Abstract
Purpose
Recurrent “driver” mutations at specific loci in BRAF, NRAS, KIT, GNAQ, and GNA11 define clinically-relevant molecular subsets of melanoma, but >30% are “pan-negative” for these recurrent mutations. We sought to identify additional potential drivers in “pan-negative” melanoma.
Experimental Design
Using a targeted next-generation sequencing (NGS) assay (FoundationOne™) and targeted RNA sequencing, we identified a novel PAPSS1-BRAF fusion in a “pan-negative” melanoma. We then analyzed NGS data from 51 additional melanomas genotyped by FoundationOne™, as well as melanoma RNA, whole genome and whole exome sequencing data in The Cancer Genome Atlas (TCGA), to determine the potential frequency of BRAF fusions in melanoma. We characterized the signaling properties of confirmed molecular alterations by ectopic expression of engineered cDNAs in 293H cells.
Results
Activation of the mitogen-activated protein kinase (MAPK) pathway in cells by ectopic expression of PAPSS1-BRAF was abrogated by MEK inhibition but not by BRAF inhibition. NGS data analysis of 51 additional melanomas revealed a second BRAF fusion (TRIM24-BRAF) in a “pan-negative” sample; MAPK signaling induced by TRIM24-BRAF was also MEK inhibitor sensitive. Through mining TCGA skin cutaneous melanoma dataset, we further identified two potential BRAF fusions in another 49 “pan-negative” cases.
Conclusions
BRAF fusions define a new molecular subset of melanoma, potentially comprising 4–8% of “pan-negative” cases. Their presence may explain an unexpected clinical response to MEK inhibitor therapy or assist in selecting patients for MEK directed therapy.
Keywords: melanoma, BRAF fusion, BRAF rearrangement, next-generation sequencing, BRAF inhibitor, MEK inhibitor, vemurafenib, trametinib
RESULTS
Identification of a PAPSS1-BRAF Fusion in a “Pan-Negative” Melanoma
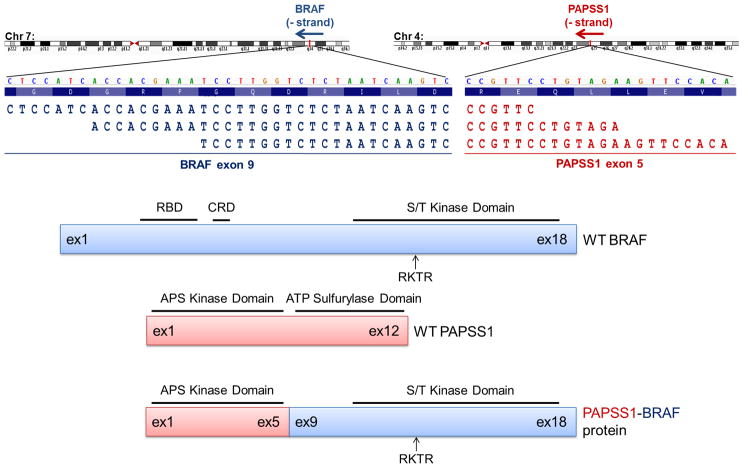
A 2.2 cm thick (Clark Level V), focally ulcerated, malignant melanoma from the left shoulder of a 27-year-old Caucasian female was screened for common melanoma driver mutations in BRAF, NRAS, KIT, GNAQ and GNA11 by the Vanderbilt melanoma SNaPshot assay (1) and determined to be “pan-negative”. Axillary dissection revealed metastatic involvement of 21 of 25 axillary lymph nodes (initial tumor staging: pT4bN3 = Stage IIIC), and the area was subsequently irradiated. Unfortunately, she developed local recurrence and lung metastasis. She was subsequently treated with experimental and standard immunotherapies. The patient’s disease progressed rapidly and she died nine months later (11 months after initial diagnosis). During the course of treatment, the primary shoulder lesion was sent to Foundation Medicine (Cambridge, MA) for comprehensive genomic profiling with the FoundationOne™ assay (2). This test involves targeted NGS of 3,320 exons in 182 cancer-related genes and 37 introns in 14 genes recurrently rearranged in cancer and simultaneously detects single nucleotide variants, insertions, deletions, copy number changes, and select rearrangements (see Supplementary Methods). Profiling revealed the presence of a large genomic deletion of approximately 350 kilobase pairs between BRAF intron 8 and an intragenic region of chromosome 7, suggesting a possible gene fusion event. Subsequent targeted RNA sequencing of tumor cDNA identified a novel, in-frame fusion between exon 5 of the sulfurylase kinase PAPSS1 (3′-phosphoadenosine 5′-phosphosulfate synthetase-1) and exon 9 of BRAF generated by a t(4;7)(q24;q34) translocation (Figure 1).
Figure 1. Detection of PAPSS1-BRAF fusion.
Three representative spanning sequence reads from targeted RNA sequencing of the “pan-negative” melanoma case shows alignment of PAPSS1 (red text) to chromosome 4 and of BRAF (dark blue text) to chromosome 7. The break-point occurs in-frame between exon 5 of PAPSS1 and exon 9 of BRAF. Below are schematics of wild-type BRAF (blue), wild-type PAPSS1 (red), and the fused PAPSS1-BRAF proteins. The adenylyl kinase domain of PAPSS1 and the serine-threonine (S/T) kinase domain of BRAF remain intact in the fused protein. WT, wild-type; ex, exon; RBD, Ras-binding domain; CRD, cysteine-rich domain; RKTR, Arg-Lys-Thr-Arg dimerization domain; APS, adenosine phosphosulfate; ATP, adenosine 5′-triphosphate.
To determine the effect of the PAPSS1-BRAF fusion on MAPK signaling in cells, we expressed cDNAs encoding FLAG-tagged WT BRAF, mutant BRAF (V600E), WT PAPSS1 or the fusion in 293H cells. Corresponding lysates were probed by immunoblotting with antibodies against phosphorylated and total forms of MEK1/2 and ERK1/2, as well as against PAPSS1, FLAG and BRAF. Ectopic expression of PAPSS1-BRAF in 293H cells led to increased levels of phosphorylated MEK1/2 and ERK1/2, similar to levels induced by BRAF V600E (Figure 2A). WT PAPSS1 did not induce MAPK pathway activation (Figure S1). These data confirm that the PAPSS1-BRAF fusion activates the MAPK signaling cascade.
Figure 2. Signaling induced by PAPSS1-BRAF is more sensitive to MEK inhibition than BRAF inhibition.
(A) Immunoblotting of lysates from 293H cells transfected with vector (empty vector) or plasmids encoding BRAF V600E-FLAG or PAPSS1-BRAF-FLAG demonstrate that the BRAF fusion activates MAPK pathway signaling similarly to BRAF V600E. (B) While MAPK pathway signaling induced by expression of BRAF V600E is sensitive to increasing doses (0, 0.1, 0.5, 1, and 5 μmol/L) of the BRAF inhibitor vemurafenib (vem) or the MEK inhibitor trametinib (tra), signaling induced by PAPSS1-BRAF is more sensitive to trametinib than vemurafenib. kDa, kilodalton.
Activation of MAPK signaling by BRAF V600E is sensitive to inhibition by both vemurafenib (a BRAF mutant-specific inhibitor) and trametinib (a MEK inhibitor) (3). To determine if signaling induced by the BRAF fusion was inhibited by these agents, we transfected 293H cells with the V600E or PAPSS1-BRAF cDNAs and treated them with vehicle control or increasing concentrations of vemurafenib or trametinib for 2 hours. Immunoblotting studies with the corresponding lysates showed that BRAF V600E-induced MEK1/2 phosphorylation was effectively reduced by vemurafenib, but MEK1/2 phosphorylation induced by PAPSS1-BRAF was not. Trametinib, however, was effective at reducing ERK1/2 phosphorylation in both V600E- and PAPSS1-BRAF-expressing cells (Figure 2B). These results suggest that downstream signaling induced by the PAPSS1-BRAF fusion could be abrogated by MEK but not mutant-specific BRAF inhibitors.
To determine whether BRAF fusions are recurrent in melanoma, we interrogated 51 additional melanomas from various institutions genotyped with the FoundationOne™ assay. This cohort was enriched with cases negative for BRAF mutations (both V600 and non-V600), likely due to referral bias. Only 8 of 52 (15%) tumors harbored V600 changes, at least less than half the expected percent in unbiased cohorts (1), and 8 of 52 (15.4%) harbored non-V600 (D594, L597, K601, etc.) changes. In addition to the PAPSS1-BRAF fusion, we identified another BRAF fusion, this time involving tripartite motif-containing 24 (TRIM24-BRAF; inv(7)(q32-34q34)) (Table 1, Supplementary Figure S2). This tumor was also “pan-negative” for other known “driver” mutations (Figure 3, Supplementary Table S1). Similar to PAPSS1-BRAF, ectopic expression of TRIM24-BRAF led to activation of the MAPK pathway which was sensitive to MEK, but not BRAF, inhibition (Figure S3). Thus, in this cohort, BRAF fusions were present in 8% [2 of 24, 95% confidence interval (C.I.) of 1.2%–27.0%] of “pan-negative” melanomas (Figure 3, Figure 4, Supplementary Table S1).
Table 1.
BRAF Rearrangements in “Pan-Negative” Melanomas
| Sample | Detection Method | BRAF Exon Break | 5′ Partner, Exon Break | Spanning Pairs (n) | Split Reads (n) |
|---|---|---|---|---|---|
| FM-Mel29 | FoundationOne™ | exon 9 | TRIM24, exon 9 | 200 | 37 |
| FM-Mel30 | FoundationOne™, RNA Kinome Seq | exon 9 | PAPSS1, exon 5 | 15 | 5 |
Figure 3. 52 melanomas genotyped on the FoundationOne™ assay.
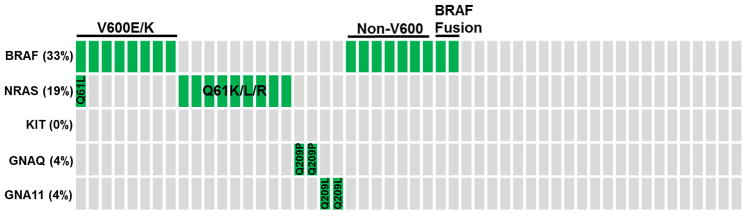
Schematic of the mutation distribution in 52 patient melanomas genotyped by the FoundationOne™ assay. Each column of boxes indicates a single patient, where green boxes indicate the presence of a mutation in BRAF, NRAS, KIT, GNAQ, and/or GNA11 and grey boxes indicate lack of mutation(s). Cases with V600E/K BRAF mutations, non-V600 BRAF mutations, BRAF fusions and certain NRAS mutations are indicated. Specific mutations for each case can be found in Supplementary Table S1. No KIT mutations were identified. Note the difference in the percent of cases positive for BRAF V600 mutations in this cohort versus those genotyped in Figure 4, demonstrating that this cohort was enriched for cases lacking BRAF V600 alterations.
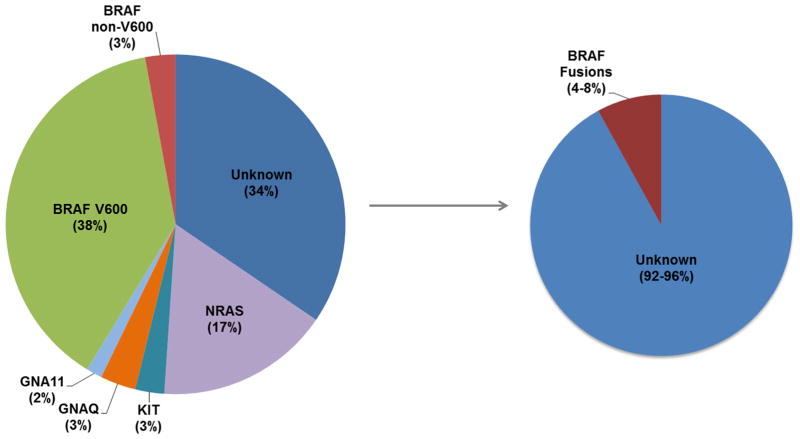
Figure 4. Molecular subsets of melanoma.
Pie chart demonstrating the percentage distribution of genes with clinically-relevant and recurrent driver mutations in individuals with melanoma, including non-V600 BRAF alterations (left), interrogated on the Vanderbilt melanoma SNaPshot assay (1). In this study, we have demonstrated that BRAF fusions occur in approximately 4–8% “pan-negative” cases (right).
To extend these findings, we also analyzed RNA, whole genome and whole exome sequencing data on an independent dataset available from The Cancer Genome Atlas (TCGA) skin cutaneous melanoma (SKCM) dataset. In two of 49 (4.1%) “pan-negative” cases, we identified sequence reads indicative of potential BRAF fusions, involving CDC27 and TAX1BP1 as 5′ partners (Figure S4). Consistent with these findings, TCGA reverse phase protein array (RPPA) data comparing levels of phosphorylated MEK1/2 in the tumors harboring fusions versus those with BRAF, NRAS, KIT, GNAQ or GNA11 mutations revealed that the fusion cases harbor phosphorylated MEK1/2 levels similar to, or greater than, levels observed in BRAF or NRAS-mutant melanomas (Figure S5). Collectively, these data suggest that BRAF fusions exist in 4–8% of “pan-negative” melanomas.
DISCUSSION
The classification and treatment of melanomas by known recurrent single-nucleotide driver mutation status in BRAF (V600), NRAS (G12/13, Q61), KIT (W557, V559, L576, K642, D816), GNAQ (Q209) and GNA11 (Q209) (1) has changed standard treatment practice by enabling rationally guided treatment. However, in our experience at Vanderbilt, using an established SNaPshot-based assay in the clinic (1), approximately one-third of melanomas are still “pan-negative” for these mutations. We recently determined that approximately 8% of cases negative for these drivers harbor other activating mutations in BRAF exon 15 (D594E/G/H/N/V, L597R/S/Q/V and K601E/I/N) rather than the better-known V600E/K/M/R/D alterations (4), and we showed in a patient harboring a BRAF L597 mutation that tumor regression could be induced by a MEK inhibitor (4). Here, we have identified another subset of potentially clinically relevant “pan-negative” melanomas defined by BRAF fusions. Specifically, we found two novel BRAF fusions (PAPSS1-BRAF and TRIM24-BRAF) in 2 of 24 (8%) “pan-negative” melanomas genotyped on an assay that examines that status of 182 cancer-related genes and 37 introns in 14 genes recurrently rearranged in cancer. Ectopic expression of either fusion activates the MAPK pathway (Figure 2A, Figure S3), and induced signaling is readily diminished by treatment with the MEK inhibitor, trametinib (Figure 2B, Figure S3). Through mining TCGA skin cutaneous melanoma dataset, we also identified two potential BRAF fusions in another 49 “pan-negative” cases, indicating a frequency of 4.1% in an independent cohort.
PAPSS1 is a bifunctional sulfurylase kinase, with an N-terminal adenosine-5′-phosphosulfate kinase domain and a C-terminal ATP sulfurylase domain (5). Only the adenylylsulfate kinase domain of PAPSS1 remains intact in the PAPSS1-BRAF fusion described herein. TRIM24 is a transcriptional co-regulator of nuclear receptors such as the retinoic acid receptor-α (RARα) (6) and is known to facilitate ubiquitination of p53 for proteasomal degradation (7). Interestingly, a version of TRIM24-BRAF fusion was identified in the early 1990s in a cDNA library derived from a model of mouse hepatocellular carcinoma (6, 8), but not identified in humans until now. In addition to BRAF, TRIM24 is also fused to the kinase domains of FGFR1 in a myeloproliferative disorder case (8p11 myeloproliferative syndrome) (9) and of RET in a case of papillary thyroid cancer (10). Like PAPSS1-BRAF, we show that expression of TRIM24-BRAF leads to activation of the MAPK pathway which is sensitive to MEK inhibition (Figure S3).
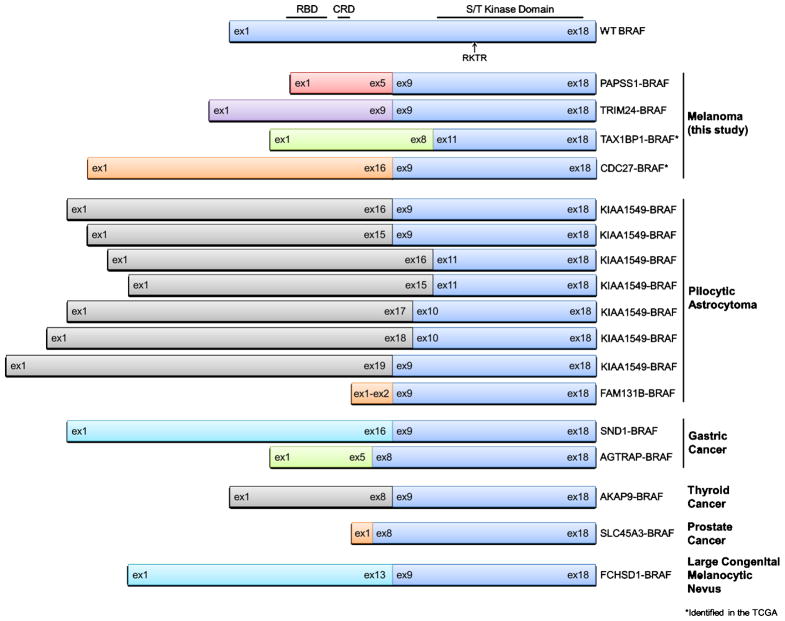
Although BRAF fusions have been found in other cancers (pilocytic astrocytoma, gastric adenocarcinoma, thyroid cancer, prostate cancer, and melanocytic nevi) (Figure 5) (11–18), to our knowledge, BRAF fusions have not yet been functionally characterized in malignant melanoma. A BRAF rearrangement was identified previously by break-apart fluorescence in situ hybridization (FISH) in a single malignant melanoma in 2010, however, insufficient sample remained for follow-up analyses that might have identified the fusion partner and allowed for its characterization (18). Additionally, a FCHSD1-BRAF fusion was identified in a large congenital melanocytic nevus (LCMN) (13). If left untreated/unresected, LCMN can be a precursor to melanoma, but this is thought to occur in fewer than 5% of LCMN cases (19). Notably, every BRAF fusion characterized to date activates MAPK pathway signaling (11–16, 18) and when interrogated, had transforming abilities (11, 12, 15, 18). Because PAPSS1-BRAF and TRIM24-BRAF are structured similarly to all other BRAF fusions (Figure 5), and because we show that both PAPSS1-BRAF and TRIM24-BRAF activate MAPK pathway signaling (Figure 2, Figure S3), we expect these melanoma BRAF fusions will also be transforming. Additional biological studies outside the scope of this manuscript are ongoing.
Figure 5. BRAF fusions identified in melanoma and other cancer types.
Schematics of wild-type BRAF (top) and all currently known BRAF fusions including those identified in this study (PAPSS1-BRAF and TRIM24-BRAF). All BRAF fusions break between exons 8 through 11, thus leaving the serine-threonine (S/T) kinase domain of BRAF intact. WT, wild-type; ex, exon; RBD, Ras-binding domain; CRD, cysteine-rich domain; RKTR, Arg-Lys-Thr-Arg dimerization domain.
In protein fusions involving receptor tyrosine kinases (RTKs), the 5′ partners usually encode coiled-coil domains which enable dimerization necessary for kinase activity (20). In the case of BRAF fusions, AKAP9 (11) and TRIM24 are the only 5′ partners that contain coiled-coil domains. BRAF harbors its own small dimerization motif (Arg-Lys-Thr-Arg, RKTR, amino acids 506–509) spanning exons 12 and 13 (21), which is intact in all currently-known BRAF fusions (Figure 5); therefore, the need for 5′ partners with dimerization ability may not be necessary for BRAF fusion function. In full-length wild-type BRAF, modulation of the RAS-binding domain (RBD) by activated RAS leads to BRAF homo-/hetero-dimerization and activation (22). This negative-regulatory RBD has been replaced by the various 5′ partners in all known BRAF fusions (Figure 5). Similarly, the recently-discovered BRAF V600E splice variants which induce vemurafenib resistance harbor N-terminal exons and mutant kinase domain exons, but RBD exons are spliced out, allowing for constitutive dimerization at the RKTR dimerization interface (23). Recently, Sievert, et al., demonstrated that KIAA1549-BRAF fusion variants can homodimerize with one another; introduction of a dimer interface mutant (R509H) disrupts this interaction (24). Future studies should ascertain the dimerization properties of the various BRAF fusions.
In summary, through NGS analysis of a “pan-negative” melanoma, we identified a novel PAPSS1-BRAF fusion. The fusion protein activates the MAPK pathway, and the induced downstream signaling is sensitive to MEK inhibition. Subsequent analysis of 51 additional melanomas (24 of which were “pan-negative”) revealed a second fusion, TRIM24-BRAF, that also activates MEK1/2 and ERK1/2. We also identified two candidate BRAF fusions in the TCGA skin cutaneous melanoma dataset. Thus, BRAF fusions may occur in 4–8% of the “pan-negative” melanoma population. Coupled with knowledge that the transforming ability of multiple BRAF fusions has already been established (11, 12, 15, 18), we believe enough evidence exists to raise awareness that BRAF fusions are present in this “pan-negative” population and have implications for MAPK pathway-targeted therapies currently in clinical trials. Their presence may explain an unexpected clinical response to MEK inhibitor therapy or assist in selecting patients for MEK-directed therapy. Collectively, these biochemical and genetic data define an additional molecular subset of melanoma that should be routinely screened for in the clinic, and knowledge about BRAF fusions in melanoma may provide insight into the mechanism of responses to treatment with an expanding list of available kinase inhibitors.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Through comprehensive molecular tumor profiling, we identified novel BRAF fusions in two of 24 melanoma patients lacking other known recurrent driver mutations in BRAF, NRAS, KIT, GNAQ, and GNA11. Surrogate kinase assays suggest that activated signaling induced by BRAF fusion proteins is sensitive to MEK inhibition. We also identified two candidate BRAF fusions in another 49 “pan-negative” cases in the TCGA skin cutaneous melanoma dataset. Thus, BRAF fusions represent a new, potentially clinically relevant target in melanomas possibly treatable with kinase inhibitors.
Acknowledgments
We would like to thank Doug Johnson, Holly Crandall and Ashlee Gecewich for coordinating patient tissue acquisition and Lynetha Verge for assisting with tissue protocol consent. For technical assistance and helpful discussions, we would like to thank Sophia Jelsma, Helen Pan, and Jordan Feigerle. Finally, we thank Kimberly B. Dahlman for critically reviewing the manuscript.
GRANT SUPPORT
Financial support was provided by the James C. Bradford Family Foundation (VICC investigators), the American Cancer Society (Mary Hendrickson-Johnson ACS Melanoma Professorship to J.A. Sosman and ACS Grant #PF-10-226-01-TBG to B.D. Lehmann), the National Institutes of Health/National Cancer Institute 5K24 CA097588-09 (J.A. Sosman) and NIH CA95131 (Specialized Program of Research Excellence in Breast Cancer to J.A. Pietenpol), and the Joanna M. Nicolay Melanoma Foundation 2013 Research Scholar Award (K.E. Hutchinson). W. Pao is supported by a Stand Up to Cancer Innovative Reseatch Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-IRG0409).
Financial Support: Financial support was provided by the James C. Bradford Family Foundation (VICC investigators), the American Cancer Society (Mary Hendrickson-Johnson ACS Melanoma Professorship to J.A. Sosman and ACS Grant #PF-10-226-01-TBG to B.D. Lehmann), the National Institutes of Health/National Cancer Institute 5K24 CA097588-09 (J.A. Sosman) and NIH CA95131 (Specialized Program of Research Excellence in Breast Cancer to J.A. Pietenpol), and the Joanna M. Nicolay Melanoma Foundation 2013 Research Scholar Award (K.E. Hutchinson). W. Pao is supported by a Stand Up to Cancer Innovative Reseatch Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-IRG0409).
Footnotes
Conflicts of Interest: I. Puzanov and J.A. Sosman have participated as a consultant and on advisory boards, respectively, for GlaxoSmithKline. All Foundation Medicine, Inc. (FMI) authors are employees and stockholders in FMI.
References
- 1.Lovly CM, Dahlman KB, Fohn LE, Su Z, Dias-Santagata D, Hicks DJ, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS One. 2012;7:e35309. doi: 10.1371/journal.pone.0035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–4. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–14. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 4.Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2012;2:791–7. doi: 10.1158/2159-8290.CD-12-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroder E, Gebel L, Eremeev AA, Morgner J, Grum D, Knauer SK, et al. Human PAPS synthase isoforms are dynamically regulated enzymes with access to nucleus and cytoplasm. PLoS One. 2012;7:e29559. doi: 10.1371/journal.pone.0029559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Douarin B, Zechel C, Garnier JM, Lutz Y, Tora L, Pierrat P, et al. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–33. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, et al. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci U S A. 2009;106:11612–6. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miki T, Fleming TP, Crescenzi M, Molloy CJ, Blam SB, Reynolds SH, et al. Development of a highly efficient expression cDNA cloning system: application to oncogene isolation. Proc Natl Acad Sci U S A. 1991;88:5167–71. doi: 10.1073/pnas.88.12.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belloni E, Trubia M, Gasparini P, Micucci C, Tapinassi C, Confalonieri S, et al. 8p11 myeloproliferative syndrome with a novel t(7;8) translocation leading to fusion of the FGFR1 and TIF1 genes. Genes Chromosomes Cancer. 2005;42:320–5. doi: 10.1002/gcc.20144. [DOI] [PubMed] [Google Scholar]
- 10.Klugbauer S, Rabes HM. The transcription coactivator HTIF1 and a related protein are fused to the RET receptor tyrosine kinase in childhood papillary thyroid carcinomas. Oncogene. 1999;18:4388–93. doi: 10.1038/sj.onc.1202824. [DOI] [PubMed] [Google Scholar]
- 11.Ciampi R, Knauf JA, Kerler R, Gandhi M, Zhu Z, Nikiforova MN, et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest. 2005;115:94–101. doi: 10.1172/JCI23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cin H, Meyer C, Herr R, Janzarik WG, Lambert S, Jones DT, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011;121:763–74. doi: 10.1007/s00401-011-0817-z. [DOI] [PubMed] [Google Scholar]
- 13.Dessars B, De Raeve LE, El Housni H, Debouck CJ, Sidon PJ, Morandini R, et al. Chromosomal translocations as a mechanism of BRAF activation in two cases of large congenital melanocytic nevi. J Invest Dermatol. 2007;127:1468–70. doi: 10.1038/sj.jid.5700725. [DOI] [PubMed] [Google Scholar]
- 14.Forshew T, Tatevossian RG, Lawson AR, Ma J, Neale G, Ogunkolade BW, et al. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218:172–81. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 15.Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–7. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee NV, Lira ME, Pavlicek A, Ye J, Buckman D, Bagrodia S, et al. A novel SND1-BRAF fusion confers resistance to c-Met inhibitor PF-04217903 in GTL16 cells though MAPK activation. PLoS One. 2012;7:e39653. doi: 10.1371/journal.pone.0039653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin A, Rodriguez FJ, Karajannis MA, Williams SC, Legault G, Zagzag D, et al. BRAF alterations in primary glial and glioneuronal neoplasms of the central nervous system with identification of 2 novel KIAA1549:BRAF fusion variants. J Neuropathol Exp Neurol. 2012;71:66–72. doi: 10.1097/NEN.0b013e31823f2cb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alikhan A, Ibrahimi OA, Eisen DB. Congenital melanocytic nevi: where are we now? Part I. Clinical presentation, epidemiology, pathogenesis, histology, malignant transformation, and neurocutaneous melanosis. J Am Acad Dermatol. 2012;67:495, e1–17. doi: 10.1016/j.jaad.2012.06.023. quiz 512–4. [DOI] [PubMed] [Google Scholar]
- 20.Chmielecki J, Peifer M, Viale A, Hutchinson K, Giltnane J, Socci ND, et al. Systematic screen for tyrosine kinase rearrangements identifies a novel C6orf204-PDGFRB fusion in a patient with recurrent T-ALL and an associated myeloproliferative neoplasm. Genes Chromosomes Cancer. 2012;51:54–65. doi: 10.1002/gcc.20930. [DOI] [PubMed] [Google Scholar]
- 21.Baljuls A, Mahr R, Schwarzenau I, Muller T, Polzien L, Hekman M, et al. Single substitution within the RKTR motif impairs kinase activity but promotes dimerization of RAF kinase. J Biol Chem. 2011;286:16491–503. doi: 10.1074/jbc.M110.194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer Cell. 2011;19:11–5. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sievert AJ, Lang SS, Boucher KL, Madsen PJ, Slaunwhite E, Choudhari N, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A. 2013;110:5957–62. doi: 10.1073/pnas.1219232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.