Abstract
Background
Family and twin studies indicate substantial overlap of genetic influences on psychotic and mood disorders. Linkage and candidate gene studies have also suggested overlap across schizophrenia (SCZ), bipolar disorder (BPD), and major depressive disorder (MDD). The objective of this study was to apply genomewide association study (GWAS) analysis to address the specificity of genetic effects on these disorders.
Method
We combined GWAS data from three large effectiveness studies of SCZ (CATIE, genotyped n = 741), BPD (STEP-BD, n = 1575) and MDD (STAR*D, n= 1938) and psychiatrically-screened controls (NIMH-GI controls, n = 1204). We applied a two-stage analytic procedure involving an omnibus test of allele frequency differences among case and control groups followed by a model selection step to identify the best-fitting model of allelic effects across disorders.
Results
The strongest result was seen for a single nucleotide polymorphism near the adrenomedullin (ADM) gene (rs6484218, p = 3.93 × 10−8), with the best-fitting model indicating that the effect is specific to bipolar II disorder. We also observed evidence suggesting that several genes may have effects that transcend clinical diagnostic boundaries including variants in NPAS3 that showed pleiotropic effects across SCZ, BPD, and MDD.
Conclusions
This study provides the first genomewide significant evidence implicating variants near the ADM gene on chromosome 11p15 in psychopathology, with effects that appear to be specific to bipolar II disorder. Although we do not detect genomewide significant evidence of cross-disorder effects, our study provides evidence that there are both pleiotropic and disorder-specific effects on major mental illness and illustrates an approach to dissecting the genetic basis of mood and psychotic disorders that can inform future large-scale cross-disorder GWAS analyses.
Introduction
Family and twin studies have established that schizophrenia (SCZ), bipolar disorder (BPD), and major depressive disorder (MDD) are familial and heritable phenotypes and that genetic factors are the most robustly validated risk factors for each disorder (1–3). However, several findings have called into question whether these disorders are etiologically distinct. First, several key clinical features, including psychosis, neurocognitive impairment and suicidality, may be observed in all three. Second, genetic epidemiologic studies have documented that SCZ, BPD, and MDD share familial and genetic determinants. Family studies have shown familial co-aggregation for SCZ and BPD (4–6) as well as BPD and MDD (1). In a population-based study of > 2 million families, Lichtenstein and colleagues (7) demonstrated increased risks of SCZ among relatives of BPD probands and increased risks of BPD among relatives of SCZ probands. Comorbidity between the disorders was mainly attributable to overlapping genetic influences. Twin studies have similarly documented substantial shared genetic variance between psychotic disorders and BPD (8) and between BPD and MDD (9).
Although family and twin studies can estimate the shared heritability across disorders, they cannot identify the genetic loci contributing to this overlap. To date, evidence implicating specific chromosomal regions and genes in the shared liability to psychotic and mood disorders has largely been limited to linkage and candidate gene association studies. Some of the regions with the strongest linkage evidence for SCZ are also among regions most strongly linked to BPD(10–12), though simulations suggest that such overlap could easily occur by chance(7). Several chromosomal microdeletions have also been associated with both mood and psychotic disorders. The balanced translocation (1;11)(q42;q14.3) that disrupts DISC1 was first identified due to its co-segregation with a broad phenotype comprising SCZ, BPD, and recurrent MDD(13). The 22q11 microdeletion responsible for velocardiofacial syndrome also appears to confer increased risk of both psychotic and mood disorders(14, 15) Candidate gene studies have also found association between specific genes and both psychotic and mood disorder phenotypes(11, 16, 17), although results have been inconsistent(18, 19).
Genomewide association studies (GWAS), which provide a survey of common genetic variation across the genome, offer a more comprehensive method for identifying risk loci at the genotypic level. Early efforts to apply this technology to major psychiatric disorders have begun to bear fruit, with GWAS studies implicating several susceptibility genes for SCZ(20), BPD(21), and MDD(22). A recent analysis examined gene-wide evidence of association using data from both a BPD and SCZ GWAS, respectively and found nominal evidence that several genes influence both disorders(23). Data from the International Schizophrenia Consortium demonstrated that common genetic variation (involving thousands of small-effect alleles) accounts for at least one-third of the total variation in liability to SCZ and that these polygenic risks are substantially shared with BPD (24). To date, however, no GWAS have been reported that examine cross-disorder analyses of the specificity of genetic influences for all three disorders. Here we report the first genomewide cross-disorder analysis incorporating samples from the three largest treatment effectiveness studies of SCZ, BPD, and MDD, respectively. To address the issue of multiple comparisons, we utilize a novel approach that examines the patterns of cross-disorder effects in a single model selection framework that controls the type I error risk at an experiment-wise level.
Methods
Clinical Samples
BPD (STEP-BD) sample
STEP-BD was a national, longitudinal public health initiative designed to examine the effectiveness of treatments and their impact on the course of BPD(25). Over a 7-year period, 4361 participants were enrolled across 20 sites and followed for up to 2 years. To maximize external validity, enrollment was offered to all eligible patients seeking outpatient treatment at one of the participating sites (26). Eligibility for STEP-BD required a consensus DSM-IV bipolar diagnosis on both the ADE and MINI-PLUS semi-structured interviews as previously described (25). From the parent STEP-BD study, 2089 individuals were enrolled in a genetic substudy.
SCZ (CATIE) sample
CATIE was a multiphase randomized controlled trial of antipsychotic medications comprising 1460 individuals with SCZ followed for up to 18 months(27, 28). Final study diagnoses of DSM-IV SCZ were established by CATIE clinicians using the Structured Clinical Interview for DSM-IV(29), including review of all available information (including psychiatric and general medical records). As detailed previously (27), exclusion criteria included: diagnosis of schizoaffective disorder, mental retardation or other cognitive disorder, single psychotic episode, and history of treatment resistance or serious adverse reaction to the study treatments. As previously described (30), the genetic sub-study included 738 cases.
MDD (STAR*D) sample
STAR*D was a multi-site, prospective, randomized multiphase clinical trial of outpatients with nonpsychotic MDD that enrolled 4041 participants over a three-year period (31). Eligibility required a single or recurrent nonpsychotic major depressive episode (by DSM-IV criteria) and a score of ≥ 14 on the 17-item Hamilton Depressive Rating Scale. Relevant exclusion criteria included history of BPD, SCZ, schizoaffective disorder or psychosis NOS. In the genetic substudy, blood samples were collected from 1953 participants.
Control (NIMH Genetics Repository) sample
As previously described (32), controls were collected by Knowledge Networks, a survey and market research company whose panel contains approximately 60,000 households representative of the US population. Subjects completed an online psychiatric screen that included questions regarding demographics, ancestry, and DSM-IV criteria for a range of psychiatric disorders. Participants who reported a history of SCZ, psychosis or BPD were excluded from the GWAS analyses as previously described(32). We also excluded individuals (N = 126) who met criteria for a history of major depressive episode.
Genotyping
Genotyping of STEP-BD and CATIE samples was performed using the Affymetrix GeneChip Human Mapping 500K Array Set, while half the STAR*D sample was genotyped with the 500K array, and half with the Affymetrix Human SNP Array 5.0. Genotyping of the STEP-BD and Control samples were performed at the Broad Institute as previously described(33). QC processing of genotypes was described in Sklar et al.(32). Genotyping of the SCZ sample was performed by Perlegen Sciences (Mountain View CA, USA) as reported elsewhere (30). Genotyping of the STAR*D sample was performed at Affymetrix (500K) or at the University of California, San Francisco (5.0), as described (34).
Quality Control and Harmonization of Genotype Data
Additional QC for the combined genotypic dataset was performed using PLINK(35) as previously described (32). In brief, individuals were excluded if they had: overall call rates <95%; for excess or insufficient heterozygosity or for apparent relatedness. We only included non-Hispanic Caucasian individuals with European ancestry based on self-reported race and ethnicity information. The PLINK nearest neighbor method (35) was used to filter out potential outliers based on the first 10 multidimensional scaling (MDS) factors. SNPs were excluded if they had a call rate <98%; minor allele frequency <1%, were inconsistent with Hardy-Weinberg Equilibrium at p<1×10−6 or showed differential rates of missingness in patient and controls (32). After QC steps, 224,395 genotyped markers were retained, with a total genotyping rate in the final sample >99%. This set of genotyped markers is smaller than that reported for the primary GWAS reports of each sample (30, 32) due to the additional QC steps described above. We used BEAGLE ver 3.1.1 (http://www.stat.auckland.ac.nz/~bbrowning/beagle/beagle.html) to impute missing genotypes, with the HapMap (CEU population, release 23, forward strand) as the reference panel. We excluded SNPs with an imputation quality R2 score less than 0.8.and obtained a total of 1,574,154 SNPs for final analysis. For each SNP, imputed dosage was then summed for each of the five phenotype groups.
Further control for confounding by population stratification was performed by calculating the first 10 quantitative ancestry indices based on the merged dataset using multidimensional scaling analysis (35). To examine the effect of each of the 10 quantitative indices (C1–C10), we calculated the genomic inflation factor (λ) from a GWA analysis of genotypes using each quantitative index as the dependent variable. We plotted the λ’s against each the 10 indices (analogous to a scree plot) and found excessive values only for C1–C4. We thus used these four indices as covariates in our GWAS of the phenotypic groups.
Statistical Analysis
Examining the pleiotropic of genetic variants across disorders in the GWAS context requires additional attention to problems of multiple testing and Type I error. One approach to the analysis is to conduct a series of pairwise comparisons of the individual disorders and their combinations. However, this effectively entails conducting multiple GWAS analyses, each of which would require correction for multiple testing. We have taken an alternative approach that involves a single genomewide omnibus test of association across disorders followed by a model selection approach that asks which configuration of phenotypes is most likely to be associated with a given variant. The sequence of analytic steps was as follows:
We first compute a likelihood ratio statistic (the omnibus test statistic) from a multinomial logistic regression in which allele frequencies can vary for each sample (SCZ, bipolar I (BPI), bipolar II (BPII), MDD, and controls) compared to a null model in which allele frequencies are the same across all groups. This 4 df test is essentially a test of whether there is any association between the variant and any of the four target disorders.
- For each SNP whose omnibus test p < α = 5× 10−5, we fit nine additional log-linear models corresponding to nine patterns of allele frequency configurations:
- Shared by all disorders: SCZ/BPD/MDD model (SCZ=BPI=BPII=MDD)≠Controls;
- Shared by psychotic disorders: SCZ/BPI model (SCZ=BP1)≠(MDD =BPII=Controls)
- Shared by mood disorders: BPD/MDD model (BPI=BPII=MDD)≠(SCZ=Controls)
- Shared by depressive disorders: BPII/MDD model (BPII=MDD)≠(BPI=SCZ=Controls)
- SCZ-specific: SCZ≠(MDD=BPI=BPII=Controls)
- BPD-specific: BPD≠(MDD=SCZ=Controls)
- BPI-specific: BPI ≠(BPII=MDD=SCZ=Controls)
- BPII-specific: BPII≠(BPI=MDD=SCZ=Controls)
- MDD-specific: MDD≠(SCZ=BP1=BPII=Controls)
Identify the best-fit model based on the Bayes Information Content (BIC)
In order to control the marker-wise type I error at α, only the omnibus test p-value (4 df) and the best-fit model are reported as primary results. Thus, the result of this analytic procedure is a p value for the single omnibus test and a best-fit model that indicates the phenotype(s) that provide the best fit for genotype-phenotype association for a given variant.
As is customary in pooled GWAS analyses, we do not adjust p values for the prior GWAS results of the individual samples.
Results
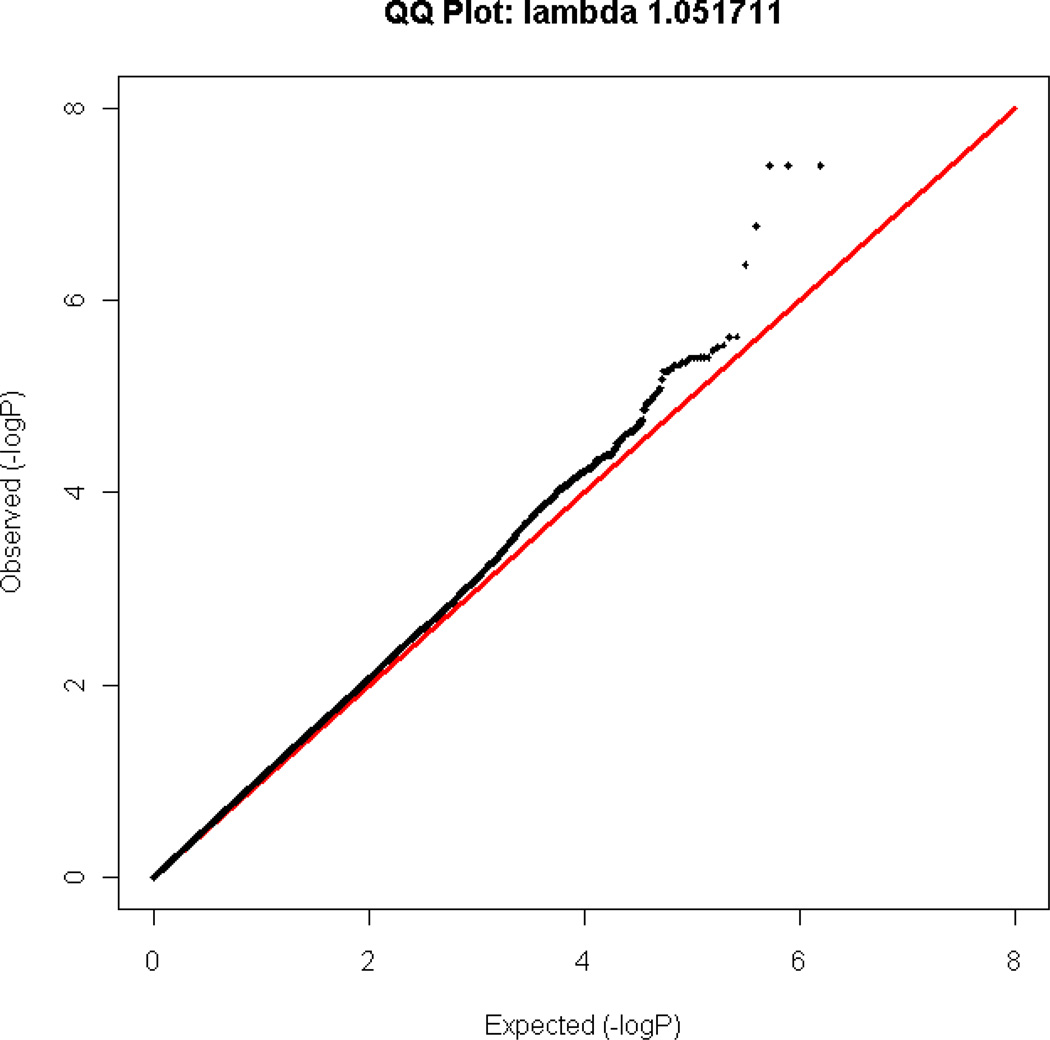
After QC, 4186 unrelated individuals of European ancestry were included in the GWAS analyses in four diagnostic groups: SCZ (n=402), BPI (n=1021), BPII (n=493), MDD (n=1210), and controls (n=1060) (Table 1). A Q-Q plot for the omnibus test is shown in Figure 1. The genomic inflation factor λ was 1.0517.
Table 1.
Demographics of the individuals of european ancestry included in GWAS analyses
| SCZ (CATIE) |
BPI (STEP-BD) |
BPII (STEP-BD) |
MDD (STAR*D) |
Controls (NIMH) |
|
|---|---|---|---|---|---|
| N | 402 | 1021 | 493 | 1,210 | 1,060 |
| % Female | 23.1 | 53.3 | 59.8 | 58.6 | 46.9 |
| Age, Mean (SD) | 41.3 (11.4) | 43 (12.7) | 42.8 (12.6) | 42.9 (13.6) | 51.7 (17.4) |
| Age at onset, Mean (SD) | 26.8 (9.2) | 17.6 (8.9) | 16.7 (8.7) | 25.5 (14.7) | N/A |
Figure 1.
Q-Q plot for the genome-wide omnibus test
Genomewide analysis and phenotypic model selection
SNPs for which the omnibus p < 5 × 10−5 are summarized in Table 2 according to the best-fitting model identified by the BIC criterion. A total of 124 SNPs in 25 independent regions exceeded this threshold. For each gene region (positions based on UCSC genome build hg18), Table 2 lists the number of SNPs in LD (r2 > 0.8) with the top SNP that exceed the p < 5×10−5 threshold (see. Supplementary Table 1 for full results).
Table 2.
Regions containing SNPs for which the Omnibus GWAS test p< 5×10−5 and best-fitting models. SNP: single nucleotide polymorphism; CHR: chromosome; freq: minor allele frequency; G/I: genotyped or imputed SNP. Genotyped P: p-value of the genotyped SNP in the region. * first allele shown is minor allele.
| # SNPs |
CHR | Top SNP | Alleles* | Gene | Position | Freq BP1 |
Freq BP2 |
Freq SCZ |
Freq MDD |
Freq CTRL |
Bes Model | BIC Diff |
OMNIBUS P | G/I | Genotyped P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 11 | rs6484218 | A/G | ADM | 61kb down | 0.13 | 0.07 | 0.13 | 0.13 | 0.14 | BP2 | 3.93E-08 | G | 3.93E-08 | |
| 3 | 22 | rs1001021 | A/G | MYO18B | intron 42 | 0.03 | 0.05 | 0.05 | 0.05 | 0.02 | SCZ_BPD_MDD | 2.39E-06 | G | 2.39E-06 | |
| 16 | 13 | rs7326068 | A/G | IFT88 | intron 16 | 0.18 | 0.23 | 0.16 | 0.23 | 0.19 | BP2_MDD | 2.92E-06 | G | 2.92E-06 | |
| 1 | 9 | rs3758354 | C/A | ANXA1 | 8kb up | 0.05 | 0.08 | 0.04 | 0.04 | 0.03 | BP2 | 3.31E-06 | G | 3.31E-06 | |
| 12 | 14 | rs4982029 | A/G | NPAS3 | intron 1 | 0.04 | 0.04 | 0.04 | 0.03 | 0.02 | SCZ_BPD_MDD | 3.96E-06 | I | 4.48E-06 | |
| 13 | 18 | rs11875674 | C/T | TXNL1 | 153kb up | 0.22 | 0.27 | 0.21 | 0.19 | 0.24 | BP2 | 3.97E-06 | I | 1.81E-05 | |
| 6 | 1 | rs4271171 | T/C | FAM20B | 8kb up | 0.53 | 0.58 | 0.49 | 0.54 | 0.5 | BPD_MDD | 4.12E-06 | I | 1.16E-05 | |
| 1 | 7 | rs12539410 | C/G | CHCHD3 | 403kb up | 0.01 | 0.03 | 0.01 | 0.02 | 0.01 | BP2_MDD | 4.74E-06 | I | -- | |
| 1 | 7 | rs4726220 | T/C | ACTR3B | 129kb down | 0.07 | 0.04 | 0.07 | 0.08 | 0.08 | BP2 | 1.16E-05 | G | 1.16E-05 | |
| 2 | 1 | rs17014011 | A/T | -- | intergenic | 0.13 | 0.15 | 0.16 | 0.15 | 0.11 | SCZ_BPD_MDD | 1.74E-05 | I | 2.36E-05 | |
| 4 | 2 | rs2372008 | G/A | CRIM1 | 782kb up | 0.44 | 0.38 | 0.45 | 0.38 | 0.41 | SCZ_BP1 | 1.78E-05 | G | 1.78E-05 | |
| 4 | 10 | rs7071307 | A/C | PPP2R2D | 360kb up | 0.43 | 0.49 | 0.46 | 0.42 | 0.42 | BP2 | 1.95E-05 | I | 1.98E-05 | |
| 1 | 5 | rs194487 | T/C | CTNND2 | intron 13 | 0.32 | 0.34 | 0.30 | 0.35 | 0.28 | BPD_MDD | 2.34E-05 | I | -- | |
| 2 | 2 | rs278865 | A/G | NR4A2 | 760kb up | 0.32 | 0.3 | 0.26 | 0.33 | 0.35 | SCZ | 2.46E-05 | I | -- | |
| 7 | 10 | rs10508451 | T/C | OPTN | 137kb up | 0.35 | 0.3 | 0.4 | 0.38 | 0.35 | BP2 | 2.87E-05 | G | 2.87E-05 | |
| 2 | 18 | rs12458992 | G/A | CCDC102B | 400kb up | 0.31 | 0.37 | 0.34 | 0.37 | 0.33 | BP2_MDD | 3.59E-05 | I | -- | |
| 1 | 15 | rs16949856 | T/C | -- | intergenic | 0.26 | 0.22 | 0.26 | 0.21 | 0.26 | BP2_MDD | 3.62E-05 | I | -- | |
| 1 | 15 | rs2117975 | A/G | EIF2AK4 | 91kb up | 0.15 | 0.11 | 0.10 | 0.15 | 0.15 | SCZ | 3.70E-05 | I | -- | |
| 10 | 20 | rs6079501 | G/T | SEL1L2 | 739kb down | 0.31 | 0.34 | 0.28 | 0.31 | 0.27 | BPD_MDD | 3.73E-05 | I | 3.82E-05 | |
| 4 | 12 | rs4140862 | A/C | TMEM16B | intron 5 | 0.3 | 0.33 | 0.33 | 0.36 | 0.36 | SCZ_BP1 | 4.03E-05 | I | 4.84E-05 | |
| 1 | 7 | rs917815 | G/A | SP8 | 270kb down | 0.4 | 0.32 | 0.35 | 0.4 | 0.4 | BP2 | 4.18E-05 | G | 4.18E-05 | |
| 1 | 3 | rs9819616 | G/A | RBMS3 | 294kb down | 0.35 | 0.34 | 0.39 | 0.31 | 0.36 | MDD | 4.46E-05 | I | -- | |
| 2 | 11 | rs11237798 | G/A | ODZ4 | 541kb down | 0.08 | 0.08 | 0.11 | 0.09 | 0.12 | BPD_MDD | 4.54E-05 | I | -- | |
| 1 | 21 | rs7280842 | T/C | SAMSN1 | 176kb down | 0.15 | 0.18 | 0.19 | 0.18 | 0.14 | SCZ_BPD_MDD | 4.56E-05 | G | 4.56E-05 | |
| 2 | 4 | rs7678609 | A/G | HAR39 | 26kb down | 0.36 | 0.32 | 0.33 | 0.34 | 0.29 | SCZ_BPD_MDD | 4.58E-05 | I | -- |
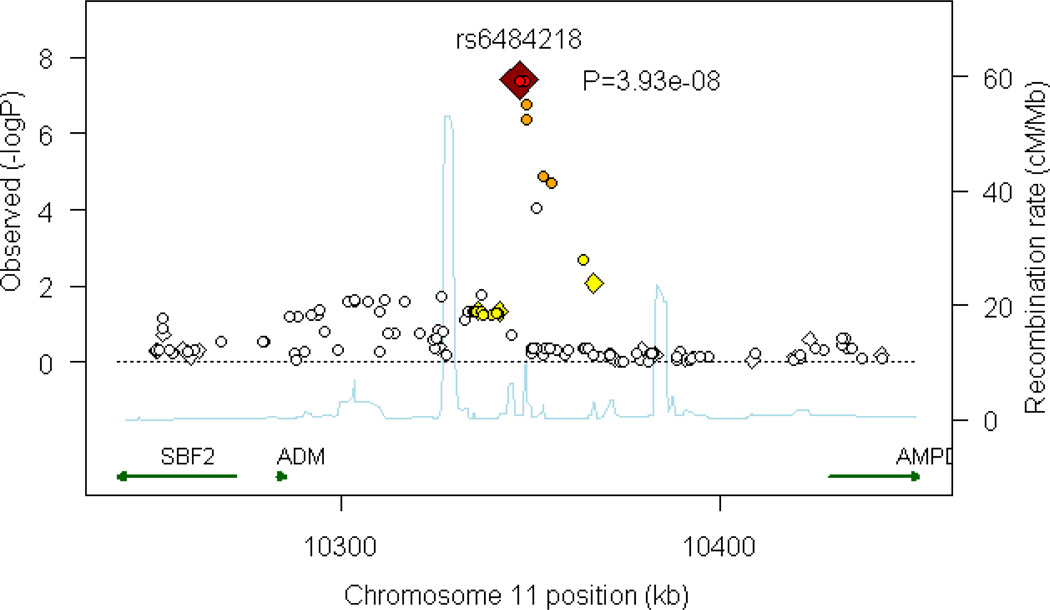
One region (comprising seven SNPs) on chromosome 11p15.4 exceeded a genomewide significant threshold for association (Figure 2). The top SNP achieving a p value of 3.93×10−8 was the genotyped SNP rs6484218; two other SNPs, rs10770107 and rs7119983 also reached a genomewide significance level of 4.07 × 10−8 and 4.22 × 10−8, respectively. These SNPs are located approximately 60 kb from the 3’ end of the nearest gene, adrenomedullin (ADM) and within an EST, EF537581, about which little is known. The best-fitting model for these SNPs indicated effects specific to bipolar II disorder. As shown in Table 2, the minor (A) allele of rs6484218 was less common among bipolar II cases compared to all other groups. Further support for the BPII-only model is depicted in Supplementary Figure 3 which plots the BIC for all 10 models at rs6484218. As shown in the Figure, the BIC for the BPII-only model is substantially lower than that of all other models.
Figure 2.
Plot of region of strongest association. Results (–log10P) are shown for directly genotyped (diamonds) and imputed (circles) SNPs. The most associated SNP is shown in dark red, and the color of the remaining markers reflects the linkage disequilibrium (r2) with the top SNP in each panel (increasing red hue associated with increasing r2). The recombination rate (second y axis) is plotted in light blue and is based on the CEU HapMap population.
The region showing the second strongest evidence of association comprised three SNPs within intron 42 of the myosin XVIIIB (MYO18B) gene. The top genotyped SNP rs1001021 showed a p-value of 2.39 × 10−6, accompanied by two imputed SNPs rs5752249 and rs16986621 with p-values < 3.12 × 10−6. The best-fitting model indicated that these SNPs have pleiotropic effects, influencing all three disorders (SCZ, BPD, and MDD). Another region showing the same pleiotropic effects across three disorders comprised 12 SNPs within intron 1 of the neuronal PAS domain 3 (NPAS3) gene (top SNP = rs4982029, p = 3.96 × 10−6), a gene previously implicated in psychotic and mood disorders (36–38). Three of the 12 SNPs were directly genotyped (rs7142052, rs10483422, and rs4982031, all p values < 9 × 10−6, supplementary Table 1).
Also of note, our model selection procedure suggested that genetic variants have effects that differ in their specificity with regard to diagnostic categories. For example, results for five of the 25 regions shown in Table 2 fit best with a model in which they have effects across SCZ, BPD and MDD, while four were most consistent with effects on the broad category of mood disorders (BPD and MDD) and 10 appeared to fit best with a single disorder.
Discussion
We combined genomewide association methods and a model selection procedure to examine the specificity of genetic influences on three major mental illnesses: SCZ, BPD and MDD. This study is the first, to our knowledge, systematically to examine genetic effects across these disorders in a genomewide framework. We observed genomewide significant evidence of association for SNPs near the ADM gene on chromosome 11p. The best-fitting model for our strongest single marker results indicated BPII disorder-specific effects. Family studies have supported the hypothesis that BPI and BPII are at least partly genetically distinct (1). Risks of BPII tend to be highest among relatives of BPII probands as opposed to those with BPI or MDD (39–44). Twin data support the heritability of BPII, but also suggest shared genetic influences with BPI (45). Linkage analyses of BPII have observed modest evidence of linkage on chromosomes 9p and 18q21(46). Interestingly, the sixth ranked region (top SNP = rs11875674, p = 3.97×10−6 in our GWAS is located on 18q21 and model-selection implicates a BPII effect, although this is 7 Mb from the reported linkage peak.
Our results are the first evidence to our knowledge implicating specific genetic variants in risk for BPII disorder and provide support for the hypothesis that the genetic etiology of this disorder is distinguishable from other DSM-IV mood disorders. The genomewide significant signals we observed are located within an EST (EF537581) that has not been well-characterized. This EST does not appear to be brain-expressed (47), making it an implausible candidate for BPII disorder. The nearest gene to the associated SNPs is ADM, the gene encoding adrenomedullin. Adrenomedullin is widely expressed in the brain, and mice lacking CNS adrenomedullin exhibit hyperactivity, increased anxiety, and increased sensitivity to the neurotoxic effects of hypobaric hypoxia (48). Elevations of serum adrenomedullin have been reported in bipolar disorder (49), and a recent study identified a functional ADM variant (rs11042725) that was associated with self-reported response to paroxetine among patients with depression(50). We were unable to examine this SNP (or proxies for it) because it is not included in our GWAS or in the HapMap database. However, the SNPs for which we found association do not appear to be in strong LD with SNPs in ADM.
Our results also provide preliminary evidence that several SNPs, including markers within several gene regions, have pleiotropic effects that cross traditional DSM-IV boundaries. Although these results did not reach genomewide thresholds for statistical significance, they are consistent with emerging evidence of shared genetic influences on psychotic and mood disorders. In particular, we note a group of SNPs in intron 1 of NPAS3 (top SNP with a p-value of 3.96 × 10−6) for which the best-fitting model indicated effects on SCZ BPD, and MDD. NPAS3 encodes a neuronal transcription factor and was identified as a candidate locus for SCZ after a balanced translocation [t(9, 14)(q34.2;q13)] disrupting the gene was observed to segregate with SCZ and learning disability (37). Mice deficient for NPAS3 display behavioral abnormalities including altered prepulse inhibition, impaired recognition memory, dysregulation of glutamatergic, dopaminergic, and serotoninergic neurotransmission, as well as diminished hippocampal neurogenesis (51–53). Pickard and colleagues (38) recently reported evidence that haplotypes of NPAS3 are associated with both SCZ and BPD, although the SNPs reported in Table 2 are not in linkage disequilibrium with those reported by Pickard et al. Nevertheless, the evidence reported here that variation in NPAS3 may have pleiotropic effects on mood and psychotic disorders is intriguing given the diverse neurobiologic functions of this gene.
Our results have several implications for future genetic studies of major mental illness. First, the analytic approach illustrated here provides a basis for identifying genes that underlie the common genetic basis of SCZ, BPD, and MDD. It is likely that genes influencing these disorders comprise a combination of disorder-specific and shared susceptibility loci. Similar evidence for cross-disorder genetic effects have emerged from GWAS studies of other medical disorders, most notably autoimmune diseases where specific genetic markers are strongly associated with multiple, clinically distinctive conditions (54, 55). For example, nearly half of the risk alleles identified as influencing either Type 1 diabetes or celiac disease have been shown to influence both disorders (55, 56). In addition, GWAS studies have demonstrated that a coding SNP in the PTPN22 gene is associated with reduced risk of Crohn’s disease but increased risk of systemic lupus erythematosis, rheumatoid arthritis, Type I diabetes, and Graves’ disease (54). Second, our results may inform case-definition in future genetic studies of SCZ, BPD, and MDD. Specifically, the power of future analyses may be enhanced by combining “affecteds” across disorders for loci that appear to have cross-disorder effects. Finally, identifying susceptibility loci that have pleiotropic effects may reveal shared etiologic mechanisms underlying multiple diseases. Biological pathways involving these loci may represent targets for the development of broad-spectrum treatments with efficacy for a wider range of psychopathology than currently available options. We note that the STAR*D sample excluded patients with psychotic depression. While this would not bias our results, it might have implications for their generalizability. To the extent that psychosis itself may be under genetic influence, this exclusion might have made the diagnostic groups more distinct than they would have been had psychosis been present in the MDD group.
Our results should be considered in light of several limitations. First, given the available sample size, our study had limited power to detect modest genetic effects or rare susceptibility variants using a strict genomewide threshold for significance. Because this is the first report of its kind and the goal was to characterize genotype-phenotype relationships, we present results down to a threshold of p < 5 × 10−5 to inform future analyses. Although our study detected only one region exceeding genomewide significant thresholds, recent pooled analyses of GWAS data have demonstrated that many variants that fall short of this threshold in a single study emerge as confirmed susceptibility loci when additional datasets are analyzed(20, 21). Thus, the “top hits” in our study provide preliminary evidence that awaits replication in future studies. A large-scale effort to use genomewide data to dissect within-disorder and cross-disorder influences has begun through the Psychiatric GWAS Consortium(57). Our results may inform these analyses which ultimately may include GWAS data on > 80,000 individuals across five disorders: SCZ, BPD, MDD, autism, and attention deficit hyperactivity disorder(57).
A second limitation is that, by design, our model fitting approach did not involve statistical testing of the each model vs. the null hypothesis. Instead, to control inflation of Type I error, we performed only the single omnibus test followed by model selection. A single test of the “true” underlying model would be expected to have greater power than the omnibus test. However, since the “true” model is unknown a priori, we would have to test multiple models to cover the range of possible true models. We have shown by simulation that the omnibus test has power comparable to a test of the “true” model, especially when the latter is corrected for multiple testing (58).
In summary, in a genomewide association analysis of three major psychiatric disorders, we identified a region of genomewide significant association on chromosome 11p15, near the ADM gene which appears to be specific to bipolar II disorder. We also found preliminary evidence for both cross-disorder and disorder-specific influences on SCZ, BPD, and MDD. Consistent with previous reports in independent samples, we observed evidence that variation in NPAS3 has effects that transcend DSM-IV diagnostic categories. The methods illustrated here may inform larger-scale efforts to examine cross-disorder genetic effects.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grant MH079799 (JWS). We thank Rutgers Cell and DNA Repository for extracting DNA and providing samples.
The STEP-BD project was funded in whole or in part with Federal funds from the National Institute of Mental Health (NIMH), National Institutes of Health, under Contract N01MH80001 to Gary S. Sachs, M.D. (PI), Michael E. Thase M.D. (Co-PI), Mark S. Bauer, M.D. (Co-PI). Active STEP-BD Sites and Principal Investigators included: Baylor College of Medicine (Lauren B. Marangell, M.D.); Case University (Joseph R. Calabrese, M.D.); Massachusetts General Hospital and Harvard Medical School (Andrew A. Nierenberg, M.D.); Portland VA Medical Center (Peter Hauser, M.D.); Stanford University School of Medicine (Terence A. Ketter, M.D.); University of Colorado Health Sciences Center (Marshall Thomas, M.D.); University of Massachusetts Medical Center (Jayendra Patel, M.D.); University of Oklahoma College of Medicine (Mark D. Fossey, M.D.); University of Pennsylvania Medical Center (Laszlo Gyulai, M.D.); University of Pittsburgh Western Psychiatric Institute and Clinic (Michael E. Thase, M.D.); University of Texas Health Science Center at San Antonio (Charles L. Bowden, M.D.). Collection of DNA from consenting participants in STEP-BD was supported by N01-MH-80001 (G Sachs, PI).
The Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study was supported by federal funds from NIMH under contract N01 MH-90003 to the University of Texas – Southwestern Medical Center at Dallas (A.J. Rush, principal investigator). Genotyping of STAR*D samples was funded by NIMH (R01 MH-072802, S.P. Hamilton, principal investigator).The CATIE project was funded by NIMH contract N01 MH90001.
Control subjects from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI), data and biomaterials were collected by the "Molecular Genetics of Schizophrenia II" (MGS-2) collaboration. The investigators and co-investigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA,MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA,MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01, MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA,MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY, MH59586, Jeremy Silverman, Ph.D. 34 (PI).
REFERENCES
- 1.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123(1):48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157(10):1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2(7):e212. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsuang MT, Winokur G, Crowe RR. Morbidity risks of schizophrenia and affective disorders among first degree relatives of patients with schizophrenia, mania, depression and surgical conditions. Br J Psychiatry. 1980;137:497–504. doi: 10.1192/bjp.137.6.497. [DOI] [PubMed] [Google Scholar]
- 5.Taylor MA, Berenbaum SA, Jampala VC, Cloninger CR. Are schizophrenia and affective disorder related? preliminary data from a family study. Am J Psychiatry. 1993;150(2):278–285. doi: 10.1176/ajp.150.2.278. [DOI] [PubMed] [Google Scholar]
- 6.Valles V, Van Os J, Guillamat R, Gutierrez B, Campillo M, Gento P, Fananas L. Increased morbid risk for schizophrenia in families of in-patients with bipolar illness. Schizophr Res. 2000;42(2):83–90. doi: 10.1016/s0920-9964(99)00117-6. [DOI] [PubMed] [Google Scholar]
- 7.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373(9659):234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardno AG, Rijsdijk FV, Sham PC, Murray RM, McGuffin P. A twin study of genetic relationships between psychotic symptoms. Am J Psychiatry. 2002;159(4):539–545. doi: 10.1176/appi.ajp.159.4.539. [DOI] [PubMed] [Google Scholar]
- 9.McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60(5):497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- 10.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7(4):405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 11.Craddock N, O'Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32(1):9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, Sklar P, Smoller JW, Abou Jamra R, Albus M, Bacanu SA, Baron M, Barrett TB, Berrettini W, Blacker D, Byerley W, Cichon S, Coryell W, Craddock N, Daly MJ, Depaulo JR, Edenberg HJ, Foroud T, Gill M, Gilliam TC, Hamshere M, Jones I, Jones L, Juo SH, Kelsoe JR, Lambert D, Lange C, Lerer B, Liu J, Maier W, Mackinnon JD, McInnis MG, McMahon FJ, Murphy DL, Nothen MM, Nurnberger JI, Pato CN, Pato MT, Potash JB, Propping P, Pulver AE, Rice JP, Rietschel M, Scheftner W, Schumacher J, Segurado R, Van Steen K, Xie W, Zandi PP, Laird NM. Combined Analysis from Eleven Linkage Studies of Bipolar Disorder Provides Strong Evidence of Susceptibility Loci on Chromosomes 6q and 8q. Am J Hum Genet. 2005;77(4) doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69(2):428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassett AS, Chow EW, AbdelMalik P, Gheorghiu M, Husted J, Weksberg R. The schizophrenia phenotype in 22q11 deletion syndrome. Am J Psychiatry. 2003;160(9):1580–1586. doi: 10.1176/appi.ajp.160.9.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassett AS, Chow EW, Husted J, Weksberg R, Caluseriu O, Webb GD, Gatzoulis MA. Clinical features of 78 adults with 22q11 Deletion Syndrome. Am J Med Genet A. 2005;138(4):307–313. doi: 10.1002/ajmg.a.30984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulze TG, Ohlraun S, Czerski PM, Schumacher J, Kassem L, Deschner M, Gross M, Tullius M, Heidmann V, Kovalenko S, Jamra RA, Becker T, Leszczynska-Rodziewicz A, Hauser J, Illig T, Klopp N, Wellek S, Cichon S, Henn FA, McMahon FJ, Maier W, Propping P, Nothen MM, Rietschel M. Genotype-phenotype studies in bipolar disorder showing association between the DAOA/G30 locus and persecutory delusions: a first step toward a molecular genetic classification of psychiatric phenotypes. Am J Psychiatry. 2005;162(11):2101–2108. doi: 10.1176/appi.ajp.162.11.2101. [DOI] [PubMed] [Google Scholar]
- 17.Williams NM, Green EK, Macgregor S, Dwyer S, Norton N, Williams H, Raybould R, Grozeva D, Hamshere M, Zammit S, Jones L, Cardno A, Kirov G, Jones I, O'Donovan MC, Owen MJ, Craddock N. Variation at the DAOA/G30 locus influences susceptibility to major mood episodes but not psychosis in schizophrenia and bipolar disorder. Arch Gen Psychiatry. 2006;63(4):366–373. doi: 10.1001/archpsyc.63.4.366. [DOI] [PubMed] [Google Scholar]
- 18.Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, Burrell GJ, Rice JP, Nertney DA, Olincy A, Rozic P, Vinogradov S, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Crowe RR, Cloninger CR, Martinez M, Gejman PV. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165(4):497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- 19.Shi J, Badner JA, Gershon ES, Liu C. Allelic association of G72/G30 with schizophrenia and bipolar disorder: a comprehensive meta-analysis. Schizophr Res. 2008;98(1–3):89–97. doi: 10.1016/j.schres.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Gejman PV, Sanders AR, Duan J, Levinson DF, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008 doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, Macintyre DJ, Maclean AW, St Clair D, Robinson M, Van Beck M, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Nicol Ferrier I, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008 doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, Arolt V, Baune BT, Blackwood D, Cichon S, Coventry WL, Domschke K, Farmer A, Fava M, Gordon SD, He Q, Heath AC, Heutink P, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hu Y, Kohli M, Lin D, Lucae S, Macintyre DJ, Maier W, McGhee KA, McGuffin P, Montgomery GW, Muir WJ, Nolen WA, Nothen MM, Perlis RH, Pirlo K, Posthuma D, Rietschel M, Rizzu P, Schosser A, Smit AB, Smoller JW, Tzeng JY, van Dyck R, Verhage M, Zitman FG, Martin NG, Wray NR, Boomsma DI, Penninx BW. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskvina V, Craddock N, Holmans P, Nikolov I, Pahwa JS, Green E, Owen MJ, O'Donovan MC. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol Psychiatry. 2009;14(3):252–260. doi: 10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachs GS, Thase ME, Otto MW, Bauer M, Miklowitz D, Wisniewski SR, Lavori P, Lebowitz B, Rudorfer M, Frank E, Nierenberg AA, Fava M, Bowden C, Ketter T, Marangell L, Calabrese J, Kupfer D, Rosenbaum JF. Rationale, design, and methods of the systematic treatment enhancement program for bipolar disorder (STEP-BD) Biol Psychiatry. 2003;53(11):1028–1042. doi: 10.1016/s0006-3223(03)00165-3. [DOI] [PubMed] [Google Scholar]
- 26.Kogan JN, Otto MW, Bauer MS, Dennehy EB, Miklowitz DJ, Zhang HW, Ketter T, Rudorfer MV, Wisniewski SR, Thase ME, Calabrese J, Sachs GS. Demographic and diagnostic characteristics of the first 1000 patients enrolled in the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) Bipolar Disord. 2004;6(6):460–469. doi: 10.1111/j.1399-5618.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- 27.Stroup TS, McEvoy JP, Swartz MS, Byerly MJ, Glick ID, Canive JM, McGee MF, Simpson GM, Stevens MC, Lieberman JA. The National Institute of Mental Health Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project: schizophrenia trial design and protocol development. Schizophr Bull. 2003;29(1):15–31. doi: 10.1093/oxfordjournals.schbul.a006986. [DOI] [PubMed] [Google Scholar]
- 28.Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, Keefe RS, Davis SM, Davis CE, Lebowitz BD, Severe J, Hsiao JK. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353(12):1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 29.Spitzer R, Williams J, Gibbon M. Structured Clinical Interview for DSM-IV, Outpatient Version (SCID-OP) New York: Biometrics Research Department, New York State Psychiatric Institute; 1996. [Google Scholar]
- 30.Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rush AJ, Fava M, Wisniewski SR, Lavori PW, Trivedi MH, Sackeim HA, Thase ME, Nierenberg AA, Quitkin FM, Kashner TM, Kupfer DJ, Rosenbaum JF, Alpert J, Stewart JW, McGrath PJ, Biggs MM, Shores-Wilson K, Lebowitz BD, Ritz L, Niederehe G. Sequenced treatment alternatives to relieve depression (STAR*D): rationale and design. Control Clin Trials. 2004;25(1):119–142. doi: 10.1016/s0197-2456(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 32.Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A, de Bakker PI, Ogdie MN, Thase ME, Sachs GS, Todd-Brown K, Gabriel SB, Sougnez C, Gates C, Blumenstiel B, Defelice M, Ardlie KG, Franklin J, Muir WJ, McGhee KA, MacIntyre DJ, McLean A, VanBeck M, McQuillin A, Bass NJ, Robinson M, Lawrence J, Anjorin A, Curtis D, Scolnick EM, Daly MJ, Blackwood DH, Gurling HM, Purcell SM. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13(6):558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 34.Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD, Reinalda MS, Slager SL, McGrath PJ, Hamilton SP. A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry. 67(2):133–138. doi: 10.1016/j.biopsych.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamnasaran D, Muir WJ, Ferguson-Smith MA, Cox DW. Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. J Med Genet. 2003;40(5):325–332. doi: 10.1136/jmg.40.5.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickard BS, Malloy MP, Porteous DJ, Blackwood DH, Muir WJ. Disruption of a brain transcription factor, NPAS3, is associated with schizophrenia and learning disability. Am J Med Genet B Neuropsychiatr Genet. 2005;136B(1):26–32. doi: 10.1002/ajmg.b.30204. [DOI] [PubMed] [Google Scholar]
- 38.Pickard BS, Christoforou A, Thomson PA, Fawkes A, Evans KL, Morris SW, Porteous DJ, Blackwood DH, Muir WJ. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14(9):874–884. doi: 10.1038/mp.2008.24. [DOI] [PubMed] [Google Scholar]
- 39.Gershon ES, Hamovit J, Guroff JJ, Dibble E, Leckman JF, Sceery W, Targum SD, Nurnberger JI, Jr, Goldin LR, Bunney WE., Jr A family study of schizoaffective, bipolar I, bipolar II, unipolar, and normal control probands. Arch Gen Psychiatry. 1982;39(10):1157–1167. doi: 10.1001/archpsyc.1982.04290100031006. [DOI] [PubMed] [Google Scholar]
- 40.Coryell W, Endicott J, Reich T, Andreasen N, Keller M. A family study of bipolar II disorder. Br J Psychiatry. 1984;145:49–54. doi: 10.1192/bjp.145.1.49. [DOI] [PubMed] [Google Scholar]
- 41.Endicott J, Nee J, Andreasen N, Clayton P, Keller M, Coryell W, Bipolar II. Combine or keep separate? J Affect Disord. 1985;8(1):17–28. doi: 10.1016/0165-0327(85)90068-0. [DOI] [PubMed] [Google Scholar]
- 42.Andreasen NC, Rice J, Endicott J, Coryell W, Grove WM, Reich T. Familial rates of affective disorder. A report from the National Institute of Mental Health Collaborative Study. Arch Gen Psychiatry. 1987;44(5):461–469. doi: 10.1001/archpsyc.1987.01800170083011. [DOI] [PubMed] [Google Scholar]
- 43.Heun R, Maier W. The distinction of bipolar II disorder from bipolar I and recurrent unipolar depression: results of a controlled family study. Acta Psychiatr Scand. 1993;87(4):279–284. doi: 10.1111/j.1600-0447.1993.tb03372.x. [DOI] [PubMed] [Google Scholar]
- 44.Simpson SG, Folstein SE, Meyers DA, McMahon FJ, Brusco DM, DePaulo JR., Jr Bipolar II: the most common bipolar phenotype? Am J Psychiatry. 1993;150(6):901–903. doi: 10.1176/ajp.150.6.901. [DOI] [PubMed] [Google Scholar]
- 45.Edvardsen J, Torgersen S, Roysamb E, Lygren S, Skre I, Onstad S, Oien PA. Heritability of bipolar spectrum disorders. Unity or heterogeneity? J Affect Disord. 2008;106(3):229–240. doi: 10.1016/j.jad.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Nwulia EA, Miao K, Zandi PP, Mackinnon DF, DePaulo JR, Jr, McInnis MG. Genome-wide scan of bipolar II disorder. Bipolar Disord. 2007;9(6):580–588. doi: 10.1111/j.1399-5618.2007.00437.x. [DOI] [PubMed] [Google Scholar]
- 47.Bettoni F, Filho FC, Grosso DM, Galante PA, Parmigiani RB, Geraldo MV, Henrique-Silva F, Oba-Shinjo SM, Marie SK, Soares FA, Brentani HP, Simpson AJ, de Souza SJ, Camargo AA. Identification of FAM46D as a novel cancer/testis antigen using EST data and serological analysis. Genomics. 2009;94(3):153–160. doi: 10.1016/j.ygeno.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez AP, Serrano J, Tessarollo L, Cuttitta F, Martinez A. Lack of adrenomedullin in the mouse brain results in behavioral changes, anxiety, and lower survival under stress conditions. Proc Natl Acad Sci U S A. 2008;105(34):12581–12586. doi: 10.1073/pnas.0803174105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savas HA, Herken H, Yurekli M, Uz E, Tutkun H, Zoroglu SS, Ozen ME, Cengiz B, Akyol O. Possible role of nitric oxide and adrenomedullin in bipolar affective disorder. Neuropsychobiology. 2002;45(2):57–61. doi: 10.1159/000048677. [DOI] [PubMed] [Google Scholar]
- 50.Glubb DM, McHugh PC, Deng X, Joyce PR, Kennedy MA. Association of a functional polymorphism in the adrenomedullin gene (ADM) with response to paroxetine. Pharmacogenomics J. 2009 Jul 28; doi: 10.1038/tpj.2009.33. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 51.Brunskill EW, Ehrman LA, Williams MT, Klanke J, Hammer D, Schaefer TL, Sah R, Dorn GW, 2nd, Potter SS, Vorhees CV. Abnormal neurodevelopment, neurosignaling and behaviour in Npas3-deficient mice. Eur J Neurosci. 2005;22(6):1265–1276. doi: 10.1111/j.1460-9568.2005.04291.x. [DOI] [PubMed] [Google Scholar]
- 52.Erbel-Sieler C, Dudley C, Zhou Y, Wu X, Estill SJ, Han T, Diaz-Arrastia R, Brunskill EW, Potter SS, McKnight SL. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci U S A. 2004;101(37):13648–13653. doi: 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pieper AA, Wu X, Han TW, Estill SJ, Dang Q, Wu LC, Reece-Fincanon S, Dudley CA, Richardson JA, Brat DJ, McKnight SL. The neuronal PAS domain protein 3 transcription factor controls FGF-mediated adult hippocampal neurogenesis in mice. Proc Natl Acad Sci U S A. 2005;102(39):14052–14057. doi: 10.1073/pnas.0506713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17(R2):R116–R121. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, Howson JM, Stevens H, McManus R, Wijmenga C, Heap GA, Dubois PC, Clayton DG, Hunt KA, van Heel DA, Todd JA. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359(26):2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plenge RM. Shared genetic risk factors for type 1 diabetes and celiac disease. N Engl J Med. 2008;359(26):2837–2838. doi: 10.1056/NEJMe0809719. [DOI] [PubMed] [Google Scholar]
- 57.A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14(1):10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- 58.Purcell S, Perlis RH, Sullivan PF, Sklar P, Smoller JW. Modifiers and subtype-specific genes in whole-genome association studies of complex disease. doi: 10.1159/000327158. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.