Abstract
Specialized metabolic enzymes biosynthesize chemicals of ecological importance, often sharing a pedigree with primary metabolic enzymes1. However, the lineage of the enzyme chalcone isomerase (CHI) remained a quandary. In vascular plants, CHI-catalyzed conversion of chalcones to chiral (S)-flavanones is a committed step in the production of plant flavonoids, compounds that contribute to attraction, defense2, and development3. CHI operates near the diffusion limit with stereospecific control4,5. While associated primarily with plants, the CHI-fold occurs in several other eukaryotic lineages and in some bacteria. Here we report crystal structures, ligand-binding properties, and in vivo functional characterization of a non-catalytic CHI-fold family from plants. A. thaliana contains five actively transcribed CHI-fold genes, three of which additionally encode amino-terminal chloroplast-transit sequences (cTP). These three CHI-fold proteins localize to plastids, the site of de novo fatty acid (FA) biosynthesis in plant cells. Furthermore, their expression profiles correlate with those of core FA biosynthetic enzymes, with maximal expression occurring in seeds and coinciding with increased FA storage in the developing embryo. In vitro, these proteins are Fatty Acid-binding Proteins (FAP). FAP knockout A. thaliana plants exhibit elevated alpha-linolenic acid levels and marked reproductive defects, including aberrant seed formation. Notably, the FAP discovery defines the adaptive evolution of a stereospecific and catalytically ‘perfected’ enzyme6 from a non-enzymatic ancestor over a defined period of plant evolution.
CHI (EC 5.5.1.6) catalyzes the intramolecular and stereospecific cyclization of chalcones to chiral flavanones via a Michael addition reaction7. The origin of CHI has been a mystery, due to the apparent absence of a related protein from primary metabolism8. CHI-like homologues in fungi and bacteria9 lack both key catalytic residues and the chalcone-binding site of bona fide CHI (Fig. 1a-c, Supplementary Fig. 1). Phylogenetic analysis shows that CHI is restricted to vascular plants, and is derived from FAP3. FAP3 forms one of two branches of FAP proteins (FAP1 together with FAP2 form the other) found in derived and basal plants and is likely homologous to CHI-fold proteins from protists and fungi (Supplementary Figs. 2,3, Supplementary Files 1,2). A. thaliana has five genes (and one pseudogene not shown) encoding CHI-fold proteins: two CHI/CHI-like (CHIL) members, AtCHI (At3g55120) and AtCHIL (At5g05270), and three FAP members, AtFAP1, AtFAP2 and AtFAP3 (At3g63170, At2g26310, and At1g53520, respectively). Pairwise sequence identity of the shared CHI-fold domain ranges from 10% to 63%. AtCHI is a bona fide enzyme, and mutations of its gene result in plants devoid of flavonoids 10. The active site of AtCHI retains residues important for catalytic activity, and AtCHI catalyzes the in vitro formation of (2S)-naringenin (Fig. 1c). In contrast, AtCHIL has substitutions of several catalytic residues (Supplementary Fig. 3). Likewise, AtFAP1, AtFAP2 and AtFAP3 encode substitutions at nearly all of the critical catalytic positions of AtCHI, and are devoid of catalytic activity (Supplementary Fig. 3). Studies with GFP fused to full-length FAP1, FAP2 and FAP3 indicate that these proteins localize in the plastid stroma (Supplementary Figs. 4a-d).
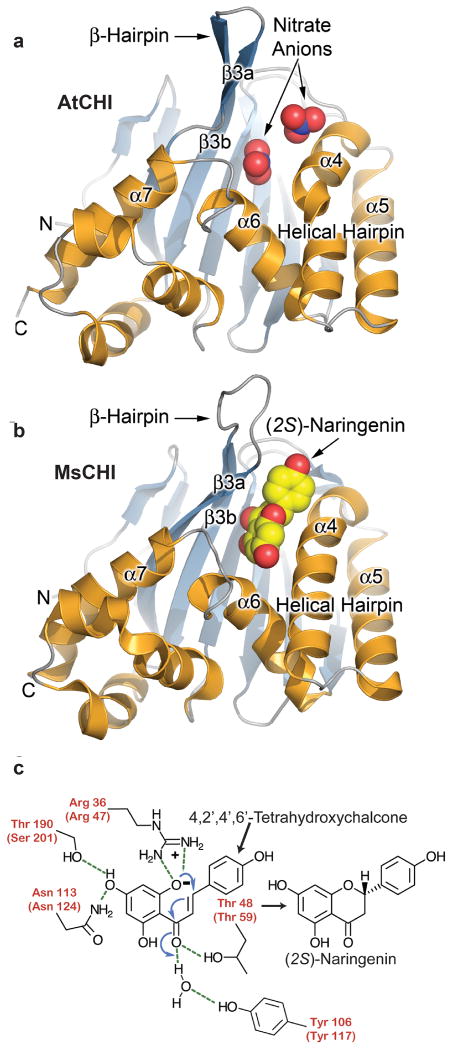
Figure 1. CHI fold, catalytic reaction and phylogeny.
a, Ribbon diagram21 of the AtCHI x-ray crystal structure, color-coded and labeled according to Jez et al.8. Two nitrate anions associate with catalytic residues in the substrate-binding site. b, Ribbon diagram21 of the MsCHI structure bound to (2S)-naringenin8. c, Chalcone is converted to (2S)-flavanone (e.g. naringenin) using a combination of electrostatic catalysis and water-mediated charge stabilization during a stereospecific Michael-type addition reaction5, 7-8. Residue numbers for AtCHI appear in parentheses. Catalytic residues are colored red.
We determined X-ray crystal structures of AtCHI, AtCHIL, and the CHI-like domains of AtFAP1 and AtFAP3, and compared these to the previously reported structure of MsCHI8. Despite low sequence conservation, all structures align within 2.3 Åroot-mean-squared deviation for the backbone atoms of equivalent residues (Supplementary Fig. 5). Ligand-binding pockets reside within the helical layer, bounded by two α-helical segments (helix-turn-helix α6-α7 and helical hairpin α4-α5), β-hairpin β3a-β3b, and the face of the core β-sheet, β3a-β3f. Notably, the largest differences in backbone conformations of MsCHI, AtCHI and AtCHIL compared to AtFAP1 and AtFAP3 are confined to the secondary structure elements surrounding the ligand-binding pockets (Supplementary Fig. 5).
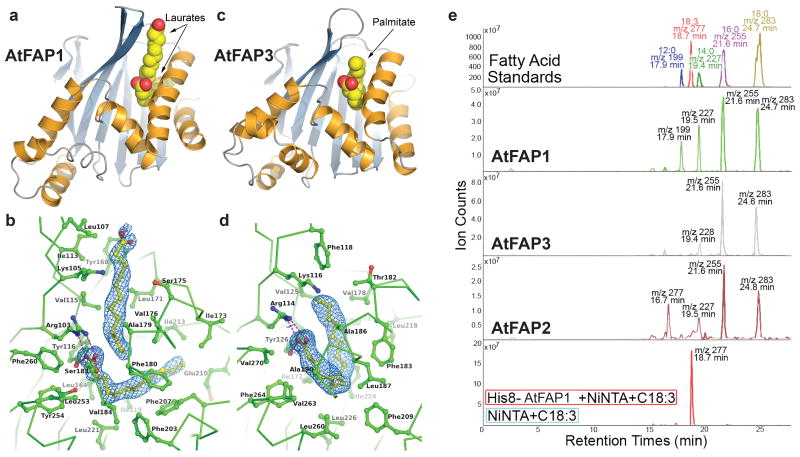
The most intriguing finding from the crystallographic structures is that the ligand-binding pockets of the recombinant FAP proteins are occupied by FAs (Fig. 2a-d). Clearly evident in electron-density maps are two abutting molecules of lauric acid (C12:0) in AtFAP1 (Fig. 2a, b), and one molecule of palmitic acid (C16:0) in AtFAP3 (Fig. 2c,d). The ligand-binding site of the FAP proteins encompasses a largely buried, non-polar cavity that sequesters the aliphatic chain(s) of the FAs, and a conserved Arg-Tyr pair that tethers a FA carboxylate group (Fig. 2b, d, and Supplementary Fig. 6). By comparison, CHI and CHIL possess more sterically restricted pockets due to the presence of larger aliphatic residues and an inward shift of the secondary structure elements surrounding their ligand-binding clefts (Figs. 1, 2, and Supplementary Fig. 5). The CHI substrate-binding pocket also includes several polar residues that form key hydrogen-bond interactions with the chalcone substrate (in MsCHI, or with polar nitrate ions in AtCHI) (Fig. 1a, b). The corresponding residues in the FAPs are non-polar. AtCHIL possesses a distinct conservation pattern of the active site pocket (Supplementary Fig. 3). Extraction of ligands with ethanol followed by high performance liquid chromatography–mass spectrometry (HPLC-MS) analyses confirms that all three FAP proteins (including AtFAP2, which was recalcitrant to crystallization) associate with saturated FAs (Fig. 2e). In contrast, with AtCHI and AtCHIL produced under similar conditions, no associated FAs were detected analytically or crystallographically. Exogenous FAs were not supplied during FAP expression in E. coli, purification, or crystallization, so the ligands bound to the FAPs were likely acquired during expression and retained during purification to homogeneity. Indeed, these saturated FAs are among the most abundant FAs in E. coli11.
Figure 2. Three-dimensional structure and ligand binding of FAPs.
a, Ribbon diagram21 of AtFAP1 oriented and color-coded as in Fig. 1. The bound C12:0 molecules are shown as van der Waals spheres where carbon is yellow and oxygen is red. b, Close-up view of the AtFAP1 FA binding sites21 with the experimental electron density of each C12:0 shown at 1σfor a SIGMAA-weighted 2Fo-Fc map23. c, Ribbon diagram21 of AtFAP3 rendered as in a. d, Close-up view of the AtFAP3 FA binding site21 with the C16:0 ligand shown as in b. e, Analysis of ligands associated with purified AtCHI-fold proteins separated and detected by reversed-phase HPLC-MS. The y-axes represent negative-ion counts for selected masses of anionic forms of FAs. The bottom panel depicts control (blue) and binding of C18:3 by His8-tagged AtFAP1 (red) coupled to Ni-affinity resin.
There are indications that the true ligands of the FAPs in planta may be different FAs. In AtFAP1, the presence of two distinct non-polar tunnels, each occupied by the aliphatic chain of a C12:0 molecule (Fig. 2b and Supplementary Fig. 6a), may indicate a proclivity for longer chain FA recognition. In AtFAP3, the horseshoe-shaped conformation of C16:0 portends a preference for binding a cis-unsaturated C16 or C18 FA, which typically assumes a bent conformation (Fig. 2d and Supplementary Fig. 6b). Notably, AtFAP1 sequesters exogenously-supplied α-linolenic acid (C18:3), which presumably displaces the bound FAs acquired during expression in E. coli (Fig. 2e). We further assessed the binding by the FAPs of a series of common FAs using thermal-stability assays12 (Supplementary Table 1). AtFAP3 interacts with most FAs tested and has maximal relative affinity for C16:0, though C18:0 induces greater thermal stabilization (Supplementary Fig. 7). AtFAP1 preferentially interacts with saturated FAs. However, the strongest stabilizing effect was observed with C18:3 (Supplementary Fig. 8), consistent with the displacement analyses (Fig. 2e) and perhaps a function of AtFAP1 in C18:3 metabolism as indicated also by phenotypic analysis. The thermal stability of AtCHI, included in these analyses as a negative control, was mostly unaffected by the addition of FAs (Supplementary Fig. 9).
We have obtained through a variety of approaches considerable direct and indirect evidence supporting the involvement of AtFAPs in FA metabolism. Network analysis of transcriptomic data13,14 demonstrates maximal expression of AtFAP1 and AtFAP3 in seeds at six days after flowering (Supplementary Fig. 10a), coinciding with the accumulation of storage lipids in the developing embryo15. AtFAP1 and AtFAP3 co-express with genes encoding FA biosynthetic enzymes (Supplementary Fig. 10b). Developmental patterns of AtFAP1, AtFAP2, and AtFAP3 expression were characterized in A. thaliana lines expressing a beta-glucuronidase (GUS) reporter under control of the respective promoters (Supplementary Figs. 11, 12, 13). The FAPs are highly expressed in developing cotyledons, young seedlings, roots, seeds, embryos, macrospores, preanthesis and tapetum. However, while AtFAP2 is expressed throughout the life of the plant, expression of AtFAP1 and especially AtFAP3 is restricted to developing and reproductive tissues.
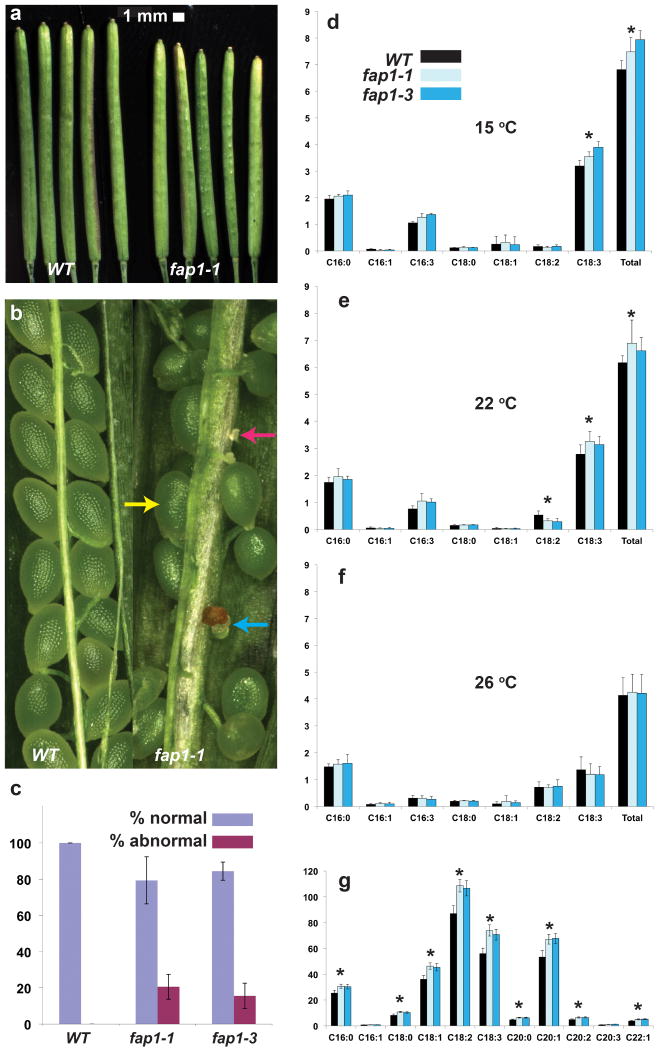
Homozygous Atfap1 and Atfap2 null plants were propagated from T-DNA insertion lines (Supplementary Figs. 14, 15). These Atfap1 and Atfap2 null plants are indistinguishable from wild-type (WT) plants during vegetative growth (Supplementary Figs. 17, 18), but the Atfap1 mutants show marked differences during reproductive stages. Specifically, Atfap1 siliques are shorter than those of wild type (∼18 mm vs. ∼20 mm) (Fig. 3a), mutant siliques contain 10-20% abnormal ovules (Fig. 3b), and the number of seeds per silique and the yield of viable seeds per plant are reduced in the Atfap1 null lines (Fig. 3c). The AtFAP3-RNAi homozygote lines also present these phenotypic alterations. In addition, the vegetative phenotype of AtFAP3-RNAi is altered, demonstrating early bolting, fast growth, increased branching, reduced apical dominance and reduced overall plant height (Supplementary Fig. 19). No AtFAP3-RNAi line with more than a ∼50% reduction in AtFAP3 RNA (relative to WT) could be identified, nor are any T-DNA tagged Atfap3 knockout lines available. These data thus indicate that a functional AtFAP3 is required for normal plant and embryo development. Double mutants bearing pairwise combinations of homozygous Atfap1, Atfap2, and AtFAP3-RNAi all show additive developmental effects (Supplementary Fig. 18).
Figure 3. Phenotypic characterization of Atfap1 null plants.
a,Length of siliques 12 days after flowering; WT siliques are longer than siliques of Atfap1 nulls (average length =17.8 mm for WT vs. 15.4 mm for Atfap1-1; 19.8 mm for WT vs. 17.1 mm for Atfap1-3 (not shown); P <0.01, n=10 siliques/plant for 10 plants/genotype). b, Atfap1 null siliques frequently contain abnormal ovules (normal, yellow arrow; aborted, blue arrow; unfertilized, pink arrow). c, Atfap1 nulls have a greater percentage of abnormal (aborted and unfertilized) ovules than WT (P <0.01 and n=10 siliques/genotype) and low seed yield mass per plant (Atfap1 average =1.3 g; WT average =1.5 g; P <0.01, n=10 plants). FA content of leaves, y-axis=mg/g fresh weight, from plants grown at d, 15 °C; e, 22 °C; f, 26 °C. d, C18:3 and total FAs is greater in leaves of Atfap1 nulls than WT. e, C18:3 and total FA is greater in leaves of Atfap1 nulls than WT. f, Atfap1 nulls and WT have similar FA content. g, In seeds of Atfap1 mutants, C16:0, C18:0, C18:1, C18:2, C18:3, C20:0, C20:1, C20:2, C22:1, and total FAs (not shown) increase relative to WT. Asterisks (*, P<0.05) represent both Atfap1 null lines; n=3 biological replicates, except for g, which has 3 experimental replicates with n=4 biological replicates; error bars represent standard deviation.
Reciprocal crosses between WT and Atfap1 or AtFAP3-RNAi lines point to the maternal inheritance of the aberrant reproductive phenotypes, as an elevated rate of ovule abortion is observed only in crosses in which the maternal parent is a null mutant (Supplementary Figs. 20-21, Supplementary Tables 2-7). The morphological phenotypes of >100 lines of heterozygotic AtFAP3-RNAi were analyzed; these consistently showed a restoration of the WT vegetative phenotype (Supplementary Fig. 22). However, the aberrant reproductive phenotype of the AtFAP3-RNAi mutant was maintained, indicating that phenotypic effects observed in the AtFAP3-RNAi lines are due to the specific knockdown of this gene. Analysis of FA composition reveals that Atfap1 null lines have elevated total fatty-acid levels in both leaves and seeds relative to WT (Fig. 3d, e and g). These phenotypic alterations in leaves are temperature dependent (apparent only in plants grown at 15°C and 22°C) and due primarily to an increased level of the lower-melting-temperature unsaturated FA, C18:3. The FA content and distribution in leaves are indistinguishable between WT and mutant plants grown at 26°C (Fig. 3f). Similarly, AtFAP3-RNAi plants and Atfap double mutants have altered FA composition and increased FA content in leaves and seeds (Supplementary Fig. 23). This temperature-dependent phenotype reflects a decrease in the more fluid acyl chain component of trienoic FAs that accompanies increasing temperature16,17.
We identified three relatives of CHI in Arabidopsis which appear to be FA-binding proteins of undetermined function. Phylogeny and sequence analyses indicate that the CHI fold occurs in most eukaryotic lineages (with the notable exception of animals) and even in bacteria. CHIL, as a divergent relative of FAP3, initially arose in mosses and may have served as the ancestor of enzymatic CHI in vascular plants (Supplementary Figs. 2 and 3). This ancestry for plant CHI clarifies the mystery surrounding the identity of the progenitor protein, and serves as a model to understand adaptation of a ligand-binding protein to a highly efficient and stereospecific enzyme. An evolutionary connection between fatty acid and flavonoid metabolism18 is also evident from the structural and mechanistic similarities between chalcone synthase (a type-III polyketide synthase (PKS)) acting one step upstream of CHI in the flavonoid pathway and ketoacyl-synthase III (KAS III) involved in chloroplast and bacterial fatty-acid biosynthesis. Together, CHI and CHS provide the foundation to examine the evolution of more recent metabolic pathways and metabolite classes from widespread biosynthetic pathways of primary metabolism.
Methods Summary
Structural biology
Crystal structures of the CHI domains of AtCHI, AtCHIL, AtFAP1, and AtFAP3 were determined as described in Supplementary Materials and Methods and summarized in Supplementary Table 8.
In vitro biochemistry
CHI activity was assayed5,7. Ligands bound to the purified AtFAP proteins were extracted with ethanol and analyzed by reversed-phase HPLC-MS. The effects of fatty acids (FAs) on the melting temperature of the AtFAP proteins were measured using a Thermofluor-based assay12.
Phylogenetic and sequence analyses
Homologs of AtCHI were identified by iterative psi-blast and profile Hidden Markov Model (HMM). Alignments were curated using protein structure superpositions and homologous groups determined manually.
Informatics
Co-expression patterns of A. thaliana genes encoding FAP proteins and genes associated with FA biosynthesis were evaluated with data compiled from 72 microarray experiments using MetOmGraph (www.metnetdb.org)13.
Plant growth and mutant analyses
Plants were grown in random block designs at 22°C under 16 hr light/8 hr dark. FAs were measured 5 hr after onset of light. Temperature effects were observed after 10-d growth at the specified temperatures. Homozygous Atfap1-1, Atfap1-3, Atfap2-1 and Atfap2-2 mutant alleles (Supplementary Fig 14 and Supplementary Table 3) were generated from SALK_130560, SALK_039829, SAIL_171_C12 and SAIL_616_D09 (Arabidopsis Biological Resource Center, ABRC) stocks, respectively. Atfap3 RNAi lines (Supplementary Fig. 16 and Supplementary Table 9) were generated from A. thaliana Col-0 plants, transformed with Agrobacterium tumefasciens strain GV3101 containing vector CATMA1a44560 (Nottingham Arabidopsis Stock Center, NASC)19. AtFAP RNA was quantified by RT-PCR (Supplementary Figs. 15 and 16). Transgenic A. thaliana lines were generated with AtFAP1, 2, 3 promoters or promoter + cTP /or + CDS fused to GUS and green fluorescent protein (GFP).
FA quantification in plants
FAs were extracted from plant tissues using barium hydroxide hydrolysis20, modified by methylation and analyzed by gas chromatography–mass spectrometry (GC-MS). Nonadecanoic acid was used as internal standard and a reference FA mixture was used for calibration and retention time determination (plantmetabolomics.org).
Supplementary Material
Acknowledgments
We thank A. Perera, B. Nikolau, H. Ilarsan and J. Peng for technical training (to M.N.N.), J. Peng for the fap1-1 homozygote mutant line, D. Nettleton & H. Wang for statistical analysis of seed FA data, and Eric Scheeff for assistance in Bayesian phylogenetic analysis. This research was supported in part by a Fulbright Fellowship (M.N.N.). This material is based in part upon work supported by the National Science Foundation under Award No. MCB-0645794 (J.P.N.), EEC-0813570 (E.S.W.), MCB-0951170 (E.S.W.), and by the National Cancer Institute Award No. CA14195 (G.M.). J.P.N. is an investigator with the Howard Hughes Medical Institute. Portions of this research were conducted at the Advanced Light Source (ALS) a national user facility operated by Lawrence Berkeley National Laboratory, on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Berkeley Center for Structural Biology is supported in part by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences. We thank the staff at the ALS for assistance with X-ray data collection. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Supplementary Information is linked to the online version at www.nature.com/nature.
Author Contributions M.N.N. R.N.P., and G.V.L. made equivalent contributions and should be considered joint first authors. M.N.N. experimentally characterized the FAP genes in planta. M.E.B., R.N.P. and E.L. expressed, purified and crystallized proteins. L.L. designed genetics experiments and constructs. G.V.L. and F.P. carried out fatty acid binding analyses and solved the x-ray crystal structures. R.N.P. carried out thermal-shift assays of fatty acid binding. G.M. and E.L. performed phylogenetic and sequence analyses. J.P.N. designed the biochemical experiments; E.S.W. designed the bioinformatics and functional genomics experiments. The manuscript was written by R.N.P., G.V.L., M.N.N., J.P.N., G.M. and E.S.W.
Author Information Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 4DOI (AtCHI), 4DOK (AtCHIL), 4DOL (AtFAP3), and 4DOO (AtFAP1).
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Hartmann T. From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochem. 2007;68:2831–2846. doi: 10.1016/j.phytochem.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Ferrer JL, Austin MB, Stewart C, Jr, Noel JP. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peer WA, Murphy AS. Flavonoids and auxin transport : modulators or regulators ? Trends Plant Sci. 2007;21:556–563. doi: 10.1016/j.tplants.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Bednar RA, Hadcock JR. Purification and Characterization of Chalcone Isomerase from Soybeans. J Biol Chem. 1988;263:9582–9588. [PubMed] [Google Scholar]
- 5.Jez JM, Noel JP. Reaction mechanism of chalcone isomerase: pH dependence, diffusion control, and product binding differences. J Biol Chem. 2002;277:1361–1369. doi: 10.1074/jbc.M109224200. [DOI] [PubMed] [Google Scholar]
- 6.Albery WJ, Knowles JR. Evolution of Enzyme Function and the Development of Catalytic Efficiency. Biochem. 1976;15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 7.Jez JM, Bowman ME, Noel JP. Role of hydrogen bonds in the reaction mechanism of chalcone isomerase. Biochem. 2002;41:5168–5176. doi: 10.1021/bi0255266. [DOI] [PubMed] [Google Scholar]
- 8.Jez JM, Bowman ME, Dixon RA, Noel JP. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat Struct Biol. 2000;7:786–791. doi: 10.1038/79025. [DOI] [PubMed] [Google Scholar]
- 9.Gensheimer M, Mushegian A. Chalcone isomerase family and fold: no longer unique to plants. Protein Sci. 2004;13:540–544. doi: 10.1110/ps.03395404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirley BW, Hanley S, Goodman HM. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell. 1992;4:333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oursel D, et al. Identification and relative quantification of fatty acids in Escherichia coli membranes by gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:3229–3233. doi: 10.1002/rcm.3177. [DOI] [PubMed] [Google Scholar]
- 12.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protocols. 2007;9:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 13.Mentzen WI, Wurtele ES. Regulon Organization of Arabidopsis. BMC Plant Biol. 2008;8:99. doi: 10.1186/1471-2229-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mentzen WI, Peng J, Ransom N, Nikolau BJ, Wurtele ES. Articulation of three core metabolic processes in Arabidopsis: fatty acid biosynthesis, leucine catabolism and starch metabolism. BMC Plant Biol. 2008;8:76–90. doi: 10.1186/1471-2229-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruuska SA, Girke T, Benning C, Ohlrogge JB. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell. 2002;14:1191–1206. doi: 10.1105/tpc.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falcone DL, Ogas JP, Somerville CR. Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition. BMC Plant Biol. 2004;4:17–31. doi: 10.1186/1471-2229-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami Y, Tsuyama M, Kobayashi Y, Kodama H, Iba K. Trienoic fatty acids and plant tolerance of high temperature. Science. 2000;287:476–479. doi: 10.1126/science.287.5452.476. [DOI] [PubMed] [Google Scholar]
- 18.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 19.Bonaventure G, Salas JJ, Pollard MR, Ohlrogge JB. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell. 2003;15:1020–1033. doi: 10.1105/tpc.008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilson P, et al. Versatile gene-specific tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Gen Res. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLano WL. DeLano Scientific; San Carlos, CA, USA: 2002. The PyMOL Molecular Graphics System. http://www.pymol.org. [Google Scholar]
- 22.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Prot Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallog. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.