Abstract
Recent studies have shown two distinct non-CIMP methylation clusters in colorectal cancer, raising the possibility that DNA methylation, involving non-CIMP genes, may play a role in the conventional adenoma–carcinoma pathway. A total of 135 adenomas (65 left colon and 70 right colon) were profiled for epigenome-wide DNA methylation using the Illumina HumanMethylation450 BeadChip. A principal components analysis was performed to examine the association between variability in DNA methylation and adenoma location. Linear regression and linear mixed effects models were used to identify locus-specific differential DNA methylation in adenomas of right and left colon. A significant association was present between the first principal component and adenoma location (P = 0.007), even after adjustment for subject age and gender (P = 0.009). A total of 168 CpG sites were differentially methylated between right- and left-colon adenomas and these loci demonstrated enrichment of homeobox genes (P = 3.0 × 10−12). None of the 168 probes were associated with CIMP genes. Among CpG loci with the largest difference in methylation between right- and left-colon adenomas, probes associated with PRAC(prostate cancer susceptibility candidate) gene showed hypermethylation in right-colon adenomas whereas those associated with CDX2(caudal type homeobox transcription factor 2) showed hypermethylation in left-colon adenomas. A subgroup of left-colon adenomas enriched for current smokers (OR = 6.1, P = 0.004) exhibited a methylation profile similar to right-colon adenomas. In summary, our results indicate distinct patterns of DNA methylation, independent of CIMP genes, in adenomas of the right and left colon.
Keywords: colon polyps, colorectal cancer, CpG island methylator phenotype, epigenetics
Colorectal cancer is the second leading cause of cancer-related deaths in the United States, with an overall lifetime risk of ∼1 in 20 (5.1%).1 Colonoscopy prevents colorectal cancer by interrupting the adenoma–carcinoma sequence through polypectomy. However, the benefit of colonoscopy may be restricted to tumors involving the left colon.2–4 This may be because detection and complete removal is more difficult for sessile polyps involving the right colon,5 or there may be underlying molecular differences between precursor lesions involving the right and left colon.
It is widely accepted that colorectal cancer arises through multiple pathways and, currently, the molecular classification of colorectal cancer is largely based on two distinct phenomena: micro-satellite instability (MSI), which subclassifies tumors into MSI-high, MSI-low, or microsatellite stable (MSS), and the CpG island methylator phenotype (CIMP), which subclassifies tumors into CIMP-high, CIMP-low, or CIMP-negative based on the methylation status of a defined set of CIMP genes.6 MSI-high and CIMP-high cancers show a predilection for the proximal colon and are believed to arise from serrated polyps with a BRAF mutation. In contrast, 47–55% of all colorectal cancers are MSS and CIMP negative, are more prevalent in the distal colon, and are thought to arise from conventional adenomas.7,8
This has led to the belief that epigenetic changes, such as DNA methylation, are important mainly in the serrated pathway of carcinogenesis (serrated polyp-dysplasia-carcinoma). It is conceivable that DNA methylation is also altered in conventional adenomas that are precursors of CIMP-negative colorectal cancers; however, these alterations have not been observed previously because they occur in genes outside the CIMP panel.9,10 Two recent studies have published data on genome-wide aberrant DNA methylation in colorectal cancer.8,10 Both studies found four distinct methylation clusters in colorectal cancer. Two of the methylation clusters correlated with CIMP-high and CIMP-low tumors, as expected, but interestingly non-CIMP tumors also segregated into two distinct methylation clusters. The presence of these two additional methylation clusters suggests that DNA methylation involving genes not included in the traditional CIMP assessment panels may also occur in conventional adenomas and likely plays an important role in the conventional pathway of colorectal carcinogenesis.
Using epigenome-wide assessment of DNA methylation, we tested the hypothesis that distinct DNA methylation patterns, independent of traditionally assessed CIMP loci, exist in conventional adenomas involving the right and left colon.
Materials and methods
Study Group and Exposure Measurement
The New Hampshire Colonoscopy Registry (NHCR) collects data on colonoscopy procedures administered in the state of New Hampshire for consenting individuals at participating facilities.11,12 Participants complete a patient information questionnaire at the time of their colonoscopy, including information on level of education, cigarette smoking (never, former, current), personal or family history of colon polyps or colorectal cancer, and known hereditary factors including familial adenomatous polyposis or Lynch syndrome. The endoscopists record detailed information in a separate form about the patient, the adequacy of bowel preparation, and the colonoscopic findings, including polyp location, size, and treatment. Histological data are retrieved from pathology reports and matching of endoscopy and pathology data is performed in a systematic manner, as described previously.11
A total of 135 conventional adenomas, from 131 consenting patients enrolled between 2005 and 2008 at one of the NHCR participating sites, were included in the study after histologic review. Case selection was dictated by presence of sufficient tissue in the paraffin block for extracting 2 μg of DNA for methylation array analysis. All adenomas selected for the study had a known anatomic location (ie, rectum, sigmoid colon, etc) and at least 75% lesional tissue. Adenomas with a significant proportion of normal colonic mucosa (>25%), adenomas with advanced histology (any villous component or high-grade dysplasia), and any serrated polyps were excluded from this study after slide review. All procedures and study materials were approved by the Committee for the Protection of Human Subjects.
Array-Based DNA Methylation Assessment and Quality Control
DNA was isolated using the QIAamp DNA formalin-fixed, paraffin-embedded tissue kit, according to the manufacturer's protocol. DNA was then ligated using the REPLI-g formalin-fixed, paraffin-embedded kit (Qiagen, Valencia, CA, USA). The ligation reaction was performed at 24°C for 90min by adding 8 μl of formalin-fixed, paraffin-embedded buffer, 1 μl formalin-fixed, paraffin-embedded enzyme, and 1 μl REPLI-g ligase to 2 μg DNA in 10ml, followed by 5min at 98°C. Samples were randomized across several plates and subsequently subjected to epigenome-wide DNA methylation assessment using the Illumina Infinium Human-Methylation450 BeadChip (Illumina, San Diego, CA, USA), which simultaneously profiles the methylation status for >485 000 CpG sites at single-nucleotide resolution, covering 96% of CpG islands, with additional coverage of island shores (<2kb from CpG islands), island shelves (2–4 kb from CpG islands), and regions flanking them. The methylation status for each CpG locus was calculated as the ratio of fluorescent signals (β = Max(M,0)/[Max(M,0) + Max(U,0) + 100]), ranging from 0 to 1, using the average probe intensity for the methylated (M) and unmethylated (U) alleles. β = 1 indicates complete methylation and β = 0 represents no methylation. The data were assembled using BeadStudio methylation software (Illumina), without normalization as per the manufacturer's instructions.
We used array control probes to assess the quality of our samples and to evaluate potential problems such as poor bisulfite conversion or color-specific issues for each array. As a quality control step, probes were removed if the median detection P-value was >0.05 across all samples, and samples were excluded if >25% of the probes associated with that sample had detection P-values >1e −5.13 All CpG loci on X and Y chromosomes were excluded from the analysis (11882 CpG loci) to avoid gender-specific methylation bias, leaving a final 473 695 CpG loci in a total of 135 samples. Technical, nonbiological noise has been shown in evaluation of samples in different batches and plates.14,15 We adjusted for the presence of plate/batch effects by applying the ComBat method16 to the logit-transformed methylation values,17 which has been shown to perform effectively and efficiently compared with competing batch/plate-adjustment methodologies18 (Supplementary Figure 1). The plate-adjusted data were then examined to ensure that plate/batch effects were successfully attenuated (Supplementary Figure 2).
Bisulfite Pyrosequencing of Target CpG Loci
As an orthogonal array validation, two CpG loci (cg01499197 for caudal type homeobox transcription factor 2 (CDX2), n = 18 and cg12374721 for prostate cancer susceptibility candidate(PRAC), n = 20) were subjected to bisulfite pyrosequencing, as these loci were found to be significantly associated with adenoma location and exhibited the largest difference in methylation between right- and left-colon adenomas. DNA samples isolated using the QIAamp DNA formalin-fixed, paraffin-embedded tissue kit (1 μg; Qiagen) were bisulfite modified using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). Bisulfite pyrosequencing was performed as previously described19 on the Pyromark MD Pyrosequencer.20 Results were analyzed using the PyroMarkCpG software version 1.0.11 (Qiagen) to obtain the methylation percent at target CpG loci. Amplification of bisulfate-converted DNA was accomplished using the PyroMark PCR kit (Qiagen) and forward and reverse biotinylated primers (IDT, Coralville, IA, USA) for each target locus. Cycling conditions were 95 °C for 15min, followed by 45 cycles of 94 °C for 30 s, 60 °C (CDX2) or 58 °C (PRAC) for 1 min, and 72 °C for 1 min, with a final extension of 10 min at 72 °C. Pyrosequencing reactions were performed in triplicate for each sample and the mean of the triplicates used in subsequent analyses. All pyrosequencing reactions included bisulfite modification controls and >93% modification efficiency was accomplished for all samples included in the analysis.
BRAF V600E Mutation Analysis
Detection of the BRAF V600E mutation was performed on isolated DNA from the adenoma tissue samples using a real-time allele-specific PCR assay. All PCR assays were run on an ABI 7500 FAST instrument with the following conditions: 95 °C for 15 min, followed by 45 cycles of 94 °C for 15 s; 65 °C for 30 s and 72 °C for 30 s. A postamplification melt curve was produced to verify the expected Tm of the wild-type (76.5 °C) and V600E (77 °C) amplicons. Cycle threshold (CT) values were used to determine the relative amount of V600E DNA detected compared with the wild-type BRAF DNA.
Statistical Analysis
Our primary analysis was aimed toward identifying differences in the underlying signature of DNA methylation between right- and left-colon adenomas. A principal components analysis was implemented to deconvolve the most prevalent sources of variability across the entire array of methylation values. Principal components analysis represents a feature extraction technique where the data are orthogonally transformed, such that the first principal component has the largest possible variance (accounting for maximal amount of variability in the data), and each succeeding component in turn has the highest variance possible. The association between the top three principal components and colon adenoma location, as well as other clinical and demographic variables, was examined using linear regression, modeling the principal component as the dependent variable and the clinical/demographic factors as the independent variables.
Locus-specific differences in DNA methylation of adenomas were examined using linear regression models fit to each CpG locus independently, modeling the logit-transformed methylation β-values as the dependent variable and adenoma location (right or left colon) as the independent variable. All models were adjusted for subject age and gender and P-values obtained from our linear regression analysis were adjusted for multiple comparisons using a stringent Bonferroni threshold (ie, 0.05/no. of CpGs tested). Because of the high frequency of missing information for adenoma size, smoking status, and alcohol consumption (11.1%, 17.0%, and 12.6%, respectively), we chose not to control for these factors in our linear regression analysis. However, given the association between adenoma size, smoking status, and alcohol consumption with adenoma location in our study and because these factors have been previously linked to alterations in DNA methylation,21–23 a sensitivity analysis was performed to ensure that differentially methylated CpG loci based on adenoma location were robust to adjustment for these factors. Although there were only four subjects for whom methylation was profiled in adenomas of both the right and left colon, we further assessed the robustness of our findings using a series of linear mixed effects models that accounted for the autocorrelation between successive measurements on the same subject.
To identify biological pathways involved in side-specific differences in DNA methylation profiles, we performed a gene set enrichment analysis on the genes that were found to be significantly associated with adenoma location using the Database for Annotation, Visualization, and Integrated Discovery tool.24
Linear regression analyses, adjusted for patient age and gender, were performed to test the linearity/nonlinearity of DNA methylation in conventional adenomas along specific anatomic location in the colon from cecum to the rectum. We created a numerical variable based on the average distance (cm) from anal verge to each anatomical location using previously published CT colonography data,25 as previously described.26 Focusing our analysis on most differentially methylated CpG loci between right and left adenomas, we fit linear regression model with logit-transformed methylation as the outcome variable and the anatomical location variable as the explanatory variable. A significant P-value based on the Wald test for the anatomical location variable indicates a linear relationship of DNA methylation along the specific colonic sites; however, this does not preclude the possibility of a nonlinear relationship. To test for nonlinearity, we used a likelihood ratio test (LRT) comparing the model with and without squared and/or cubic anatomic location variable.26 A significant P-value by the Wald test and a nonsignificant LRT P-value would indicate a linear relationship, whereas a significant LRT P-value would suggest a nonlinear relationship between DNA methylation and anatomic location in conventional adenomas.
All analyses were carried out using the R statistical package, version 2.13 (www.r-project.org/).
Results
Patient Characteristics and Adenoma Location
A detailed description of the characteristics of the patients with adenomas included in the study is presented in Table 1. Patient characteristics were generally similar between subjects with right- and left-sided adenomas, except that right-sided adenomas tended to be smaller in size (P = 0.004), were less likely to be present in current smokers (P = 0.01), and more likely to be present in heavy drinkers (P = 0.004). There were no statistically significant differences between subjects with right and left adenomas for any of the other study factors, including age, gender, race/ethnicity, body mass index (BMI), and education status.
Table 1. Demographic and clinical factors for the study group.
| Characteristic | Total number of adenomas (N = 135) | Left adenomas (N = 65, 48.1%) | Right adenomas (N = 70, 51.9%) |
|---|---|---|---|
| Age at report (years) | 59.7 (8.8) | 58.58 (8.9) | 60.64 (8.6) |
| Polyp size (mm) | |||
| <5 | 59 [43.7] | 20 [30.8] | 39 [55.7] |
| 5–9 | 43 [31.9] | 27 [41.5] | 16 [22.9] |
| 10–20 | 8 [5.9] | 5 [7.7] | 3 [4.3] |
| 420 | 2 [1.5] | 1 [1.5] | 1 [1.4] |
| Unknown | 23 [17.0] | 12 [18.5] | 11 [15.7] |
| Anatomic location | |||
| Rectum | 20 [14.8] | 20 [30.8] | 0 [0.0] |
| Sigmoid colon | 36 [26.7] | 36 [55.4] | 0 [0.0] |
| Descending colon | 8 [5.9] | 8 [12.3] | 0 [0.0] |
| Splenic flexure | 3 [2.2] | 0 [0.0] | 3 [4.3] |
| Transverse colon | 18 [13.3] | 0 [0.0] | 18 [25.7] |
| Hepatic flexure | 4 [3.0] | 0 [0.0] | 4 [5.7] |
| Ascending colon | 25 [18.5] | 0 [0.0] | 25 [35.7] |
| Cecum | 20 [14.8] | 0 [0.0] | 20 [28.6] |
| Unknown | 1 [0.7] | 1 [1.5] | 0 [0.0] |
| Smoking status | |||
| Never | 55 [40.7] | 24 [36.9] | 31 [44.3] |
| Former | 48 [35.6] | 22 [33.8] | 26 [37.1] |
| Current | 17 [12.6] | 14 [21.5] | 3 [4.3] |
| Unknown | 15 [11.1] | 5 [7.7] | 10 [14.3] |
| Gender | |||
| Male | 82 [60.7] | 36 [55.4] | 46 [65.7] |
| Female | 53 [39.3] | 29 [44.6] | 24 [34.3] |
| Education | |||
| Less than high school | 6 [4.4] | 2 [3.1] | 4 [5.7] |
| High school or equivale | nt 21 [15.6] | 11 [16.9] | 10 [14.3] |
| Some college/technical school | 24 [10.4] | 15 [23.1] | 9 [12.9] |
| College or post-college graduate | 65 [48.1] | 29 [44.6] | 36 [51.4] |
| Unknown | 19 [14.1] | 8 [12.3] | 11 [15.7] |
| Body mass index (BMI) | 28.2 (5.0) | 27.3 (5.0) | 29.1 (4.9) |
| Alcohol consumption (drinks/week) | |||
| 0 | 36 [26.7] | 22 [33.8] | 14 [20.0] |
| 1–4 | 38 [28.1] | 21 [32.3] | 17 [24.3] |
| 5–8 | 24 [17.8] | 11 [16.9] | 13 [18.6] |
| 9–12 | 12 [8.9] | 5 [7.7] | 7 [10.0] |
| 13–16 | 4 [3.0] | 0 [0.0] | 4 [5.7] |
| 17–20 | 3 [2.2] | 0 [0.0] | 3[4.3] |
| 21–25 | 1 [0.7] | 0 [0.0] | 1 [1.4] |
| >26 | 0 [0.0] | 0 [0.0] | 0 [0.0] |
| Unknown | 17 [12.6] | 6 [9.2] | 11 [15.7] |
Values are presented as mean (s.d.) for continuous variables and count [percentage] for categorical variables.
Adenoma Location is Associated with Epigenome-Wide Variability in DNA Methylation
Of the 135 conventional adenomas included in the study, 70 were located in the right and 65 in the left colon. Using plate-adjusted methylation data, we examined the extent to which epigenome-wide variability in DNA methylation was associated with adenoma location. A principal components analysis demonstrated that the first principle component (PC1), which accounts for 8.2% of the variation in methylation across the array, was significantly associated with adenoma location (P = 0.007; Table 2). This association remained significant even after adjustment for subject age and gender (P = 0.009). Principal components 2 and 3 (PC2 and PC3), which only explained 2.9% and 2.6% of the variation in methylation across the array, were not significantly associated with adenoma location.
Table 2. Results examining the association between the top three principal components (PCs) and various clinical and demographic factors.
| Associations components with principle (P-value) | |||
|---|---|---|---|
|
|
|||
| Variable | PC1(8.2%) | PC2(2.9%) | PC3(2.6%) |
| Colon adenoma side | 0.007 | 0.118 | 0.476 |
| Colon polyp size | 0.068 | 0.102 | 0.041 |
| Gender | 0.351 | 0.056 | 0.151 |
| Education level | 0.455 | 0.014 | 0.146 |
| Smoking status | 0.135 | 0.035 | 0.340 |
| Alcohol consumption | 0.690 | 0.620 | 0.309 |
| Body mass index (BMI) | 0.561 | 0.976 | 0.870 |
PCp (X%) denotes the pth principal component, which explained X% of epigenome-wide variability in DNA methylation across the array. P-values, derived from a linear model, are presented for each clinical and demographic factor across each of the top three PCs.
Locus-Specific Patterns of DNA Methylation by Adenoma Location
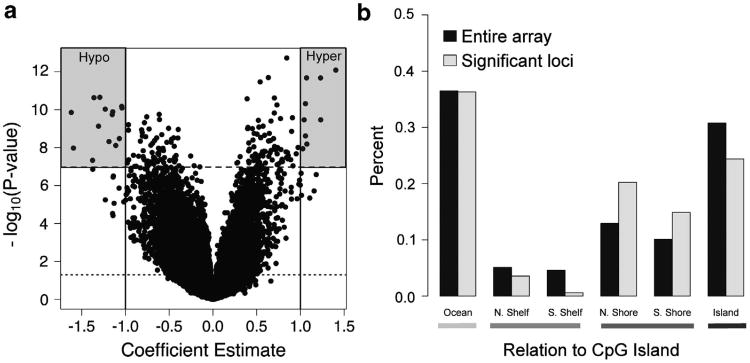
We next attempted to identify locus-specific patterns of differential methylation between right- and left-colon adenomas. This analysis (Figure 1a) revealed 57 795 (12.2%) differentially methylated CpG loci (P ≤ 0.05) and 168 of these remained significant after correcting for multiple comparisons (Supplementary Table 1). This list was robust to adjustment for age, gender, smoking status, adenoma size, and weekly alcohol consumption, with 77% of the adjusted coefficient estimates being within one standard error of the unadjusted coefficient estimate and all 168 loci remaining statistically significant (Supplementary Table 1). Linear mixed effects modeling to account for between- and within-subject variation in methylation measurements demonstrated that all 168 CpGs still remained statistically significant (Supplementary Table 1). The CpG loci that remained statistically significant after multiple adjustments were disproportionately located in CpG island shores (35% versus 23% frequency on the entire array; Fisher's exact P = 0.0004; Figure 1b) and demonstrated enrichment of genes involved in homeobox (4.97-fold enrichment, P = 3.0 × 10 -12), developmental protein (4.97-fold enrichment, P = 8.3 × 10−8), DNA binding (4.97-fold enrichment, P = 3.4 × 10−6), and transcription (4.97-fold enrichment, P = 2.3 × 10−3).
Figure 1.
Results from the locus-by-locus examination of differential methylation between right- and left-colon adenomas. (a) Volcano plot based on the locus-by-locus analysis. Points represent the −log10(P-value) obtained from testing the difference in methylation between right- and left-colon polyps, adjusted for subject age and gender. Dotted line indicates −log10(0.05) and the dashed line represents the Bonferroni threshold (ie, −log10(0.05/no. of tests)). (b) Distribution of significant probes (those significant at the Bonferroni threshold) across CpG islands, shores, shelves, and oceans.
Probes that were significantly associated with adenoma location, and those that exhibited the largest difference in DNA methylation level (probes with a > 1 or < −1 average difference in methylation on the logit scale) were then selected as candidates for technical validation via pyrosequencing. We identified 23 candidates for pyrosequencing, of which 9 were hypermethylated in right-colon adenomas and 14 were hypermethy lated in left-colon adenomas (Figure 1a and Table 3). The 23 target probes were predominantly located in the CpG island shore regions (70%), and the remaining 30% in CpG islands (Table 3). Median differences in percentage methylation between right- and left-colon adenomas ranged from 13.8 to 40.3% (Supplementary Figures 3 and 4). Examining the linearity/nonlinearity of the DNA methylation by specific anatomic location showed that the Wald P-value was statistically significant for all 23 target CpG loci (Supplementary Table 2). However, for all but four of these loci, linear regression models with a squared (quadratic) term for anatomic location resulted in a significantly improved model fit, suggesting a nonlinear relationship between anatomic location and DNA methylation for the majority of these 23 CpG loci (Supplementary Table 2 and Supplementary Figures 5 and 6).
Table 3. Candidate CpG loci for pyrosequencing.
| CpG Name | Coef Est. | P-value | Relation to CpG island | Gene |
|---|---|---|---|---|
| cg00347757 | 1.23 | 3.44E − 10 | S. Shore | GJB2 |
| cg00849267 | 1.06 | 2.43E − 09 | S. Shore | LMF1 |
| cg04574034 | 1.07 | 2.11E − 12 | N. Shore | PRAC;PRAC2 |
| cg09866983 | 1.06 | 4.84E 11 | N. Shore | PRAC;PRAC2 |
| cg12374721 | 1.40 | 8.24E − 13 | N. Shore | PRAC2;PRAC |
| cg14326989 | 1.23 | 2.13E 12 | N. Shore | PRAC2;PRAC |
| cg20643952 | 1.03 | 1.15E − 08 | N. Shore | — |
| cg20945566 | 1.07 | 6.54E − 09 | N. Shore | PRAC;PRAC2 |
| cg27170782 | 1.04 | 3.45E − 10 | N. Shore | PRAC;PRAC2 |
| cg01499197 | −1.15 | 1.30E-10 | Island | CDX2 |
| cg05222982 | −1.36 | 2.34E − 11 | S. Shore | |
| cg06137852 | −1.04 | 6.75E − 11 | S_Shore | |
| cg07984133 | −1.23 | 9.42E − 11 | Island | CDX2 |
| cg08248516 | −1.19 | 4.82E − 09 | Island | HOXA7 |
| cg09024126 | −1.38 | 4.68E − 08 | Island | PROM1 |
| cg10393811 | −1.29 | 2.24E − 11 | S. Shore | HOXD10 |
| cg11283429 | −1.07 | 3.35E − 09 | N. Shore | |
| cg12938068 | −1.59 | 1.06E−08 | N Shore | |
| cg13677149 | −1.11 | 7.74E − 09 | Island | EVX1 |
| cg15254559 | −1.04 | 8.18E − 11 | Island | |
| cg16201038 | −1.62 | 1.40E − 10 | Island | CDX2 |
| cg22210145 | −1.15 | 1.82E −10 | S. Shore | — |
| cg26708100 | −1.31 | 7.45E − 10 | S. Shore | HOXD10 |
Coef Est. represents the coefficient estimate that was estimated from a linear regression model examining the difference in methylation between right- and left-colon adenomas. Positive values of Coef Est. indicate probes that are hypermethylated in right-colon adenomas and negative values indicate probes that are hypermethylated in left-colon adenomas. CpG loci highlighted in gray and genes highlighted in bold represent those that were eventually selected for subsequent pyrosequencing validation.
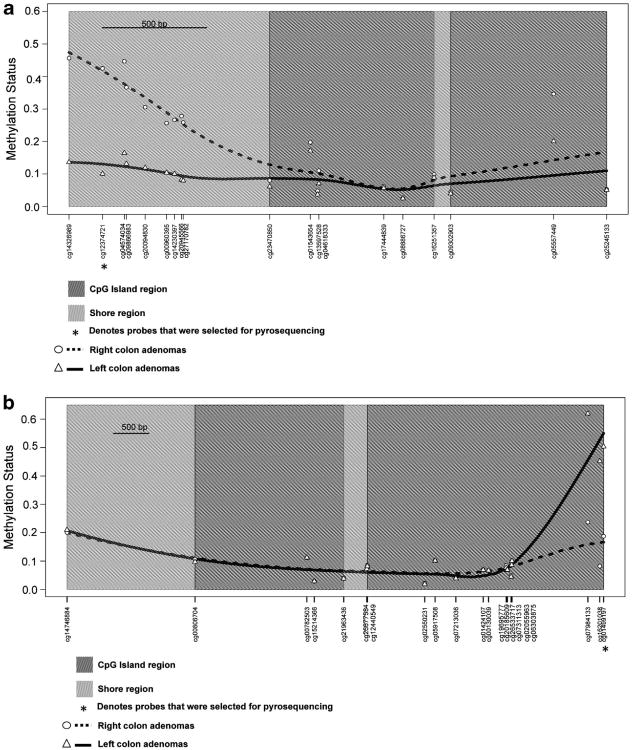
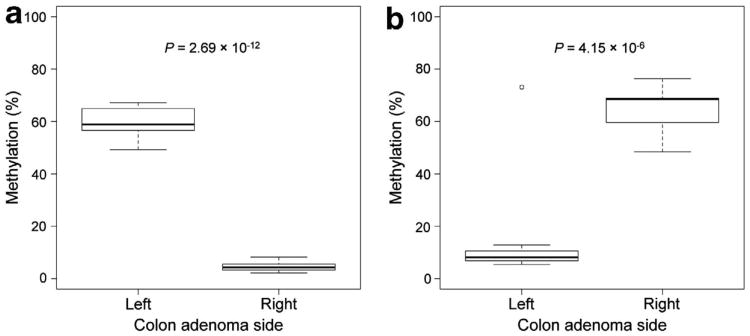
Among the nine probes that were differentially hypermethylated in right-colon adenomas, six were located in a CpG island shore region associated with PRAC and PRAC2—prostate cancer susceptibility candidate genes (Figure 2a). Of the 14 probes that were hypermethylated in left-colon adenomas, 3 were located in a CpG island region associated with the CDX2 gene, a homeobox transcription factor critical for intestinal differentiation. The methylation status for most of the CpG loci associated with CDX2 was fairly stable between right- and left-colon adenomas, but showed distinctly higher levels of methylation in left adenomas within a CpG island associated with this gene (Figure 2b). Therefore, the loci associated with PRAC and CDX2 were ultimately selected as targets for pyrosequencing to verify our methylation assessment. The results from pyrosequencing of two target probes cg01499197 (CDX2) and cg12374721 (PRAC) are shown in Figure 3 and show a high degree of concordance with the extent of methylation obtained from the Illumina methylation array. The mean percent methylation difference between right and left adenomas was −54.3% and 49.5% for cg01499197 and cg12374721, respectively, thereby validating the array results.
Figure 2.
Methylation status between right- and left-colon adenomas for PRAC(a) and CDX2(b). Points represent the median methylation status for left- and right-colon polyps and lines represent the locally weighted polynomial smoothed methylation status across the profiled CpGs associated with that gene.
Figure 3.
Difference in percent methylation between right- and left-colon adenomas based on the two target probes, (a) cg01499197, associated with gene CDX2, and (b) cg12374721, associated with PRAC, selected for pyrosequencing. P-values were adjusted for subject age and gender and were obtained from linear regression models fit to percent methylation measurements obtained from pyrosequencing.
Differentially Methylated Loci are Under Represented among CIMP Genes
None of the 168 differentially methylated probes in our study were associated with CIMP genes (Supplementary Table 3). Broadening our examination to include probes with a significant P-value before adjustment for multiple comparisons (57 795 with P ≤ 0.05) demonstrated that the differentially methylated CpG loci based on adenoma location were not only independent of CIMP genes, but that differential methylation of CIMP genes was observed less frequently among differentially methylated CpG loci than would be expected by random chance (P = 0.021).
DNA methylation involving CIMP-associated genes shows a strong correlation with BRAF muta-tion.6 To further confirm that the differential methylation seen in right- and left-colon adenomas was independent of the CIMP phenotype, we performed BRAF mutation analysis on all adenomas. Only 1 of 135 adenomas was positive for BRAF V600E mutation, whereas the rest were wild type.
DNA Methylation Pattern Also Reveals a Distinct Subset among Left-Colon Adenomas
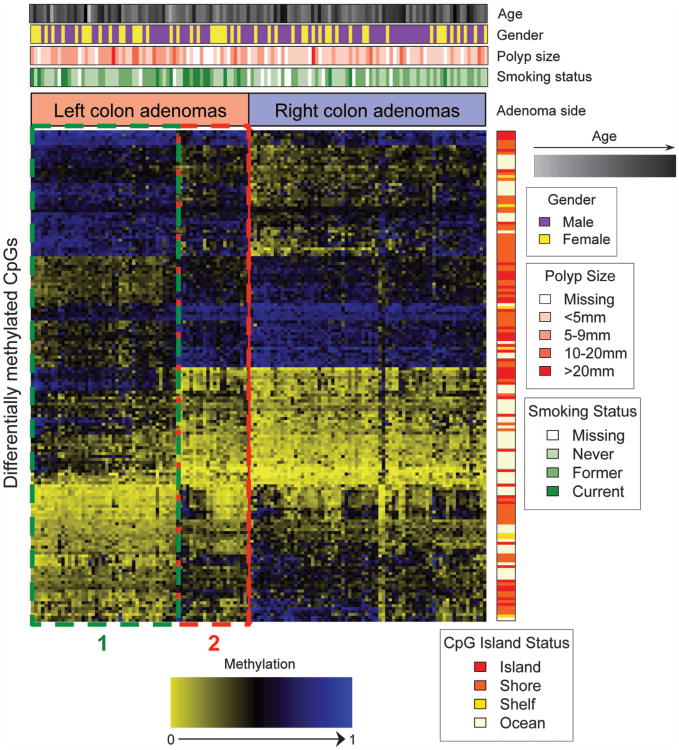
In an effort to visualize the methylation status of all 168 differentially methylated probes, we generated a heat map that depicts the methylation status for each of these loci across right- and left-colon adenomas (Figure 4). Unexpectedly, we found two subgroups within left-colon adenomas, hereafter referred to as subgroups 1 and 2. Interestingly, the methylation profile for subgroup 2 left-colon adenomas was similar to that of the right-colon adenomas. We next sought to understand if there were underlying differences in the demographic and clinical characteristics among the subjects with adenomas in these two subgroups. The subjects associated with adenomas in subgroup 2 were more frequently current smokers (OR = 6.1, P = 0.004), as indicated by the green bar above the heat map (Figure 4). However, there were no statistically significant associations between subjects with adenomas in subgroup 2 and other clinical and demographic factors in our study.
Figure 4.
Heat map of the 168 significantly differentially methylated CpG loci between right- and left-colon adenomas. Methylation values are presented as unmethylated (yellow) to methylated (blue). Rows are represented by the 168 differentially methylated loci and columns represent subjects who are grouped based on adenoma side. On the right-hand side of the heat map is a color side bar, which indicates the island status for the 168 CpG loci. Above the heat map are color side bars that indicate smoking status, polyp size, gender, and age for the study subjects. Red and green dotted boxes indicate two subgroups of left-colon adenoma subjects, referred to as subgroups 1 and 2.
Discussion
Colorectal carcinogenesis occurs through two distinct pathways: the conventional adenoma– carcinoma pathway involving APC, KRAS, and TP53 mutations, leading to MSS, aneuploid colorectal cancer, and the serrated polyp-carcinoma pathway involving BRAF mutation and promoter hypermethylation, which leads to CIMP-high and MSI-high colorectal cancer.9 There may be some overlap between these two pathways and the concept of a ‘fusion pathway’ to colorectal cancer has also been proposed.27 DNA methylation is currently believed to be an important pathogenetic mechanism in the serrated but not the conventional pathway of colorectal carcinogenesis. However, demonstration of distinct methylation clusters in CIMP-negative colorectal cancer raises the possibility that epigenetic changes may also play a role in the development and progression of conventional adenomas.8,10
The findings in our study suggest that epigenetic changes not only occur in conventional adenomas but also show distinctive methylation patterns in adenomas located in the right and left colon. A total of 168 CpG loci were differentially methylated and these loci not only included CpG islands (24.4%) but also CpG island shores (35.1%), CpG island shelves (4.2%), and CpG oceans (36.3%) (Figure 1b). None of the differentially methylated genes in these conventional adenomas involved a CIMP panel gene (Supplementary Table 1), suggesting that epigenetic changes in conventional adenomas represent a unique methylation signature that is independent of the traditional CIMP gene panel. Unlike the linear association between MSI-high and CIMP-high phenotypes and anatomic location of colorectal cancer shown in prior studies,26 our results indicate that a nonlinear relationship exists for the most differentially methylated CpG loci between right- and left-colon conventional adenomas.
Although the role of promoter CpG island methylation in epigenetic gene control and disease states is well established,28,29 the biologic significance of methylation involving CpG island shores, which are DNA regions up to 2kb adjacent to CpG islands, is uncertain, but recent data suggest that CpG island shores may be critical for normal development and somatic cell reprogramming.30 Aberrant DNA hypomethylation and hypermethylation involving CpG island shores has also been shown recently in colorectal cancer31 and to be concordant with gene expression by 5-aza-2′-deoxycytidine and DNA methyltransferase knockout experiments, highlighting the importance of these regions in epigenetic gene control. Whether DNA methylation outside of the CpG island merely sets up a field effect that increases the likelihood of developing an adenoma or is a critical pathogenic mechanism needs to be addressed in future studies.
The location-specific, differentially methylated CpG loci in our study were enriched for homeobox genes. The HOX family is the largest among homeo-box gene groups and encodes transcription factors critical for controlling cell growth and differentiation during embryonic and normal tissue development. Deregulated expression of HOX genes has been previously described in many solid tumors, including colorectal cancer, and derivative cell lines.32–36 There is also variation in HOX expression between colorectal cancer and normal colon mucosa. Among the 39 genes in the HOX group, 14 (HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXB5, HOXB6, HOXB9, HOXC4, HOXC5, HOXC6, HOXC9, HOXD9, and HOXD10) were represented in our list of differentially methylated loci (Supplementary Table 1). Of the 30 CpG loci associated with these 14 HOX genes, 10 were located in CpG islands and 20 in the CpG island shore regions. Of the 30 loci, 28 showed hypermethylation in left-sided adenomas. Expression levels of HOXB6 and HOXB9 are elevated in normal right colon as compared with the left, whereas HOXB13 and D13 show the reverse pattern, suggesting a role in normal colonic differentiation for these proteins.37 Expression of HOX genes, with the exception of HOXB13, has been shown to vary with the location of colorectal cancer, with the highest expression seen in proximal colon tumors.38 CpG island shore hypermethylation of PRAC and PRAC2 genes—prostate cancer susceptibility candidates—in adenomas of the right colon is also an unexpected and interesting finding in our study. PRAC2 lies between the HOXB13 and PRAC, and both PRAC2 and PRAC are predominantly expressed in prostate and distal colon and encode small nuclear proteins.39 Whereas little is known about PRAC and PRAC2 in the context of colorectal cancer, hypermethylation of PRAC has been described in solid tumors other than prostate carcinoma.40
CDX2 is a transcription factor that plays a major role in development and maturation of intestinal epithelium.41 Recent work has demonstrated that CDX2 activates expression of the proto cadherin Mucdhl, which interacts with β-catenin and regulates activities of intestinal cells, and loss of CDX2 expression can, therefore, reduce expression of Mucdhl and lead to malignancy.42 We have shown differential CpG island hypermethylation of the CDX2 gene in adenomas from the left colon as compared with the right colon. CDX2-negative colorectal cancers have been described as high-grade tumors, with a preferential distribution in the right colon that trend toward increased relapse and reduced survival.43,44 However, others have found CDX2-negative colorectal cancer to have no association with tumor location and survival.41 Regardless, the phenotype of CDX2-negative colorectal cancer is probably set very early in the course of tumor development, as our study shows CpG island hypermethylation of CDX2 gene at the earliest conventional adenoma stage.
CDX2 expression was evaluated in a selected sample of six conventional adenomas that showed DNA hypermethylation by array analysis and by pyrosequencing. Expression of CDX2 was reduced, to varying degrees, in four of six cases analyzed and diffuse strong staining was present in the remaining two. Interestingly, however, aberrant nuclear and/or cytoplasmic localization of β-catenin was present in all six adenomas, suggesting suppressed CDX2 activity (Supplementary Figure 7). These results suggest that the level of hypermethylation seen in these adenomas is insufficient to cause complete loss of protein expression, but may still have a functional impact on the Wnt signaling pathway.
Unexpectedly, a subset of patients with left-colon adenomas, who were more likely to be current smokers, exhibited a methylation profile that closely resembled that of right-colon adenomas (Figure 4). Smoking has been shown to alter the underlying signature of DNA methylation across a variety of different tissue types.20,22,45 Genomic methylation in normal rectal mucosa of smokers has been shown to be significantly lower than in nonsmokers, and in patients with adenoma compared with those without adenoma.46 This suggests that aberrant methylation of key genes in normal mucosa may be responsible for setting the stage for subsequent adenoma development. Heavy smokers (>20 cigarettes per day) have been shown to be at high risk for CIMP-high and BRAF-mutated colorectal cancer.47 Our findings suggest that smoking may also modulate the DNA methylation pattern of conventional adenomas in a site-specific manner.
In summary, our study provides evidence that aberrant DNA methylation occurs in conventional adenomas and is likely to be of pathogenetic significance. The side-specific methylation observed in conventional adenomas is independent of the CIMP-related genes and instead involves the intestinal transcription factor CDX2, the prostate cancer susceptibility genes PRAC and PRAC2, and a subset of homeobox genes, notably the HOX group. Smoking modulates the site-specific methylation signature, whereby left-colon adenomas may acquire a methylation profile similar to those of right-colon adenomas. The clinical impact of these findings and the interplay between epigenetic and genetic changes in the conventional pathway of carcinogen-esis needs to be explored in future studies.
Supplementary Material
Acknowledgments
We thank the New Hampshire Colonoscopy Registry staff, providers, and participants for their valuable contributions to this work. This work was supported by: CA078609, ES00002, 5 P42 ES05947, RR018787, R25CA134286, and LM009012 from the National Cancer Institute, NIH, National Institute of Environmental Health Sciences, NIH, and the National Library of Medicine, NIH.
Footnotes
Disclosure/conflict of interest: The authors declare no conflict of interest.
Supplementary Information accompanies the paper on Modern Pathology website (http://www.nature.com/modpathol)
References
- 1.American Cancer Society Cancer Facts and Figures 2012. Atlanta: 2012. [Google Scholar]
- 2.Brenner H, Hoffmeister M, Arndt V, et al. Protection from right- and left-sided colorectal neoplasms after colonoscopy: population-based study. J Natl Cancer Inst. 2010;102:89–95. doi: 10.1093/jnci/djp436. [DOI] [PubMed] [Google Scholar]
- 3.Baxter NN, Goldwasser MA, Paszat LF, et al. Association of colonoscopy and death from colorectal cancer. Ann Intern Med. 2009;150:1–8. doi: 10.7326/0003-4819-150-1-200901060-00306. [DOI] [PubMed] [Google Scholar]
- 4.Brenner H, Chang-Claude J, Rickert A, et al. Risk of colorectal cancer after detection and removal of adenomas at colonoscopy: population-based case-control study. J Clin Oncol. 2012;30:2969–2976. doi: 10.1200/JCO.2011.41.3377. [DOI] [PubMed] [Google Scholar]
- 5.Pohl H, Srivastava A, Bensen SP, et al. Incomplete polyp resection during colonoscopy-results of the complete adenoma resection (CARE) study. Gastroenterology. 2013;144:74–80. e1. doi: 10.1053/j.gastro.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 6.Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawasaki T, Ohnishi M, Nosho K, et al. CpG island methylator phenotype-low (CIMP-low) colorectal cancer shows not only few methylated CIMP-high-specific CpG islands, but also low-level methylation at individual loci. Mod Pathol. 2008;21:245–255. doi: 10.1038/modpathol.3800982. [DOI] [PubMed] [Google Scholar]
- 8.Hinoue T, Weisenberger DJ, Lange CP, et al. Genomescale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 10.Muzny DM, Bainbridge MN, Chang K, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butterly LF, Goodrich M, Onega T, et al. Improving the quality of colorectal cancer screening: assessment of familial risk. Dig Dis Sci. 2010;55:754–760. doi: 10.1007/s10620-009-1058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carney PA, Goodrich ME, Butterly LF, et al. The design and development of a population-based colonoscopy registry. J Registry Manag. 2006;33:91–99. [Google Scholar]
- 13.Marsit CJ, Christensen BC, Houseman EA, et al. Epigenetic profiling reveals etiologically distinct patterns of DNA methylation in head and neck squamous cell carcinoma. Carcinogenesis. 2009;30:416–422. doi: 10.1093/carcin/bgp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teschendorff AE, Zhuang J, Widschwendter M. Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies. Bioinformatics. 2011;27:1496–1505. doi: 10.1093/bioinformatics/btr171. [DOI] [PubMed] [Google Scholar]
- 16.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 17.Kuan PF, Wang S, Zhou X, et al. A statistical framework for Illumina DNA methylation arrays. Bioinformatics. 2010;26:2849–2855. doi: 10.1093/bioinformatics/btq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Grennan K, Badner J, et al. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS One. 2011;6:e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilhelm-Benartzi CS, Houseman EA, Maccani MA, et al. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ Health Perspect. 2012;120:296–302. doi: 10.1289/ehp.1103927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joubert BR, Haberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kakar S, Deng G, Cun L, et al. CpG island methylation is frequently present in tubulovillous and villous adenomas and correlates with size, site, and villous component. Hum Pathol. 2008;39:30–36. doi: 10.1016/j.humpath.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Mani S, Szymanska K, Cuenin C, et al. DNA methylation changes associated with risk factors in tumors of the upper aerodigestive tract. Epigenetics. 2012;7:270–277. doi: 10.4161/epi.7.3.19306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan ES, Qiu W, Baccarelli A, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012;21:3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barker DJ. Fetal and Infant Origins of Disease. BMJ Books; London: 1992. [Google Scholar]
- 25.Khashab MA, Pickhardt PJ, Kim DH, et al. Colorectal anatomy in adults at computed tomography colonography: normal distribution and the effect of age, sex, and body mass index. Endoscopy. 2009;41:674–678. doi: 10.1055/s-0029-1214899. [DOI] [PubMed] [Google Scholar]
- 26.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jass JR, Baker K, Zlobec I, et al. Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a ‘fusion’ pathway to colorectal cancer. Histopathology. 2006;49:121–131. doi: 10.1111/j.1365-2559.2006.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira HJ, Heyn H, Moutinho C et al. CpG island hypermethylation-associated silencing of small nucleolar RNAs in human cancer. RNA Biol. 2012;9 doi: 10.4161/rna.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenzen JM, Martino F, Thum T. Epigenetic modifications in cardiovascular disease. Basic Res Cardiol. 2012;107:245. doi: 10.1007/s00395-012-0245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aberdam D, Negreanu V, Sachs L, et al. The oncogenic potential of an activated Hox-2 4 homeobox gene in mouse fibroblasts. Mol Cell Biol. 1991;11:554–557. doi: 10.1128/mcb.11.1.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cillo C, Barba P, Freschi G, et al. HOX gene expression in normal and neoplastic human kidney. Int J Cancer. 1992;51:892–897. doi: 10.1002/ijc.2910510610. [DOI] [PubMed] [Google Scholar]
- 34.De Vita G, Barba P, Odartchenko N, et al. Expression of homeobox-containing genes in primary and metastatic colorectal cancer. Eur J Cancer. 1993;29A:887–893. doi: 10.1016/s0959-8049(05)80432-0. [DOI] [PubMed] [Google Scholar]
- 35.Maulbecker CC, Gruss P. The oncogenic potential of deregulated homeobox genes. Cell Growth Differ. 1993;4:431–441. [PubMed] [Google Scholar]
- 36.Peverali FA, D'Esposito M, Acampora D, et al. Expression of HOX homeogenes in human neuroblastoma cell culture lines. Differentiation. 1990;45:61–69. doi: 10.1111/j.1432-0436.1990.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 37.Kanai M, Hamada J, Takada M, et al. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol Rep. 2010;23:843–851. [PubMed] [Google Scholar]
- 38.Sanz-Pamplona R, Cordero D, Berenguer A, et al. Gene expression differences between colon and rectum tumors. Clin Cancer Res. 2011;17:7303–7312. doi: 10.1158/1078-0432.CCR-11-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsson P, Motegi A, Bera TK, et al. PRAC2: a new gene expressed in human prostate and prostate cancer. Prostate. 2003;56:123–130. doi: 10.1002/pros.10185. [DOI] [PubMed] [Google Scholar]
- 40.Arai E, Chiku S, Mori T, et al. Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis. 2012;33:1487–1493. doi: 10.1093/carcin/bgs177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baba Y, Nosho K, Shima K, et al. Relationship of CDX2 loss with molecular features and prognosis in colorectal cancer. Clin Cancer Res. 2009;15:4665–4673. doi: 10.1158/1078-0432.CCR-09-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinkel I, Duluc I, Martin E, et al. Cdx2 controls expression of the protocadherin Mucdhl, an inhibitor of growth and beta-catenin activity in colon cancer cells. Gastroenterology. 2012;142:875–885 e3. doi: 10.1053/j.gastro.2011.12.037. [DOI] [PubMed] [Google Scholar]
- 43.Knosel T, Chen Y, Hotovy S, et al. Loss of desmocollin 1-3 and homeobox genes PITX1 and CDX2 are associated with tumor progression and survival in colorectal carcinoma. Int J Colorectal Dis. 2012;27:1391–1399. doi: 10.1007/s00384-012-1460-4. [DOI] [PubMed] [Google Scholar]
- 44.Bauer KM, Hummon AB, Buechler S. Right-side and left-side colon cancer follow different pathways to relapse. Mol Carcinog. 2012;51:411–421. doi: 10.1002/mc.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wangsri S, Subbalekha K, Kitkumthorn N, et al. Patterns and possible roles of LINE-1 methylation changes in smoke-exposed epithelia. PLoS One. 2012;7:e45292. doi: 10.1371/journal.pone.0045292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paun BC, Kukuruga D, Jin Z, et al. Relation between normal rectal methylation, smoking status, and the presence or absence of colorectal adenomas. Cancer. 2010;116:4495–4501. doi: 10.1002/cncr.25348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samowitz WS, Albertsen H, Sweeney C, et al. Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst. 2006;98:1731–1738. doi: 10.1093/jnci/djj468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.