, Brittain T, Wright J, Boycheva S, Mortuza GB, Fulop V, Watmough NJ. Redox-linked structural changes associated with the formation of a catalytically competent form of the diheme cytochrome c peroxidase from Pseudomonas aeruginosa . Biochemistry. 2008;47:1947–1956. doi: 10.1021/bi702064f. [DOI] [PubMed] [Google Scholar]
- 17.De Smet L, Pettigrew GW, Van Beeumen JJ. Cloning, overproduction and characterization of cytochrome c peroxidase from the purple phototrophic bacterium Rhodobacter capsulatus . European Journal of Biochemistry. 2001;268:6559–6568. doi: 10.1046/j.0014-2956.2001.02610.x. [DOI] [PubMed] [Google Scholar]
- 18.Pulcu GS, Frato KE, Gupta R, Hsu HR, Levine GA, Hendrich MP, Elliott SJ. The Diheme Cytochrome c Peroxidase from Shewanella oneidensis Requires Reductive Activation. Biochemistry. 2012;51:974–985. doi: 10.1021/bi201135s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellfolk N, Ronnberg M, Aasa R, Andreasson LE, Vanngard T. Properties and function of the two hemes in Pseudomonas cytochrome c peroxidase. Biochim Biophys Acta. 1983;743:23–30. doi: 10.1016/0167-4838(83)90413-2. [DOI] [PubMed] [Google Scholar]
- 20.Pettigrew GW, Prazeres S, Costa C, Palma N, Krippahl L, Moura I, Moura JJ. The structure of an electron transfer complex containing a cytochrome c and a peroxidase. Journal of Biological Chemistry. 1999;274:11383–11389. doi: 10.1074/jbc.274.16.11383. [DOI] [PubMed] [Google Scholar]
- 21.Pettigrew GW, Pauleta SR, Goodhew CF, Cooper A, Nutley M, Jumel K, Harding SE, Costa C, Krippahl L, Moura I, Moura J. Electron transfer complexes of cytochrome c peroxidase from Paracoccus denitrificans containing more than one cytochrome. Biochemistry. 2003;42:11968–11981. doi: 10.1021/bi034829c. [DOI] [PubMed] [Google Scholar]
- 22.Arciero DM, Hooper AB. A di-heme cytochrome c peroxidase from Nitrosomonas europaea catalytically active in both the oxidized and half-reduced states. Journal of Biological Chemistry. 1994;269:11878–11886. [PubMed] [Google Scholar]
- 23.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of Molecular Microbiology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Schutz B, Seidel J, Sturm G, Einsle O, Gescher J. Investigation of the electron transport chain to and the catalytic activity of the diheme cytochrome c peroxidase CcpA of Shewanella oneidensis . Applied Environmental Microbiology. 2011;77:6172–6180. doi: 10.1128/AEM.00606-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philo JS. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 26.Rai N, Nollmann M, Spotorno B, Tassara G, Byron O, Rocco M. SOMO (SOlution MOdeler) differences between X-Ray- and NMR-derived bead models suggest a role for side chain flexibility in protein hydrodynamics. Structure. 2005;13:723–734. doi: 10.1016/j.str.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 27.Schuetz B, Schicklberger M, Kuermann J, Spormann AM, Gescher J. Periplasmic electron transfer via the c-type cytochromes MtrA and FccA of Shewanella oneidensis MR-1. Applied Environmental Microbiology. 2009;75:7789–7796. doi: 10.1128/AEM.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore GR, Pettigrew GW. Cytochromes c : evolutionary, structural, and physicochemical aspects. 1990 [Google Scholar]
- 29.Gilmour R, Goodhew CF, Pettigrew GW, Prazeres S, Moura I, Moura JJ. Spectroscopic characterization of cytochrome c peroxidase from Paracoccus denitrificans . Biochemical Journal. 1993;294:745–752. doi: 10.1042/bj2940745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foote N, Peterson J, Gadsby PM, Greenwood C, Thomson AJ. Redox-linked spin-state changes in the di-haem cytochrome c-551 peroxidase from Pseudomonas aeruginosa . Biochemistry Journal. 1985;230:227–237. doi: 10.1042/bj2300227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foote N, Peterson J, Gadsby PM, Greenwood C, Thomson AJ. A study of the oxidized form of Pseudomonas aeruginosa cytochrome c-551 peroxidase with the use of magnetic circular dichroism. Biochemistry Journal. 1984;223:369–378. doi: 10.1042/bj2230369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias JM, Alves T, Bonifacio C, Pereira AS, Trincao J, Bourgeois D, Moura I, Romao MJ. Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617. Structure. 2004;12:961–973. doi: 10.1016/j.str.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 33.Fulop V, Ridout CJ, Greenwood C, Hajdu J. Crystal structure of the di-haem cytochrome c peroxidase from Pseudomonas aeruginosa . Structure. 1995;3:1225–1233. doi: 10.1016/s0969-2126(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 34.Jaffe EK. Morpheeins--a new structural paradigm for allosteric regulation. Trends Biochem Sci. 2005;30:490–497. doi: 10.1016/j.tibs.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 36.Selwood T, Jaffe EK. Dynamic dissociating homo-oligomers and the control of protein function. Arch Biochem Biophys. 2012;519:131–143. doi: 10.1016/j.abb.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He MM, Smith AS, Oslob JD, Flanagan WM, Braisted AC, Whitty A, Cancilla MT, Wang J, Lugovskoy AA, Yoburn JC, Fung AD, Farrington G, Eldredge JK, Day ES, Cruz LA, Cachero TG, Miller SK, Friedman JE, Choong IC, Cunningham BC. Small-molecule inhibition of TNF-alpha. Science. 2005;310:1022–1025. doi: 10.1126/science.1116304. [DOI] [PubMed] [Google Scholar]
- 38.Hayouka Z, Rosenbluh J, Levin A, Loya S, Lebendiker M, Veprintsev D, Kotler M, Hizi A, Loyter A, Friedler A. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc Natl Acad Sci U S A. 2007;104:8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 40.Jeffery CJ. Moonlighting proteins--an update. Mol Biosyst. 2009;5:345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.