Abstract
The goal of this study was to characterize how depleted uranium (DU) causes DNA damage. Procedures were developed to assess the ability of organic and inorganic DNA adducts to convert to single strand breaks (SSB) in pBR322 plasmid DNA in the presence of heat or piperidine. DNA adducts formed by methyl methanesulfonate (MMS), cis-platin (cis-Pt), and chromic chloride were compared to those formed by reaction of uranyl acetate (UA) and ascorbate (Asc). Uranyl ion in the presence of Asc produced U-DNA adducts that converted to SSB upon heating. Piperidine, which acted on DNA methylated by MMS to convert methyl-DNA adducts to SSB, served in the opposite fashion with U-DNA adducts by decreasing SSB. The observation that piperidine also decreased the gel shift for metal-DNA adducts formed by monofunctional cis-Pt and chromic chloride was interpreted to suggest that piperidine served to remove U-DNA adducts. Radical scavengers did not affect formation of U-induced SSB, suggesting that SSB arose from the presence of U-DNA adducts and not from free radicals. A model is proposed to predict how U-DNA adducts may serve as initial lesions that convert to SSB or AP sites. Results suggest that DU can act as a chemical genotoxin that does not require radiation for its mode of action. Characterizing the DNA lesions formed by DU is necessary to assess the relative importance of different DNA lesions in the formation of DU-induced mutations. Understanding mechanisms of formation of DU-induced mutations may contribute to identification of biomarkers of DU exposures in humans.
Keywords: depleted uranium, DNA damage, gel electrophoresis, heavy metal, pBR322 plasmid DNA
Introduction
Uranium is an emerging toxicant whose links to cancer have been established, but whose interactions with biological molecules have not been fully characterized. It is a unique toxic metal because it displays both radiological and chemical toxicity. The radiological toxicity of uranium, ascribed to the alpha and beta emissions produced through the decay of the 238U, 235U and 234U isotopes, has been well established through human exposures occurring during mining and processing of the natural ore, with lung cancer being a major outcome [1, 2]. Possible associations between uranium mining and other cancers such as leukemia, malignant melanoma, and gallbladder cancer have also been noted [3, 4]. Communities impacted by uranium mining over the past several decades remain challenged by health risks into the present day [5–7]. In addition, concerns are expanding to include uranium’s chemical toxicity as a heavy metal, largely due to the expanding use of depleted uranium (DU) in the military [8–10].
The depleted form of uranium, consisting mostly of 238U after the fissionable 235U isotope has been extracted from natural uranium, is used by the military in armor-piercing projectiles and tank armor. The extensive use of DU in recent global conflicts has created questions regarding possible health risks for both soldiers and civilians. Exposure to DU has been cited as a major concern for veterans reporting a range of physical or mental health issues [11, 12]. Results from epidemiological studies are sparse; however, the Royal Society did conclude that a 2-fold increase in lung cancer risk could exist for soldiers with the highest exposures to DU on the battlefield [13]. Continued surveillance and monitoring of DU-exposed veterans has found detectable increases in uranium concentrations in urine and sperm; however, no significant adverse health effects have been reported [14–16]. Birth defects and cancers including leukemia, lymphoma, lung, breast, stomach, bladder and skin, have been alleged for civilians exposed to DU [17–20], but correlation of observed illnesses to DU exposure specifically is confounded by exposures to other genotoxic agents including chemical warfare agents, and polycyclic aromatic hydrocarbons generated by the burning of oil fields [19].
The major exposure routes for DU are inhalation of uranium oxides, predominantly U3O8, UO2, and UO3, that are formed from the burning of metallic DU [21, 22], and dermal absorption of uranium from DU-containing shrapnel embedded in the skin [23]. DU munitions, DU shrapnel, and the uranium oxides that form upon burning will dissolve in the environment [9, 24], in simulated gastrointestinal fluid [25], and in vivo [14, 15, 26]. Therefore, understanding the toxicology of soluble uranium may provide relevant insight into all long-term uranium exposures. Furthermore, the chemistry of uranium is identical for the 238U, 235U and 234U isotopes; therefore, DU is the best model to explore the chemical toxicity that could occur from any type of uranium exposure, natural or depleted.
In terms of its chemical properties, U(VI) is the most stable oxidation state under aqueous biological conditions, and it exists in the form of uranyl ion UO22+. Uranyl ion is a hard Lewis acid [27] preferring to coordinate with oxygen-containing ligands, for example, carboxylate and phosphate, which explains its preference for targeting proteins [28] and nucleic acids [29]. Thus uranium has significant potential to interfere with biological processes in many different ways, and at high enough exposures is expected to have adverse health effects. However, information that is still needed to fully assess these health affects includes an understanding of the exposure levels necessary to observe adverse effects, and of the biomarkers of exposure that point to a uranium-relevant endpoint.
The purported link between DU and either cancers or birth defects is supported by the results of numerous genotoxicity studies. Soluble uranyl ion has been found to be clastogenic, transforming, and aneugenic in cell culture [30–32]. Mutations at the hprt locus have been reported for soluble uranyl ion in Chinese Hamster Ovary (CHO) and V79 cells [8, 33], as well as in peripheral T-cells of gulf war veterans exposed to DU containing shrapnel [34]. Characterization of the mutagenic spectrum in CHO EM9 cells exposed to uranyl acetate (UA) showed more small multiexon 1–22 bp deletions and 1–2 bp insertions than were observed in either spontaneously-generated or H2O2-induced hprt mutants [35]. This mutation spectrum was also distinct from that observed for radon (i.e., alpha irradiation) in CHO cells, in which whole gene deletions were more prevalent [36]. It is not yet understood which specific DNA lesions are responsible for which, if any, of the clastogenic or mutagenic endpoints.
This increasing body of evidence illustrating the genotoxicity of DU points to a need to understand interactions between uranium and DNA at the molecular level. Several types of DNA lesions have been reported from DU exposures, including DNA strand breaks [33, 37], U-DNA adducts [33], abasic sites [Yellowhair et al., in preparation], and 8-oxodG in the combined presence of H2O2 [38]. The mechanism for formation of these different DNA lesions is the focus of the current work.
Ascorbate (vitamin C, Asc) has previously been shown to activate UA toward DNA damage [37]. This activation is similar to that observed for chromium(VI), another genotoxic, mutagenic, clastogenic and carcinogenic heavy metal [39]; however, the reaction between chromate and Asc is one of electron transfer under physiologically relevant conditions, producing reactive Cr(V), Cr(IV) and organic free radicals [40]. Conversely, the electron transfer reaction between uranyl ion and Asc is favored only at a pH < 2 [41], whereas at neutral pH Asc predominantly acts as a ligand for uranyl ion [42]. In the presence of DNA, reactions of uranyl ion and Asc produced DNA strand breaks that were inferred to be formed through binding of an Asc-bound uranyl ion to the DNA phosphate backbone, which subsequently promotes DNA backbone hydrolysis [37]. Thus the reaction of uranyl ion with Asc serves as one model system for the formation of U-DNA adducts and DNA single strand breaks.
Based on the above observations, that redox chemistry for uranyl ion is limited at neutral pH, that oxidative DNA damage is not observed in the absence of added H2O2, and that uranyl ion is known to have a strong association with the DNA phosphate backbone, we are exploring the general hypothesis that uranyl-DNA adducts may serve as a “parent lesion” for other types of secondary DNA damage such as abasic sites and single strand breaks. The purpose of the current study was to further characterize the stability of the DNA lesions formed by reactions of Asc with DU as UA in vitro, specifically by exploring the presence of heat-labile or piperidine- labile adducts in plasmid DNA. The hypothesis being tested was that if uranium reacts with DNA in the presence of Asc to form labile U-DNA adducts, then exposure of U-DNA adducts to heat could convert those adducts to DNA strand breaks that can be visualized by gel electrophoresis. The stability of these adducts with respect to formation of alkali-labile sites was further explored by exposing them to warm piperidine in the manner used to analyze alkylated guanine bases by the Maxam-Gilbert chemical method of DNA sequencing [43]. A protocol was developed in which unreacted DNA was not affected by heat or warm piperidine exposure, and this protocol was used to compare damage in plasmid DNA exposed to methyl methanesulfonate (MMS), to the DNA-reactive metals chromic chloride and cis-platin (cis-Pt), and to reactions of UA and Asc. Results supported our hypothesis in that the U-DNA lesions were heat labile; however, piperidine treatment did not result in an increase in DNA strand breaks for any of the metals tested.
Materials and Methods
Materials
Uranyl acetate dihydrate (CAS 6159–44–0, UA) was obtained from Spectrum Chemical Mfg. Corp. (Gardena, CA) and used as received. The 234U/238U atom ratios were determined by ICP-MS, and found to be 7.6 ± 2×10−6, consistent with DU (Dr. M. E. Ketterer, unpublished). The 234U/238U atom ratios expected for natural uranium would be on the order of 55×10−6. [44]. Methyl methane sulfonate was obtained from Spectrum Chemical (Gardena, CA). Chromic chloride was obtained from JT Baker (Phillipsburg, NJ). Cis-diamminedichloroplatinum(II) (cis-Pt) was obtained from Selleck Chemical (Houston, TX). Sodium ascorbate (Asc) was obtained from Alfa Aesar (Ward Hill, MA). N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) buffer (VWR International, Radnor, PA), was prepared as 33.3 mM stock solutions, and the desired pH was obtained by addition of NaOH or HCl at 37 °C. Buffers and Asc solutions in buffer were treated with Chelex 100 resin (Bio-Rad Laboratories, Hercules, CA) followed by 0.2 μm filtration to remove trace metals. The pBR322 plasmid was obtained from New England Biolabs (Ipswich, MA) and diluted in water to 0.7 mM DNA-P and reacted at concentrations of 0.2 mM DNA-P. All metal solutions were prepared immediately before use. The final pHs of reaction solutions were measured in mock 2 mL reaction solutions. All reactions with MMS, cis-Pt, and UA in pH 7.4 ACES buffer yielded a final reaction pH of 7.40 ± 0.05 at 37 °C. Reactions with chromic chloride yielded a final pH of 6.60 ± 0.01 at 37 °C.
DNA incubations with MMS or metal complexes
Reactions of pBR322 plasmid DNA (0.2 mM DNA-P) were carried out with MMS, chromic chloride or cis-Pt in 25.0 mM ACES buffer for 30 min or 24 hr at 37 °C. Concentrations of MMS were 1.0, 5.0, or 10 mM, concentrations of chromic chloride were 100, 200, or 300 μM, and concentrations of cis-Pt were 4.0, 6.0, and 8.0 μM.
Reactions of UA (500 μM), Asc (500 μM) and DNA (0.2 mM DNA-P) were carried out in 25.0 mM ACES buffer for 30 min at 37 °C. Control reactions consisted of untreated pBR322 DNA, and pBR322 DNA reacted individually with UA and Asc. Experiments were also repeated in the presence of 500 μM mannitol and 91 U/mL catalase in order to evaluate the involvement of free radicals and hydrogen peroxide.
Heat-induced and piperidine-induced DNA degradation
Reactions of pBR322 DNA with MMS, chromic chloride, cis-Pt, or UA and Asc were carried out as described above, in triplicate. After the 37 °C incubations, three different post-treatment exposures were carried out to measure the affect of heat and piperidine on the reaction-induced DNA lesions. One set of samples serving as the control was placed in an unheated (RT) heat block for 30 min after receiving a volume of water equivalent to that for the piperidine exposures. The second set of samples, representing the heat treatment, was incubated with an equal volume of water in the absence of piperidine for 30 min in a heat block at 60 °C. The third set of samples was incubated for 30 min at 60 °C with 30 μM piperidine. Samples were then treated with 2 μL loading dye and analyzed by gel electrophoresis.
Gel Electrophoresis
Relaxation of supercoiled pBR322 plasmid DNA was observed by gel electrophoresis on 1% agarose gels with 0.5× TBE running buffer (45 mM Tris base, 45 mM boric acid, and 1 mM EDTA) at 120 V for 90 min. Gels were stained with ethidium bromide, destained with water, and images were digitally captured on a FluoroChem SP Camera (Cell Biosciences, Inc., Santa Clara, CA). The percent plasmid relaxation relative to supercoiled DNA was quantified from digital images using UN-SCAN-IT gel software, Macintosh version 5.3 (Silk Scientific Inc., Orem, UT). Data are presented as mean ± SEM for n = 4–14 independent experiments.
Statistics
Multiple comparisons among group means were accomplished by paired ANOVA followed by a Tukey HSD post hoc test. The statistical significance of the effect of piperidine, mannitol or catalase on plasmid relaxation was evaluated for differences of ± treatment by a paired Student’s t-test. Differences were considered significant at p < 0.05 for both tests. Statistical outliers were verified by Grubbs’ test (extreme studentized deviate method), p < 0.05.
Results and Discussion
Protocol optimization
The first aim of this work was to verify the conditions that would allow for detection of MMS and metal-induced DNA lesions that were heat- or piperidine-sensitive, but would not produce interference from degradation of untreated DNA. This was accomplished by determining the maximum temperature and piperidine concentration that would not result in observable effects in untreated plasmid DNA.
Samples of untreated pBR322 DNA were incubated in buffer for 30 min at a series of temperatures from 37 – 80 °C. A representative gel illustrating the effect of temperature on degradation of untreated plasmid DNA is provided in Fig. S1A, and the quantification of differences in amounts of form II plasmid for heated samples vs. RT samples for multiple independent experiments is provided in Fig. S1B. Heating plasmid DNA at 37, 50, or 60 °C for 30 min did not produce a significant change in relative amounts of relaxed (form II) or supercoiled (form I) plasmid relaxation relative to DNA incubated at RT for 30 min; however, incubation of DNA at 70 or 80 °C resulted in a 10-fold, and 3-fold loss of relaxed DNA relative to DNA incubated at RT, respectively. These results were consistent with the previous reports that the melting temperature of supercoiled pBR322 plasmid was greater than 70 °C [45]. These data showed that for the proposed experiments an incubation temperature of 60 °C for 30 min should not affect untreated plasmid DNA.
The affect of piperidine on the degradation of untreated plasmid DNA was determined at 60 °C. A representative gel illustrating the effect of 15 – 60 μM aqueous piperidine on degradation of untreated plasmid DNA at RT vs. 60 °C is provided in Fig. S2A, and the quantification of differences in amounts of form II plasmid for samples with and without piperidine is provided in Fig. S2B. Data showed that incubations of plasmid DNA with 15 – 60 μM aqueous piperidine for 30 min without heating (i.e., RT) did not affect relaxation of supercoiled plasmid. Upon heating at 60 °C, the concentrations of 15 and 30 μM aqueous piperidine had no statistically significant effect on plasmid relaxation after a 30 min; however, the 60 μM concentration of piperidine decreased relaxed plasmid DNA levels of 1.4-fold relative to DNA heated at 60 °C for 30 min in the absence of piperidine. The nonspecific degradation of DNA by piperidine has been noted by others [46]. The current data were interpreted to suggest that incubation of untreated plasmid DNA with ≤30 μM piperidine for 30 min at 60 °C would not result in degradation of DNA in the absence of chemically-induced lesions that could be converted to single strand breaks by hydrolysis.
Effects of heat and piperidine on DNA adducts induced by MMS
Methyl methanesulfonate (MMS) was used as a positive control for the formation of adducts at DNA bases. MMS is known to form N7-methyl guanine (N7-MeG) as the major DNA adduct, as well as minor amounts of N3-MeG and O6-MeG, and N1-, N3-, and N7-methyl adenine (N7-MeA) [47]. Although the N7-MeG adduct may be less relevant than the O6-MeG adduct in terms of mutagenesis, it is proposed to serve as a biomarker for DNA alkylation in vivo, with the caveat that the N7-MeG lesion itself is unstable [48]. The instability of the N7- MeG lesion toward depurination and strand break formation, which can be enhanced in the presence of aqueous piperidine, serves as the basis for Maxam-Gilbert DNA sequencing [43].
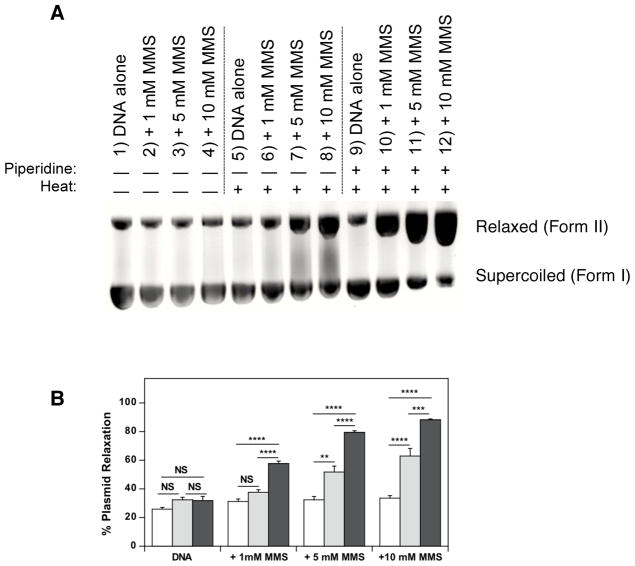
MMS was reacted with pBR322 DNA in 25.0 mM ACES (pH 7.4, 37 °C) at concentrations of 0, 1.0, 5.0, and 10 mM for 30 min, 37 °C in triplicate. One set of reactions was then incubated for 30 min with 30 μM aqueous piperidine at 60 °C, one set was incubated with an equivalent volume of water at 60 °C, or an equivalent volume of water at RT. All samples were then subjected to gel electrophoresis (Fig. 1). MMS had no affect on the gel migration of plasmid DNA for the 30 min incubation at 37 °C followed by a 30 min incubation at RT (Fig. 1A, lanes 2–4 vs. lane 1, quantified in Fig. 1B). As expected, the methyl-DNA adducts were slightly heat labile (Fig. 1A, lanes 6–8 vs. 2–4, quantified in Fig. 1B), and the methyl-DNA adducts were strongly piperidine sensitive (Fig. 1A, lanes 10–12 vs. 2–4, quantified in Fig. 1B). Post-incubation treatments with heat or heat + piperidine had no significant effect on plasmid relaxation for unreacted DNA (Fig. 1B). Heat in the absence of piperidine caused a significant increase in plasmid relaxation for the 5.0 mM and 10 mM MMS treatments of 1.6-fold and 1.9- fold, respectively. Heat and piperidine caused significant increases in plasmid relaxation for the 1.0 mM, 5.0 mM and 10 mM doses of 1.8-fold, 2.4-fold and 2.6-fold, respectively. Thus this modified Maxam-Gilbert method produces the expected result for conversion of a methyl-DNA adduct to a strand break.
Figure 1.
Effects of heat and piperidine incubations on pBR322 plasmid DNA degradation after exposure to methyl methanesulfonate (MMS). (A) Representative gel illustrating reaction of MMS (0 – 10 mM) with pBR322 DNA (0.2 mM DNA-P, 25 mM ACES, pH 7.4, 37 °C, 30 min) followed by post-treatment exposure to water (30 min, RT) (lanes 1–4), water and heat (30 min, 60 °C) (lanes 5–8) or 30 μM piperidine and heat (30 min, 60 °C) (lanes 9–12). (B) Quantification of DNA degradation as % DNA migrating as Form II for post-treatment exposure to either water (30 min, RT) (open bars); water (30 min, 60 °C) (grey bars); or 30 μM piperidine (30 min, 60 °C) (black bars). Data represent mean ± SEM for n = 5 independent experiments. Statistical significance of the effect of ± heat or ± piperidine was determined by ANOVA (NS not significant, **p<0.01, ***p<0.001, ****p<0.0001).
Effects of heat and piperidine on DNA adducts induced by cis-platin and chromic chloride
The next aim of this work was to establish the effects of heat and piperidine on plasmid DNA exposed to metal compounds whose interactions with DNA are relatively better characterized than interactions of DNA with uranyl ion. Cis-Pt and chromic chloride are examples of metals that are known to form DNA adducts in the absence of single-strand breaks. Cis-Pt interactions with DNA have been well characterized. The initial interaction between Pt and DNA produces a monofunctional purine adduct that converts to a bifunctional adduct over time [49, 50]. The major bifunctional adduct is a 1,2-intrastrand crosslink at the N7 positions of two adjacent guanines (Pt-GG, 65%), with minor adducts including a 1,2-intrastrand crosslink (Pt-AG, 25%), and a 1,3-intrastrand crosslink (Pt-GNG, 10%) [49]. The bifunctional Pt-dGpG adduct in a duplex DNA oligonucleotide has been structurally characterized [51]. Interaction of cis-Pt with duplex DNA produces distorting lesions at purine bases with an unwinding angle of 13°C [52].
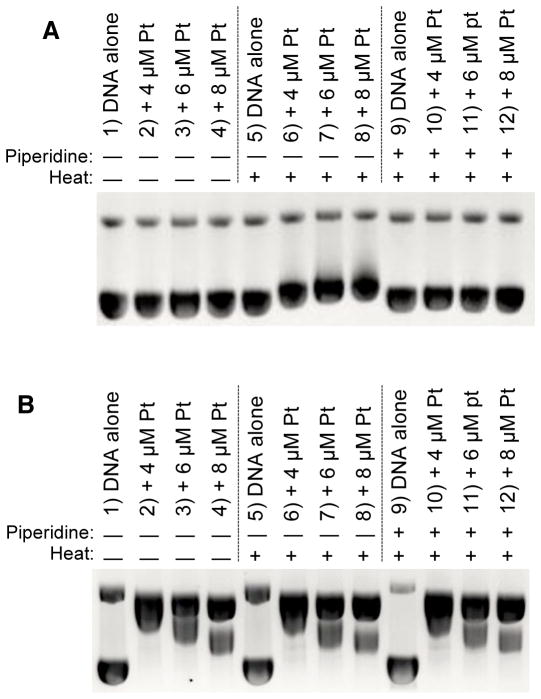
The affect of heat and piperidine on Pt-DNA lesions was determined in the current experimental system at two different reaction times, to favor generation of either monofunctional or bifunctional Pt-DNA adducts. Samples of pBR322 plasmid DNA were incubated in triplicate at 37 °C with 4.0, 6.0, and 8.0 μM cis-Pt in 25.0 mM ACES (pH 7.4, 37 °C) for either 30 min or 24 hr. Each set of reactions was then further incubated for 30 min as described above, at RT without piperidine, at 60 °C without piperidine, or at 60 °C with 30 μM aqueous piperidine, followed by gel electrophoresis. Results showed that heat and piperidine had different effects on platinated DNA depending on the time allowed for reaction of Pt with DNA (Fig. 2).
Figure 2.
Effects of heat and piperidine incubations on pBR322 plasmid DNA gel shifts after exposure to cis-platin for 30 min or 24 h. (A) Representative gel illustrating reaction of cis-platin (0 – 8 μM) with pBR322 DNA (0.2 mM DNA-P, 25 mM ACES, pH 7.4, 37 °C, 30 min) followed by post-treatment exposure to water (30 min, RT) (lanes 1–4), water and heat (30 min, 60 °C) (lanes 5–8) or 30 μM piperidine and heat (30 min, 60 °C) (lanes 9–12). (B) Representative gel illustrating reaction of cis-platin (0 – 8 μM) with pBR322 DNA (0.2 mM DNA-P, 25 mM ACES, pH 7.4, 37 °C, 24 h) followed by post-treatment exposure to water (30 min, RT) (lanes 1–4), water and heat (30 min, 60 °C) (lanes 5–8) or 30 μM piperidine and heat (30 min, 60 °C) (lanes 9–12).
For the 30 min reactions (Fig. 2A), samples post-treated at RT or post-treated with heat and piperidine showed no changes in amounts of relaxed DNA and no gel shift of supercoiled DNA (Fig. 2A, lanes 2–4 vs. lane 1, and 10–12, vs. lane 9). However, a gel shift was observed for supercoiled DNA bands in samples incubated at 60 °C in the absence of piperidine (Fig. 2A, lanes 6–8 vs. lane 5). These results were interpreted to suggest that the 30 min incubation with cis-Pt produced predominantly monofunctional (nondistorting) Pt-DNA adducts that were to some extent converted to bifunctional (distorting) Pt-DNA adducts during the 60 °C post-treatment incubation. The presence of 30 μM piperidine in heated samples was interpreted to be interfering with the conversion of monofunctional adducts to bifunctional adducts, either by removing the Pt from DNA, acting as a competing ligand with the N7-purine positions, or substituting for H2O in the coordination sphere to block bifunctional coordination at the N7-purine positions.
For the 24 hr reactions of cis-Pt with DNA, none of the three post-treatment exposures had an affect on the gel shift band patterns (Fig. 2B). Samples incubated with cis-Pt then post-treated at RT showed a dose response for the generation of distorting Pt-DNA adducts that unwound supercoiled DNA past the coalescence point, which is the point at which DNA adducts unwind supercoiled DNA to the extent that it co-migrates with relaxed DNA [53], to create a positive supercoil that increased with increasing dose of Pt (Fig. 2B, lanes 2–4 vs. 1) [see also 54]. Post-treatment incubations at 60 °C in the absence or presence of piperidine had no affect on gel shifts (Fig. 2B lanes 6–8 and 10–12 vs. lanes 2–4). Data were interpreted to suggest that the 24 hr incubation of cis-Pt with DNA was sufficient to saturate the DNA with stable bifunctional Pt-DNA adducts. The coordination of Pt in a bifunctional DNA adduct was not further changed by additional heating at 60 °C nor by the presence of piperidine. This lack of ability for piperidine to induce a strand break at Pt-DNA intrastrand N7-G adducts has been noted by others [55].
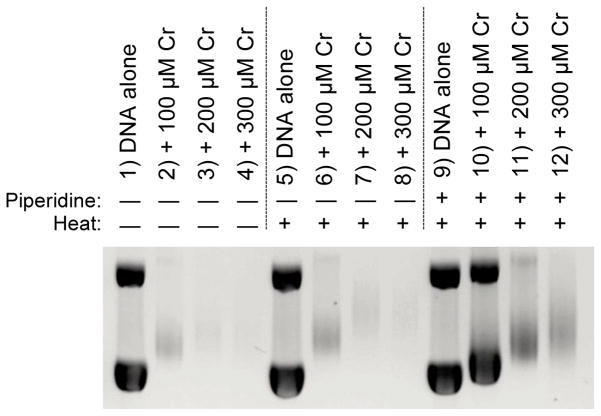
Chromic chloride forms Cr(III)-DNA adducts that have not been structurally characterized, but have been proposed to form predominantly at the DNA phosphate backbone [56, 57]. The interaction of Cr(III) with plasmid DNA has been found to be a relatively non-distorting lesion, with an unwinding angle of ~1°, consistent with an interaction at the phosphate backbone [56]. Samples of pBR322 plasmid DNA were incubated with 100, 200, and 300 μM CrCl3-6H2O in 25.0 mM ACES buffer (pH 6.6, 37 °C) for 24 hr at 37 °C. Each set of reactions was then further incubated for 30 min as described above, at RT without piperidine, at 60 °C without piperidine, or at 60 °C with 30 μM aqueous piperidine, followed by gel electrophoresis.
Similar to results with cis-Pt, reactions of plasmid DNA with 100–300 μM CrCl3 produced a gel shift of the supercoiled DNA bands (Fig. 3) that was consistent with previous work [56]. The requirement for a relatively high concentration range of Cr(III) to produce the observed gel shift is consistent with the interpretation that the resulting Cr(III)-DNA adducts are less distorting than those produced by cis-Pt. Without the addition of post-treatment heating at 60°C or piperidine, increasing concentrations of Cr(III) produced an increasing gel shift (Fig. 3A, lanes 2–4). The faintness of the bands was interpreted as being due to Cr-DNA binding interfering with ethidium bromide intercalation. Post-incubation of samples at 60 °C produced no discernable difference in the gel band shift (Fig. 3A, lanes 6–8 vs. lanes 2–4). This experimental system is constrained by the concentrations of Cr(III) necessary to produce a gel shift, 100–300 μM, and the concentration of piperidine that did not produce affects on untreated DNA, 30 μM, which result in Cr(III) being present in excess of piperidine. Nevertheless, post-treatment exposure of Cr-treated DNA to heat and piperidine decreased the loss of DNA bands in lanes containing Cr-treated plasmid, and lessened the extent of the gel shift of the supercoiled bands (Fig. 3A, lanes 10–12 vs. lanes 2–4 and 6–8). This observation was consistent with the interpretation that, similar to that for the Pt-DNA monofunctional adducts, piperidine acted as a competing ligand for Cr(III), removing it from the DNA phosphate backbone. If the Cr-DNA adducts produced unwinding of the supercoiled plasmid due to distortion of the double helix, then adduct removal due to Cr-piperidine chelation would result in a lower amount of modified DNA observed as a gel shift during electrophoresis. It was inferred that piperidine’s effect on Cr(III)-DNA unwinding was less than its effect on Pt(II)-DNA unwinding, observed as a less than complete abrogation of the gel shift, because Cr(III) was present in excess relative to piperidine.
Figure 3.
Effects of heat and piperidine incubations on pBR322 plasmid DNA gel shifts after exposure to chromic chloride for 24 h. Representative gel illustrating reaction of CrCl3•6H2O (0 – 300 μM) with pBR322 DNA (0.2 mM DNA-P, 25.0 mM ACES, pH 6.5, 37 °C, 24 hr) followed by post-treatment exposure to water (30 min, RT) (lanes 1–4), water and heat (30 min, 60 °C) (lanes 5–8) or 30 μM piperidine and heat (30 min, 60 °C) (lanes 9–12).
Thus the effects of heat and piperidine on modified plasmid DNA were verified with relatively well-characterized chemicals that form direct DNA lesions. Methyl-DNA adducts formed from MMS were resistant to heat but converted to strand breaks in the presence of piperidine. Monofunctional Pt-DNA adducts were converted to bifunctional adducts in the presence of heat but were removed from DNA by piperidine. Bifunctional Pt-DNA adducts were resistant to heat and piperidine. Chromium-DNA adducts were resistant to heat but were removed by piperidine. Therefore, although metal-DNA adducts do not behave as organic-DNA adducts under these modified Maxam-Gilbert methods, i.e., warm piperidine does not convert a purine-bound metal-DNA adduct nor a phosphate-bound metal-DNA adduct to a strand break, the presence of piperidine may be used to probe the presence of metal-DNA adducts, especially those that are relatively non-distorting to the DNA double helix.
Effect of heat and piperidine on DNA adducts induced by uranyl ion and ascorbate
Uranyl ion has been reported to cause U-DNA adducts [37] efficiently repaired AP sites [Yellowhair et al., in preparation], and single strand breaks [37]; however, the relationship among these lesions, if any, is not understood. It was hypothesized that if U-DNA adducts could serve as initial lesions that could convert to AP sites or strand breaks, then U-DNA adducts in plasmid DNA should be heat or piperidine sensitive.
Samples of pBR322 plasmid DNA were incubated in triplicate at 37 °C with 500 μM UA, 500 μM Asc, or combined exposures in 25.0 mM ACES (pH 7.4, 37 °C) for 30 min. Each set of reactions was then further incubated for 30 min as described above, at RT without piperidine, at 60 °C without piperidine, or at 60 °C with 30 μM aqueous piperidine, followed by gel electrophoresis (Fig. 4).
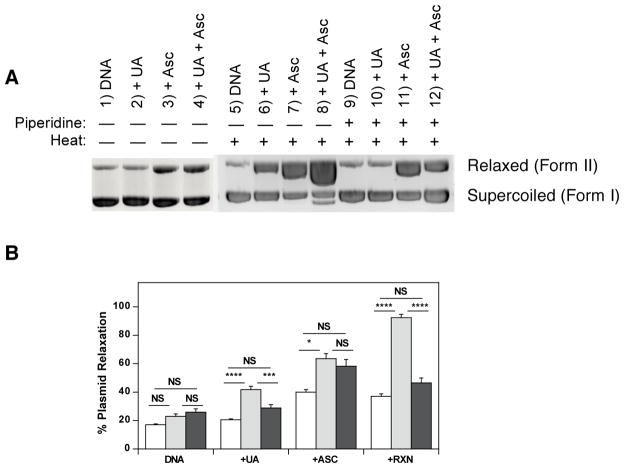
Figure 4.
Effects of heat and piperidine post-treatment incubations on pBR322 plasmid DNA degradation measured as % DNA plasmid relaxation (Form II) after exposure to uranyl acetate and ascorbate. (A) Representative gel illustrating reactions of UA (0.50 mM) and ascorbate (0.50 mM) with pBR322 DNA (0.2 mM DNA-P, 25.0 mM ACES, pH 7.4, 37 °C, 30 min) followed by post-treatment exposure to either water (30 min, RT) (left); water (30 min, 60 °C) (center); or 30 μM piperidine (30 min, 60 °C) (right). (B) Quantification of DNA degradation as % DNA migrating as Form II for post-treatment exposure to either water (30 min, RT) (open bars); water (30 min, 60 °C) (grey bars); or 30 μM piperidine (30 min, 60 °C) (black bars). Data represent mean ± SEM for n = 4–14 independent experiments. Statistical significance of the effect of ± heat or ± piperidine was determined by ANOVA (NS not significant, *p<0.05, **p<0.01, ****p<0.0001).
Reactions of UA and Asc that were not subjected to further heat were consistent with previous results [37]; however, less plasmid relaxation was observed at the lower reactant concentrations in the current study (500 μM vs. 1 mM) (Fig. 4A, lanes 1–4, quantified in Fig. 4B). No significant plasmid relaxation was observed for UA exposure alone, but DNA exposed to Asc alone or to Asc and uranyl ion produced 2-fold higher plasmid relaxation relative to untreated DNA (p < 0.0001). The plasmid relaxation induced by Asc alone was attributed to the presence of trace iron and copper in either the buffer or Asc solutions that were not completely removed by Chelex-100 resin [58].
Reactions of UA and Asc that were further subjected to heating at 60 °C showed an increase in plasmid relaxation for all reactants relative to the corresponding untreated DNA control (Fig. 4A, lanes 5–8, quantified in Fig. 4B). Reactions of UA alone produced almost 2-fold more plasmid relaxation than untreated DNA (p < 0.0001), whereas reactions of Asc alone or UA and Asc together produced 2.8-fold and 4.0-fold higher plasmid relaxation than was measured in untreated DNA, respectively (p < 0.0001).
Reactions of UA and Asc that were subjected to heat and piperidine showed no significant plasmid relaxation from exposure to UA alone, but increased relaxation for reactions of Asc alone or UA and Asc together relative to the corresponding untreated DNA control (Fig. 4A, lanes 9–12, quantified in Fig. 4B). Reactions of Asc alone or UA and Asc together produced over 2-fold higher plasmid relaxation than was measured in untreated DNA (p < 0.0001).
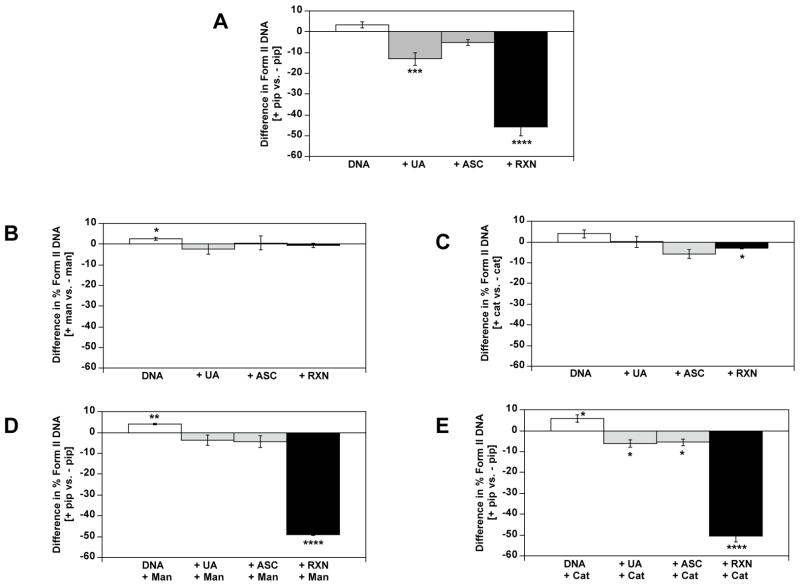
Comparison of the effects of heat and piperidine on each reaction sample showed that piperidine had the strongest influence on DNA samples that contained uranium. There were no statistical differences in levels of plasmid relaxation among untreated DNA controls for any of the three post-treatment exposures (Fig. 4A, lanes 1, 5, and 9, quantified in Fig. 4B). Reactions of DNA with Asc alone (Fig. 4A, lanes 3, 7, and 11) produced 1.6-fold more plasmid relaxation with 60 °C heating (p < 0.05, Fig. 4B); however, 60 °C piperidine exposures did not significantly change plasmid relaxation levels relative to 60 °C heat exposure in the absence of piperidine (p > 0.07, Fig. 4B). In contrast, reactions of plasmid DNA with UA alone (Fig. 4A, lanes 2, 6, and 10) or UA and Asc (Fig. 4A, lanes 4, 8, and 12) produced 2.1-fold and 2.5-fold more plasmid relaxation, respectively with 60 °C post-incubation heating (p < 0.0001, Fig. 4B). Furthermore, samples that were exposed to heat and piperidine showed decreased plasmid relaxation levels that were statistically equivalent to samples that were not subjected to 60 °C heating (Fig. 4B). Thus the presence of piperidine prevented the heat-induced plasmid relaxation in samples exposed to either UA alone or UA and Asc. Results are consistent with the interpretation that uranyl binding to DNA precedes the generation of a DNA strand break. Two modes of action are possible: either piperidine competes with DNA for binding to uranyl ion or the addition of piperidine to open coordination sites on uranium in a uranyl-DNA adduct prevents further processing of a uranyl-DNA adduct to a strand break.
Effect of mannitol and catalase on DNA damage induced by uranyl ion and ascorbate
The last set of experiments explored the effects of mannitol and catalase on the generation of DNA lesions during the reactions of uranyl and Asc with DNA. It was hypothesized that if the reaction of uranyl and Asc produced reactive oxygen species through Fenton-type chemistry, then the addition of mannitol or catalase should decrease the levels of plasmid relaxation observed. Previous work had shown that the levels of strand breaks induced by reactions or UA and Asc were not affected by co-incubation with the free radical scavengers mannitol, sodium azide, or 5,5-dimethyl-1-pyrroline-N-oxide, but were slightly decreased in the presence of the H2O2 scavenger catalase [37]. Reactions of UA and Asc with plasmid DNA were repeated in the presence of 500 μM mannitol or 91 U/mL catalase, and subjected to post-treatment exposure to either heat, or heat and piperidine, followed by gel electrophoresis. Amounts of relaxed (form II) DNA were quantified from gels as described above, and comparisons were made for ± mannitol, ± catalase, + mannitol ± piperidine, and + catalase ± piperidine (Fig. 5). The effects of piperidine are re-graphed from Figure 4 to allow direct comparisons (Fig. 5A). Mannitol had no affect on the heat-induced production of form II DNA, other than a slightly significant increase in strand breaks measured in the untreated plasmid (Fig. 5B, p = 0.04). Thus it was inferred that diffusible free radicals were not involved in the generation of frank strand breaks arising from reactions of uranyl ion and Asc with DNA. This observation is consistent with a previous report that the radical scavenger glycerol had no effect on uranyl photocleavage of DNA [59]. Reactions of UA and Asc carried out in the presence of catalase had slightly lowered levels of heat-induced DNA strand breaks (Fig. 5C, p = 0.01), an effect that was not seen for control reactions with untreated DNA or DNA exposed to uranyl ion or Asc alone. This is consistent with the previous interpretation that a redox reaction between uranyl ion and Asc is not completely absent at pH 7.4 although it is favored at pH < 2 [41]; however, the affect was minor so this cannot be inferred to be a major pathway under the current conditions.
Figure 5.
Effect of mannitol and catalase on % DNA plasmid relaxation (Form II) in the absence and presence of piperidine. pBR322 DNA was reacted with UA and ascorbate as described in Figure 4. (A) For comparison purposes, the reactions from Figure 4B are shown as difference in % Form II DNA for [piperidine vs. no piperidine]. Reactions were also carried out in the added presence of: (B) 500 μM mannitol, followed by addition of water or 30 μM piperidine and incubation for 30 min at 60 °C. (C) 70 U/mL catalase, followed by addition of water and incubation for 30 min at 60 °C. (D) 500 μM mannitol, followed by addition of 30 μM piperidine and incubation for 30 min at 60 °C. (E) 70 U/mL catalase, followed by addition of 30 μM piperidine and incubation for 30 min at 60 °C. In all cases data represent mean ± SEM for differences in % Form II for [treatment vs. no treatment], for n = 5 independent experiments. Differences were significantly different than 0 by Student’s t-test (*p<0.05, **p<0.01, ****p<0.0001).
For both the mannitol and catalase co-incubations, post-treatment exposure to piperidine at 60 °C caused the same loss of relaxed DNA that was observed without mannitol or catalase treatment (Fig. 5D, 5E vs. Fig. 5A). Results do not support an interpretation that is different from that proposed above, that piperidine interacts with the uranyl-DNA adducts before they can produce heat-mediated hydrolysis of the DNA backbone.
The above results support the ability of uranyl ion to act as a chemical genotoxic agent that does not require radiation effects for its mode of action. Cellular damage from ionizing radiation is known to manifest through two mechanisms: as “direct damage” when radiation interacts with biological molecules, and as “indirect damage” mediated by oxygen radicals that result from interactions of radiation with water [60]. The major DNA lesions from ionizing radiation include double strand breaks and oxidative clustered DNA lesions [61], with a major signature being locally multiply damaged sites [60]. Neither of those pathways is consistent with the DNA lesions observed in this study, which were heat-induced DNA single strand breaks that were decreased by the presence of piperidine but not strongly affected by scavengers of reactive oxygen species. Nor are those radiological pathways consistent with the DNA lesions observed in cell culture, which were uranyl-DNA adducts, DNA single strand breaks, and AP sites [33, Yellowhair et al., in preparation].
Growing evidence, at least in isolated DNA, supports a proposal for formation of uranyl-DNA adducts at the phosphate backbone of the minor groove, with possible involvement of the N3 position of adenine. Nielsen’s elegant work harnessing the photoactivation of uranyl ion as a tool for DNA footprinting found that uranyl photocleavage of DNA in vitro was strongest at the minor groove of AT tracts [62, 63], which was inferred to be due to the presence of a more narrow minor groove and thus higher electronegative potential for cation binding relative to the wider minor groove in GC tracts [62]. Mass spectrometry studies of uranyl bonding to oligonucleotides found bifunctional binding of uranyl to two phosphodiester moieties of the phosphate backbone [64], although the oligonucleotides in that study did not contain adenine and the experimental system did not contain competing ligands for uranyl ion such as Asc, or other small molecules or peptides that may be present in biological systems. Metal complexes that interact with the minor groove of DNA, including those of the d-block divalent metals Cu(II), Zn(II), and Cd(II), have been shown to favor association with the N3-position of adenine [65].
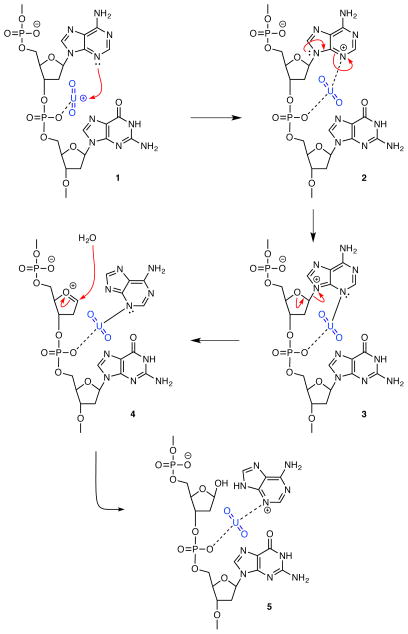
The working model that emerges from our results and related studies is that the uranyl-DNA adduct at the phosphate backbone may serve as a parental lesion that may further convert to strand breaks, AP sites, or possibly uranyl-mediated DNA crosslinks. The ability of uranyl ion to facilitate hydrolysis of the DNA phosphate backbone has been described [37]. A mechanism that could serve as one example for the conversion of a uranyl-DNA adduct to an AP site is proposed in Scheme I. The in vitro results summarized above suggest an interaction with the minor groove, and the current study does not present any results to the contrary; therefore, Scheme I depicts an association of uranyl ion with the phosphate backbone (1) that is stabilized, if only transiently, by binding at N3-adenine (2). The resonance transfer of charge from N3 to N9 (2 to 3) facilitates labilization of the hemiaminal ether moiety, thus leading to cleavage of the glycosidic bond (3 to 4). Addition of water to the oxonium ion followed by proton transfer to N9 of adenine leads to formation of the deoxyribose hemiacetal and the uranyl-coordinated purine.
Scheme I.
Proposed mechanism for conversion of a uranyl-DNA adduct to an AP site
Structural support for this model is drawn from the literature. As one example, the Dickerson Drew dodecamer d(CGCGAATTCGCG)2, which represents a structurally characterized example of B-form DNA [66], has a minor groove of 3.2 Å in the 5’AATT segment [67]. Calculations of bond distances with PyMOL molecular visualization open source software [68] give N3-adenine to O-phosphate distances of 6.9 and 7.7 Å (Dr. Suman Siramulla, personal communication). Estimating Van der Waals radii of nitrogen, uranium, and oxygen as 155, 240, and 152 pm [69], gives a sum of Van der Waals interactions of ~7.9 Å. These estimates are within bonding range without taking into consideration the flexibility of the phosphate backbone that would be increased by the charge neutralization brought on by uranyl binding [70]. One example in the literature demonstrates the spanning of cobalt hexamine between phosphate oxygen and both N7 and O6 of guanine in the d(CGCGCG)2 oligonucleotide crystal structure [71]. That structure and the current working model are consistent with the proposal, first put forth by Eichhorn in the 1960’s, that divalent metals may manipulate DNA structure through their ability to interact with both the bases and the phosphate backbone [72, 73].
There is growing evidence for this type of a mechanism to occur in culture cells, although it is not assumed to require such site-specificity. UA has been shown to be more cytotoxic in the repair-deficient CHO EM9 line than the repair-proficient CHO AA8 line [33]. This line is XRCC1 deficient, and thus is impaired in its ability to carry out base excision repair (BER). Conversely, UA is not more cytotoxic in the nucleotide excision repair (NER) deficient CHO UV5 line than the CHO AA8 line, although a slightly higher relative toxicity emerges with high doses and longer exposure times [Wilson et al., in preparation]. These observations are consistent with the interpretation that an initial uranyl adduct at the phosphate backbone is nondistorting and targeted by BER, whereas lesions formed with longer incubation times and higher concentrations are more distorting and targeted by NER.
Conclusions
The purpose of this study was to develop a protocol to investigate the presence of uranium-induced DNA damage in order to understand the mechanism of formation of relevant DNA lesions. In summary, uranyl ion, in the presence of Asc, produced uranium-DNA adducts that converted to single strand breaks upon heating. Piperidine, which acted on DNA methylated by MMS to convert a methyl-DNA adduct to a strand break, served in the opposite fashion to decrease the formation of uranyl-induced strand breaks. The observation that piperidine also decreased the gel shift for metal-DNA adducts formed by monofunctional cis-Pt and chromic chloride was interpreted to suggest that piperidine served to remove uranyl-DNA adducts. This interpretation, combined with the observation that radical scavengers did not significantly affect formation of strand breaks, suggested that DNA strand breaks arose from the presence of uranium-DNA adducts and not from diffusible free radicals. Results are consistent with previous data showing the formation of adducts, single strand breaks and AP sites in cultured cells. The likely dominant interaction of uranyl ion with the DNA phosphate backbone predicts a relatively non-distorting DNA lesion that should be targeted by BER rather than NER. Hydrolysis of the DNA phosphate backbone would produce strand breaks, and weaker associations of the uranyl-phosphate adduct with the DNA bases may provide a mechanism for the formation of AP sites. These data further support the interpretation that DU as uranyl ion can display chemical genotoxicity independent of its radioactivity.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R15 ES019703. Ms. Wilson was supported by the STEP program within the Partnership for Native American Cancer Prevention (NACP), NIH grant U54 CA143925, and the John & Sophie Ottens Native American Student Research Program. Ms. Young was supported by the NAU Bridges Program, NIH grant R25 GM102788.
Abbreviations
- ACES
N-(2- acetamido)-2-aminoethanesulfonic acid
- Asc
ascorbate
- BER
base excision repair
- CHO
Chinese hamster ovary
- Cis-Pt, cis-platin
cis-diamminedichloroplatinum(II)
- DU
depleted uranium
- MMS
methyl methanesulfonate
- NER
nucleotide excision repair
- SSB
single strand breaks
- TBE
tris-boric acid-EDTA
- UA
uranyl acetate dihydrate
References
- 1.Boice JD, Jr, Mumma MT, Blot WJ. Radiat Res. 2010;174(5):624–636. doi: 10.1667/RR2180.1. [DOI] [PubMed] [Google Scholar]
- 2.Tirmarche M, Harrison J, Laurier D, Blanchardon E, Paquet F, Marsh J. Ann ICRP. 2012;41(3–4):368–377. doi: 10.1016/j.icrp.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 3.Möhner M, Gellissen J, Marsh JW, Gregoratto D. Health Phys. 2010;99(3):314–321. doi: 10.1097/HP.0b013e3181cd6536. [DOI] [PubMed] [Google Scholar]
- 4.Kulich M, Rericha V, Rericha R, Shore DL, Sandler DP. Environ Res. 2011;111(3):400–405. doi: 10.1016/j.envres.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.deLemos JL, Brugge D, Cajero M, Downs M, Durant JL, George CM, Henio-Adeky S, Nez T, Manning T, Rock T, Seschillie B, Shuey C, Lewis J. Environ Health. 2009;8:29. doi: 10.1186/1476-069X-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson SE, Madsen GE. Health Phys. 2011;101(5):618–625. doi: 10.1097/HP.0b013e3182243a7a. [DOI] [PubMed] [Google Scholar]
- 7.de Dinis ML, Fiúza A. J Environ Radioact. 2012;119:63–69. doi: 10.1016/j.jenvrad.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Miller AC, McClain D. Rev Environ Health. 2007;22(1):75–89. doi: 10.1515/reveh.2007.22.1.75. [DOI] [PubMed] [Google Scholar]
- 9.Handley-Sidhu S, Keith-Roach MJ, Lloyd JR, Vaughan DJ. Sci Total Environ. 2010;408(23):5690–5700. doi: 10.1016/j.scitotenv.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Briner W. Int J Environ Res Public Health. 2010;7(1):303–313. doi: 10.3390/ijerph7010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmer DA, Rossignol M, Blatt M, Agarwal R, Teichman R, Lange G. J Occup Environ Med. 2007;49(5):475–480. doi: 10.1097/JOM.0b013e318042d682. [DOI] [PubMed] [Google Scholar]
- 12.Melanson MA, Geckle LS, Davidson BA. US Army Med Dep J. 2012 Jul-Sep;:82–87. [PubMed] [Google Scholar]
- 13.The Royal Society. Policy document 6/01. London, UK: The Royal Society; 2001. [Google Scholar]
- 14.Squibb KS, Gaitens JM, Engelhardt S, Centeno JA, Xu H, Gray P, McDiarmid MA. J Occup Environ Med. 2012;54(6):724–732. doi: 10.1097/JOM.0b013e31824fe138. [DOI] [PubMed] [Google Scholar]
- 15.Todorov TI, Ejnik JW, Guandalini G, Xu H, Hoover D, Anderson L, Squibb K, McDiarmid MA, Centeno JA. J Trace Elem Med Biol. 2013;27(1):2–6. doi: 10.1016/j.jtemb.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Hines SE, Gucer P, Kligerman S, Breyer R, Centeno J, Gaitens J, Oliver M, Engelhardt S, Squibb K, McDiarmid M. J Occup Environ Med. 2013;55(8):937–944. doi: 10.1097/JOM.0b013e31829176c7. [DOI] [PubMed] [Google Scholar]
- 17.Fathi RA, Matti LY, Al-Salih HS, Godbold D. Med Confl Surviv. 2013;29(1):7–25. doi: 10.1080/13623699.2013.765173. [DOI] [PubMed] [Google Scholar]
- 18.Shelleh HH. Saudi Med. 2012;33(5):483–488. [PubMed] [Google Scholar]
- 19.Al-Hadithi TS, Al-Diwan JK, Saleh AM, Shabila NP. Confl Health. 2012;6(1):3. doi: 10.1186/1752-1505-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aitken M. BMJ. 1999;319(7207):401. doi: 10.1136/bmj.319.7207.401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papastefanou C. Health Phys. 2002;83(2):280–282. doi: 10.1097/00004032-200208000-00013. [DOI] [PubMed] [Google Scholar]
- 22.Krupka KM, Parkhurst MA, Gold K, Arey BW, Jenson ED, Guilmette RA. Health Phys. 2009;96(3):276–291. doi: 10.1097/01.HP.0000298229.60229.10. [DOI] [PubMed] [Google Scholar]
- 23.Craft E, Abu-Qare A, Flaherty M, Garofolo M, Rincavage H, Abou-Donia M. J Toxicol Environ Health B Crit Rev. 2004;7(4):297–317. doi: 10.1080/10937400490452714. [DOI] [PubMed] [Google Scholar]
- 24.Heffernan TE, Lodwick JC, Spitz H, Neton J, Soldano M. Health Phys. 2001;80(3):255–262. doi: 10.1097/00004032-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Lind OC, Salbu B, Skipperud L, Janssens K, Jaroszewicz J, De Nolf W. J Environ Radioact. 2009;100(4):301–307. doi: 10.1016/j.jenvrad.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Eidson AF. Health Phys. 1994;67(1):1–14. doi: 10.1097/00004032-199407000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Pearson RG. J Am Chem Soc. 1963;85(22):3533–3539. [Google Scholar]
- 28.Van Horn JD, Huang H. Coord Chem Rev. 2006;250:765–775. [Google Scholar]
- 29.Zobel CR, Beer M. J Biophys Biochem Cytol. 1961;10:335–346. doi: 10.1083/jcb.10.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin RH, Wu LJ, Lee CH, Lin-Shiau SY. Mutat Res. 1993;319(3):197–203. doi: 10.1016/0165-1218(93)90079-s. [DOI] [PubMed] [Google Scholar]
- 31.Miller AC, Blakely WF, Livengood D, Whittaker T, Xu J, Ejnik JW, Hamilton MM, Parlette E, John TS, Gerstenberg HM, Hsu H. Environ Health Perspect. 1998;106(8):465–471. doi: 10.1289/ehp.98106465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darolles C, Broggio D, Feugier A, Frelon S, Dublineau I, De Meo M, Petitot F. Toxicol Lett. 2010;192(3):337–348. doi: 10.1016/j.toxlet.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Stearns DM, Yazzie M, Bradley AS, Coryell VH, Shelley JT, Ashby A, Asplund CS, Lantz RC. Mutagenesis. 2005;20(6):417–423. doi: 10.1093/mutage/gei056. [DOI] [PubMed] [Google Scholar]
- 34.McDiarmid MA, Albertini RJ, Tucker JD, Vacek PM, Carter EW, Bakhmutsky MV, Oliver MS, Engelhardt SM, Squibb KS. Environ Mol Mutagen. 2011;52(7):569–581. doi: 10.1002/em.20658. [DOI] [PubMed] [Google Scholar]
- 35.Coryell VH, Stearns DM. Mol Carcinog. 2006;45(1):60–72. doi: 10.1002/mc.20155. [DOI] [PubMed] [Google Scholar]
- 36.Jostes RF, Fleck EW, Morgan TL, Stiegler GL, Cross FT. Radiat Res. 1994;137(3):371–379. [PubMed] [Google Scholar]
- 37.Yazzie M, Gamble SL, Civitello ER, Stearns DM. Chem Res Toxicol. 2003;16(4):524–530. doi: 10.1021/tx025685q. [DOI] [PubMed] [Google Scholar]
- 38.Miller AC, Stewart M, Brooks K, Shi L, Page N. J Inorg Biochem. 2002;91(1):246–252. doi: 10.1016/s0162-0134(02)00391-4. [DOI] [PubMed] [Google Scholar]
- 39.Zhitkovich A. Chem Res Toxicol. 2011;24(10):1617–1629. doi: 10.1021/tx200251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stearns DM, Wetterhahn KE. Chem Res Toxicol. 1994;7(2):219–230. doi: 10.1021/tx00038a016. [DOI] [PubMed] [Google Scholar]
- 41.Taqui Khan MM, Martell AE. J Am Chem Soc. 1969;91(17):4668–4672. doi: 10.1021/ja01045a011. [DOI] [PubMed] [Google Scholar]
- 42.Comyns AE. Chem Rev. 1960;60(2):115–146. [Google Scholar]
- 43.Mattes WB, Hartley JA, Kohn KW. Biochim Biophys Acta. 1986;868(1):71–76. doi: 10.1016/0167-4781(86)90088-6. [DOI] [PubMed] [Google Scholar]
- 44.Karpas Z, Lorber A, Sela H, Paz-Tal O, Hagag Y, Kurttio P, Salonen L. Health Phys. 2005;89(4):315–321. doi: 10.1097/01.hp.0000165450.76676.10. [DOI] [PubMed] [Google Scholar]
- 45.Víglaský V, Antalík M, Adamcík J, Podhradský D. Nucleic Acids Res. 2000;28(11):e51. doi: 10.1093/nar/28.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treiber DK, Chen Z, Essigmann JM. Nucleic Acids Res. 1992;20(21):5805–5810. doi: 10.1093/nar/20.21.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beranek DT, Heflich RH, Kodell RL, Morris SM, Casciano DA. Mutat Res. 1983;110(1):171–180. doi: 10.1016/0027-5107(83)90026-x. [DOI] [PubMed] [Google Scholar]
- 48.Boysen G, Pachkowski BF, Nakamura J, Swenberg JA. Mutat Res. 2009;678(2):76–94. doi: 10.1016/j.mrgentox.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eastman A. Biochemistry. 1986;25(13):3912–3915. doi: 10.1021/bi00361a026. [DOI] [PubMed] [Google Scholar]
- 50.Schaller W, Reisner H, Holler E. Biochemistry. 1987;26(3):943–950. doi: 10.1021/bi00377a039. [DOI] [PubMed] [Google Scholar]
- 51.Takahara PM, Frederick CA, Lippard SJ. J Am Chem Soc. 1996;118:12309–12321. [Google Scholar]
- 52.Bellon SF, Coleman JH, Lippard SJ. Biochemistry. 1991;30(32):8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- 53.Bauer WR. Annu Rev Biophys Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- 54.Keck MV, Lippard SJ. J Am Chem Soc. 1992;114:3386–3390. [Google Scholar]
- 55.Ponti M, Forrow SM, Souhami RL, D’Incalci M, Hartley JA. Nucleic Acids Res. 1991;19(11):2929–2933. doi: 10.1093/nar/19.11.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blankert SA, Coryell VH, Picard BT, Wolf KK, Lomas RE, Stearns DM. Chem Res Toxicol. 2003;16(7):847–854. doi: 10.1021/tx034007g. [DOI] [PubMed] [Google Scholar]
- 57.Reynolds M, Peterson E, Quievryn G, Zhitkovich A. J Biol Chem. 2004;279(29):30419–30424. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- 58.Buettner GR, Jurkiewicz BA. Radiat Res. 1996;145(5):532–541. [PubMed] [Google Scholar]
- 59.Nielsen PE, Hiort C, Sönnichsen SH, Buchardt O, Dahl O, Norden B. J Am Chem Soc. 1992;114(13):4967–4975. [Google Scholar]
- 60.Ward JF. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 61.Hada M, Georgakilas AG. J Radiat Res. 2008;49(3):203–210. doi: 10.1269/jrr.07123. [DOI] [PubMed] [Google Scholar]
- 62.Nielsen PE, Møllegaard NE, Jeppesen C. Nucleic Acids Res. 1990;18(13):3847–3851. doi: 10.1093/nar/18.13.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sönnichsen SH, Nielsen PE. J Mol Recognit. 1996;9(3):219–227. doi: 10.1002/(sici)1099-1352(199605)9:3<219::aid-jmr242>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 64.Wu Q, Cheng X, Hofstadler SA, Smith RD. J Mass Spectrom. 1996;31(6):669–675. doi: 10.1002/(SICI)1096-9888(199606)31:6<669::AID-JMS340>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 65.Galindo MA, Amantia D, Martinez AM, Clegg W, Harrington RW, Martinez VM, Houlton A. Inorg Chem. 2009;48(21):10295–10303. doi: 10.1021/ic9013448. [DOI] [PubMed] [Google Scholar]
- 66.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Proc Natl Acad Sci USA. 1981;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neidle S. DNA Structure and Recognition. Oxford University Press; Oxford, UK: 1994. p. 47. [Google Scholar]
- 68.PyMol molecular visualization open source software, version 1.6. http://pymol.org.
- 69.Los Alamos National Laboratory. Periodic Table of the Elements. http://periodic.lanl.gov/list.shtml.
- 70.Okonogi TM, Alley SC, Harwood EA, Hopkins PB, Robinson BH. Proc Natl Acad Sci USA. 2002;99(7):4156–4160. doi: 10.1073/pnas.072067799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gessner RV, Quigley GJ, Wang AH, van der Marel GA, van Boom JH, Rich A. Biochemistry. 1985;24(2):237–240. doi: 10.1021/bi00323a001. [DOI] [PubMed] [Google Scholar]
- 72.Eichhorn GL, Shin YA. J Am Chem Soc. 1968;90(26):7323–7328. doi: 10.1021/ja01028a024. [DOI] [PubMed] [Google Scholar]
- 73.Shin YA, Eichhorn GL. Biochemistry. 1968;7(3):1026–1032. doi: 10.1021/bi00843a022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.