Abstract
Lines of mice were created by selective breeding for the purpose of identifying genetic mechanisms that influence magnitude of the selected trait and to explore genetic correlations for additional traits thought to be influenced by shared mechanisms. DNA samples from high and low methamphetamine drinking (MADR) and high and low methamphetamine sensitization lines were used for quantitative trait locus (QTL) mapping. Significant additive genetic correlations between the two traits indicated common genetic influence, and a QTL on chromosome X was detected for both traits, suggesting one source of this commonality. For MADR mice, a QTL on chromosome 10 accounted for more than 50% of the genetic variance in that trait. Microarray gene expression analyses were performed for 3 brain regions for methamphetamine-naïve MADR line mice: nucleus accumbens, prefrontal cortex and ventral midbrain. Many of the genes that were differentially expressed between the high and low MADR lines were shared in common across the 3 brain regions. A gene network highly enriched in transcription factor genes was identified as being relevant to genetically-determined differences in methamphetamine intake. When the mu opioid receptor gene (Oprm1), located on chromosome 10 in the QTL region, was added to this top ranked transcription factor network, it became a hub in the network. These data are consistent with previously published findings of opioid response and intake differences between the MADR lines and suggest that Oprm1 or a gene that impacts activity of the opioid system, plays a role in genetically–determined differences in methamphetamine intake.
Keywords: QTL, selected lines, gene expression, network analysis, opioid
Introduction
Quantitative trait locus (QTL) analysis allows dissection of genetic variation underlying a trait into its component loci by mapping each one to a chromosomal region. To generate the needed data, a population is measured for a trait (phenotype) of interest, then genotyped at marker loci distributed genome-wide. There are two basic approaches to carrying out a genome-wide search for QTLs, either marker-based or trait-based (Lebowitz et al., 1987). For the marker-based approach, the question asked is, for each marker locus (or an interval between markers), do animals of one genotype score differently on a trait than do animals of other genotypes? Because of linkage, the genotypes at a QTL can be predicted accurately from those at a nearby marker or flanking markers. Alternatively, in the trait-based approach, extreme high and low scoring animals for the trait (e.g., selected lines) are examined and the question is asked, do they differ in allele or genotype frequencies for any of the markers? If the answer is yes, and the difference is significant, then a QTL has been detected and mapped to the chromosomal region of the marker(s).
This paper describes QTL analyses for two separate trait-based experiments, using bidirectionally selected lines of mice. Each set of selected lines was focused on one of two traits, either two-bottle choice methamphetamine (MA) drinking (MADR lines) or MA-induced sensitization to locomotor activating effects of MA (MASENS lines). Each independent selection began with the testing of a population of F2 intercross mice from the C57BL/6J (B6) and DBA/2J (D2) inbred mouse strains. For the MADR lines, the F2 mice that consumed the most MA, when it was offered versus water, were chosen as breeders to establish the high MA drinking (MAHDR) line, whereas those that consumed the least were used to establish the low MA drinking (MALDR) line (Wheeler et al., 2009; Shabani et al., 2011). For the MASENS lines, a separate set of F2 mice was tested for magnitude of locomotor sensitization to MA over the course of 5 total MA treatments. Mice with the highest sensitization scores (those with the largest increase in locomotor response after the final MA treatment, compared to the response after the initial treatment) were interbred to create the MA high sensitization (MAHSENS) line and those with the lowest sensitization scores (with the smallest change in locomotor response after the final MA treatment, compared to the response after the initial treatment) established the MA low sensitization (MALSENS) line (Scibelli et al., 2011). The lines were tested and perpetuated across multiple generations using an individual or mass selection procedure (Falconer and Mackay, 1996). This bidirectional selection method resulted in divergent lines for each of the two MA traits, with the extent of divergence dependent on the proportion of genetic variation underlying each trait (aka, the heritability; Falconer and Mackay, 1996). The details of how each selection project was carried out are given in Wheeler et al. (2009) and Scibelli et al. (2011), respectively. The current paper addresses the question of whether QTLs were located in common genetic regions for these two MA traits.
In addition, for the MADR trait, differences in gene expression that are related to greater and lesser genetic risk for MA intake were examined using brain samples from MA naive MADR line mice. The brain regions chosen for analysis were the prefrontal cortex, nucleus accumbens and ventral midbrain (including the ventral tegmental area), because of their roles in effects of MA that are related to MA use (e.g., Keleta and Martinez, 2012; Zhu et al., 2012; Lin et al., 2007). Although there have been gene expression profiling studies examining the effects of MA (e.g., Yang et al., 2008; Martin et al., 2012), genetic risk for MA use has not been the focus of investigation.
Materials and methods
Genotyping
QTL analyses were performed using DNA samples obtained prior to MA exposure from all of the breeders of the S2 and S5 generation offspring (N = 40–48 per generation). These animals represent the most extreme scoring mice for high and low MADR of those tested in S1 and S4. Initial genotyping used custom single nucleotide polymorphism (SNP) arrays with 95 markers and the Illumina Golden Gate Assay (San Diego, CA). These informative SNPs were spaced at approximately 21 Mb intervals throughout the genome. The SNPs were validated using DNA from C57BL/6J (B6) and DBA/2J (D2) parental strains. The samples were analyzed locally using the Illumina Bead Station 500 X (Illumina, Inc, San Diego, CA) genotyping platform, and procedures were performed exactly as recommended by the manufacturer. Subsequently, in an effort to increase map resolution for one significant QTL location, four additional microsatellite markers were added to proximal chromosome (Chr) 10 where SNP marker coverage was initially less than optimal.
Selection line QTL analysis
The genetic basis for bidirectional selective breeding (or selection) is the change in allele frequencies in opposite directions, when comparing one line with the other. All allele frequencies for markers begin at close to 0.5 for each of the two possible alleles (B6 or D2) in the founding F2. Allele frequencies will diverge as a function of generations of selection, approaching 1.0 and zero in the two oppositely-selected lines, for markers close to QTLs influencing the MA trait under selection. In contrast, markers in those portions of the genome not in close proximity to QTLs will not change in allele frequency within the limits of sampling error (e.g., random or genetic drift). The B6D2F2 has been used as the founding population for several previous short-term selective breeding projects for drug-related traits (Belknap et al., 1997; Metten et al., 1998; Phillips et al., 2005; Kamens et al., 2006), and this was one factor in our decision to use them for these MA traits. In addition, multiple QTL mapping projects for traits relevant to addictive drug effects have been based on populations derived from this F2 (e.g., Belknap et al., 1997; Grisel et al., 1997; Phillips et al., 1998; Janowsky et al., 2001; Hitzemann et al., 2004; Boone et al., 2008). We have recently published data showing that D2 mice have greater intake and preference for MA, compared to B6 mice (Eastwood and Phillips, 2013). Both B6 and D2 mice develop locomotor sensitization to MA, but D2 mice show sensitization after fewer MA treatments (Phillips et al., 1994).
The selected lines were tested for QTLs in the second and fifth selected generations, when random drift was relatively small. The QTL analysis method of Belknap et al. (1997) was used, which is based on allele frequency changes due to selection and is thus explicitly designed for selection line data. The null hypothesis is that the observed divergence in allele frequencies was due to random drift and allele frequency sampling error, but not selection. We refer to the D2 allele frequency as q and the B6 allele frequency as p, such that p + q = 1. In the F2, both p and q = 0.5 (approximately). Selection for a trait will cause the D2 allele frequency (q) for all QTL-associated markers to diverge away from 0.5, and approach 1.0 in one line and 0 in the other line. Because the expected outcome for p is the mirror image of q, we need focus only on q. Evidence for the presence of a QTL was gained from the difference in relative allele frequencies between the high and low lines at a nearby marker (δ = qH − qL) significantly exceeding that expected from genetic drift and sampling error. The value of Z, the normal deviate, was calculated as follows for each marker and used to test for QTL significance:
| (Equation 1) |
where the first term in the denominator is the expected genetic drift variance (Falconer and Mackay, 1996), the second and third terms are the variances due to sampling error in the high and low selected lines, respectively, nL and nH are the sample sizes in each line, pH, qH, pL, qL are the allele frequencies in each line, F is the inbreeding coefficient at a given selection generation (Falconer and Mackay, 1996) and p0 and q0 are the initial allele frequencies in the founding F2 population (both about 0.5). F was calculated as [1− (1−1/2Ne)]t where Ne is the effective number of breeders, which averaged 24 in both short-term selection experiments, and t is the generation of selection (Falconer and Mackay, 1996). The QTL results are presented as logarithm of the odds (LOD) scores (df=2) calculated as the negative logarithm (base 10) of the p value associated with the Z value from Equation 1. For example, LOD = 3.0 when p =0.001. We used directional LOD scores, which were positive if the D2 allele was associated with higher MA trait scores and negative if the B6 allele was associated with higher MA trait scores.
Two selected generations, S2 and S5, were chosen because, for large effect QTLs, selection causes allele frequencies to approach fixation (q → 0 or 1) rapidly in both lines in the S2, thus limiting further allele frequency divergence in subsequent generations. In contrast, smaller effect QTLs will show moderate allele divergence at the S2 and continue to show further divergence approximately linearly out to the S5. For this reason, it was important to assess both S2 and S5 selected generations to more optimally detect a broader range of QTL effect sizes for the two MA behavioral traits.
All procedures using animals were approved by the Institutional Animal Care and Use Committee and were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Genetic correlation between the two MA behavioral traits
Using LOD scores generated by the trait-based allele frequency method, a genetic correlation can be estimated by constructing a vector of LOD scores from Chr 1 to Chr X (i.e., genome-wide) for each of the two traits and correlating the two vectors using Pearson’s product moment correlation. This approach was greatly facilitated by the use of the same SNP markers in both short-term selection experiments and the use of a B6D2F2 population to initiate both selection experiments. For this purpose, directional LOD scores were used, where the original LOD scores were multiplied by −1 if the B6 allele was associated with higher trait scores and by +1 (left unchanged) if the D2 allele was associated with higher trait scores. Because the selection response relies primarily on additive genetic variation, the genetic correlation estimated in this way is primarily additive, or rA (Falconer and Mackay, 1996).
Microarray Gene Expression studies in MADR lines
Expression data have been collected, thus far, only in the MADR lines. In part, this decision was based on available resources, but more importantly on our more promising results for genetic influences on the MADR trait; i.e., calculated heritability at the end of selection for the MADR lines was 0.35 (Wheeler et al., 2009), whereas it was 0.08 for MA sensitization (Scibelli et al., 2011). Heritability was somewhat higher at 0.17 for the sensitization lines earlier in selection, when most of the response to selection occurred; however, it was still half that found for MADR.
For the array data, we used a 2 × 2 design in which one factor was line (MAHDR or MALDR) and the second factor was treatment (2 mg/kg MA or saline, both given i.p.) for each of three brain areas, the nucleus accumbens (NAC), prefrontal cortex (PFC) and ventral midbrain (VMB; substantia nigra and ventral tegmental area). Two-way ANOVA was used to test for significance for each brain area implemented in the MeV 4.6 software package (www.tm4.org). We also performed a three-way factorial ANOVA (line × treatment × brain area) using a full (or saturated) model implemented in the LIMMA package in R. However, in this paper we are reporting only the results for the main effect of line, because it reflects genetic differences in the predisposition to MA drinking – the focus of this paper; the treatment factor and its interaction with line will be the focus of a subsequent paper. S5 generation mice contributed brain tissue for expression profiling. A total of 20 Affymetrix 430 2.0 mouse GeneChips were used per brain area, with 5 chips per cell of the 2 × 2 factorial design per brain area, for a total of 60 arrays overall. RNA samples from four male mice, aged 60–67 days, were pooled per chip. Each pool was hybridized to one 430 2.0 chip, resulting in a total of 45,101 probe sets (transcripts) for analysis, representing about 19,000 distinct genes.
Tissue preparation and analysis
Four hours after injection, mice were killed by cervical dislocation, followed promptly by brain removal, chilled saline rinse, and dissection. Dissection was performed using RNAse free treated tools and an aluminum dissection stage, and dissected regions were placed into RNAse free microcentrifuge tubes containing Trizol (Invitrogen, Carlsbad, CA, USA) that were immediately placed on dry ice. Samples were then stored at −80°C for less than 2 months prior to RNA extraction. The prefrontal cortex was removed as the medial third of a 1.5 mm slice from the anterior part of the brain just behind the olfactory bulbs. The next 1.4 mm slice was then taken, and the NAC was collected from this slice, using the anterior commissure as a landmark and a micropunch fashioned from a 16 gauge blunt cut needle. Then, using the superior colliculus as a landmark, the most anterior portion of the brain was removed and discarded, and the next 1 mm slice was taken. The ventral third of that slice and the cerebral cortex were removed, and the remaining tissue was collected as the ventral midbrain.
Total RNA was isolated with TRIZOL, using a modification of the single-step acid guanidinium thiocyanate-phenol-chloroform extraction method (Chomczynski and Sacchi, 1987), according to the manufacturer’s guidelines. The extracted RNA was then purified using RNAeasy (Qiagen, Valencia, CA, USA). RNA purity and concentration were evaluated using a Nanodrop spectrophotometer (ND-1000 ThermoScientific, Wilmington, DE USA) and all samples met our criterion of a 260/280 nm absorption ratio >1.8. Samples containing less than 10 μg of total RNA were vacufuged using a DNA120 SpeedVac (ThermoScientific, Wilmington, DE USA) until the desired concentration was obtained (as measured by the spectrophotometer). RNA samples that met these standards were sent to the Oregon Health & Science University Affymetrix Microarray Core for analysis. The Affymetrix Microarray Core at Oregon Health & Science University runs additional quality control checks, according to Affymetrix recommendations, including determination of RNA quality by fluorescence-based capillary electrophoresis, using the Agilent RNA 6000 Pico LabChip kit and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA USA). All of the samples used in this study passed all quality control measures. The procedures used at the Affymetrix Microarray Core facility faithfully follow the manufacturer’s recommended procedures as detailed at the Affymetrix Microarray Core web site: http://www.ohsu.edu/gmsr/amc. This core facility performed all of the subsequent steps of RNA labeling, hybridization and subsequent intensity scanning using Affymetrix scanners.
Probe (low) level data analysis
We used robust multiarray analysis (RMA; Irizarry et al., 2003), using the Bioconductor Affy package (www.bioconductor.org) running in the R statistical computing environment (v. 2.9.1) with default settings (Gautier et al., 2004).
Testing for selection line differences in expression
The multiple comparison significance threshold was determined as the 5% false discovery rate (FDR), which is based on the proportion of all declared significant findings that are expected to be false positives (Benjamini and Hochberg, 1995; Storey and Tibshirani, 2003). This differs from the conventional multiple comparison correction approach (e.g., Bonferroni), which is based on the proportion of all tests that are expected to be false positives. False discovery rates (FDR) were calculated using the q value software package within the R statistical program package (Storey and Tibshirani, 2003). FDR values adjust the observed p values to correct for the effects of multiple testing.
Comparative network analysis
This approach seeks to move beyond looking at genes one-at-a-time and instead seeks to identify groups of genes showing coordinated gene function as the unit of analysis, such as those in an interacting pathway or pathways. We used Metacore (www.genego.com) which is a bioinformatics package that relies on an extensive database of mostly interacting protein information gleaned from the literature on about 700 known networks. Differentially expressed (DE) genes from a microarray experiment are entered along with their p values and fold-change values to be used as weights. The program identifies those pathways from among the 700 in the database that are statistically significantly overrepresented with DE genes from a microarray experiment (Nikolsky et al., 2009). Note that this approach does not involve any clustering of DE genes in the statistical sense, and thus does not require the construction of a correlation or other similarity matrix as does the clustering algorithm approach.
Comparative Network analysis was conducted on the DE candidates (up-regulated, FDR adjusted p-value <0.05) for all 3 brain regions (N=103 for NAC; N=439 for PFC and N=191 for VMB) for the MAHDR versus MALDR comparison. To discover common regulatory networks, for every transcription factor (TF) with direct target(s) in the overlapping candidate list, the shortest path algorithm generates a sub-network consisting of all shortest paths to this TF from the closest receptor with direct ligand(s) in the candidate list using high confidence interaction data from the Metacore Knowledgebase. The p-value for the sub-network is based on a hypergeometric distribution in which the p-value essentially represents the probability of a particular mapping arising by chance, givne the number of genes in the candidate list and the distribution of pathways and membership from the knowledgebase.
Gene Ontology database to identify overrepresented functional categories among differentially expressed genes between the MAHDR and MALDR selection lines
The NIH DAVID online database and analytical tools (https://david.abcc.ncifcrf.gov) were used to infer functional characteristics of DE genes between the high and low selection lines in all three brain areas (Huang et al., 2009a,b). All three broad GO categories of BP (Biological Processes), MF (Molecular Function) and CC (Cellular Component) were examined for over representation of DE genes in all subcategories within each of these three broad categories compared to all genes represented on the microarray (Huang et al., 2009a). This program was also used to identify significant over representation among the twenty Chr in the mouse genome by these same DE genes.
Results
Genetic correlation between the two behavioral traits
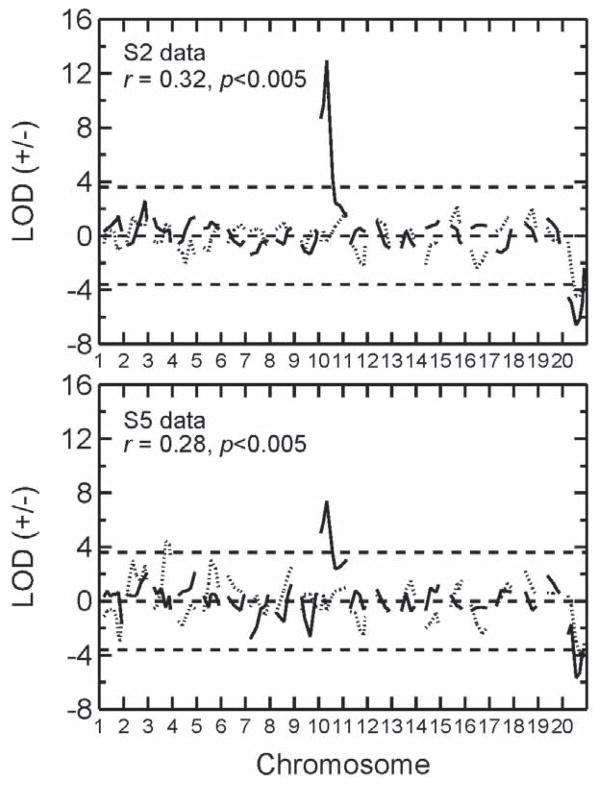
The correlation of the two vectors of LOD scores, derived from the directional LOD scores for each trait, indicated significant genetic codetermination of the MA drinking and sensitization traits. The additive genetic correlation between the two MA traits was rA = 0.32 for the S2 (p<0.001) and rA = 0.28 for the S5 (p<0.005), indicating significant common genetic influences for the two traits. These results are shown in Figure 1.
Figure 1.
Directional genome-wide LOD scores resulting from QTL analysis for the MADR (solid line) and MASENS (dotted line) short-term selected lines in the S2 and S5 generations. Positive LOD scores indicate that the D2 strain allele confers higher trait scores, while negative LOD scores indicate that the B6 allele confers higher trait scores. The genetic correlation between the two traits is also shown, indicating a statistically significant degree of genetic codetermination between the two behavioral traits in both generations. Horizontal dashed lines indicate genome-wide significance threshold based on permutation tests (Doerge and Churchill, 1996).
QTL detection in the MADR and MASENS selection lines
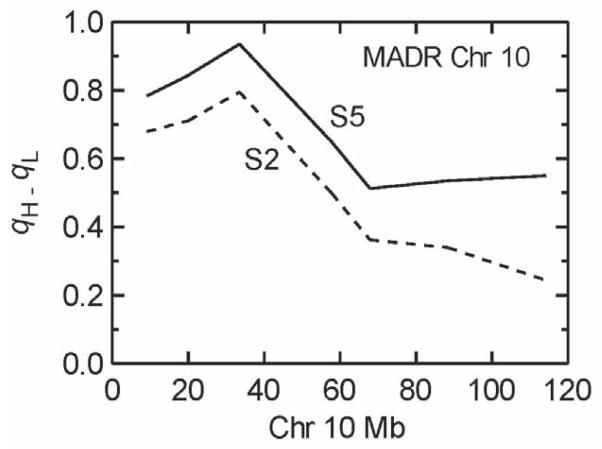
The QTL results for both traits are also shown in Figure 1 for the S2 and S5 selected generations, with results presented as directional LOD scores (df=2). For the MADR lines, two QTLs emerged as significant, one on proximal Chr 10 (LOD 12.9) and another mid Chr X (LOD 6.6), with 2-LOD support intervals of 10–40 and 60–140 Mb, respectively; the 2-LOD support interval approximates a 95% confidence interval. The Chr 10 QTL was by far the largest detected for either trait, accounting for 24% of the phenotypic variance and 60% of the genetic variance in the S2. As expected for such a large effect QTL, the divergence in allele frequencies (q) due to selection was unusually rapid, attaining in the S2 a value of q=0.96 in the MAHDR line compared to q=0.17 in the oppositely-selected MALDR line. By S5, this QTL approached fixation, with q=0.97 in the MAHDR and q=0.02 in the MALDR line. This is clearly evident in Figure 2 which plots the difference in allele frequencies (qH –qL) for all markers on Chr 10. In contrast, no evidence for a Chr 10 QTL was seen for the MASENS lines, so this QTL is unique to the MADR lines. Suggestive QTLs (Lander and Kruglyak, 1995), which are those where one QTL is expected to be a false positive in a genome-wide search, were detected on distal Chr 2 (LOD 2.6) and mid Chr 9 (LOD 2.6). The Lander and Kruglyak (1995) criteria for attaining significant or suggestive status are considered to be unusually stringent compared to other widely accepted criteria (Doerge and Churchill, 1996; Broman and Sen, 2009).
Figure 2.
Difference in allele frequencies between the two oppositely-selected MADR lines, qH - qL, for all markers on Chr 10 where the largest QTL was found. The S2 and S5 generations are shown indicating increasing allele frequency divergence as a function of generation of selection, especially for the proximal portion of this chromosome where the QTL resides. There is a suggestion of a second QTL at the distal end of Chr 10, but this did not meet our statistical criteria for being significant.
For the MASENS lines, only one QTL emerged as significant which was located on mid Chr X (LOD 4.5 in S2). The maximum LOD score was seen at SNP marker mm13483927 at 97 Mb (2-LOD support interval 60–140 Mb), the same X-Chr marker showing the maximum LOD score on this Chr in the MADR S2 and in the same direction; i.e., the B6 allele conferred higher trait scores for both MA behavioral traits. Thus, this mid Chr X QTL, present in both MADR and MASENS sets of lines, is a prime candidate for being the major cause of the positive additive genetic correlation (rA) between these two traits when examined genome-wide. For the MASENS trait, four suggestive QTLs emerged on distal Chr 3 (LOD 3.3), distal Chr 1 (LOD 3.0), distal Chr 2 (LOD 2.6) and mid Chr 5 (LOD 3.0). Only for the distal Chr 2 suggestive QTL for MASENS was there a corresponding suggestive QTL for the MADR trait at the same location and in the same direction. Thus, this suggestive chromosomal region could also contribute to the genome-wide positive genetic correlation, although likely to a lesser extent than the common (between the two traits) significant QTL on mid Chr X.
MAHDR vs MALDR expression differences
For the NAC, 195 probe sets were found to be DE at an FDR < 0.05, which in terms of unadjusted p values was p<0.0005. For the PFC, 787 probe sets were detected as significantly DE at this same FDR threshold, corresponding to an unadjusted p<0.002. For the VMB, 399 probe sets were significantly DE at this same threshold, corresponding to p<0.001.
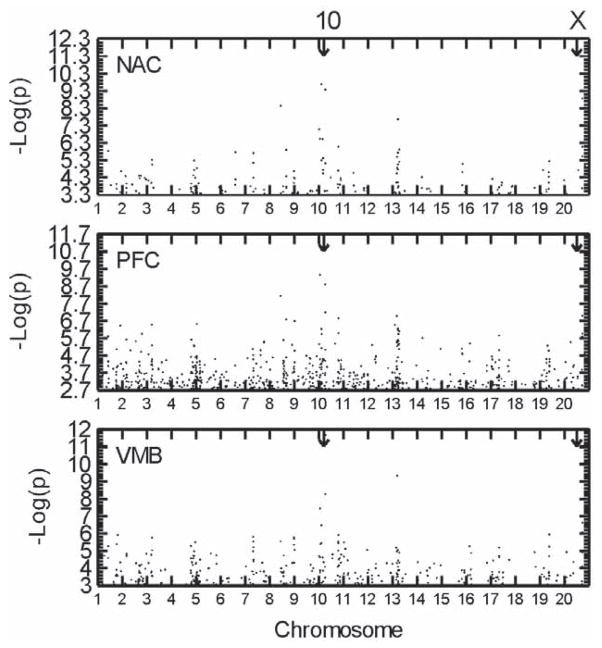
The genome-wide distributions of DE probe sets for all three brain areas are shown in Figure 3 for all probe IDs showing FDR < 0.05. The same threshold was adopted for the purpose of data plotting to put all three brain areas on an equal footing. As can be seen, there was an overrepresentation of DE probe sets (IDs) in the proximal Chr 10 QTL region in all three brain areas, especially in terms of the magnitude of the -log(p) values which were highest in this chromosomal region compared to any other. The mid Chr X QTL region showed a diminished degree of enrichment of DE probe sets compared to the genome-wide average in all three brain regions, suggesting that this QTL is predominantly based, not on differences in transcript abundance, but on protein variants differing in functional capacity. However, the distal Chr 2 and mid Chr 9 suggestive QTL regions did show an enrichment of DE probe IDs in all three brain regions compared to genome-wide averages.
Figure 3.
Genome-wide distribution of differentially expressed (DE) probe sets between the MAHDR and MALDR lines that attained FDR < 0.05 for each of the three brain areas, either nucleus accumbens (NAC), prefrontal cortex (PFC) or ventral midbrain (VMB). As can be seen, the clustering of DE probe sets in particular chromosomal locations was closely similar for all three brain areas although the probe sets making up each cluster often were not the same. The arrows show the locations of the two significant QTLs for the MA preference drinking trait on Chr 10 and X. The X axis in all cases is the negative logarithm of the p value for the line difference.
There were clusters of DE genes at the same chromosomal locations in all three brain regions not associated with any MADR QTL, either suggestive or significant. Because QTLs always map to the causative polymorphisms, these non-QTL clusters of DE genes are likely caused indirectly by QTLs residing elsewhere in the genome. These non-QTL DE clusters were strongly apparent on distal Chr 4, proximal 5, proximal 7 and mid 19 (Figure 3). These findings suggest that these non-QTL regions of DE gene clusters reflect the secondary expression effects of QTLs on downstream pathways or reflect the widespread effects of one or more transcription factors. Because these DE gene clusters are also DE between the two lines despite the absence of coincident QTLs, they may well play a role in the influence QTLs have on the MADR trait via non-QTL portions of the genome. An obvious mechanism for this is QTL-mediated trans regulation effects on other portions of the genome which in turn may also influence the MADR behavioral trait.
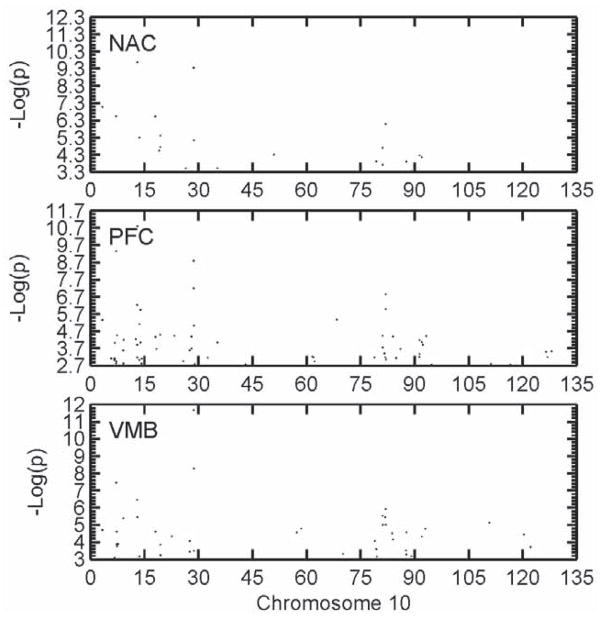
Figure 4 shows the distribution of DE probe sets on Chr 10 where the largest effect QTL was found for the MADR trait. All three brain areas showed a closely similar clustering of DE probe sets in the QTL region within the 2-LOD support interval. These are tabulated in Table 1S of the Supplementary Materials. Note that another DE cluster at 80–85 Mb is apparent where there is no known QTL, again suggesting trans regulation emanating from the proximal Chr 10 (or other) QTL causing differential expression of genes on distal portions of Chr 10.
Figure 4.
The distribution of differentially expressed (DE) probe sets on Chr 10 between the MAHDR and MALDR lines that attained FDR < 0.05 for each of the three brain areas, either nucleus accumbens (NAC), prefrontal cortex (PFC) or ventral midbrain (VMB). As can be seen, the clustering of DE probe sets in particular chromosomal locations was closely similar for all three brain areas although the probe sets making up each cluster often did not agree. The QTL influencing the MADR trait was on proximal Chr 10, with a 2-LOD support interval ranging from 10 to 40 Mb; however, the lack of markers at the most proximal end (centromere) impacts the ability to exclude this region. The X axis in all cases was the negative logarithm of the p value for the line difference.
While all three brain areas showed closely similar clusters in terms of chromosomal location, the probe sets making up these clusters differed considerably among the brain areas. For example, within the Chr 10 QTL region, 62 probe sets were DE for at least one brain area, 12 in the NAC, 30 in the PFC and 20 in the VMB (Figure 4). Of these 62, 28 of them (45%) were DE for only one brain area and not the other two. Only 18 of these DE probe sets (29%) were DE for all three brain areas (See Supplemental Table 1S). Similar findings emerged for the clusters of DE probe sets in non-QTL regions seen on distal Chr 4, proximal 5, proximal 7 and mid 19 (Figure 3). While the clustering was closely similar in all three brain areas, the probe sets making up each cluster often differed substantially among the brain areas.
The NIH DAVID website (Huang et al., 2009a, b) was used to detect over representation of DE genes between the two lines by Chr at p<0.01 (compared to all 20 mouse Chr) for each of the three brain areas subjected to microarray analysis. For those DE genes showing higher expression values in the high MADR compared to the low MADR selected line, Chr 10 was over represented in the NAC (p=9×10−4), PFC (p=4×10−4) and VMB (p=7×10−7); all FDR < 0.05. Chr 4 and 12 were over represented, but only in the PFC at p=0.007 and p=0.0034, respectively, both FDR < 0.05. For those DE genes showing lower expression values in the high MADR compared to the low MADR selection line, Chr 10 was over represented in the NAC (p=5×10−3) and PFC (p=0.01), both FDR < 0.10. Chr 4 was over represented in the PFC (p=1×10−7) and VMB (p=2×10−4), both FDR < 0.05, and the NAC (p=0.01), FDR < 0.10. To summarize for all three brain areas, the strongest and most consistent over representation by Chr of DE genes was seen for Chr 10, followed by Chr 4. While Chr 10 has a large effect and highly significant QTL, Chr 4 does not have even a single suggestive QTL for any brain area.
Network Analysis
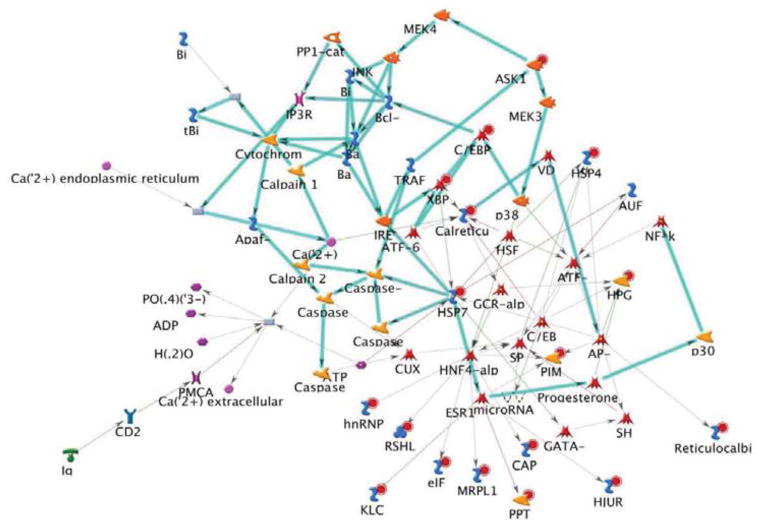
A highly significant transcription-factor centric sub-network was identified based on the shared DE genes (P=1.87e-40, Figure 5). Examination of annotation of the subnetwork members also revealed that this network was functionally enriched for regulation of apoptotic processes (P=2.728e-25), consistent with our previous data (Wheeler et al., 2009). Transcription Factor hubs for this network included HNF4-alpha (12 edges), AP-1 (11 edges), ATF-3 (9 edges), C/EDBPalpha (7 edges), c-Jun (6 edges), ESR1 (6 edges), HSF1 (6 edges), NF-KB1 (6 edges), SP2 (6 edges) and ATF-6 (5 edges). There was enrichment for genes from the dorsal neural tube (Calr, Klc1, Hspa5, Hjurp, Pcmt1, Xbp1, Serpinh1, Cap1, Ddit3, Hspa1b, Mrpl15, Map3k5, Rcn1, Hpgd; P=7.532e-26) and brain (P=3.031e-21).
Figure 5.
Results of comparative network analysis conducted on differentially expressed genes in 3 brain regions (nucleus accumbens, prefrontal cortex and ventral midbrain) identifies a network rich in transcription factor genes (p = 1.87e-40). Shown are inferred functional characteristics of genes that were differentially expressed between the high and low methamphetamine drinking selected lines.
Relation of Oprm1 to the network
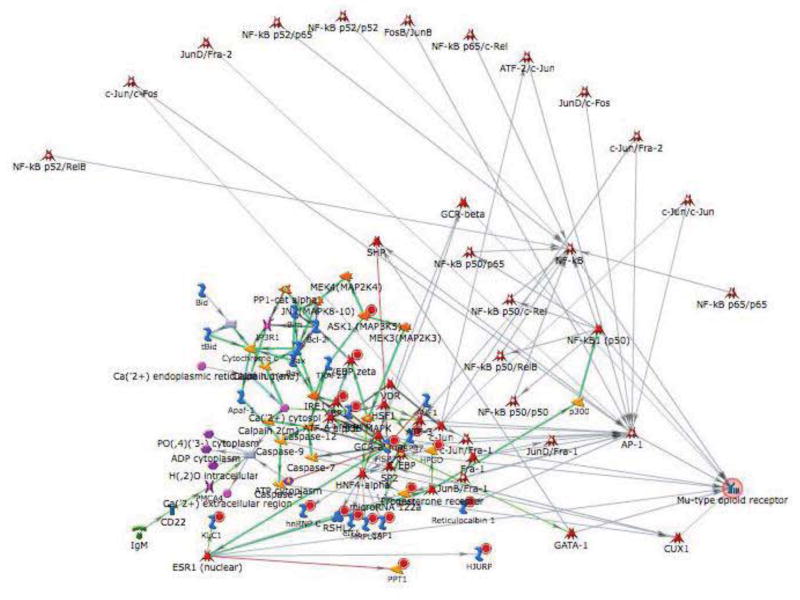
A potential candidate in the Chr 10 QTL region is Oprm1, the mu-opioid receptor (MOP-r) gene. We have determined that the MADR lines differ in sensitivity to MOP-r agonist drugs (Eastwood and Phillips, 2013) and that the low MADR line voluntarily consumes more morphine than the high MADR line (Eastwood and Phillips, unpublished). When Oprm1 was added to this network to determine which genes it interacted with and identify its location in the network, it became a hub, with 9 edges or interactions in the network (with AP1, C/EBPbeta, NF-kB, NF-kB(nuclear), GCR-alpha, ESR1, GCR-beta, CUX1, and GATA-1), all of which are with transcription factors (Figure 6).
Figure 6.
Candidate gene Oprm1 (mu-type opioid receptor) becomes a hub when added to the top ranked transcription factor network of genes that show differential expression in the nucleus accumbens, prefrontal cortex and ventral midbrain of methamphetamine naïve high and low methamphetamine drinking line mice.
Discussion
Bidirectionally selected lines have a number of advantages as QTL detection and mapping tools. Because of the greatly increased frequency of extreme phenotypes and associated genotypes generated by selection, the effects of QTLs will be greatly leveraged, facilitating their detection. An advantage of selection from an F2 of two inbred strains stems from the fact that there are only two possible alleles at each locus, each with frequencies initially of about 0.5 per allele, which is close to the optimum for each QTL to contribute maximally to the additive genetic variance upon which selection progress depends (Falconer and Mackay, 1996). This, in turn, is conducive to a rapid selection response compared to other foundation populations (Belknap et al., 1996). Further, because the unit of genetic analysis is the selected line, rather than individual mice, smaller anatomical brain areas can be used by pooling multiple mice from the same selected line for expression analysis. This is not possible in an F2, where each mouse is a unique genotype and must be analyzed as such. In the current paper, we demonstrate a large effect QTL on Chr 10 for the strongly heritable trait, MA drinking. Not surprisingly, QTLs were more difficult to detect for the less heritable trait, MA-induced sensitization. Based on stronger genetic contribution, we carried out gene expression analysis in the MADR lines and identified a highly significant transcription factor centric sub-network associated with risk for MA intake. Oprm1, a gene located in the region of the Chr 10 QTL serves as a hub with multiple interactions within this transcription factor centric network.
As previously reported, selective breeding produced a rapid divergence between the oppositely-selected MADR lines, resulting in a realized heritability of 0.34 in the S4 generation (Wheeler et al., 2009). These heritability estimates represent the proportion of the total trait (phenotypic) variation due to genetic variation, and are based on the observed divergence between the lines (selection response) as a fraction of the cumulated total selection pressure applied, estimated by the divergence in the breeder trait means for each generation (Falconer and Mackay 1996). The realized heritability calculated through S2 for the MADR lines was somewhat higher at 0.40, indicating that divergence occurred early in selection and began to wane in later generations as fixation was approached for trait relevant genes (QTLs), thus reducing the genetic variance upon which further selection progress depends.
We have recently replicated the selection for MA drinking, following the same procedures as for the first, and we obtained virtually identical selection and heritability results; realized heritability was 0.35 in the S4 generation (Shabani et al., 2011). In addition, QTL analysis in the second set of MADR lines produced a remarkably similar pattern of mapping results, including the identification of a large effect QTL in the same region on Chr 10 as reported here (see Supplementary materials Figure 1S). When mapping results are combined for the two selections (Figure 1S), the QTLs on both Chr 10 and X are more strongly supported and more modest support for QTLs on other Chr appear; however, the mapping results for the independent and combined replicates are remarkably similar. On the other hand, the MASENS selection experiment showed a realized heritability of only 0.17 in the S2 and 0.08 in the S5 generation (Scibelli et al., 2011), roughly half that of the MADR trait. We have therefore decided not to replicate this selection.
One drawback of the current approach to QTL mapping is that each generation of selection increases the possibility of genetic drift (also known as random drift), which is a change in allele frequencies, resulting from sampling error accumulating over generations. In part, this is due to the use of a finite number of breeders that are used to generate offspring in each generation. Thus, drift can cause a divergence in allele frequencies between the selected lines by random events alone, unrelated to the effects of selection, which is a bidirectional process. The variance due to drift is a simple function of the inbreeding coefficient, F, as follows: vardrift = p0q0F (Falconer and Mackay, 1996), where p0 and q0 are the initial allele frequencies of each of the two possible alleles per locus. The way to minimize the effect of drift is to minimize F, either by using a larger number of breeders, avoiding the mating of relatives, and/or restricting selection to only a few generations when F is relatively low compared to later generations. In the MADR and MASENS selection projects, we selected for only five generations (to S5) to minimize inbreeding and thus genetic drift, and we maintained reasonably large numbers of breeding pairs for each line (Wheeler et al., 2009; Scibelli et al., 2011). This approach can be described as short-term selective breeding, because at the outset there was no plan to perpetuate these lines indefinitely. However, for QTL detection purposes, earlier generations may be optimal for study because this practice often maximizes the effect of selection compared to random drift as contributors to allelic frequency differences between the lines. The genetic data from the second selection generation (S2) proved to be optimal for the largest-effect QTLs in both of the selection experiments reported here, although the S5 data provided evidence of smaller-effect QTLs which took longer to show their effects, as allele frequency differences accumulated.
For the purpose of gene discovery, QTL analysis of a behavioral trait and microarray analysis of gene expression have several complementary strengths that address some of their independent weaknesses and make their combination especially attractive when applied to the same population of mice. When a statistically significant QTL is first detected, three important facts become known. First, the QTL influences a behavioral trait such as MA drinking in the sense that it controls (accounts for) a portion of the trait variation. In other words, in our two-allele population, substituting one allele for the other at a QTL is associated with a significant change in the behavioral trait score. This substitution effect (or QTL effect) is the basis for detecting QTLs and is thus the cornerstone of all types of QTL analyses (Falconer and Mackay, 1996). Second, a QTL will map to the region of the causative polymorphism, thus providing important clues as to which polymorphisms are trait-relevant. Third, the QTL effect allowing detection is unidirectional; the QTL influences trait variation and not the reverse. What is not known initially is the identity of the gene underlying the QTL due to insufficient mapping precision. Finding the responsible gene is challenging, but new technologies now make this task easier; gene expression microarrays have been one of these (Belknap et al., 2001; Phillips and Belknap, 2002; Hitzemann et al., 2004; Schadt et al., 2003; 2005; Han et al., 2008; Yeung et al., 2011), although the analytic tools have continued to evolve.
When variation in mRNA (transcript) abundance is found in a microarray study, the identity of the gene involved is known, in most cases, as is map location. What is not known is whether there is any influence of that gene on a behavioral trait, because variation in mRNA abundance may not translate to a protein function difference or effects on the behavioral trait. Also, differential expression of a gene may be due to a polymorphism in or near the gene in the case of cis regulation (e.g., the promoter), but it may be elsewhere in the genome in the case of trans regulation (e.g, a transcription factor). Another frequent concern is that observed variation in expression could be a cause of variation in the behavioral trait, or it could be the consequence of such trait variation–the direction of effect is often difficult to disentangle unless the experiment is designed to differentiate these possibilities (e.g., see Slonim, 2002). This is a problem when the process of measuring the trait (e.g., consumption of MA for several days) might alter the expression of many genes causing them to be DE between the selected lines. However, short-term selective breeding experiments provide an effective way to take advantage of both approaches in a complementary fashion. For example, here, differences in gene expression were examined in mice that had been bred for differences in susceptibility to MA consumption, without having been exposed to MA themselves. In this way, risk genes and mechanisms are identified. With regard to the Chr 10 QTL region, the genes that we have found to be DE in one or more of the 3 brain regions include, Oprm1, Esr1 (estrogen receptor 1), pcmt1 (protein-L-isoaspartate [D-aspartate] O-methyltransferase 1), ppil4 (peptidylprolyl isomerase [cyclophilin]-like 4), Hivep2 (human immunodeficiency virus type 1 enhancer binding protein 2), Nhsl1 (NHS-like 1), Pex7 (peroxisome biogenesis factor 7), Map3k5 (mitogen activated protein kinase kinase kinase 5), Amd1 (S-adenosylmethionine decarboxylase 1), Cirbp (cold inducible RNA binding protein), Rfx4 (regulatory factor S, 4), Dmt2 (dorso-medial telencephalon gene 2) and Fgd6 (FYVE, RhoGEF and PH domain containing 6). Partly because good pharmacological tools exist to manipulate mu opioid receptors, but also because several polymorphisms and linkage disequilibrium blocks in human OPRM1 have been associated with MA dependence/psychosis (Ide et al., 2006) and because buprenorphine, a mu-opioid receptor partial agonist, attenuated the effects of MA on dopaminergic neuro-transmission in rats (Pereira et al., 2011), we have pursued a line of research that has demonstrated highly significant differences between MALDR and MAHDR mice in both opioid sensitivity (Eastwood and Phillips, 2013) and intake (Eastwood and Phillips, submitted). However, any of the genes in the QTL region could be relevant. For example, a human ESR1 functional polymorphism has been associated with MA-induced psychosis (Kishi et al., 2009); mice lacking Map3k5 (aka, Ask1) exhibited hyperactivity and altered dopamine levels which would be expected to affect responses to MA (Kumakura et al., 2010); Fgd6 regulates Rho protein signal transduction and Rho-associated kinase activity has been associated with dopamine levels and behavioral effects of MA (Narita et al., 2003). Before pursuing other candidates, we are taking steps to narrow the QTL region and thus, the list of potential candidates for further study.
The current network analysis based on DE genes for three different brain regions allowed a more global analysis of interactions and identified a highly significant sub-network that is enriched in transcription factor genes. This expression study is unique in its focus on genetic risk for MA intake. However, there has been a series of studies examining initial MA sensitivity, which may (or may not) predict future use (see de Wit and Phillips, 2012). The first of these studies used an approach similar to the one used here, in that QTL and expression analyses were conducted in selected lines bred for sensitivity to a single acute administration of MA, and suggested casein kinase 1 epsilon (Csnk1e) as a candidate gene in one QTL region (Palmer et al., 2005). These results were translationally applied to a human study that obtained evidence of a role for the human CSNK1E gene region in subjective response to d-amphetamine (Veenstra-Vanderweele et al., 2006). Subsequent work has verified involvement of Csnk1e in sensitivity to both psychostimulants and opioids (Bryant et al., 2012), which is of interest given the interconnectedness of Oprm1 in the current network analysis. However, the QTL for MA sensitivity is on mouse Chr 15, whereas the major QTL for MA drinking is on Chr 10, indicating that this is not a pleiotropic effect.
Uhl et al. (2008) performed a genome-wide association study for human MA dependence and reported associations of genes involved in cell adhesion, enzymatic functions, transcription, and other cellular functions, using samples from MA dependent individuals and matched controls from Japan and Taiwan. Several genetic association studies have focused on specific genes; e.g., Oprm1 (Ide et al., 2006). Bousman et al. (2009) reviewed the genetic association studies for MA use disorders and noted that 18 genes within 38 studies identified significant associations. However, they also noted that replication, greater statistical power and development of more rigorous methods for classification and reporting were needed. Another recent paper reports the failure to replicate results for 12 candidate genes for subjective and physiological responses to amphetamine. This included CSNK1E (discussed above), and the authors drew similar conclusions about power, and also cautioned that allelic effect sizes may not be accurately predicted by genome-wide association studies (Hart et al., 2013). There are certain advantages to pursuing drug-related genetic factors in genetic animal models, including the ability to test large numbers of subjects, allowing for increased power, and the ability to control drug history. The network approach described here, as well as approaches that identify transcriptional modules and coexpression patterns (Li et al., 2005; Piechota et al., 2010; Mulligan et al., 2011; Iancu et al., 2013), attempt to identify genetic interactions and hub genes with significant effects on a relevant network that could be manipulated to produce a desired effect on the important trait. Perhaps this systems approach to genetic effects will provide a more functional application to human MA addiction.
Supplementary Material
Acknowledgments
This work was supported by NIDA grant P50 DA018165 (JKB, TJP), a grant from the Department of Veterans Affairs (TJP) and two Senior Research Career Scientist Awards (JKB, TJP) from the Department of Veterans Affairs.
References
- Belknap JK, Hitzemann R, Crabbe JC, Phillips TJ, Buck KJ, Williams RW. QTL analysis and genomewide mutagenesis in mice: complementary genetic approaches to the dissection of complex traits. Behav Genet. 2001;31:5–15. doi: 10.1023/a:1010249607128. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Mitchell SR, O’Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav Genet. 1996;26:149–160. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Richards SP, O’Toole LA, Helms ML, Phillips TJ. Short-term selective breeding as a tool for QTL mapping: Ethanol preference drinking in mice. Behav Genet. 1997;27:55–66. doi: 10.1023/a:1025615409383. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Boone EM, Hawks BW, Li W, Garlow SJ. Genetic regulation of hypothalamic cocaine and amphetamine-regulated transcript (CART) in BxD inbred mice. Brain Res. 2008;1194:1–7. doi: 10.1016/j.brainres.2007.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousman CA, Glatt SJ, Everall IP, Tsuang MT. Genetic association studies of methamphetamine use disorders: A systematic review and synthesis. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:1025–1049. doi: 10.1002/ajmg.b.30936. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. New York, NY: Springer; 2009. [Google Scholar]
- Bryant CD, Parker CC, Zhou L, Olker C, Chandrasekaran RY, Wager TT, Bolivar VJ, Loudon AS, Vitaterna MH, Turek FW, Palmer AA. Csnk1e is a genetic regulator of sensitivity to psychostimulants and opioids. Neuropsychopharmacology. 2012;37:1029–1035. doi: 10.1038/npp.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- de Wit H, Phillips TJ. Do initial responses to drugs predict future use or abuse? Neurosci Biobehav Rev. 2012;36:1565–1576. doi: 10.1016/j.neubiorev.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood EC, Phillips TJ. Opioid sensitivity in mice selectively bred to consume or not consume methamphetamine. Addict Biol. 2013 doi: 10.1111/adb.12003. epub ahead of print Nov 12 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood EC, Phillips TJ. Opioid intake and role in the acquisition of methamphetamine intake. Genes Brain Behav. doi: 10.1111/gbb.12100. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4. Essex, England: Longman Group Ltd; 1996. [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Grisel JE, Belknap JK, O’Toole LA, Helms ML, Wenger CD, Crabbe JC. Quantitative trait loci affecting methamphetamine responses in BXD recombinant inbred mouse strains. J Neurosci. 1997;17:45–54. doi: 10.1523/JNEUROSCI.17-02-00745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Altman NS, Mong JA, Klein LC, Pfaff DW, Vandenbergh DJ. Comparing quantitative trait loci and gene expression data. Adv Bioinformatics article. 2008:719818, 6. doi: 10.1155/2008/719818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AB, de Wit H, Palmer AA. Candidate gene studies of a promising intermediate phenotype: failure to replicate. Neuropsychopharmacology. 2013;38:802–816. doi: 10.1038/npp.2012.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Reed C, Malmanger B, Lawler M, Hitzemann B, Cunningham B. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcohol Clin Exp Res. 2004;28:1437–1448. doi: 10.1097/01.alc.0000139827.86749.da. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Oberbeck D, Darakjian P, Metten P, McWeeney S, Crabbe JC, Hitzemann R. Selection for drinking in the dark alters brain gene coexpression networks. Alcohol Clin Exp Res. 2013;37:1295–1303. doi: 10.1111/acer.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Kobayashi H, Ujike H, Ozaki N, Sekine Y, Inada T, Harano M, Komiyama T, Yamada M, Iyo M, Iwata N, Tanaka K, Shen H, Iwahashi K, Itokawa M, Minami M, Satoh M, Ikeda K, Sora I. Linkage disequilibrium and association with methamphetamine dependence/psychosis of mu-opioid receptor gene polymorphisms. Pharmacogenomics J. 2006;6:179–188. doi: 10.1038/sj.tpj.6500355. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Gautier L, Cope L. An R package for analysis of Affymetrix oligonucleotide arrays. In: Parmigiani RIG, Garrett ES, Ziegler S, editors. The Analysis of Gene Expression Data: Methods and Software. Springer; Berlin: 2003. pp. 102–119. [Google Scholar]
- Janowsky A, Mah C, Johnson RA, Cunningham CL, Phillips TJ, Crabbe JC, Eshleman AJ, Belknap JK. Mapping genes that regulate density of dopamine transporters and correlated behaviors in recombinant inbred mice. J Pharmacol Exp Ther. 2001;298:634–643. [PubMed] [Google Scholar]
- Kamens HM, Burkhart-Kasch S, McKinnon CS, Li N, Reed C, Phillips TJ. Sensitivity to psychostimulants in mice bred for high and low stimulation to methamphetamine. Genes Brain Behav. 2005;4:110–125. doi: 10.1111/j.1601-183X.2004.00101.x. [DOI] [PubMed] [Google Scholar]
- Keleta YB, Martinez JL. Brain circuits of methamphetamine place reinforcement learning: the role of the hippocampus-VTA Loop. Brain Behav. 2012;2:128–141. doi: 10.1002/brb3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Ikeda M, Kitajima T, Yamanouchi Y, Kinoshita Y, Kawashima K, Okochi T, Tsunoka T, Okumura T, Inada T, Ujike H, Yamada M, Uchimura N, Sora I, Iyo M, Ozaki N, Iwata N. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:895–898. doi: 10.1016/j.pnpbp.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Kumakura K, Nomura H, Toyoda T, Hashikawa K, Noguchi T, Takeda K, Ichijo H, Tsunoda M, Funatsu T, Ikegami D, Narita M, Suzuki T, Matsuki N. Hyperactivity in novel environment with increased dopamine and impaired novelty preference in apoptosis signal-regulating kinase 1 (ASK1)-deficient mice. Neurosci Res. 2010;66:313–320. doi: 10.1016/j.neures.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lebowitz RJ, Soller M, Beckmann JS. Trait-based analyses for the detection of linkage between marker loci and quantitative trait loci in crosses between inbred strains. Theor Appl Genet. 1987;73:556–562. doi: 10.1007/BF00289194. [DOI] [PubMed] [Google Scholar]
- Li H, Lu L, Manly KF, Chesler EJ, Bao L, Wang J, Zhou M, Williams RW, Cui Y. Inferring gene transcriptional modulatory relations: a genetical genomics approach. Hum Mol Genet. 2005;14:1119–1125. doi: 10.1093/hmg/ddi124. [DOI] [PubMed] [Google Scholar]
- Lin SK, Pan WH, Yeh PH. Prefrontal dopamine efflux during exposure to drug-associated contextual cues in rats with prior repeated methamphetamine. Brain Res Bull. 2007;71:365–371. doi: 10.1016/j.brainresbull.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Martin TA, Jayanthi S, McCoy MT, Brannock C, Ladenheim B, Garrett T, Lehrmann E, Becker KG, Cadet JL. Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens. PLoS One. 2012;7:e34236. doi: 10.1371/journal.pone.0034236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, Erwin VG, Belknap JK. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome. 1998;9:983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Harris RA, Ponomarev I. Molecular profiles of drinking alcohol to intoxication in C57BL/6J mice. Alcohol Clin Exp Res. 2011;35:659–670. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Takaqi M, Aoki K, Kuzumaki N, Suzuki T. Implication of Rho-associated kinase in the elevation of extracellular dopamine levels and its related behaviors induced by methamphetamine in rats. J Neurochem. 2003;86:273–282. doi: 10.1046/j.1471-4159.2003.01784.x. [DOI] [PubMed] [Google Scholar]
- Nikolsky Y, Kirillov E, Zuev R, Rakhmatulin E, Nikolskaya T. Functional analysis of OMICs data and small molecule compounds in an integrated “knowledge-based” platform. Methods Mol Biol. 2009;563:177–196. doi: 10.1007/978-1-60761-175-2_10. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Verbitsky M, Suresh R, Kamens HM, Reed CL, Li N, Burkhart-Kasch S, McKinnon CS, Belknap JK, Gilliam TC, Phillips TJ. Gene expression differences in mice divergently selected for methamphetamine sensitivity. Mammal Genome. 2005;16:291–305. doi: 10.1007/s00335-004-2451-8. [DOI] [PubMed] [Google Scholar]
- Pereira FC, Gough B, Macedo TR, Ribeiro CF, Ali SF, Binienda ZK. Buprenorphine modulates methamphetamine-induced dopamine dynamics in the rat caudate nucleus. Neurotox Res. 2011;19:94–101. doi: 10.1007/s12640-009-9143-9. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Belknap JK. Complex-trait genetics: emergence of multivariate strategies. Nat Rev Neurosci. 2002;3:478–485. doi: 10.1038/nrn847. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Broadbent J, Burkhart-Kasch S, Henderson C, Wenger CD, McMullin C, McKinnon CS, Cunningham CL. Genetic correlational analyses of ethanol reward and aversion phenotypes in short-term selected mouse lines bred for ethanol drinking or ethanol-induced conditioned taste aversion. Behav Neurosci. 2005;119:892–910. doi: 10.1037/0735-7044.119.4.892. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Dickinson S, Burkhart-Kasch S. Behavioral sensitization to drug stimulant effects in C57BL/6J and DBA/2J mice. Behav Neurosci. 1994;108:789–803. doi: 10.1037//0735-7044.108.4.789. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Huson MG, McKinnon CS. Localization of genes mediating acute and sensitized locomotor responses to cocaine in BXD/Ty recombinant inbred mice. J Neurosci. 1998;18:3023–3034. doi: 10.1523/JNEUROSCI.18-08-03023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechota M, Korostynski M, Solecki W, Gieryk A, Slezak M, Bilecki W, Ziolkowska B, Kostrzewa E, Cymerman I, Swiech L, Jaworski J, Przewlocki R. The dissection of transcriptional modules regulated by various drugs of abuse in the mouse striatum. Genome Biol. 2010;11:R48. doi: 10.1186/gb-2010-11-5-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Scibelli AC, McKinnon CS, Reed C, Burkhart-Kasch S, Li N, Baba H, Wheeler JM, Phillips TJ. Selective breeding for magnitude of methamphetamine-induced sensitization alters methamphetamine consumption. Psychopharmacology. 2011;214:791–804. doi: 10.1007/s00213-010-2086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabani S, McKinnon CS, Reed C, Cunningham CL, Phillips TJ. Sensitivity to rewarding or aversive effects of methamphetamine determines methamphetamine intake. Genes Brain Behav. 2011;10:625–636. doi: 10.1111/j.1601-183X.2011.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32(Suppl):502–508. doi: 10.1038/ng1033. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Liu QR, Johnson C, Walther D, Komiyama T, Harano M, Sekine Y, Inada T, Ozaki N, Iyo M, Iwata N, Yamada M, Sora I, Chen CK, Liu HC, Ujike H, Lin SK. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry. 2008;65:345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Qaadir A, Palmer AA, Cook EH, Jr, de Wit H. Association between the casein kinase 1 epsilon gene region and subjective response to D-amphetamine. Neuropsychopharmacology. 2006;31:1056–1063. doi: 10.1038/sj.npp.1300936. [DOI] [PubMed] [Google Scholar]
- Wheeler JM, Reed C, Burkhardt-Kasch S, Li N, Cunningham CL, Janowsky AJ, Franken FH, Wiren KM, Hashimoto JG, Scibelli AC, Phillips TJ. Genetically correlated effects of selective breeding for high and low methamphetamine consumption. Genes Brain Behav. 2009;8:758–771. doi: 10.1111/j.1601-183X.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Jung MS, Lee MJ, Yoo KH, Yook YJ, Park EY, Choi SH, Suh YJ, Kim KW, Park JH. Gene expression profiling of the rewarding effect caused by methamphetamine in the mesolimbic dopamine system. Mol Cells. 2008;26:121–130. [PubMed] [Google Scholar]
- Yeung KY, Dombek KM, Lo K, Mittler JE, Zhu J, Schadt EE, Bumgarner RE, Raftery AE. Construction of regulatory networks using expression time-series data of a genotyped population. Proc Natl Acad Sci U S A. 2011;108:19436–19441. doi: 10.1073/pnas.1116442108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen Y, Zhao N, Cao G, Dang Y, Han W, Xu M, Chen T. Distinct roles of dopamine D3 receptors in modulating methamphetamine-induced behavioral sensitization and ultrastructural plasticity in the shell of the nucleus accumbens. J Neurosci Res. 2012;90:895–904. doi: 10.1002/jnr.22821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.