Abstract
p21-activated kinases (PAKs) are effectors of RhoGTPases. PAK4 contributes to regulation of cofilin at the leading edge of migrating cells through activation of Lin-11/Isl-1/Mec-3 kinase (LIMK). PAK4 activity is regulated by an autoinhibitory domain that is released upon RhoGTPase binding as well as phosphorylation at serine 474 in the activation loop of the kinase domain. We here add another level of complexity to PAK4 regulation by showing that phosphorylation at serine residue 99 is required for its targeting to the leading edge. This phosphorylation is mediated by protein kinase D1 (PKD1). Phosphorylation of PAK4 at S99 also mediates binding to 14-3-3 protein, and is required for the formation of a PAK4/LIMK/PKD1 complex that regulates cofilin activity and directed cell migration.
Keywords: PKD, protein kinase D, p21-activated kinase 4, 14-3-3, leading edge, localization
Introduction
p21-activated kinases (PAKs) are effectors of RhoGTPases including Cdc-42, Rac1 and RhoA and play essential roles in controlling many cellular functions such as cell survival and motility [1, 2]. PAK4 together with PAK5 and PAK6 belong to the group II of PAK kinases [2]. Of these, PAK4 and PAK5 are localized at the leading edge of migrating cells, whereas PAK6 is mainly localized to the nucleus [1, 3]. At the leading edge PAK4 is a member of a multi-protein complex comprising Lin-11/Isl-1/Mec-3 kinase (LIMK), Slingshot 1L (SSH1L) and 14-3-3 proteins [4]. Activation of LIMK by PAK4 mediates phosphorylation and inactivation of cofilin, and this is part of the “cofilin cycle” that facilitates actin reorganization processes required for directed cell migration [4-7]. In many cancers PAK4 was shown to be overexpressed or mutated, and its inhibition by pharmacological means was suggested to target cancer metastases [2, 8].
PAK4 requires activation loop phosphorylation at S474 for its activity [9, 10]. Phosphorylation of this site can be mediated by autophosphorylation and by protein kinase D1 (PKD1) [10, 11]. However, S474-phosphorylated PAK4 only acquires full activity upon binding to RhoGTPases and release of its autoinhibitory domain (AID) [12]. The mechanisms by which PAK4 is directed to the leading edge however are unknown.
PKD1 is downregulated in expression or activity in invasive breast, prostate, and gastric cancer [13-15]. In response to active RhoA, PKD1 negatively regulates cofilin activity at the leading edge by phosphorylating key enzymes in both of its regulatory pathways. PKD1 phosphorylates PAK4 at S474 mediating its activation and downstream signaling to LIMK [11]. It also phosphorylates the cofilin phosphatase slingshot 1L (SSH1L) at multiple sites mediating its inactivation [16-18]. Net effect of such signaling is a shift of the cofilin pool at the leading edge to the phosphorylated inactive state, resulting in inhibition of barbed end formation and directed cell migration [19].
Here we identify a previously undescribed mechanism that is required to target PAK4 to the leading edge. We show that PKD1-mediated phosphorylation of PAK4 at serine residue 99 (S99) generates a binding site for 14-3-3 proteins. S99 phosphorylation and 14-3-3 binding is necessary for localization of PAK4 to the leading edge and mutation of this residue to an alanine leads to its retention in the cytosol.
Methods
Cell Lines, Antibodies, and Reagents
All cell lines were obtained from ATCC (Manassas, VA). Hek293T and HeLa cells were maintained in DMEM with 10% FBS. Anti-PKD1 and anti-14-3-3 antibodies were from Santa Cruz (Santa Cruz, CA), anti-PKD3 from Bethyl Laboratories (Montgomery, TX), anti-HA, anti-FLAG (M2) and anti-β-actin from Sigma-Aldrich (St. Louis, MO), anti-PKD2 and anti-HIS from Millipore (Billerica, MA), anti-pMOTIF (PKD substrate antibody), anti-LIMK, anti-PAK4, anti-pS474-PAK4, anti-cofilin, and anti-pS3-cofilin from Cell Signaling Technology (Danvers, MA). Secondary HRP-linked antibodies were from Millipore (Billerica, MA). Secondary antibodies for immunofluorescence (Alexa Fluor 568 F(ab’)2 fragment of goat-anti-mouse IgG or Alexa Fluor 546 F(ab’)2 fragment of goat-anti-rabbit) were from Invitrogen. HeLa-Monster was used for transient transfection of HeLa, and TransIT 293 for transient transfection of Hek293T cells, both from Mirus (Madison, WI). The PKD inhibitor CID755673 was from TOCRIS (Ellisville, MO) and the PAK4 inhibitor from MedKoo Biosciences (Chapel Hill, NC). Purified recombinant PKD1 was from Millipore, PKD2 and PKD3 both were from Enzo Life Sciences (Farmingdale, NY).
DNA Constructs
The expression plasmids for HA-tagged constitutively-active PKD1 (PKD1.CA, PKD1.S738E/S742E mutation) and HA-tagged kinase-inactive PKD1 (PKD1.KW, PKD1.K612W mutation) were described before [20]. The expression plasmid for FLAG-tagged kinase-inactive PKD2 (PKD2.KW, PKD2.K580W mutation), was generated by site-directed mutagenesis using a previously-described plasmid as template [21] and the following primer pair: 5′-GGCCGGGACGTGGCAGTTTGGGTCATTGACAAACTGCGC-3′ and 5′-GCGCAGTTTGTCAATGACCCAAACTGCCACGTCCCGGCC-3′. The expression plasmids for FLAG-tagged or GST-tagged (GST fusion proteins) human wildtype, K350M- and S474A-mutated PAK4 were described before [11]. To obtain S99A-mutated PAK4 site directed mutagenesis was carried out using the QuikChange kit (Stratagene, La Jolla, CA), wildtype, K350M- and/or S474A-mutated PAK4 expression constructs as template, and 5′-GTGACACGCTCCAACGCCCTGCGGAGAGACAGC-3′ and 5′-GCTGTCTCTCCGCAGGGCGTTGGAGCGTGTCAC-3′ as primers.
In Vitro Kinase Assays
Kinase assays with GST fusion proteins were carried out by adding 250 ng of active, purified PKD enzyme to 2 μg of purified GST or GST-fusion protein in a volume of 40 μl kinase buffer (50 mM Tris pH 7.4, 10 mM MgCl2 and 2 mM DTT) supplemented with 100 μM ATP (cold assay) or 100 μM ATP containing 10 μCi of [γ-32P]ATP in kinase buffer (hot assay). The kinase reaction (30 min, RT) was stopped by adding 2× Laemmli buffer.
Immunoprecipitation, PAGE, Far-Western blot and Immunoblotting
Cells were washed twice with 4 °C cold phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.2) and then lysed using lysis buffer I (50 mM Tris-HCl pH7.4, 1% Triton X-100, 150 mM NaCl, 5 mM EDTA pH 7.4) plus Protease Inhibitor Cocktail (PIC, Sigma-Aldrich, St. Louis, MO). After vortexing and incubating for 30 min on ice, lysates were centrifuged (15 min, 13,000 rpm, 4 °C). To obtain the actin fraction, pellets were resolved in lysis buffer II (50 mM Tris-HCl, pH7.6, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 1% Nonindet P-40, 10% glycerol) plus Protease Inhibitor Cocktail and boiled (details described in [17]). For all other experiments, the supernatant was collected and either subjected directly to SDS-PAGE (Western blotting) or proteins of interest were immunoprecipitated by incubating for 1 hour at 4 °C with a specific antibody (2 μg) followed by another 30 min incubation with protein G-Sepharose (Amersham Biosciences). After washing the immune-complexes 3 times with TBS (50 mM Tris-HCl pH 7.4, 150 mM NaCl), 20 μl TBS and 20 μl 2× Laemmli buffer were added and the samples were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes and visualized by immunostaining or subjected to Far-Western blotting. For Far-Western blotting, nitrocellulose membranes were blocked with 5% PhosphoBLOCKER Blocking Reagent (Cell Biolabs, San Diego, CA) in TBST (TBS + 0.2% Tween-20) for 1 hour and overlayed with HIS-tagged 14-3-3ζ protein (4 μg/ml) in 5% PhosphoBLOCKER Blocking Reagent solution for 16 hours at 4 °C. Membranes then were washed 3 times briefly with TBST and 14-3-3ζ binding to proteins was visualized by immunostaining with α-HIS antibody.
Immunofluorescence
Cells were either seeded and transfected in 8 well ibiTreat μ-Slides (ibidi, Integrated BioDiagnostics, Martinsried, Germany), or grown in 6 cm plates, transfected as indicated and re-seeded onto coverslips in a 24 well plate at a density of 2 × 104 cells per well. The following day, cells were washed with phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde (15 min, 37 °C). Following two washes with PBS, cells were permeabilized with 0.1% Triton X-100 for 10 min at room temperature (RT) and blocked with 3% bovine serum albumin and 0.05% Tween 20 in PBS (blocking solution) for 30 min at RT. The samples were then incubated with primary antibody (α-FLAG 1:4000 in blocking solution) overnight at 4 °C. Samples were washed five times with PBS and incubated with secondary antibody (goat-anti-mouse IgG Alexa Fluor 568 F(ab’)2 or goat-anti-rabbit Alexa Fluor 488 F(ab’)2 (both Invitrogen) at a dilution of 1:1600 in blocking solution for 2 hours at RT. F-actin and nuclei were stained together with secondary antibodies by incubating with phalloidin (Alexa Fluor 633-Phalloidin, Invitrogen) and DAPI (Sigma-Aldrich), respectively, in blocking solution. Following 3 washes with PBS, the coverslips were mounted onto slides using Fluoromount-G (SouthernBiotech) and examined using a IX81 DSU Spinning Disc Confocal from Olympus with a 40× objective. Images were processed using NIH ImageJ.
Analysis of Free Actin Filament Barbed Ends
Cells were transfected and reseeded on 8 well ibiTreat μ-Slides (ibidi, Integrated BioDiagnostics, Martinsried, Germany) and serum-starved for 16 hours. Then the medium was removed, cells were washed with pre-warmed PBS, permeabilized and labeled with 0.4 μM Actin-Alexa Flour594 in permeabilization buffer (20 mM HEPES, 138 mM KCl, 4 mM MgCl2, 3 mM EGTA, 0.2 mg/ml saponin, 1 % BSA) plus 1 mM ATP for 30 seconds at 37 °C. The cells were fixed with 4 % paraformaldehyde in PBS (RT, 10 min). Samples were examined using a IX81 DSU Spinning Disc Confocal from Olympus.
Impedance-based Real-time Cell Migration and Proliferation Assays
Cells were transfected as indicated and after 24 hours were seeded on Transwell CIM-plate 16 (motility assays) or E-plates (proliferation assays) from Roche (Indianapolis, IN). For proliferation assays, after attachment of cells on the E-plate, impedance was continuously monitored in real-time over a period of 16 hours using the xCELLigence RTCA DP instrument (Roche). For migration assays, after attachment of cells, cell migration towards NIH-3T3 conditioned media was continuously monitored in real-time (over 16 hours) using the xCELLigence RTCA DP instrument (Roche). Error bars (grey) represent three experiments.
Transwell Assays
Cells were transfected and after 24 hours suspended in 0.1% BSA in serum-free media and seeded into transwells containing a polycarbonate membrane with pores of 8 μm (Corning, Corning NY). After allowing cells to migrate towards NIH 3T3-conditioned media (for 16 hours), the cells on top of the transwell were removed. The remaining cells were fixed with 3.7 % formaldehyde (30 min, RT), stained with β-gal solution (40 μM K3Fe(CN)6, 40 μM K4Fe(CN)6, 16 μM MgCl2, 20 μM 5-bromo-4-chloro-indolyl-β-D-galactopyranoside (X-Gal) in PBS) and counted manually.
Statistical Analysis
Data are presented as mean ± SD. P values were acquired with the student’s t-test using Graph Pad software, and p < 0.05 is considered statistically significant.
Results
PKD1 phosphorylates PAK4 at serine residue 99
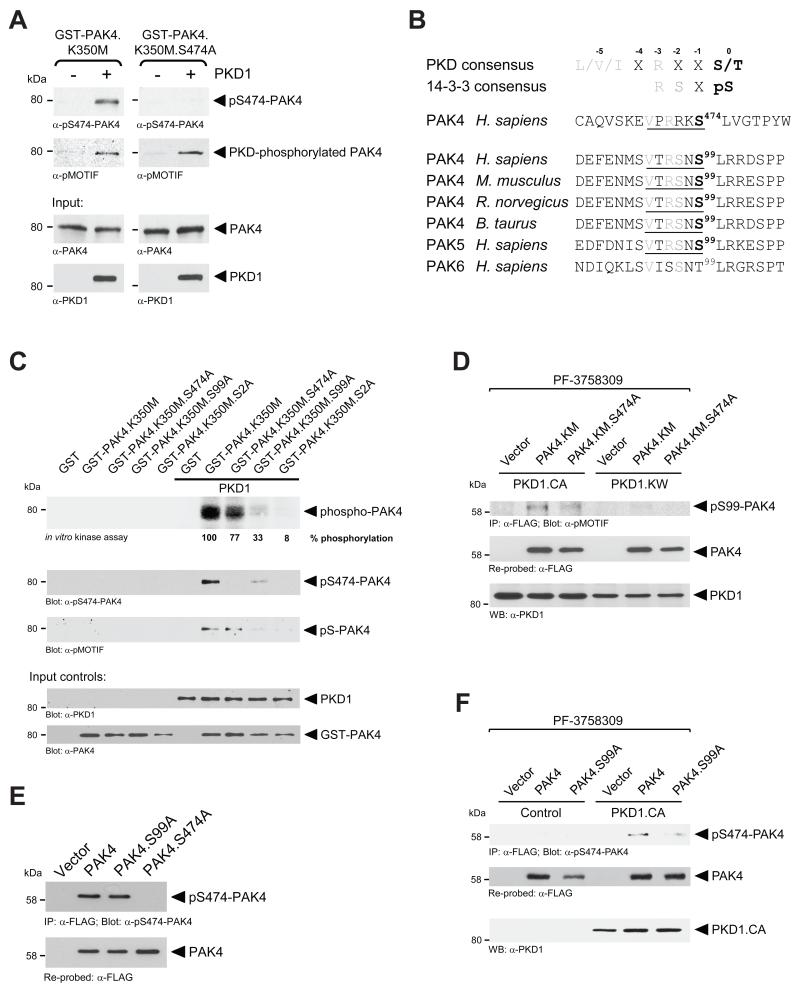
PAK4 can autophosphorylate at S474, a key residue in the activation loop of the kinase domain that contributes to activity [10]. Using a kinase-dead PAK4 (PAK4.K350M), PKD1 also was shown to directly phosphorylate this residue in vitro and in vivo [11]. In vitro kinase assays with PKD1 and GST-tagged purified kinase-dead PAK4 additionally mutated at S474 (GST-PAK4.K350M.S474A mutant) expectedly led to a loss of PAK4 phosphorylation at S474. However, probing PKD-phosphorylated GST-PAK4.K350M.S474A with the pMOTIF antibody that is designed to recognize the phosphorylated PKD consensus motif [22, 23] still picked up a GST-PAK4.K350M.S474A mutant, indicating additional PKD1 phosphorylation sites in PAK4 (Fig. 1A).
Figure 1. S99 is a novel PKD phosphorylation site on PAK4.
A In vitro kinase assays were performed with purified recombinant PKD1 and either purified GST-tagged PAK4 with only an inactivating K350M mutation (GST-PAK4.K350M) or additional mutation at S474 (GST-PAK4.K350M.S474A) as substrates. To analyze substrate phosphorylation, Western blots of resolved proteins were probed with α-pS474-PAK4 and α-pMOTIF (recognizes a phosphorylated PKD substrate motif). Additional blots (α-PAK4 and α-PKD1) were performed to control equal input of purified proteins. B. Analysis of group II p21-activated kinases (PAKs 4-6) for PKD phosphorylation sites other than S474 showed that serine residue 99 (S99) in PAK4 and PAK5 lies within a PKD1 phosphorylation consensus motif. This motif is conserved through species and its phosphorylation generates a 14-3-3 consensus motif. C. In vitro kinase assays were performed with purified recombinant PKD1 and bacterially-expressed GST-tagged kinase-dead PAK4 (GST-PAK4.K350M) additionally mutated in S99A, S474A, or both, as indicated. Top panel shows a hot kinase assay, other panels show cold kinase assays. Following the kinase reactions, samples were separated on SDS-PAGE and transferred to nitrocellulose. PKD1-mediated phosphorylation of PAK4 was determined by autoradiography (top panel) or by probing with α-pS474-PAK4 and α-pMOTIF antibodies. Additional staining of nitrocellulose blots was performed (α-PAK4 and α-PKD1) to control equal input of purified proteins. D. HeLa cells were co-transfected with vector control, FLAG-tagged PAK4.KM or PAK4.KM.S474A mutant and active PKD1 (PKD1.CA) or kinase-dead PKD1 (PKD1.KW), as indicated. Immediately after transfection, samples were treated with the PAK4 inhibitor PF-3758309 (400 nM) for 16 hours. Cells were lysed and PAK4 immunoprecipitated (α-FLAG). Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for PKD1-mediated phosphorylation of PAK4 (α-pMOTIF) and total PAK4 (α-FLAG). Additionally, lysates were analyzed by Western blotting for PKD1 expression. E. HeLa cells were transfected with vector control, FLAG-tagged PAK4, or indicated PAK4 mutants. PAK4 was immunoprecipitated from cell lysates using α-FLAG antibody. Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for PAK4 activation loop phosphorylation (with α-pS474-PAK4) and total PAK4 (α-FLAG). F. HeLa cells were co-transfected with vector control, FLAG-tagged PAK4 or PAK4.S99A mutant and vector control or active PKD1 (PKD1.CA), as indicated. Immediately after transfection, samples were treated with the PAK4 inhibitor PF-3758309 (400 nM) for 16 hours. Cells were lysed and PAK4 immunoprecipitated (α-FLAG). Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for PAK4 activation loop phosphorylation (with α-pS474-PAK4) and total PAK4 (α-FLAG). Additionally, lysates were analyzed by Western blotting for PKD1.CA expression.
Analysis of the amino-acid sequence of PAK4 indicated only one additional PKD phosphorylation consensus motif (VTRSNS99) that is conserved between species and is also present in PAK5, but not in PAK6 (Fig. 1B). Moreover, Ser99 phosphorylation of PAK4 has been previously detected by masspec [24] and was reported in phosphorylation databases such as Phosphosite (www.phosphosite.org). Therefore, we tested whether PKD1 can phosphorylate PAK4 at S99. We performed in vitro kinase assays with a series of bacterially-expressed GST-PAK4 fusion proteins encompassing kinase-dead PAK4 (K350M mutation) combined with S474A, S99A or both mutations (Fig. 1C). In vitro kinase assays using radioactive ATP as well as “cold” kinase assays and probing with pS474-PAK4 and pMOTIF antibodies suggested that PKD1 indeed can phosphorylate PAK4 at both residues, S99 and S474. For example, a kinase-dead and S474A-mutated PAK4 was still phosphorylated by PKD1 (77% of control), whereas a S99A mutant was phosphorylated significantly less (33% of control). Only mutation of both sites fully diminished PKD1-mediated phosphorylation. Probing “cold” kinase assays with pMOTIF indicated that this antibody primarily recognizes PAK4 phosphorylated at S99. Moreover, probing with pS474-PAK4 suggested that the phosphorylation at S99 may prime for PKD1-mediated phosphorylation at S474 in vitro (In Fig. 1C compare PKD1-phosphorylated GST-PAK4.K350M to PKD1-phosphorylated GST-PAK4.K350M.S99A, pS474-PAK4 blot). Of the two other PKD family members, only PKD2 is a S99 kinase, whereas PKD2 and PKD3 are S474 kinases (Supplemental Figure S1 and [11]).
We then tested if active PKD1 mediates phosphorylation of PAK4 at S99 in cells. Therefore, we expressed a kinase-dead PAK4 mutant (PAK4.KM) or a kinase-dead S474A double mutant (PAK4.KM.S474A) in combination with constitutively-active PKD1 (PKD1.CA) or kinase-dead PKD1 (PKD1.KW). Additionally, all cells were treated with the PAK4 inhibitor PF-3758309 [25] to block phosphorylations by endogenous PAK4. We found that active PKD1 phosphorylated PAK4.KM as well as PAK4.KM.S474A, but to a less extent. A kinase-dead PKD1 blocked both phosphorylations. This indicates that PKD1 can contribute to phosphorylation of both residues in cells (Fig. 1D).
Next we tested whether S99 phosphorylation can prime for S474 phosphorylation in cells. Therefore, we first expressed PAK4, PAK4.S99A, or PAK4.S474A in HeLa cells and probed for S474 phosphorylation. However, a PAK4.S99A mutant showed phosphorylation at S474 at a level comparable to wildtype PAK4 (Fig. 1E). This may be explained by previous reports demonstrating a high autophosphorylation activity of PAK4 towards this residue [10]. We therefore applied the PAK4 inhibitor PF-3758309 to dampen PAK4 autophosphorylation. Under such conditions, constitutively-active PKD1 (PKD1.CA) induced PAK4 phosphorylation at S474 in wildtype PAK4 but significantly less in the PAK4.S99A mutant (Fig. 1F). In summary, our data indicate that S99 phosphorylation to some extent can prime for PKD1-mediated S474 phosphorylation but is not necessary for PAK4 autophosphorylation at this serine residue.
S99 is necessary for the localization of PAK4 to the leading edge
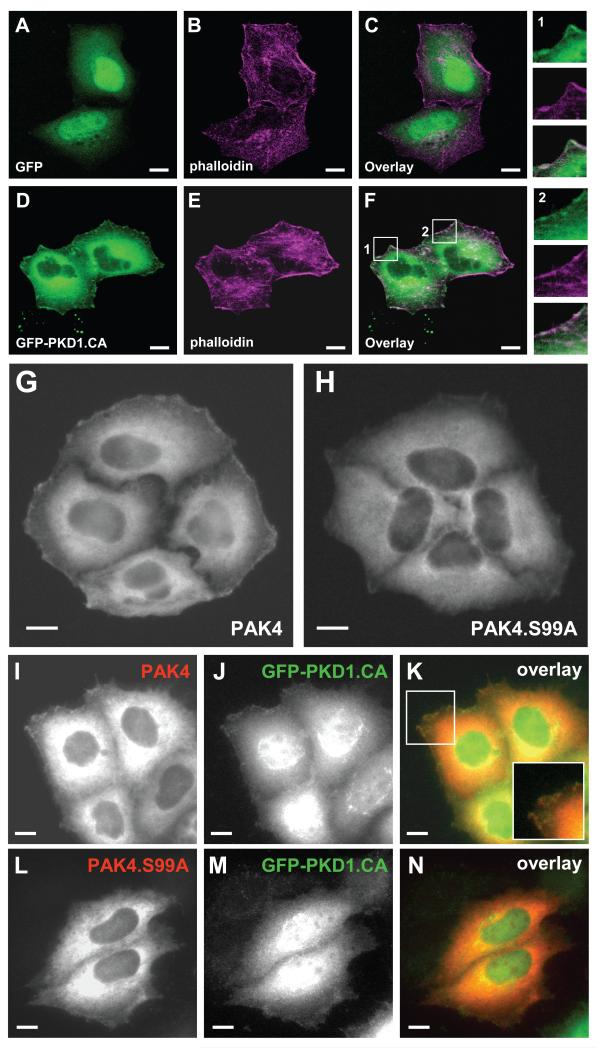
The group II PAK kinases PAK4 and PAK5 which contain the S99 phosphorylation motif locate to the leading edge, whereas PAK6 which does not contain this motif is a nuclear protein (Supplemental Figure S2). This prompted us to test if the phosphorylation of this residue by PKD1 can affect the cellular localization of PAK4. In HeLa cells endogenous and overexpressed PKD1 is localized to the leading edge where it co-localizes with F-actin filaments [13, 17]. Similarly, a GFP-tagged version of the active PKD1 protein co-localized with F-actin filaments at the leading edge (Fig. 2 D-F). Further, PAK4 localized to the leading edge of cells (Fig. 2G), but a PAK4.S99A mutant was retained in the cytosol (Fig. 2H). Interestingly, the localization of active PKD1 to the leading edge was linked to PAK4 phosphorylation at S99, since wildtype PAK4 co-localized with active PKD1 at the leading edge (Fig. 2 I-K) and a PAK4.S99A mutant led to loss of PKD1 localization to the leading edge (Fig. 2 L-N). This suggests that the translocation of both proteins to the leading edge is coupled to the PAK4 phosphorylation status at S99.
Figure 2. S99 is necessary for the localization of PAK4 to the leading edge.
A-F. HeLa cells were transfected with GFP vector control (A-C) or GFP-PKD1.CA (D-F). 24 hours after transfection, cells were fixed and F-actin was stained with phalloidin. In the overlay in F, two areas are enhanced (1, 2). Both show areas of PKD1.CA and F-actin co-localization at the leading edge of cells. The bar represents 10 μm. G-N. HeLa cells were transfected with either wild-type FLAG-tagged PAK4 (PAK4) or FLAG-tagged PAK4.S99A and either empty vector (G, H) or GFP-tagged constitutively-active PKD1 (PKD1.CA) (I-N). Samples were subjected to immunofluorescence analysis in which PAK4 was detected by using α-FLAG antibody and Alexa Fluor 568 as secondary antibody. Scale bars indicate 10 μm.
S99 phosphorylation mediates binding of PAK4 to 14-3-3 proteins
Phosphorylation of proteins in the PKD substrate motif has been shown to generate a binding site for 14-3-3 proteins [17, 26-29]. In PAK4 phosphorylation of S99 with RSNpS99 generates an ideal (RSXpS) 14-3-3 binding motif (Fig. 1B and [30]).
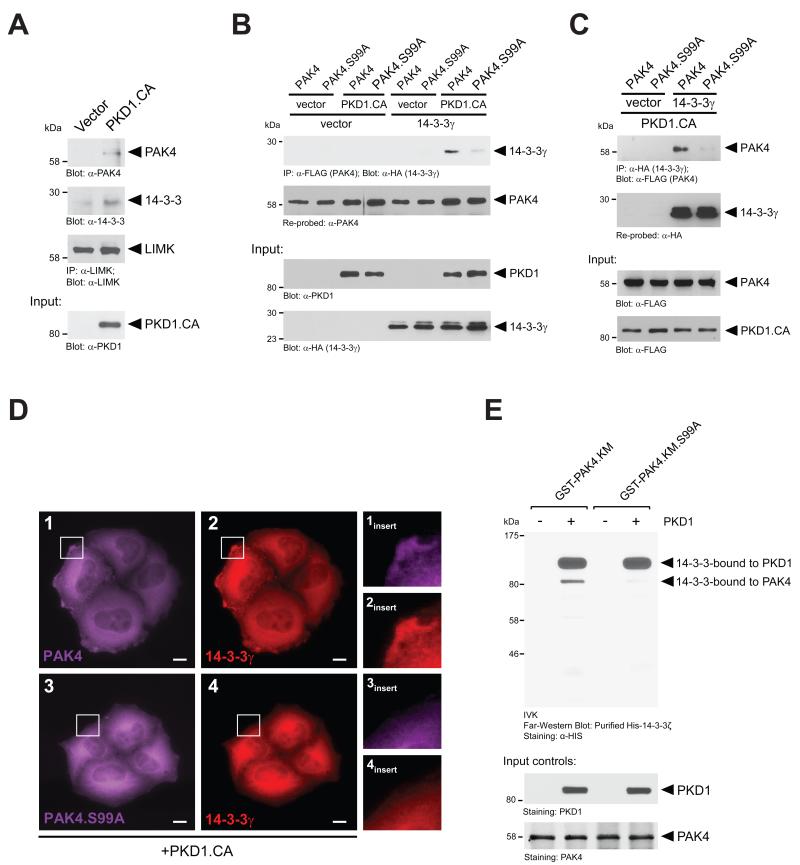
Previously, a multi-protein complex consisting of PAK4, actin, and 14-3-3 scaffolding proteins along with the PAK4 substrate LIMK was shown to regulate ADF/cofilin activity at the leading edge [4]. Therefore, we determined if the formation of such a complex is dependent on active PKD1. To test this, we introduced a constitutively-active PKD1 into cells, immunoprecipitated endogenous LIMK and probed samples for co-immunoprecipitated endogenous PAK4 and 14-3-3. We found that both proteins associated with LIMK in presence of active PKD1 (Fig. 3A). Since 14-3-3 scaffold proteins may function as the ‘connectors’ between signaling proteins of this complex, we next tested if PAK4, when phosphorylated by PKD1, can bind to 14-3-3 proteins and if this is dependent on S99 phosphorylation. We found that active PKD1 induces the interaction of PAK4 with 14-3-3γ, and this is blocked when PAK4 is mutated at S99. These effects were obtained when PAK4 or PAK4.S99A were immunoprecipitated and samples probed for co-immunoprecipitated 14-3-3γ (Fig. 3B), but also vice versa when 14-3-3γ was immunoprecipitated and samples probed for PAK4 or PAK4.S99A mutant (Fig. 3C). Moreover, in presence of active PKD1, both PAK4 and 14-3-3γ co-localized at the leading edge, whereas when a PAK4.S99A mutant was combined with 14-3-3γ both did not localize to the leading edge (Fig. 3D). The effects observed were not 14-3-3 isoform specific since similar results were obtained when 14-3-3ζ was expressed instead of the γ isoform (Supplemental Figure S3 and not shown).
Figure 3. S99 phosphorylation mediates binding of PAK4 to 14-3-3 proteins.
A. Hek293T cells were transfected with HA-tagged constitutively-active PKD1 (PKD1.CA). Endogenous LIMK was immunoprecipitated (α-LIMK). Following SDS-PAGE and transfer to nitrocellulose, samples were analyzed for co-immunoprecipitated endogenous PAK4 (α-PAK4) or 14-3-3 (α-14-3-3) and then probed for LIMK (α-LIMK). Additionally, lysates were analyzed by Western blotting for PKD1.CA expression (input control). B. Hek293T cells were co-transfected with FLAG-tagged PAK4 or PAK4.S99A mutant combined with vector or HA-tagged 14-3-3γ and vector or constitutively-active PKD1 (PKD1.CA), as indicated. PAK4 was immunoprecipitated from cell lysates using α-FLAG antibody. Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for co-immunoprecipitated 14-3-3γ (α-HA antibody). Samples were re-probed for total PAK4 (α-PAK4). Additionally, lysates were analyzed by Western blotting for PKD1 expression (α-PKD1) and 14-3-3γ (α-HA) expression. C. Hek293T cells were co-transfected with FLAG-tagged PAK4 or PAK4.S99A mutant combined with vector or HA-tagged 14-3-3γ and constitutively-active PKD1 (PKD1.CA), as indicated. 14-3-3γ was immunoprecipitated from cell lysates using α-HA antibody. Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for co-immunoprecipitated PAK4 (α-FLAG antibody). Samples were re-probed for total 14-3-3γ (α-HA). Additionally, lysates were analyzed by Western blotting for PKD1 expression (α-FLAG) and PAK4 (α-FLAG) expression. D. HeLa cells were co-transfected with FLAG-tagged PAK4 or PAK4.S99A mutant, HA-tagged 14-3-3γ and constitutively-active PKD1 (PKD1.CA), as indicated. Samples were subjected to immunofluorescence analysis in which PAK4 was detected by using α-FLAG antibody and Alexa Fluor 647 as secondary antibody and 14-3-3α was detected using an α-HA antibody and Alexa Fluor 568 as secondary antibody. E. In vitro kinase assays were performed with purified recombinant PKD1 and bacterially-expressed GST-tagged kinase-dead PAK4 (GST-PAK4.K350M), or GST-tagged kinase-dead PAK4 additionally mutated at S99A (GST-PAK4.K350M.S99A), as indicated. Following the kinase reactions, samples were separated on SDS-PAGE and transferred to nitrocellulose. A Far-Western blot was performed using recombinant HIS-tagged 14-3-3ζ. 14-3-3 protein bound to PKD1 and PAK4 was determined by immunostaining with anti-HIS antibody. Additional staining was performed to control equal input of purified PAK4 or PKD1 proteins.
In order to test if 14-3-3 directly binds to PAK4 phosphorylated at S99, we performed an in vitro kinase assay, in which we phosphorylated wildtype PAK4 or the PAK4.S99A mutant with recombinant PKD1. We then performed a Far-Western blot to detect binding of recombinant 14-3-3. We found that 14-3-3 can directly bind to PAK4, when phosphorylated by PKD1 (Fig. 3E). Recombinant 14-3-3 also bound to autophosphorylated PKD1, as previously shown by Hausser et al [31].
S99 phosphorylation affects cell motility
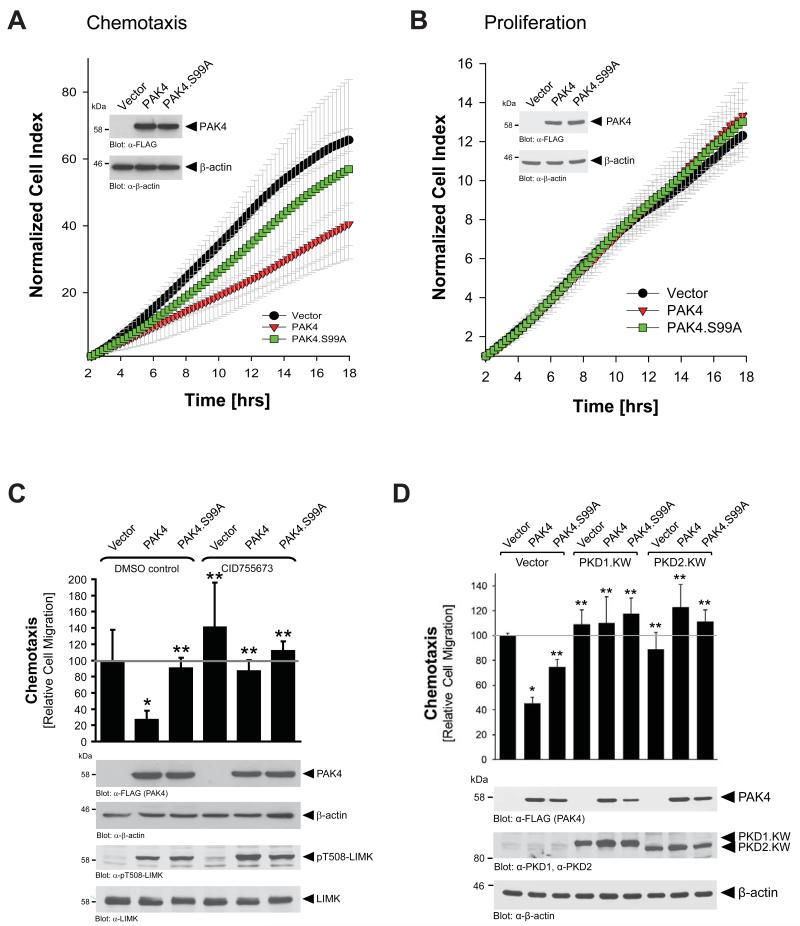
At the leading edge PAK4 has been shown to activate LIMK, but also to inactivate SSH1L [4]. Net effect of such signaling is an increase in (inactive) pS3-cofilin and decreased directed cell migration [11]. Consequently, ectopic introduction of wildtype PAK4 decreased directed cell migration, whereas a PAK4.S99A mutant that does not localize to the leading edge did not significantly decrease directed cell migration when compared with the vector control (Fig. 4A). Effects observed were not caused by differences in cell proliferation, since neither ectopic expression of wildtype PAK4, nor the PAK4.S99A mutant had an impact on cell proliferation in the time period of analysis (Fig. 4B). The calculated doubling of the cell index in the logarithmic growth phase were 5.2 −/+ 0.24 hours for vector control, 4.84 −/+ 0.19 hours for PAK4 wildtype and 4.97 −/+ 0.21 hours for PAK4.S99A mutant. Taken together, these data suggest that blocking PAK4 localization to the leading edge can favor directed cell migration. The negative effects of wildtype PAK4 on directed cell migration are blocked when cells are treated with the PKD inhibitor CID755673 (Fig. 4C). Inhibiting PKD1 led to a reversion of PAK4-mediated inhibitory effects on cell migration (compare PAK4 expressing cells in absence and presence of CID755673). These results are also consistent with previous reports showing that introduction of active PKD1 into motile cells decreases directed cell migration by inhibiting cofilin-induced free actin barbed end formation (Supplemental Figures S4, S5 and [11, 17, 18]). Since both PKD isoforms PKD1 and PKD2 can phosphorylate PAK4 at S99, we next tested if kinase-dead versions of these kinases can block PAK4-mediated decrease in cell migration. We found that both mutants, PKD1.KW and PKD2.KW fully restored cell migration (Fig. 4D), indicating that both kinases are redundant in their function towards PAK4.
Figure 4. S99 phosphorylation affects cell motility, and not cell proliferation.
A, B. HeLa cells were transfected with control vector, wildtype PAK4 or PAK4.S99A mutant, and reseeded in Transwell CIM-plate 16 plates (chemotaxis assays, A), or E-plates (proliferation assays, B). After attachment of cells, cell migration or proliferation was continuously monitored in real-time using the xCELLigence RTCA DP instrument. Error bars (grey) represent five experiments. Protein expression was controlled by Western blot (α-FLAG for PAK4 expression, α-β-actin for loading control). C, D. HeLa cells were transfected with control vector, wildtype PAK4 or PAK4.S99A mutant, and treated with either DMSO (control) or the PKD inhibitor CID755673 (20 μM, during time of the assay) as indicated (C), or were additionally transfected with PKD1.KW or PKD2.KW as indicated (D). Transwell assays were performed to assess cell migration towards NIH-3T3-conditioned media at 16 hours. In C: Protein expression was controlled by Western blot (α-FLAG for PAK4 expression, α-β-actin for loading control). Additionally, samples were analyzed by Western blot for expression and activity of LIMK (α-LIMK, α-pT508-LIMK). In D: Protein expression was controlled by Western blot for PAK4 (α-FLAG), PKD1 (α-PKD1), PKD2 (α-PKD2), or β-actin (for loading control). * indicates statistical significance as compared to vector control (first bar), ** indicates statistical significance as compared to wildtype PAK4 (second bar).
S99 phosphorylation affects PAK4 localization and downstream signaling at the leading edge
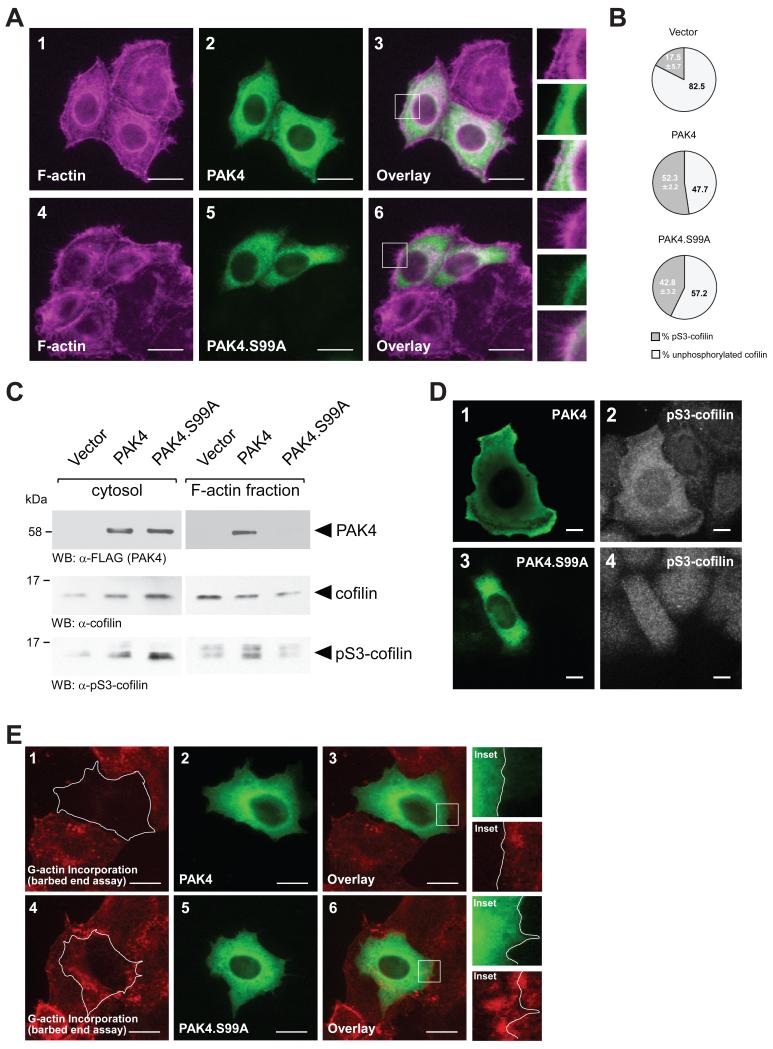
Since the PAK4.S99A mutant did not localize to the cell periphery (Fig. 3D), we next analyzed if this affects downstream signaling towards cofilin at the leading edge. First, we determined F-actin structures in cells expressing wildtype PAK4 or PAK4.S99A. Cells expressing the S99A mutant showed a more motile phenotype as judged by organization of the F-actin structures (Fig. 5A) and this correlated with our findings on directed cell migration (Fig. 4A). However, when we quantitated cofilin phosphorylation in cells expressing vector control, PAK4 or PAK4.S99A, we found only marginal differences between the two versions of PAK4 (Fig. 5B). In control transfected cells 17.5 −/+ 5.7 percent of the total cofilin was phosphorylated at S3. In cells expressing wildtype PAK4, cofilin phosphorylation was at 52.3 −/+ 2.2 percent, and in cells expressing PAK4.S99A we detected slightly-decreased phospho-cofilin levels at 42.8 −/+ 3.2 percent. This correlated with data on LIMK activity shown in Fig. 4C, and indicates that the S99A mutated form of the kinase is functional. However, when we isolated the cytosolic and F-actin fractions, we found that while PAK4 occurs in both fractions, PAK4.S99A is exclusively localized to the cytosol (Fig. 5C). Moreover, localization of PAK4 or mutant correlated with cofilin phosphorylation. This indicates that the mechanism by which S99 phosphorylation affects PAK4 function is that it regulates its function at the lamellipodium. Indeed, expression of wildtype PAK4 led to increased cofilin phosphorylation at the lamellipodia and cytosol (Fig. 5D, top row), whereas expression of a PAK4.S99A mutant that is not localized at the leading edge also led to cofilin phosphorylation but only in the cytosol (Fig. 5D, bottom row). This was further confirmed by performing barbed end formation (G-actin incorporation) assays, in which we show that PAK4 completely blocks barbed-end formation, whereas cells expressing PAK4.S99A can still form barbed ends and show G-actin incorporation (Fig. 5E). Taken together, this indicates that the ability of PAK4 to induce cofilin phosphorylation at S3 is not affected by its mutation at S99 but that alone the lack of localization to the leading edge may be responsible for its effects on directed cell migration.
Figure 5. Loss of S99 phosphorylation decreases cofilin phosphorylation at the leading edge.
A. HeLa cells were transfected with wildtype PAK4 or PAK4.S99A mutant. 24 hours after transfection, cells were fixed and PAK4 was stained by immunofluorescence (α-FLAG). F-actin was stained with phalloidin. The insert shows an area at the leading edge of cells. The bar represents 20 μm. B. HeLa cells were transfected with control vector, wildtype PAK4 or PAK4.S99A mutant. Total cell lysates (n=5 experiments) were analyzed by Western blot for cofilin phosphorylation (α-pS3-cofilin antibody) or total cofilin (α-cofilin). Percentage of S3-phosphorylated cofilin and unphosphorylated cofilin was calculated and depicted in a pie graph. C. HeLa cells were transfected with control vector, wildtype PAK4 or PAK4.S99A mutant. Cytosolic and F-actin fractions were prepared and analyzed by Western blot for presence of PAK4 (α-FLAG), or presence of endogenous cofilin or pS3-cofilin. D. HeLa cells were transfected with wildtype PAK4 or PAK4.S99A mutant as indicated. Samples were subjected to immunofluorescence analysis in which PAK4 was detected by using α-FLAG antibody and Alexa Fluor 488 as secondary antibody and endogenous pS3-cofilin by using α-pS3-cofilin antibody and Alexa Fluor 546 as secondary antibody. E. Free barbed end-induced actin incorporation at the leading edge is blocked when PAK4 is overexpressed, but not when PAK4.S99A is overexpressed. HeLa cells (5 × 104 cells, 8 well ibiTreat μ-slide) were transfected as indicated. To analyze cofilin-mediated actin incorporation F-actin free barbed ends were performed as described in materials and methods. PAK4-expressing cells were stained by immunofluorescence using primary antibodies directed against their tag as well as Alexa Fluor 488 as secondary antibody. Bar is 20 μm.
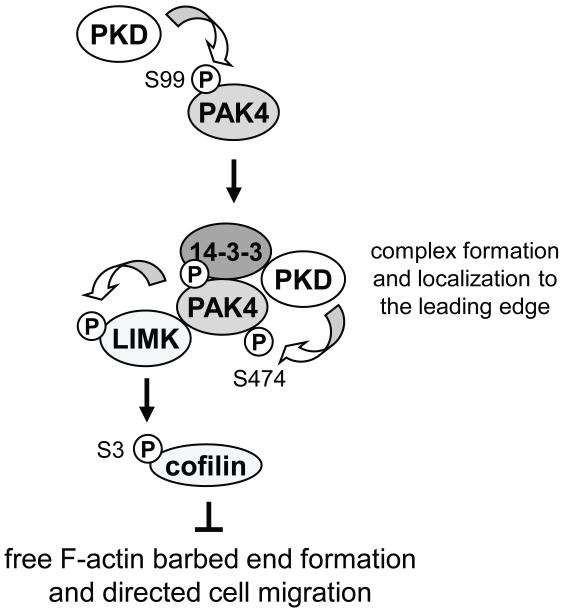
In summary, with S99 we describe a novel PKD1 phosphorylation site in PAK4 that regulates PAK4 localization and complex formation with LIMK and 14-3-3 proteins. We suggest that lack of phosphorylation of PAK4 at S99 prevents inactivation of cofilin at the leading edge, thus affecting directed cell migration (Fig. 6).
Figure 6. Scheme of how S99 phosphorylation of PAK4 is mediated and may contribute to directed cell migration.
S99A phosphorylation of PAK4 is mediated by PKD1. This leads to complex formation of PAK4 with 14-3-3, LIMK, and active PKD1 at the leading edge, allowing activation loop phosphorylation of PAK4 at S474 and activation of its substrate LIMK. LIMK then phosphorylates and inactivates cofilin at S3, blocking actin barbed end formation at the leading edge and directed cell migration.
Discussion
The group II p21-activated kinases are effectors of RhoGTPases and play essential roles in regulating cell motility [1, 2]. Similarly, Protein Kinase D isoforms are regulated by RhoGTPases [17, 32-34]. We have previously shown that PKD1, downstream of RhoA, can directly phosphorylate PAK4 at serine residue 474 (S474) in its activation loop, increasing its activity towards its substrate LIMK [11]. Functional consequences of such signaling are an increase of the phospho-cofilin pool and decreased actin reorganization at the leading edge and directed cell migration [11]. Here, with serine residue 99 we identify an additional PKD1 phosphorylation site in PAK4 that contributes to directed cell migration.
After blocking S99 phosphorylation with a mutational approach (S to A mutation), we found that the presence of S99 phosphorylation had a slight but reproducible effect on PKD1-mediated activation loop phosphorylation at S474 in vitro and in vivo (Fig. 1C, 1F). But this was only observed when PAK4 autophosphorylation was inhibited either by using kinase-dead PAK4 mutants or PF-3758309 (PAK4 inhibitor). However, in cells with PAK4 autophosphorylation activity, phosphorylation at S474 most likely is independent of PKD1-mediated phosphorylation of S99 (Fig. 1E).
Of the group II PAK enzymes, only PAK4 and PAK5, but not PAK6 contain a PKD-consensus motif surrounding S99 (Fig. 1B). As PAK4 and PAK5 are localized at the leading edge of migrating cells, and PAK6 is mainly localized to compartments in the nucleus [Supplemental Figure S2 and [1, 3]], we tested if S99 phosphorylation is required to translocate PAK4 to the leading edge. We found loss of PAK4 at the leading edge when S99 cannot be phosphorylated (Fig. 2). This translated to altered cell migration. Overexpression of wildtype PAK4 decreased directed cell migration, whereas a S99A mutant that was not localized at the leading edge showed migration behavior similar to wildtype cells (Figs. 4A, 4C, 4D).
Actin reorganization at the leading edge, a key process in directed cell migration, is facilitated by the cofilin cycle [35]. PKD1 can regulate activities of both LIMK and SSH1L, thereby being a key regulator of cofilin activity [11, 17, 18, 36]. Activation of LIMK by PAK4 mediates phosphorylation and inactivation of cofilin [4-7]. Although showing significant differences in their effects on cell migration, the expression of PAK4.S99A similar as it was described for wildtype PAK4, increased cofilin phosphorylation (Fig. 5B). This is not surprising since residue S99 is neither located in the p21-Rho-binding domain (amino acids 10-66), nor in the protein kinase domain (amino acids 322-572), suggesting that binding to RhoGTPases still can occur and that the kinase activity is intact. However, we found that both PAK4 and PAK4.S99A localized in the cytosol, but that only wildtype PAK4 localized to the F-actin fraction (Fig. 5C). In addition we found that cofilin in PAK4.S99A expressing cells is mainly phosphorylated in the cytosol and that in wildtype PAK4 expressing cells phosphorylation occurs in the cytosol and the F-actin fraction. Consequently, expression of wildtype PAK4 led to increased cofilin phosphorylation at the lamellipodia and cytosol, whereas expression of a PAK4.S99A mutant that is not localized at the leading edge also led to cofilin phosphorylation but only in the cytosol (Fig. 5D). To demonstrate decreased cofilin activity when cells express PAK4, we performed G-actin incorporation (barbed end) assays. Cofilin-initiated G-actin incorporation at the cell periphery was blocked when cells were expressing wildtype PAK4, but not when PAK4.S99A was expressed (Fig. 5E). This indicates that the ability of PAK4 to induce cofilin phosphorylation at S3 is not affected by its mutation at S99 but that alone the lack of localization to the leading edge may be responsible for its effects on directed cell migration.
Phosphorylation of the PKD substrate motifs can generate a 14-3-3 protein consensus motif (Fig. 1B). For example, PKD1-mediated phosphorylation of SSH1L at S978 has been shown to lead to binding of 14-3-3 [17]. Similarly, PKD1 phosphorylation of PAK4 at S99 created a 14-3-3 binding motif (Figs. 3B, 3C) and mediated direct binding of 14-3-3 in Far-Western assays (Fig. 3E). Although both PKD1-phosphorylated PAK4 and PKD1 directly bind 14-3-3, this does not prove the formation of a complex between these three proteins. It was shown previously that PAK4 at the leading edge, along with LIMK, interacts with SSH1L and 14-3-3 to form a multi-protein complex which regulates the activity of cofilin [4]. We found that PKD1 may contribute to such complex formation, since both PAK4 and 14-3-3 associated with LIMK in presence of active PKD1 (Fig. 3A). The mechanisms by which such a complex is formed are not yet fully elucidated. One possibility of how 14-3-3 proteins could contribute to complex formation is due to their ability to form dimers or multimers [37]. Moreover, the binding of 14-3-3 proteins to PKD1, as well as to LIMK1-phosphorylated cofilin has been described previously [31, 38], suggesting that 14-3-3 may function as scaffold to keep this complex at the leading edge. However, our data do not exclude additional levels of regulation of the PAK4/SSH1L/LIMK complex through 14-3-3 proteins or other scaffolding proteins.
To conclude, in this study we identified an additional mechanism by which PAK4 function and directed cell migration can be regulated. We describe PKD1-mediated phosphorylation of S99 as a requirement for complex formation of PAK4 with 14-3-3 proteins, a mechanism that targets a PAK4/LIMK complex to the leading edge and facilitates cofilin phosphorylation at this location (Fig. 6).
Supplementary Material
Acknowledgements
This work was supported by a Bankhead-Coley grant (10BG11) from the Florida Department of Health and the NIH grants GM86435 and CA140182, all to PS.
Abbreviations
- LIMK
Lin-11/Isl-1/Mec-3 kinase
- PAK4
p21-activated kinase 4
- PKD
protein kinase D
- RT
room temperature
- SSH
slingshot
Footnotes
Author Contributions
Conceived and designed the experiments: LIB, HD, SEP, PS. Performed the experiments: LIB, HD, SEP, ND, SJS. Analyzed the data: LIB, HD, SEP, SJS, PS. Wrote the paper: PS.
References
- 1.Bokoch GM. Biology of the p21-activated kinases. Annu. Rev. Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 2.Eswaran J, Soundararajan M, Knapp S. Targeting group II PAKs in cancer and metastasis. Cancer Metastasis Rev. 2009;28:209–217. doi: 10.1007/s10555-008-9181-4. [DOI] [PubMed] [Google Scholar]
- 3.Yang F, Li X, Sharma M, Zarnegar M, Lim B, Sun Z. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J. Biol. Chem. 2001;276:15345–15353. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- 4.Soosairajah J, Maiti S, Wiggan O, Sarmiere P, Moussi N, Sarcevic B, Sampath R, Bamburg JR, Bernard O. Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. EMBO J. 2005;24:473–486. doi: 10.1038/sj.emboj.7600543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 6.Mouneimne G, DesMarais V, Sidani M, Scemes E, Wang W, Song X, Eddy R, Condeelis J. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 2006;16:2193–2205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Oser M, Condeelis J. The cofilin activity cycle in lamellipodia and invadopodia. J. Cell. Biochem. 2009;108:1252–1262. doi: 10.1002/jcb.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whale AD, Dart A, Holt M, Jones GE, Wells CM. PAK4 kinase activity and somatic mutation promote carcinoma cell motility and influence inhibitor sensitivity. Oncogene. 2013;32:2114–2120. doi: 10.1038/onc.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu J, Cammarano MS, Shi Q, Ha KC, de Lanerolle P, Minden A. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol. Cell. Biol. 2001;21:3523–3533. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callow MG, Clairvoyant F, Zhu S, Schryver B, Whyte DB, Bischoff JR, Jallal B, Smeal T. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J. Biol. Chem. 2002;277:550–558. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- 11.Spratley SJ, Bastea LI, Doppler H, Mizuno K, Storz P. Protein kinase D regulates cofilin activity through p21-activated kinase 4. J. Biol. Chem. 2011;286:34254–34261. doi: 10.1074/jbc.M111.259424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baskaran Y, Ng YW, Selamat W, Ling FT, Manser E. Group I and II mammalian PAKs have different modes of activation by Cdc42. EMBO Rep. 2012;13:653–659. doi: 10.1038/embor.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eiseler T, Doppler H, Yan IK, Goodison S, Storz P. Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 2009;11:R13. doi: 10.1186/bcr2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M, Jang HR, Kim JH, Noh SM, Song KS, Cho JS, Jeong HY, Norman JC, Caswell PT, Kang GH, Kim SY, Yoo HS, Kim YS. Epigenetic inactivation of protein kinase D1 in gastric cancer and its role in gastric cancer cell migration and invasion. Carcinogenesis. 2008;29:629–637. doi: 10.1093/carcin/bgm291. [DOI] [PubMed] [Google Scholar]
- 15.Mak P, Jaggi M, Syed V, Chauhan SC, Hassan S, Biswas H, Balaji KC. Protein kinase D1 (PKD1) influences androgen receptor (AR) function in prostate cancer cells. Biochem. Biophys. Res. Commun. 2008;373:618–623. doi: 10.1016/j.bbrc.2008.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barisic S, Nagel AC, Franz-Wachtel M, Macek B, Preiss A, Link G, Maier D, Hausser A. Phosphorylation of Ser 402 impedes phosphatase activity of slingshot 1. EMBO Rep. 2011;12:527–533. doi: 10.1038/embor.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eiseler T, Doppler H, Yan IK, Kitatani K, Mizuno K, Storz P. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat. Cell Biol. 2009;11:545–556. doi: 10.1038/ncb1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 2009;69:5634–5638. doi: 10.1158/0008-5472.CAN-09-0718. [DOI] [PubMed] [Google Scholar]
- 19.Storz P. Protein kinase D1: a novel regulator of actin-driven directed cell migration. Cell Cycle. 2009;8:1975–1976. [PubMed] [Google Scholar]
- 20.Storz P, Doppler H, Johannes FJ, Toker A. Tyrosine phosphorylation of protein kinase D in the pleckstrin homology domain leads to activation. J. Biol. Chem. 2003;278:17969–17976. doi: 10.1074/jbc.M213224200. [DOI] [PubMed] [Google Scholar]
- 21.Sturany S, Van Lint J, Muller F, Wilda M, Hameister H, Hocker M, Brey A, Gern U, Vandenheede J, Gress T, Adler G, Seufferlein T. Molecular cloning and characterization of the human protein kinase D2. A novel member of the protein kinase D family of serine threonine kinases. J. Biol. Chem. 2001;276:3310–3318. doi: 10.1074/jbc.M008719200. [DOI] [PubMed] [Google Scholar]
- 22.Doppler H, Storz P, Li J, Comb MJ, Toker A. A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J. Biol. Chem. 2005;280:15013–15019. doi: 10.1074/jbc.C400575200. [DOI] [PubMed] [Google Scholar]
- 23.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 24.Wissing J, Jansch L, Nimtz M, Dieterich G, Hornberger R, Keri G, Wehland J, Daub H. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol. Cell. Proteomics. 2007;6:537–547. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Murray BW, Guo C, Piraino J, Westwick JK, Zhang C, Lamerdin J, Dagostino E, Knighton D, Loi CM, Zager M, Kraynov E, Popoff I, Christensen JG, Martinez R, Kephart SE, Marakovits J, Karlicek S, Bergqvist S, Smeal T. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc. Natl. Acad. Sci. U S A. 2010;107:9446–9451. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hausser A, Link G, Hoene M, Russo C, Selchow O, Pfizenmaier K. Phospho-specific binding of 14-3-3 proteins to phosphatidylinositol 4-kinase III beta protects from dephosphorylation and stabilizes lipid kinase activity. J. Cell Sci. 2006;119:3613–3621. doi: 10.1242/jcs.03104. [DOI] [PubMed] [Google Scholar]
- 27.Watkins JL, Lewandowski KT, Meek SE, Storz P, Toker A, Piwnica-Worms H. Phosphorylation of the Par-1 polarity kinase by protein kinase D regulates 14-3-3 binding and membrane association. Proc. Natl. Acad. Sci. U S A. 2008;105:18378–18383. doi: 10.1073/pnas.0809661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziegler S, Eiseler T, Scholz RP, Beck A, Link G, Hausser A. A novel protein kinase D phosphorylation site in the tumor suppressor Rab interactor 1 is critical for coordination of cell migration. Mol. Biol. Cell. 2011;22:570–580. doi: 10.1091/mbc.E10-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valente C, Turacchio G, Mariggio S, Pagliuso A, Gaibisso R, Di Tullio G, Santoro M, Formiggini F, Spano S, Piccini D, Polishchuk RS, Colanzi A, Luini A, Corda D. A 14-3-3gamma dimer-based scaffold bridges CtBP1-S/BARS to PI(4)KIIIbeta to regulate post-Golgi carrier formation. Nat. Cell Biol. 2012;14:343–354. doi: 10.1038/ncb2445. [DOI] [PubMed] [Google Scholar]
- 30.Tzivion G, Avruch J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 31.Hausser A, Storz P, Link G, Stoll H, Liu YC, Altman A, Pfizenmaier K, Johannes FJ. Protein kinase C mu is negatively regulated by 14-3-3 signal transduction proteins. J. Biol. Chem. 1999;274:9258–9264. doi: 10.1074/jbc.274.14.9258. [DOI] [PubMed] [Google Scholar]
- 32.Mullin MJ, Lightfoot K, Marklund U, Cantrell DA. Differential requirement for RhoA GTPase depending on the cellular localization of protein kinase D. J. Biol. Chem. 2006;281:25089–25096. doi: 10.1074/jbc.M603591200. [DOI] [PubMed] [Google Scholar]
- 33.Yuan J, Rey O, Rozengurt E. Activation of protein kinase D3 by signaling through Rac and the alpha subunits of the heterotrimeric G proteins G12 and G13. Cell Signal. 2006;18:1051–1062. doi: 10.1016/j.cellsig.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Yuan J, Slice LW, Rozengurt E. Activation of protein kinase D by signaling through Rho and the alpha subunit of the heterotrimeric G protein G13. J. Biol. Chem. 2001;276:38619–38627. doi: 10.1074/jbc.M105530200. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell. Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olayioye MA, Barisic S, Hausser A. Multi-level control of actin dynamics by protein kinase D. Cell Signal. 2013;25:1739–1747. doi: 10.1016/j.cellsig.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Shen YH, Godlewski J, Bronisz A, Zhu J, Comb MJ, Avruch J, Tzivion G. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol. Biol. Cell. 2003;14:4721–4733. doi: 10.1091/mbc.E02-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gohla A, Bokoch GM. 14-3-3 regulates actin dynamics by stabilizing phosphorylated cofilin. Curr. Biol. 2002;12:1704–1710. doi: 10.1016/s0960-9822(02)01184-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.