Abstract
Atypical antipsychotics have become a common therapeutic option in both schizophrenia and bipolar disorder. However, these medications come with a high risk of metabolic side effects, particularly dyslipidemia and insulin resistance. Therefore, identification of patients who are at increased risk for metabolic side effects is of great importance. The genetics of fatty acid metabolism is one area of research that may help identify such patients. Therefore, in this present study, we aimed to determine the effect of one commonly studied genetic polymorphism from both fatty acid desaturase 1 (FADS1) and FADS2 gene on a surrogate measure of insulin resistance and lipid levels in a metabolically high-risk population of patients largely exposed to atypical antipsychotics. This study used a cross-sectional design, fasting blood draws, and genetic analysis to investigate associations between polymorphisms, haplotypes, and metabolic measures. A total of 320 subjects with schizophrenia (n = 226) or bipolar disorder (n = 94) were included in this study. The mean age of the population was 42.5 years and 45% were male. A significant association between FADS1 and FADS2 haplotypes was found with insulin resistance while controlling for confounders. Further investigation is required to replicate this finding.
1. Background
The use of antipsychotics, particularly the atypical antipsychotics (AAPs), is considered the standard of care in schizophrenia symptom management and is becoming a common therapeutic choice in the management of bipolar disorder [1–3]. The fact that AAPs are commonly used in both of these populations may be due to the overlapping symptomatology that is seen as well as the genetic overlap that has been identified in several disease linkage studies [4–7]. Although many studies support the use of AAPs in the severely mentally ill, these medications come with a high risk of metabolic side effects. This risk requires careful monitoring and management as the cardiometabolic side effects have been shown to increase the cost of care, decrease adherence, and, most severely, have negative consequences on length and quality of life [8–11]. Therefore, investigation into lifestyle, diet, and genetic factors that may increase or attenuate the risk of metabolic side effects in patients taking AAPs is important and of high interest. One current line of research within the area of AAP metabolic side effects concentrates on fatty acid metabolism and its influence on metabolic measures.
Fatty acids (FAs) serve many important physiological functions including energy reserves, structural components of cell membranes, precursors of eicosanoids, and regulators of gene expression. The role FAs play in cell membranes is of particular interest as they influence translocation of glucose transporters and insulin receptor binding and signaling in addition to cell membrane fluidity and permeability. This indicates that FAs may play an important role in the development of insulin resistance and type 2 diabetes mellitus [12–14]. FA levels in the blood are determined by both dietary FA intake and to a larger extent endogenous FA metabolism. Endogenous FA metabolism is mediated by a series of elongation and desaturation steps controlled by two rate-limiting enzymes called delta-5 desaturase (D5D) and delta-6 desaturase (D6D). These enzymes, which are expressed at high levels in the liver, brain, heart, and lungs, are responsible for conversion of linoleic acid to n-6 polyunsaturated fatty acids (PUFA) and α-linolenic acid to n-3 PUFAs. Furthermore, it has been determined that D5D is encoded by the fatty acid desaturase 1 (FADS1) gene and D6D is encoded by the fatty acid desaturase 2 (FADS2) gene which are located on chromosome 11. Minor alleles of polymorphisms within the FADS1/2 gene cluster are commonly associated with lower D5D and D6D activities and thus are used as a surrogate marker for desaturase activity [15]. Many studies in various populations have shown correlations with FADS1 and/or FADS2 polymorphisms and FA or lipid levels [16, 17]. Additionally, there is also a growing body of literature showing correlations between D5D and D6D activity, insulin resistance, and risk for type 2 diabetes mellitus. The two desaturases have an opposite effect on the risk of developing diabetes. Specifically, an increased D5D activity is associated with lower risk of developing diabetes, whereas increased D6D activity is associated with a higher risk. A few studies have indicated possible differences in FA desaturase activity and FADS gene expression due to the influence of antipsychotics; however this data primarily comes from animal populations and postmortem brain studies. Thus, to our knowledge no studies have investigated FADS genetic variants and metabolic biomarkers from the mental health population taking antipsychotics [18–23].
Therefore, in this present study, we aimed to determine the effect of one commonly studied genetic polymorphism (SNP) from both the FADS1 and FADS2 gene on a surrogate measure of insulin resistance and lipid levels in a metabolically high-risk population of patients diagnosed with schizophrenia or bipolar disorder and largely exposed to AAPs. We also aimed to use a haplotype analysis to evaluate the combined effects of each gene's variant on insulin resistance and lipid levels.
2. Subjects and Methods
2.1. Study Population
Male and female participants were recruited from outpatient mental health clinics in the Southeastern Michigan area. Subjects were considered for inclusion if they met the following criteria: (1) aged 18–80 and diagnosed with schizophrenia, schizoaffective disorder, or bipolar disorder, (2) currently taking an antipsychotic, and (3) no medication changes for the previous 6 weeks. Subjects were excluded based on the following criteria: (1) having an active substance abuse or dependence diagnosis, (2) currently taking a medication for diabetes (to avoid bias in the insulin resistance measure), or (3) unwilling or unable to participate. The study was approved by the University of Michigan Institutional Review Board.
2.2. Clinical and Metabolic Measurements
Participants came to the University of Michigan Clinical Research Unit (MCRU) for a single visit. Study visits were completed in the morning, within 2 hours of the participants' usual wakening time. Participants were required to fast overnight for the visit. After obtaining an informed consent, participants underwent the structured clinical interview for DSM-IV-TR diagnoses (SCID) performed by a trained research assistant in order to confirm their psychiatric diagnosis. Psychiatric diagnoses were also confirmed by medical chart review when possible. Subjects were asked about basic demographic information (e.g., age, race, and gender), current medications (also confirmed by pharmacy records), and current or past cigarette smoking. A registered nurse took height and waist measurements along with a blood pressure measurement and a blood draw. Body mass index (BMI, kg/m2) was calculated from this information. The blood draw was collected for genetic analysis and fasting labs such as lipids (which included total cholesterol (TC), triglycerides (TG), high-density lipoproteins (HDL), and low-density lipoprotein (LDL)), blood glucose, and insulin measurements were measured. Insulin resistance was calculated using the homeostatic model assessment of insulin resistance [24] (HOMA-IR, [fasting insulin (μIU/mL) × fasting glucose (mmol/L)]/22.5) which has been correlated to more invasive measures of insulin resistance [25–28].
2.3. Selection of SNPs and Genotyping Methods
Whole blood was used for DNA extraction using the salt precipitation method [29]. We aimed to conduct a candidate-gene study by using one polymorphism from both the FADS1 and FADS2 genes. The variants, FADS1 rs174537 and FADS2 rs174570, were chosen based on a literature review in which these variants were associated with metabolic measures in various large cohorts with replicated results [17, 29–34]. The FADS1 rs174537 (G/T) variant is located 14 kb upstream of the FADS1 gene and has a minor allele frequency (MAF) of approximately 33% (from the 1000 genomes project) while the FADS2 rs174570 (C/T) is located in intron 1 of the FADS2 gene and has a MAF of approximately 24% (from the 1000 genomes project). Genotyping was completed by polymerase chain reaction followed by pyrosequencing [35] (specific assay conditions available upon request).
2.4. Haplotype Analyses
Linkage analysis between the FADS1 and FADS2 variants and evaluation of a haplotype block were conducted by using Haploview 4.2 [36]. Pairwise haplotypes were inferred using the PHASE 2.1 program and only haplotypes with a frequency >1% were used in statistical analysis [37, 38]. PHASE 2.1 uses a Bayesian statistical method for determining haplotypes from population data. The computational algorithms used in the PHASE 2.1 program have been shown to be superior compared to other commonly employed haplotype inference methods like the expectation-maximization (EM) algorithm.
2.5. Statistical Analysis
Statistical analyses were performed with JMP Pro 9.0 software (JMP, Version 9.0. SAS Institute Inc., Cary, NC, 1989–2012). Hardy-Weinberg equilibrium (HWE) was evaluated using Haploview 4.2. One-way analysis of variance (ANOVA) was used to assess differences in mean values of clinical and metabolic variables within psychiatric diagnosis and genetic variant groups (FADS1 SNP, FADS2 SNP, and FADS1/2 haplotypes) for continuous variables (age, BMI, blood pressure, lipid levels, glucose, insulin, and HOMA-IR). Chi-squared analysis was used to compare dichotomous variables (gender, race, AAP status, and smoking status) by psychiatric diagnosis and genetic variant groups. A two-tailed value of P < 0.05 was considered statistically significant for these tests. To examine our main hypothesis, the relationship between either SNP or haplotype variants and lipid or HOMA-IR, a regression model was constructed using the metabolic measures (TC, TG, HDL, LDL, or HOMA-IR) as the dependent variable and psychiatric diagnosis, age, race, gender, bmi, smoking status, and AAP status as the independent variables. In order to account for multiple testing and reduce the rate of false positives, a Bonferroni correction was applied for our regression analyses. Given tests were conducted for each SNP and haplotype for the four lipid values (TC, TG, LDL, and HDL) and HOMA-IR; only P values <0.003 were considered statistically significant (0.05/15 tests analyzed in regression). Results are expressed as means ± standard deviation (S.D.) or %.
3. Results
3.1. Study Population Characteristics
A total of 320 subjects with schizophrenia (n = 226) or bipolar disorder (n = 94) were included in this study. The mean age of the population was 42.5 years, with 45% male and 70% identifying themselves as Caucasian. Table 1 represents the demographic and metabolic parameters of our study population. The schizophrenia spectrum and bipolar disorder cohorts were compared and significant differences were found for gender, race, current smokers, AAP use, and three lipid measures (TC, HDL, and LDL). Therefore, in our analyses of the combined cohort regarding our main hypotheses, psychiatric diagnosis was used as a confounder in order to account for differences between the two populations taking antipsychotics.
Table 1.
Demographic and metabolic characteristics of the schizophrenia, bipolar, and combined samples.
| Schizophrenia spectrum (n = 226) | Bipolar disorder (n = 94) | Combined (n = 320) | |
|---|---|---|---|
| Age (year) | 42.7 ± 11.6 | 41.8 ± 12.0 | 42.5 ± 11.7 |
| Gender (% female) | 37 | 63 | 44.7¥ |
| Race (% Caucasian/% African American/% other*) | 65/26/9 | 81/12/7 | 70/21/9¥ |
| Current smokers (%) | 53 | 32 | 47¥ |
| Currently on AAP (%) | 85 | 74 | 82¥ |
| BMI (kg/m2) | 31.4 ± 7.24 | 31.8 ± 8.89 | 31.5 ± 7.74 |
| SBP (mmHg) | 122 ± 16.0 | 123 ± 17.7 | 122 ± 16.5 |
| DBP (mmHg) | 74.2 ± 11.4 | 72.9 ± 11.0 | 73.8 ± 11.3 |
| TC (mg/dL) | 179 ± 41.2 | 191 ± 45.7 | 183 ± 42.9¥ |
| TG (mg/dL) | 130 ± 88.9 | 142 ± 108 | 134 ± 95.0 |
| HDL (mg/dL) | 53.1 ± 17.3 | 57.6 ± 15.1 | 54.4 ± 16.8¥ |
| LDL (mg/dL) | 108 ± 34.8 | 118 ± 38.8 | 111 ± 36.2¥ |
| Glucose (mg/dL) | 99.0 ± 18.5 | 95.2 ± 10.7 | 97.9 ± 16.6 |
| Insulin (µIU/mL) | 20.3 ± 14.6 | 24.3 ± 21.6 | 21.5 ± 17.0 |
| HOMA-IR | 5.07 ± 4.23 | 5.90 ± 5.66 | 5.32 ± 4.70 |
Means ± S.D. or percentage.
AAP: atypical antipsychotic, BMI: body mass index, SBP: systolic blood pressure, DBP: diastolic blood pressure, TC: total cholesterol, TG: triglycerides, HDL: high-density lipoprotein, LDL: low-density lipoprotein, and HOMA-IR: homeostasis model assessment-insulin resistance.
*Other includes self-defined race categories of Asian, Hispanic, latino, and others.
¥Significant difference based on a P value cutoff of 0.05.
3.2. Genetic Distribution and Analysis for FADS Variants
The FADS1 and FADS2 variants used in this study satisfied HWE within the schizophrenia and bipolar populations as well as in the combined sample (all P > 0.1). Table 2 illustrates the distribution of the variants within our population based on a dichotomized, dominant genetic model (e.g., wildtype = 0 and heterozygote/homozygote = 1). This model was used due to the low frequency of homozygote variants seen with the FADS variants as well as previous studies establishing this model as predictive of FA desaturase enzyme activity. Of note, two samples were unable to be genotyped for the FADS1 variant due to sample fatigue. Variant distribution did not significantly differ between schizophrenia and bipolar diagnoses (P > 0.4). Within the combined sample, the two variants were found to be highly linked from a linkage disequilibrium test (D′ = 0.92) using Haploview version 4.2 [36] and within a haplotype block using the conservative four-gamete rule (Figure 1). Pairwise haplotypes were calculated and empirical haplotype frequencies are presented in Table 3.
Table 2.
FADS genetic distribution for the schizophrenia, bipolar, and combined samples.
| Schizophrenia spectrum | Bipolar disorder | Combined | ||
|---|---|---|---|---|
| FADS1 rs174537 | GG genotype | 54.5 (122) | 48.9 (46) | 52.8 (168) |
| T allele | 45.5 (102) | 51.2 (58) | 47.2 (150) | |
| FADS2 rs174570 | CC genotype | 75.2 (170) | 71.3 (67) | 74.1 (237) |
| T allele | 24.8 (56) | 28.7 (27) | 26.0 (83) |
% (n); distribution based on dominant genetic model did not significantly differ between schizophrenia and bipolar diagnoses.
Figure 1.
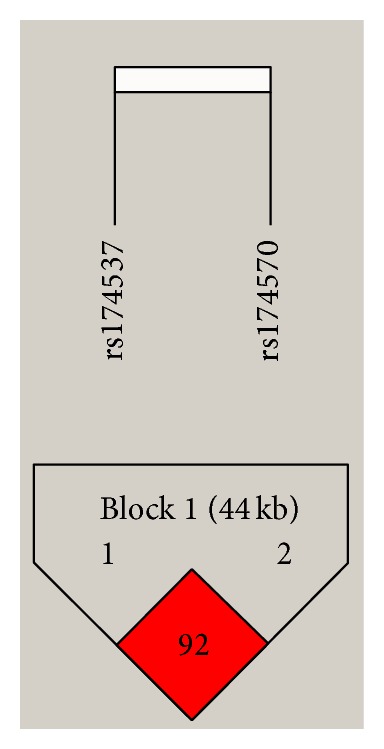
FADS haplotype block. Haplotype block analysis using the four-gamete rule for the FADS1 and FADS2 SNPS investigated in this study.
Table 3.
Haplotype frequencies for combined sample.
| Haplotype number | FADS1 rs174537 | FADS2 rs174570 | Frequency |
|---|---|---|---|
| 1 | G | C | 0.708 |
| 2 | G | T | 0.006 |
| 3 | T | C | 0.159 |
| 4 | T | T | 0.127 |
Empirical haplotype frequencies. Gives total count for all haplotypes inferred.
3.3. Clinical and Metabolic Characteristics according to Genotype and Haplotype
Table 4 represents the metabolic characteristics according to the FADS1 genotype, FADS2 genotype, and FADS1/2 haplotypes, respectively, for the combined sample. Significant differences are indicated within the table. Of the metabolic measurements, significant differences were identified with fasting insulin and HOMA-IR based on haplotype.
Table 4.
Demographic and metabolic characteristics based on FADS variants or FADS haplotype.
| SNP/haplotypes | FADS1 rs174537 | FADS2 rs174570 | 11 | 13 | 14 | 33 | 34 | ||
|---|---|---|---|---|---|---|---|---|---|
| Genotype/allele/haplotype genotypes | GG genotype (n = 167) | T allele (n = 150) | CC genotype (n = 236) | T allele (n = 83) | GC/GC (n = 163) | GC/TC (n = 61) | GC/TT (n = 56) | TC/TC (n = 11) | TC/TT (n = 18) |
|
| |||||||||
| Age (years) | 42.8 ± 11.6 | 42.1 ± 11.9 | 43.2 ± 11.8 | 40.4 ± 11.2 | 43.0 ± 11.5 | 43.0 ± 11.6 | 39.3 ± 11.8 | 46.6 ± 16.7 | 44.3 ± 9.30 |
| Gender (% female) ¥# | 37 | 52 | 44 | 48 | 37 | 54 | 50 | 73 | 33 |
| Race (% Caucasian/% African American/% other*)¥ψ# | 57/34/9 | 83/8/9 | 65/27/8 | 81/8/11 | 57/35/8 | 82/10/8 | 78/11/11 | 100/0/0 | 94/6/0 |
| Current smokers (%) | 48 | 47 | 47 | 47 | 48 | 51 | 43 | 27 | 61 |
| Currently on AAP (%) | 78 | 85 | 81 | 84 | 78 | 88 | 85 | 91 | 73 |
| BMI (kg/m2)ψ | 31.3 ± 7.63 | 31.7 ± 7.91 | 31.0 ± 7.62 | 33.0 ± 7.92 | 31.4 ± 7.71 | 30.5 ± 7.61 | 33.5 ± 8.68 | 27.7 ± 5.99 | 31.2 ± 6.34 |
| SBP (mmHg) | 123 ± 16.1 | 121 ± 16.9 | 122 ± 16.4 | 123 ± 16.9 | 123 ± 15.9 | 119 ± 16.9 | 122 ± 14.4 | 121 ± 21.5 | 127 ± 21.7 |
| DBP (mmHg) | 74.3 ± 11.0 | 73.2 ± 11.6 | 74.0 ± 10.7 | 73.1 ± 12.7 | 74.4 ± 10.9 | 73.0 ± 10.7 | 73.2 ± 11.5 | 71.9 ± 6.95 | 76 ± 15.5 |
| TC (mg/dL) | 184 ± 41.7 | 181 ± 44.6 | 183 ± 43.1 | 182 ± 42.6 | 183 ± 41.3 | 184 ± 48.9 | 180 ± 41.6 | 181 ± 40.5 | 171 ± 42.3 |
| TG (mg/dL) | 131 ± 94.0 | 138 ± 96.4 | 136 ± 101.5 | 126 ± 72.1 | 130 ± 93.8 | 162 ± 124 | 121 ± 65.9 | 104 ± 45.6 | 127 ± 85.2 |
| HDL (mg/dL) | 54.0 ± 17.0 | 54.6 ± 16.3 | 54.4 ± 17.4 | 54.4 ± 15.1 | 54.0 ± 17.2 | 52.7 ± 16.0 | 55.2 ± 16.5 | 65.2 ± 20.3 | 51.8 ± 13.0 |
| LDL (mg/dL) | 112 ± 37.6 | 109 ± 34.9 | 110 ± 37.8 | 112 ± 31.5 | 111 ± 37.5 | 110 ± 39.9 | 110 ± 31.0 | 103 ± 32.5 | 105 ± 32.9 |
| Glucose (mg/dL) | 98.3 ± 17.9 | 97.2 ± 15.1 | 98.1 ± 17.1 | 97.2 ± 15.4 | 98.1 ± 18.0 | 98.6 ± 15.5 | 96.4 ± 16.2 | 94.2 ± 12.9 | 95.8 ± 12.4 |
| Insulin (µIU/mL)# | 20.8 ± 12.3 | 22.4 ± 21.2 | 22.1 ± 18.4 | 19.7 ± 12.2 | 20.8 ± 12.3 | 27.3 ± 29.2 | 20.4 ± 13.9 | 15.7 ± 11.1 | 16.7 ± 6.51 |
| HOMA-IR# | 5.18 ± 3.74 | 5.49 ± 5.63 | 5.52 ± 5.13 | 4.72 ± 3.16 | 5.166 ± 3.74 | 6.90 ± 7.86 | 4.83 ± 3.54 | 3.89 ± 3.18 | 3.94 ± 1.61 |
*Other includes self-defined race categories of Asian, Hispanic, latino, and others.
¥Significant difference based on a P value cutoff of 0.05 for the FADS1 dominant genetic model.
ψSignificant difference based on a P value cutoff of 0.05 for the FADS2 dominant genetic model.
#Significant difference based on a P value cutoff of 0.05 for haplotype comparisons.
3.4. Regression Analysis of Lipid and Insulin Resistance Measures Based on Genotype or Haplotype
Our candidate gene hypotheses regarding the influence of FADS1 and FADS2 variants on lipid measures and insulin resistance were tested on the combined sample using generalized linear regression analysis adjusting for the following confounders: psychiatric diagnosis, age, gender, race, BMI, smoking status, and AAP status. This model was conducted for the FADS1 dominant model, FADS2 dominant model, and the inferred haplotypes. Due to multiple testing, only P values below 0.003 were considered statistically significant.
No statistically significant associations were found for the FADS1 dominant model. An association was found for the FADS2 dominant model with HOMA-IR (whole model F(9,301) = 6.77, P < 0.0001). However, this effect was mainly due to BMI (P < 0.0001) since the effect of FADS2 variant (P = 0.02) did not meet the more conservative cutoff value.
When using haplotypes as the independent variable in our regression analysis, a trend with triglycerides was found that did not meet multiple testing cutoff (whole model F(12, 290) = 2.57, P = 0.003) due to the effect of BMI (P = 0.002) and haplotype (P = 0.004). Finally, of note, the significant association found above for HOMA-IR and haplotypes remained significant after controlling for confounders. The whole model was significant (F(12, 292) = 5.86, P < 0.0001) due to the effect of haplotype (P = 0.0004) and BMI (P < 0.0001). The HOMA-IR values for this significant association can be found in Figure 2.
Figure 2.
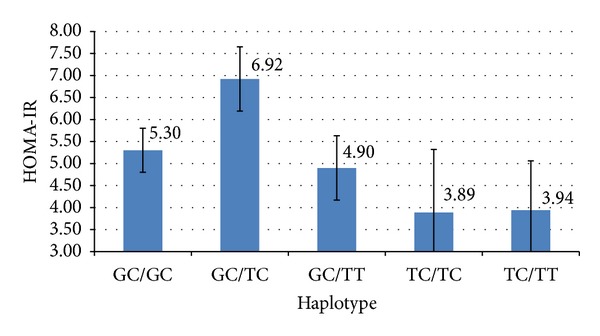
Association between HOMA-IR and FADS haplotype. Insulin resistance according to haplotype of FADS1 and FADS2 polymorphisms adjusting for psychiatric diagnosis, age, gender, race, BMI, smoking status, and AAP use. Means ± s.e. bars.
4. Discussion
Within our study, we found that HOMA-IR was associated with the haplotype of two investigated FADS gene variants. This is the first report of such an association found in a population of schizophrenia and bipolar patients largely exposed to AAPs. However, associations with insulin measures have been reported previously in healthy Caucasian and Korean populations. Associations between FADS1 polymorphisms and insulin resistance in a population with European descent arose from a GWAS meta-analysis conducted by Dupuis and colleagues [39]. This finding was subsequently confirmed via direct insulin resistance measures by Ingelsson and colleagues [40]. More recently, a study found correlations between HOMA-IR and variants in both the FADS1 and FADS2 genes in a population of healthy Korean men [41]. All of these studies used different polymorphisms found within the FADS1 and FADS2 genes, and thus, our results reflect gene cluster associations previously found but not necessarily individual variant effects.
HapMap CEU data has shown that the FADS1 gene and a large portion of the FADS2 gene are known to be in a linkage disequilibrium block; therefore, haplotype analyses are a natural extension of FADS genetic investigations. However, there have been relatively few studies to date incorporating FADS haplotype in the analyses [32, 33, 42] which could be explained by the ability only to infer haplotypes when familial genetic data is unavailable. Haplotype reconstruction may be of particular importance in studies looking at insulin resistance measures or diabetes risk as D5D (FADS1) and D6D (FADS2) activities have been shown to have an opposite relationship with diabetes risk. Thus, examining polymorphisms from only one FADS gene region at a time does not take into account the opposing effects of the other FADS region. Our data demonstrates that FADS1 and FADS2 variants had opposite relationships between HOMA-IR and the minor allele (nonsignificant, Table 4), but when the variants are combined into a haplotype, a relationship with HOMA-IR is exposed. This finding may indicate the importance of simultaneously considering variants from FADS1 and FADS2 when exploring metabolic measures and particularly diabetes biomarkers in studies.
A nonsignificant trend after a multiple testing adjustment (P = 0.003) was identified for triglycerides and haplotype which mirrored the results of the statistically significant haplotype and HOMA-IR relationship. The same haplotype identified to have the highest HOMA-IR value, GC/TC, was found to have the highest TG level as well (147 mg/dL). The closest TG level was that of the GC/GC haplotype (116 mg/dL). This finding, although not statistically significant with the multiple testing correction, may be of clinical significance given the link between insulin resistance and TGs. Hypertriglyceridemia is considered a defining feature of insulin resistance and metabolic syndrome. TG-mediated changes in very-low-density lipoprotein and lipoprotein lipase activity ultimately lead to increased expression of angiopoietin-like protein 4, and thus, insulin resistance [43]. An association between the GC/TC haplotype and both HOMA-IR and triglyceride levels adds epidemiologic and mechanistic plausibility to our findings.
Several limitations of our study need to be addressed. First, this is a cross-sectional study and causal associations cannot be drawn from the data; prospective, randomized studies are needed in populations treated with AAPs to draw further conclusions. Second, our study had differences in demographics between the bipolar disorder and the schizophrenia subjects. While these are important differences to consider, we used psychiatric diagnosis as a confounder in our main hypothesis testing; therefore, these differences are a natural reflection of the diagnosis and recruitment area. Third, our subject population was largely exposed to AAPs with known metabolic side effects which makes translation of our results to other disease populations or populations exposed to other medications challenging. Fourth, we used a surrogate measure of insulin resistance, which, although not a direct measure of insulin resistance, has been highly correlated to more invasive measures such as the glucose clamps and the oral glucose tolerance test [25, 26]. HOMA-IR was used to make our findings more translatable to the clinic setting but more direct measures of insulin resistance would be useful in supporting our study's results. Our study did not collect dietary data or individually measure D5D and D6D fatty acid indices and these important measures should be taken into account in future studies. Finally, we only investigated one variant from each gene based on a literature search. Although this was done to strengthen our candidate gene approach, one cannot rule out the many polymorphisms found in this gene cluster including the FADS1 rs174550 variant, which has been previously implicated in insulin resistance.
Despite these limitations, our study is the first to identify a relationship between a FADS1/2 haplotype and insulin resistance in a severely mentally ill population taking antipsychotics. Our population is unique in that it is at higher risk for insulin resistance and dyslipidemia due to the side effects associated with atypical antipsychotics. Indeed, those taking AAPs had higher HOMA-IR and lipid values compared to subjects taking typical antipsychotics (data not shown). AAP status was used as a confounder in our pharmacogenetic analysis; however, it adds evidence to the importance of discovering factors that may increase the risk for metabolic side effects including insulin resistance as this can lead to higher rates of cardiovascular disease and life years lost. The ability to personalize antipsychotic treatment based on a patient's metabolic risk profile, including pharmacogenetic analysis, may be one step to minimizing the damaging side effects of these drugs while maintaining their efficacy.
5. Conclusion
HOMA-IR was associated with a FADS1/2 haplotype in a population with severe mental illness taking antipsychotics. This study is the first to identify such an association; however the results need to be repeated and further haplotype analyses with other high-risk FADS variants would substantiate the current findings.
Conflict of Interests
The authors have no conflict of interests to disclose in regard to this paper.
Acknowledgments
The following sources were utilized for this publication: NIMH (R01 MH082784, K08 MH64158), NIH-NCCR, GCRC Program (UL1RR024986), the Chemistry Core of the Michigan Diabetes Research and Training Center (NIH5P60 DK 20572) (all Bethesda, Maryland) and the Washtenaw Community Health Organization (WCHO, Ann Arbor, Michigan), The Brain and Behavior Research Foundation (formerly NARSAD, Great Neck, New York), and the Prechter Longitudinal Study of Bipolar Disorder (Ann Arbor, Michigan).
References
- 1.Stahl SM, Morrissette DA, Citrome L, et al. ‘Meta-guidelines’ for the management of patients with schizophrenia. CNS Spectrums. 2013;18(3):150–162. doi: 10.1017/S109285291300014X. [DOI] [PubMed] [Google Scholar]
- 2.Geddes JR, Miklowitz DJ. Treatment of bipolar disorder. The Lancet. 2013;381(9878):1672–1682. doi: 10.1016/S0140-6736(13)60857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connolly KR, Thase ME. The clinical management of bipolar disorder: a review of evidence-based guidelines. Primary Care Companion to the Journal of Clinical Psychiatry. 2011;13(4) doi: 10.4088/PCC.10r01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SH, Ripke S, Neale BM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Genetics. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smoller JW, Craddock N, Kendler K, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. The Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green EK, Grozeva D, Jones I, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Molecular Psychiatry. 2010;15(10):1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peerbooms OLJ, van Os J, Drukker M, et al. Meta-analysis of MTHFR gene variants in schizophrenia, bipolar disorder and unipolar depressive disorder: evidence for a common genetic vulnerability? Brain, Behavior, and Immunity. 2011;25(8):1530–1543. doi: 10.1016/j.bbi.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Colton CW, Manderscheid RW. Congruencies in increased mortality rates, years of potential life lost, and causes of death among public mental health clients in eight states. Preventing Chronic Disease. 2006;3(2, article A42) [PMC free article] [PubMed] [Google Scholar]
- 9.Fagiolini A, Chengappa KNR, Soreca I, Chang J. Bipolar disorder and the metabolic syndrome: causal factors, psychiatric outcomes and economic burden. CNS Drugs. 2008;22(8):655–669. doi: 10.2165/00023210-200822080-00004. [DOI] [PubMed] [Google Scholar]
- 10.Laursen TM, Munk-Olsen T, Vestergaard M. Life expectancy and cardiovascular mortality in persons with schizophrenia. Current Opinion in Psychiatry. 2012;25(2):83–88. doi: 10.1097/YCO.0b013e32835035ca. [DOI] [PubMed] [Google Scholar]
- 11.Ward A, Quon P, Abouzaid S, Haber N, Ahmed S, Kim E. Cardiometabolic consequences of therapy for chronic schizophrenia using second-generation antipsychotic agents in a medicaid population: clinical and economic evaluation. P and T. 2013;38(2):109–115. [PMC free article] [PubMed] [Google Scholar]
- 12.Boden G, Shulman GI. Free fatty acids in obesity and type 2 diabetes: defining their role in the development of insulin resistance and β-cell dysfunction. European Journal of Clinical Investigation. 2002;32(supplement 3):14–23. doi: 10.1046/j.1365-2362.32.s3.3.x. [DOI] [PubMed] [Google Scholar]
- 13.Boden G. Effects of free fatty acids (FFA) on glucose metabolism: significance for insulin resistance and type 2 diabetes. Experimental and Clinical Endocrinology and Diabetes. 2003;111(3):121–124. doi: 10.1055/s-2003-39781. [DOI] [PubMed] [Google Scholar]
- 14.Boden G. Obesity, insulin resistance and free fatty acids. Current Opinion in Endocrinology, Diabetes and Obesity. 2011;18(2):139–143. doi: 10.1097/MED.0b013e3283444b09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merino DM, Ma DW. Genetic variation in lipid desaturases and its impact on the development of human disease. Lipids in Health and Disease. 2010;9, article 63 doi: 10.1186/1476-511X-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser C, Lattka E, Rzehak P, Steer C, Koletzko B. Genetic variation in polyunsaturated fatty acid metabolism and its potential relevance for human development and health. Maternal and Child Nutrition. 2011;7(supplement 2):27–40. doi: 10.1111/j.1740-8709.2011.00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lattka E, Illig T, Heinrich J, Koletzko B. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes? Clinical Nutrition. 2010;29(3):277–287. doi: 10.1016/j.clnu.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 18.McNamara RK, Jandacek R, Rider T, et al. Abnormalities in the fatty acid composition of the postmortem orbitofrontal cortex of schizophrenic patients: gender differences and partial normalization with antipsychotic medications. Schizophrenia Research. 2007;91(1–3):37–50. doi: 10.1016/j.schres.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vik-Mo AO, Birkenaes AB, Fernø J, Jonsdottir H, Andreassen OA, Steen VM. Increased expression of lipid biosynthesis genes in peripheral blood cells of olanzapine-treated patients. International Journal of Neuropsychopharmacology. 2008;11(5):679–684. doi: 10.1017/S1461145708008468. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Jandacek R, Rider T, Tso P, McNamara RK. Elevated delta-6 desaturase (FADS2) expression in the postmortem prefrontal cortex of schizophrenic patients: relationship with fatty acid composition. Schizophrenia Research. 2009;109(1–3):113–120. doi: 10.1016/j.schres.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNamara RK, Jandacek R, Rider T, Tso P, Cole-Strauss A, Lipton JW. Differential effects of antipsychotic medications on polyunsaturated fatty acid biosynthesis in rats: relationship with liver delta6-desaturase expression. Schizophrenia Research. 2011;129(1):57–65. doi: 10.1016/j.schres.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNamara RK, Jandacek R, Rider T, Tso P, Cole-Strauss A, Lipton JW. Atypical antipsychotic medications increase postprandial triglyceride and glucose levels in male rats: relationship with stearoyl-CoA desaturase activity. Schizophrenia Research. 2011;129(1):66–73. doi: 10.1016/j.schres.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, McNamara RK. Elevated Delta-6 desaturase (FADS2) gene expression in the prefrontal cortex of patients with bipolar disorder. Journal of Psychiatric Research. 2011;45(2):269–272. doi: 10.1016/j.jpsychires.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews DR, Hosker JP, Rudenski AS. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 25.Muniyappa R, Lee S, Chen H, Quon MJ. Current approaches for assessing insulin sensitivity and resistance in vivo: advantages, limitations, and appropriate usage. The American Journal of Physiology. 2008;294(1):E15–E26. doi: 10.1152/ajpendo.00645.2007. [DOI] [PubMed] [Google Scholar]
- 26.Rössner SM, Neovius M, Mattsson A, Marcus C, Norgren S. HOMA-IR and QUICKI: decide on a general standard instead of making further comparisons. Acta Paediatrica, International Journal of Paediatrics. 2010;99(11):1735–1740. doi: 10.1111/j.1651-2227.2010.01911.x. [DOI] [PubMed] [Google Scholar]
- 27.Ascaso JF, Pardo S, Real JT, Lorente RI, Priego A, Carmena R. Diagnosing insulin resistance by simple quantitative methods in subjects with normal glucose metabolism. Diabetes Care. 2003;26(12):3320–3325. doi: 10.2337/diacare.26.12.3320. [DOI] [PubMed] [Google Scholar]
- 28.Borai A, Livingstone C, Kaddam I, Ferns G. Selection of the appropriate method for the assessment of insulin resistance. BMC Medical Research Methodology. 2011;11, article 158 doi: 10.1186/1471-2288-11-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabatti C, Service SK, Hartikainen A-L, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nature Genetics. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka T, Shen J, Abecasis GR, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI study. PLoS Genetics. 2009;5(1) doi: 10.1371/journal.pgen.1000338.e1000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aulchenko YS, Ripatti S, Lindqvist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 european population cohorts. Nature Genetics. 2009;41(1):47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaeffer L, Gohlke H, Müller M, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Human Molecular Genetics. 2006;15(11):1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 33.Malerba G, Schaeffer L, Xumerle L, et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43(4):289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 34.Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nature Genetics. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh S, King CR, Garsa AA, McLeod HL. Pyrosequencing of clinically relevant polymorphisms. Methods in Molecular Biology. 2005;311:97–114. doi: 10.1385/1-59259-957-5:097. [DOI] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. The American Journal of Human Genetics. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingelsson E, Langenberg C, Hivert M-F, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59(5):1266–1275. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim OY, Lim HH, Yang LI, Chae JS, Lee JH. Fatty acid desaturase (FADS) gene polymorphisms and insulin resistance in association with serum phospholipid polyunsaturated fatty acid composition in healthy Korean men: cross-sectional study. Nutrition and Metabolism. 2011;8, article 24 doi: 10.1186/1743-7075-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bokor S, Dumont J, Spinneker A, et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. Journal of Lipid Research. 2010;51(8):2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bays HE, Toth PP, Kris-Etherton PM, et al. Obesity, adiposity, and dyslipidemia: a consensus statement from the national lipid association. Journal of Clinical Lipidology. 2013;7(4):304–383. doi: 10.1016/j.jacl.2013.04.001. [DOI] [PubMed] [Google Scholar]


