Abstract
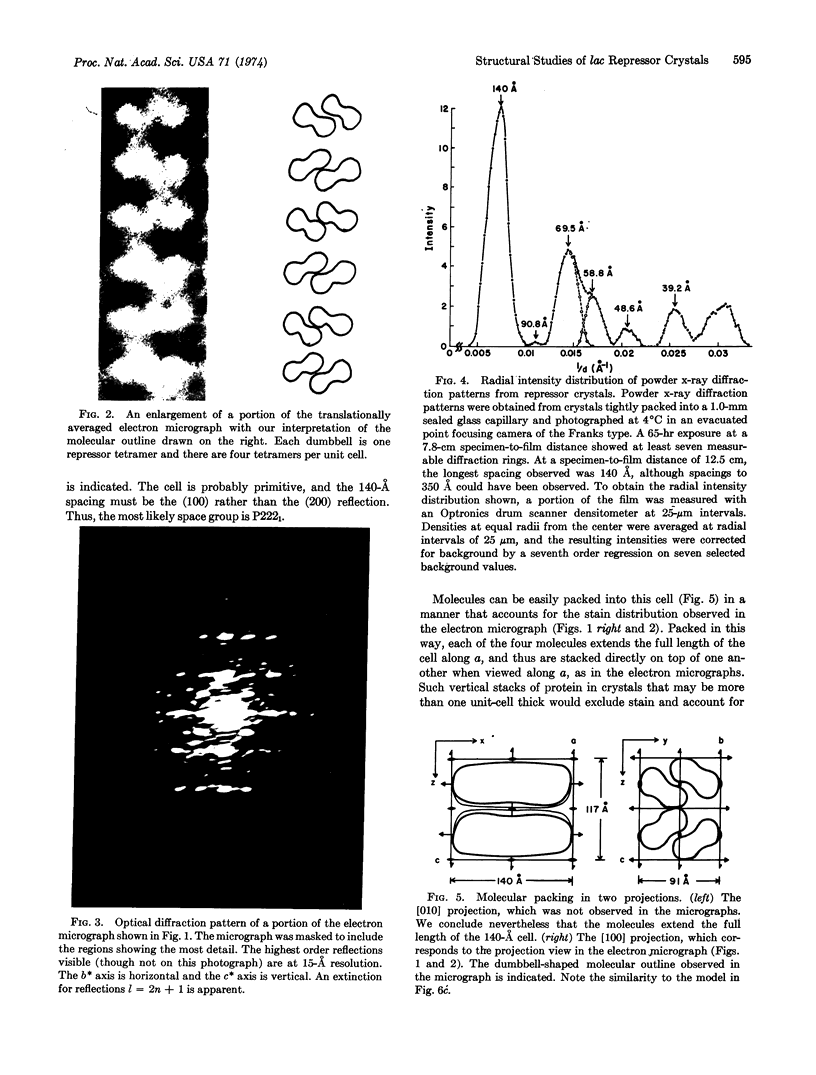
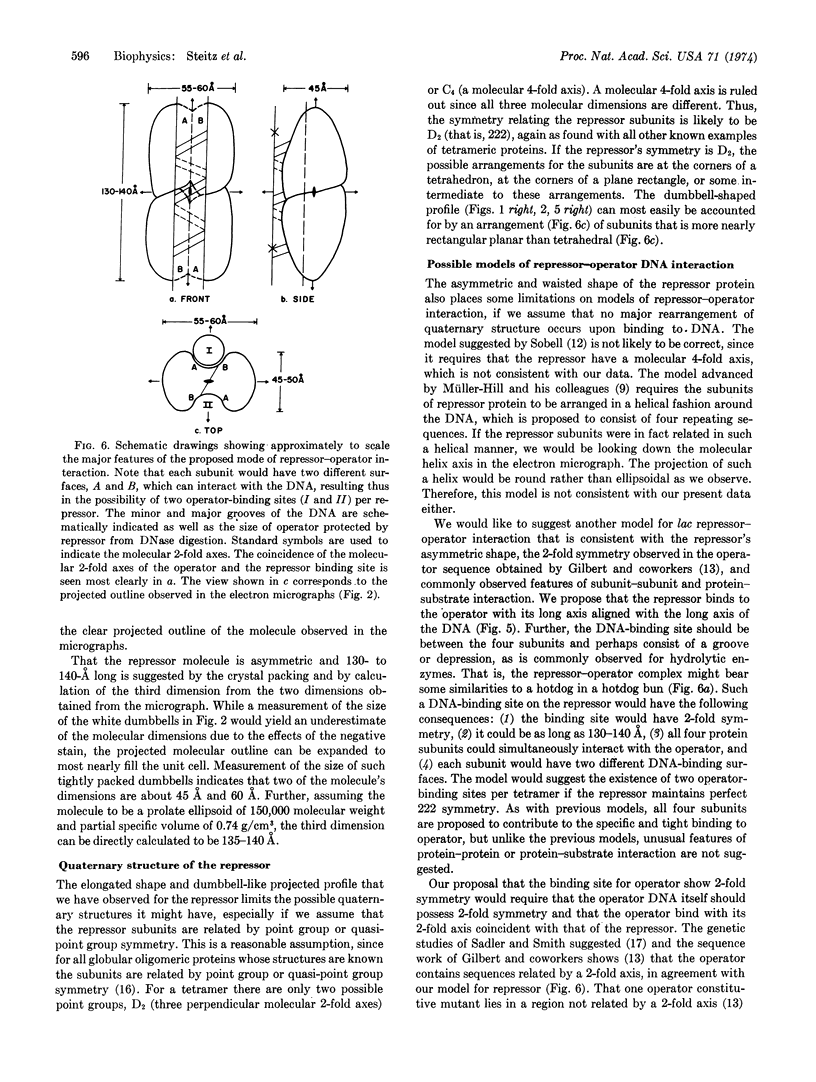
Electron microscopic and powder x-ray diffraction studies of small crystals of the lac repressor protein provide evidence on its molecular shape and subunit structure which in turn suggests a possible mode of repressor—operator interaction. The crystals are probably orthorhombic space group P2221 with unit cell dimensions of a = 140, b = 91, c = 117 Å. This tetrameric protein appears rather asymmetric, having approximate molecular dimensions of 140 Å by 60 Å by 45 Å. The dumbbell shape of the projected molecular outline observed in the electron micrographs can be explained by assuming that the subunits are related by 222 symmetry and are placed at the corners of a plane rectangle. We propose a model for repressor—operator interaction in which the DNA binds to the repressor with its long axis aligned with that of the repressor and with its 2-fold axis coincident with a twofold axis of the repressor.
Keywords: electron microscopy, x-ray diffraction, protein-DNA interaction
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler K., Beyreuther K., Fanning E., Geisler N., Gronenborn B., Klemm A., Müller-Hill B., Pfahl M., Schmitz A. How lac repressor binds to DNA. Nature. 1972 Jun 9;237(5354):322–327. doi: 10.1038/237322a0. [DOI] [PubMed] [Google Scholar]
- Crick F. General model for the chromosomes of higher organisms. Nature. 1971 Nov 5;234(5323):25–27. doi: 10.1038/234025a0. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Müller-Hill B. Isolation of the lac repressor. Proc Natl Acad Sci U S A. 1966 Dec;56(6):1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Müller-Hill B. The lac operator is DNA. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2415–2421. doi: 10.1073/pnas.58.6.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACOB F., MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961 Jun;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Labaw L. W., Davies D. R. The molecular outline of human gamma G1 immunoglobulin from an EM study of crystals. J Ultrastruct Res. 1972 Aug;40(3):349–365. doi: 10.1016/s0022-5320(72)90106-2. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J Mol Biol. 1968 Apr 28;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B. Lac repressor. Angew Chem Int Ed Engl. 1971 Mar;10(3):160–172. doi: 10.1002/anie.197101601. [DOI] [PubMed] [Google Scholar]
- Platt T., Weber K., Ganem D., Miller J. H. Translational restarts: AUG reinitiation of a lac repressor fragment. Proc Natl Acad Sci U S A. 1972 Apr;69(4):897–901. doi: 10.1073/pnas.69.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Newby R. F., Cohn M. DNA binding of the lac repressor. J Mol Biol. 1968 Jul 14;34(2):365–368. doi: 10.1016/0022-2836(68)90261-1. [DOI] [PubMed] [Google Scholar]
- Sadler J. R., Smith T. F. Mapping of the lactose operator. J Mol Biol. 1971 Nov 28;62(1):139–169. doi: 10.1016/0022-2836(71)90136-7. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. Molecular mechanism for genetic recombination. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2483–2487. doi: 10.1073/pnas.69.9.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Fletterick R. J., Hwang K. J. Structure of yeast hexokinase. II. A 6 angstrom resolution electron density map showing molecular shape and heterologous interaction of subunits. J Mol Biol. 1973 Aug 15;78(3):551–561. doi: 10.1016/0022-2836(73)90475-0. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structure of yeast hexokinase-B. I. Preliminary x-ray studies and subunit structure. J Mol Biol. 1971 Nov 14;61(3):695–700. doi: 10.1016/0022-2836(71)90073-8. [DOI] [PubMed] [Google Scholar]