Abstract
Aberrant cholesterol/lipid homeostasis is linked to a number of diseases prevalent in the developed world including metabolic syndrome, type II diabetes, and cardiovascular disease. We have previously uncovered gene regulatory mechanisms of the Sterol Regulatory Element-Binding Protein (SREBP) family of transcription factors, which control the expression of genes involved in cholesterol and lipid biosynthesis and uptake. Intriguingly, we recently discovered conserved microRNAs (miR-33a/b) embedded within intronic sequences of the human SREBF genes that act in a concerted manner with their host gene products to regulate cholesterol/lipid homeostasis. Indeed, miR-33a/b control the levels of ABCA1, a cholesterol efflux pump critical for high-density lipoprotein (HDL) synthesis and reverse cholesterol transport from peripheral tissues. Importantly, antisense inhibition of miR-33 in mice results in elevated HDL and decreased atherosclerosis. Intriguingly, miR-33a/b also act in the fatty acid/lipid homeostasis pathway by controlling the fatty acid β-oxidation genes CROT, HADHB and CPT1A, as well as the energy sensor AMPK, the NAD+-dependent sirtuin SIRT6, and the insulin signaling intermediate IRS-2, key regulators of glucose and lipid metabolism. These results have revealed a highly integrated microRNA-host gene circuit governing cholesterol/lipid metabolism and energy homeostasis in mammals that may have important therapeutic implications for the treatment of cardiometabolic disorders.
Introduction
Cholesterol and other lipids are crucial for metazoan life since they are essential components of membranes as well as precursors for hormone and signaling molecules. Furthermore, imbalance in cholesterol and lipids are strongly associated with diseases prevalent in the developed world such as metabolic syndrome, type II diabetes and cardiovascular disease. To improve treatment of these diseases we need to expand our knowledge of how cholesterol/lipid homeostasis is maintained, and elucidate the underlying molecular mechanisms contributing to metabolic abnormalities. Members of the sterol regulatory element-binding protein (SREBP) family of basic-helix-loop-helix-leucine zipper transcription factors are crucial regulators of lipid and cholesterol homeostasis. SREBPs activate the complete program (>30 genes) of fatty acid, triglyceride, phospholipid and cholesterol biosynthesis and uptake (Horton et al. 2002), as well as genes involved in the production of NADPH, an essential cofactor for lipid and cholesterol synthesis. A recent study from our lab has revealed that SREBPs also activate many genes in the one-carbon cycle to provide S-adenosylmethionine (SAMe), the major cellular methyl donor, and a key cofactor for the synthesis of the membrane phospholipid phosphatidylcholine (Walker et al. 2011). The mammalian SREBP gene family contains 2 members: SREBF1 (which codes for 2 isoforms, SREBP-1c and SREBP-1a) and SREBF2, coding for the SREBP-2 protein. SREBP-1 isoforms have been shown to preferentially activate genes involved in fatty acid, triglyceride, and phospholipid biosynthesis, such as fatty acid synthase (FASN) and stearoyl Co-A desaturase (SCD). By contrast, SREBP-2 is more important for the expression of genes involved in cholesterol biosynthesis, such as 3-hydroxy-3-methylglutaryl- coenzyme A reductase (HMGCR), and uptake (i.e., low density lipoprotein receptor (LDLR)) (Horton et al. 2002).
SREBPs are regulated extensively on multiple levels in order to coordinate cellular and physiologic metabolic processes for optimal animal growth and homeostasis (Osborne and Espenshade 2009). An important and well-studied level of SREBP regulation is the post-transcriptional control of SREBP proteolytic processing/maturation by cholesterol. This regulation is essential for proper cholesterol homeostasis and constitutes a feedback loop involving cholesterol levels and SREBP-2. When sufficient levels of cholesterol are present in the cell, SREBP-2 precursors are tethered in the endoplasmic reticular membrane in complex with the cholesterol-sensing chaperones SCAP and INSIG (Osborne and Espenshade 2009). When cholesterol levels fall below a threshold, the complex dissociates and SREBP-2 is trafficked together with SCAP to the Golgi for proteolytic processing to its active form and released to the nucleus to activate genes for cholesterol biosynthesis and uptake. While SREBP-2 is controlled by cholesterol in a negative feedback loop, we have recently found that proteolytic maturation of metazoan SREBP-1 precursors are controlled by levels of phosphatidylcholine (PC), a downstream product of SREBP-1-regulated pathways (fatty acid synthesis, one-carbon cycle and PC biosynthesis genes) (Walker et al. 2011).
In addition to proteolytic processing, SREBPs are controlled at the transcriptional level. For example, the expression of the SREBP-1c isoform is regulated by the liver × receptor (LXR) nuclear receptor in response to oxysterols (Repa et al. 2000; Schultz et al. 2000). Both SREBP-1 and -2 isoforms also activate their own expression, ensuring higher levels of SREBPs through a feed-forward mechanism. In the liver, regulation of SREBP-1c by insulin is of great physiological importance. Post-prandial elevation in circulating insulin levels leads to increased hepatic glucose and fatty acid uptake, and production of triglycerides for storage. Pathological insulin levels associated with insulin resistance, a hallmark of metabolic syndrome, are linked to increased hepatic expression and enhanced nuclear accumulation of SREBP-1c. This in turn results in elevated expression of lipogenic genes and hepatic steatosis. The insulin-dependent increase in SREBP-1c mRNA is mediated through the LXR binding sites in the SREBF1c promoter (Chen et al. 2004). The mechanisms of post-transcriptional regulation of SREBPs by insulin are not completely understood, but have been suggested to involve both the INSIG/SCAP complex as well as the function of the serine/threonine protein kinase AKT. Indeed, SREBPs are known to be extensively controlled by phosphorylation. Acting downstream of insulin, AKT can directly phosphorylate SREBP, as well as inhibit GSK3, a kinase that promotes SREBP turnover, thereby stimulating SREBP-dependent gene regulation (Sundqvist et al. 2005; Yellaturu et al. 2009). In addition, insulin activation of the mTOR signaling pathway can also augment SREBP function through an mTORC1-dependent mechanism (Lewis et al.; Porstmann et al. 2008; Peterson et al. 2011).
The nutrient sensor AMPK (5' AMP-activated protein kinase) is a master regulator of cellular energy homeostasis in response to energy stress (e.g., low ATP/AMP ratio), negatively regulating energy-consuming cellular processes such as the synthesis of proteins, fatty acids and cholesterol, while simultaneously promoting mitochondrial biogenesis and activating fatty acid β-oxidation for the production of ATP (Hardie 2011a). Activated AMPK directly inactivates several SREBP downstream targets such as acetyl-CoA carboxylase (ACC) and HMGCR by phosphorylation, but can also inhibit SREBPs themselves on multiple levels (Zhou et al. 2001; Foretz et al. 2005). This latter mechanism includes inhibition of LXR (Yap et al. 2011), as well as direct phosphorylation of SREBPs (Li et al. 2011). Increased AMPK activity has also been shown to promote the activity of the NAD+-dependent deacetylase SIRT1 (Schug and Li 2011). We and others recently showed that SIRT1 is a key negative regulator of SREBPs in metazoans in response to nutrient deprivation, such as during fasting (Ponugoti et al. 2010; Walker et al. 2010). Under such conditions, AMPK is active, NAD+ levels are elevated, and SIRT1 protein is increased, together promoting SIRT1-dependent deacetylation of nuclear SREBPs, which then results in their ubiquitylation and proteasomal degradation, and a reduction in lipogenic gene expression and fat storage (Ponugoti et al. 2010; Walker et al. 2010; Schug and Li 2011). These findings suggest that SREBPs sit at the nexus of a highly integrated and interconnected metabolic regulatory network governing cholesterol/lipid and energy homeostasis.
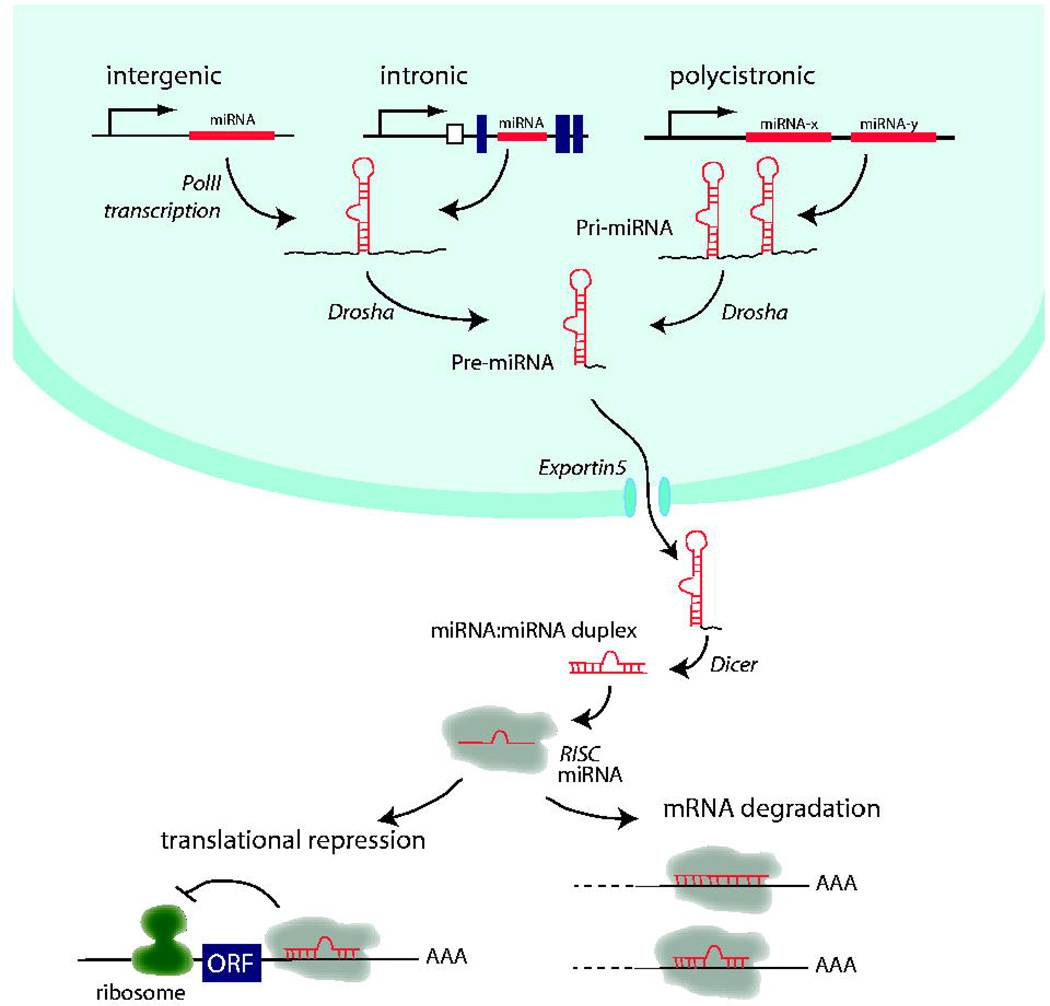
MicroRNAs have recently been shown to represent yet another layer of regulation of metabolism and cholesterol/lipid homeostasis (Najafi-Shoushtari; Fernandez-Hernando et al. 2011). MicroRNAs (miRNAs) are short (22–25 nucleotides) single stranded non-coding RNAs with important functions in most biological processes. They are transcribed by RNA polymerase II and subsequently processed by the miRNA biogenesis pathway including Drosha, DGCR8, and Dicer and incorporated into the RNA-Induced Silencing Complex (RISC) (Fig. 1). Mature miRNAs then act to inhibit mRNA expression by binding to partially complementary target sequences in their 3' UTRs and either reducing their translation or causing mRNA degradation (Bartel 2009). Individual miRNAs typically have a number of predicted targets based on complementarity with their seed base sequences, as well as evolutionary conservation of target sites (Lewis et al. 2005; Grimson et al. 2007; Friedman et al. 2009).
Figure 1.
MicroRNA biogenesis pathway.
We and others have recently discovered that the human SREBP-encoding genes harbor intronic microRNAs (miR-33a/b) that appear to act in concert with their host gene products to control cholesterol/lipid homeostasis (Gerin et al. 2010; Horie et al. 2010; Marquart et al. 2010; Najafi-Shoushtari et al. 2010; Rayner et al. 2010). These studies show that miR-33, through its regulation of the ABCA1 cholesterol transporter, cooperates with SREBP to regulate cholesterol homeostasis. In addition, we and others have found that miR-33 controls the expression of fatty acid β-oxidation genes such as CROT, CPT1A and HADHB, as well as the energy homeostasis regulators AMPKα1, SIRT6 and IRS-2. These results uncover a broader, concerted regulation of metabolic networks by the SREBP and miR-33 transcription factor/miRNA circuit. Moreover, it hints at the therapeutic value of antisense-based targeting of miR-33 for diseases such as metabolic syndrome and atherosclerosis that are associated with dysregulation of cholesterol/lipid metabolism and insulin signaling.
Results
SREBPs are host genes to conserved microRNAs, miR-33a/b
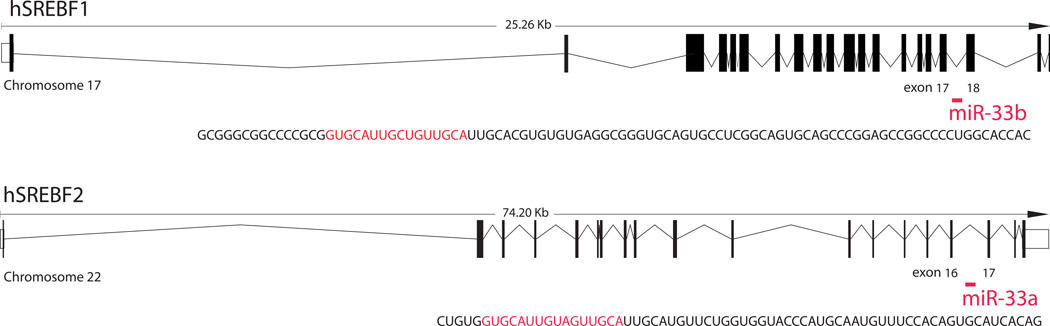
During our analysis of the mechanism of transcriptional regulation by the SREBP transcription factors we noticed the presence of conserved miRNAs (miR-33b and miR-33a) within the intronic sequence of both SREBF1 and SREBF2 genes, respectively (Najafi-Shoushtari et al. 2010) (Fig. 2). Database searches revealed that miR-33 is conserved inside the SREBP genomic sequences from insects such as Drosophila to humans. Interestingly, miR-33b is present within the SREBF1 gene in mammals, including humans and primates, but is absent in rodents; the physiological significance of this difference will be further touched upon in the discussion section. To explore the potential functional link of the SREBP transcription factors and miR-33a/b, we initial investigated the expression of the SREBF genes and their embedded miRNAs. Consistent with the possibility of coordinated functions, we found that miR-33a/b are extensively co-expressed with their SREBF host genes in human (miR-33a/b) and mouse (miR-33) tissues (Najafi-Shoushtari et al. 2010). To gain an understanding of the possible regulatory roles of miR-33a/b, we used several target prediction programs such as TargetScan (Grimson et al. 2007) Pictar (Krek et al. 2005) and miRanda (Betel et al. 2008). This analysis revealed that miR-33a and miR-33b (which differ by only two nucleotides) share a number of conserved targets predicted by several algorithms (Table 1). Intriguingly, the list of predicted targets includes genes involved in cholesterol transport such as the ATP-binding cassette (ABC) transporter ABCA1 and the endolysosomal transport protein Niemann-Pick C1 (NPC1). This suggested that miR-33a/b may indeed be functionally linked with their host genes.
Figure 2.
SREBF gene structure with their intronic microRNAs.
The human SREBF1 and 2 genes contain the intronic miR-33b and miR-33a, respectively. miR-33a is conserved from insects (e.g., Drosophila) to humans whereas miR-33b is conserved in all mammals, except in rodents.
Table 1.
Other selected predicted conserved targets of miR-33a/b involved in lipid homeostasis.
| Target gene |
Gene name | # sites |
Target scan |
PICTAR | miRanda | targeted in |
|---|---|---|---|---|---|---|
| ABCA1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 3 | x | x | x | Hsa, Ptr, Mmu, Rno, Cfa, Mdo, Oan, Gga, Xtr |
| CROT | Carnitine O-octanoyltransferase | 2 | x | (x) | x | Hsa, Ptr, Mmu, Rno, Fca, Aca, Gga |
| MRPS25 | Mitochondrial ribosomal protein S25 | 2 | x | x | x | Hsa, Ptr, Mmu, Rno, Cfa, Mdo, Oan, Gga |
| NPC1 | Niemann-Pick disease, type C1 | 2 | x | x | Hsa, Ptr, Mmu, Rno, Cfa, Gga | |
| SIRT6 | Sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) | 1 | x | x | Hsa, Ptr, Mmu, Cfa | |
| PRKAA1 | Protein kinase, AMP-activated, alpha 1 catalytic subunit | 2 | x | x | x | Hsa, Ptr, Mmu, Rno, Cfa, Mdo, Gga, Xtr |
| HADHB | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 1 | x | x | x | Hsa, Ptr, Mmu, Rno, Cfa, Mdo, Oan, Gga |
| SNF1LK | SNF1-like kinase | 1 | x | x | x | Hsa, Ptr, Mdo, Oan, Gga |
| IRS-2 | Insulin receptor substrate 2 | 1 | x | x | x | Hsa, Ptr, Mmu, Rno, Cfa, Mdo, Oan |
| HMGCR | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | 1 | x | x | Hsa, Ptr, Mmu, Oan | |
| CS | Citrate synthase | 1 | x | x | Hsa, Ptr, Cfa, Oan, Xtr | |
| CPT1A | Carnitine palmitoyltransferase 1A (liver) | 1 | x | Hsa, Mmu, Rno, Cfa, Gga, Dme |
Selected conserved miR-33a/b targets involved in cholesterol/lipid and energy homeostasis as predicted by TargetScan (Grimson et al. 2007), PICTAR, and miRanda.
Hsa = human; Ptr = chimpanzee; Mmu = mouse; Rno = rat; Cfa = dog; Fca = Cat; Mdo = opossum; Oan = platypus; Gga = chicken Xtr = frog; Dme = fruitfly
MiR-33 targets the ABCA1 cholesterol transporter
We were particularly intrigued by the possibility of ABCA1 as a target of miR-33a/b. ABCA1 ranks highest among predicted miR-33 targets and encodes a cholesterol transporter. ABCA1 is a major regulator of cellular cholesterol and phospholipid homeostasis by mediating the efflux of cholesterol and phospholipids to apolipoprotein A1, which then form nascent high-density lipoproteins (HDL) (Tall et al. 2008). It is also critical for the reverse cholesterol transport from peripheral tissues to the liver. Hence, whereas SREBPs promote the intracellular accumulation of cholesterol/lipids, ABCA1 acts to decrease their levels. If miR-33a/b would indeed negatively regulate ABCA1 expression, it would thus represent the first example of miRNA-host gene cooperativity.
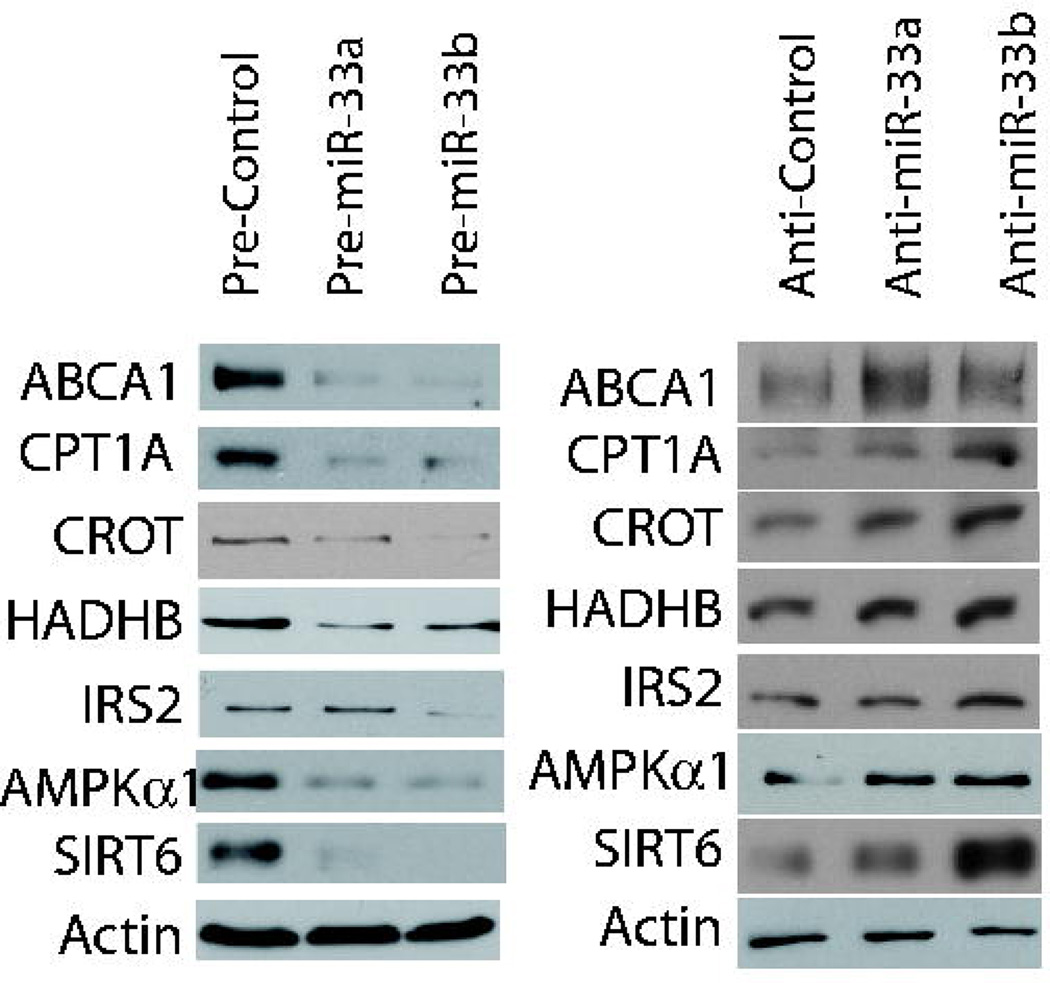
To confirm ABCA1 as a bona fide miR-33 target we first showed that ABCA1 protein levels are reduced upon introduction into cells of exogenous miR-33a/b precursors; conversely, ABCA1 levels are increased after miR-33a/b antisense inhibition (Fig. 3) (Najafi-Shoushtari et al. 2010). MicroRNAs typically act to lower protein expression by targeting the 3’UTR of mRNAs. The ABCA1 3’UTR harbors three closely spaced/overlapping predicted miR-33a/b target sites, and fusion of the ABCA1 3’UTR fragment containing these sites to a Luciferase reporter confirmed that miR-33a/b can indeed regulate the ABCA1 3'UTR through the miR-33 binding sites, as shown by site-directed mutagenesis (Najafi-Shoushtari et al. 2010). In accord with the notion that SREBPs and miR-33 may be functionally linked, we showed that SREBP-2 and miR-33 are co-regulated by cholesterol in J774 mouse macrophage cells (Najafi-Shoushtari et al. 2010). Importantly, manipulation of miR-33 also affected cholesterol efflux in these cells (Najafi-Shoushtari et al. 2010), suggesting that miR-33 could negatively impact reverse cholesterol transport from peripheral cells, including atherogenic macrophages/foam cells, thereby promoting atherosclerosis. Importantly, we found that injection of mice on a Western-type diet with Locked Nucleic Acid (LNA) antisense inhibitors of miR-33 led to a significant 25–30% elevation in circulating HDL levels (Najafi-Shoushtari et al. 2010). Similar results were reported by others (Gerin et al. 2010; Horie et al. 2010; Marquart et al. 2010; Najafi-Shoushtari et al. 2010; Rayner et al. 2010). Additionally, in support of a potential pathological role for miR-33 in atherosclerosis as suggested by our data (Najafi-Shoushtari et al. 2010), it was recently reported that injecting LDLR−/− mice on an atherogenic diet with an anti-miR-33 oligonucleotide reduces atherosclerotic plaque size (Rayner et al. 2011). Our ongoing studies in non-human primates fed a Western-type diet have confirmed that injection of LNA antisense oligonucleotides directed against miR-33a/b elicits significant elevation of HDL levels, without adverse effects (unpublished data), suggesting the feasibility of therapeutic targeting of miR-33. Taken together, the targeting of ABCA1 by miR-33a/b and the apparent cooperativity of miR-33 and SREBP adds an elegant layer of regulation to the cholesterol homeostasis mechanism, and supports the idea that miR-33a/b may represent attractive therapeutic targets to raise HDL and ameliorate atherosclerosis.
Figure 3.
MiR-33a/b targets involved in cholesterol/lipid and energy homeostasis
A. Excess miR-33a/b (through introduction of precursors, Pre-miR-33a/b) in human HepG2 hepatoma cells reduces ABCA1, CROT, HADHB, CPTA1, IRS2, AMPKα1 and SIRT6 protein levels. B. Transfection of miR-33a/b antisense oligonucleotides (Anti-miR-33a/b)into HepG2 cells increases ABCA1, CROT, HADHB, CPTA1, IRS2, AMPKα1 and SIRT6 protein levels.
MiR-33 targets fatty acid β-oxidation genes
While the results clearly show concerted regulation of cholesterol by miR-33a and the SREBP-2 host gene product, the question then arose whether a similar cooperation is found between SREBP-1 and miR-33b in regulating lipogenesis. This could be an important point from a physiological and clinical view because of the known transcriptional upregulation of SREBP-1c in the liver in response to insulin signaling and in cases of insulin resistance. Analogous to the SREBP-2/ABCA1 connection, miR-33 could also target genes that decrease intracellular levels of fatty acids and triglycerides. Indeed, the list of predicted miR-33a/b targets also include genes that promote fatty acid β-oxidation, such as carnitine O-octanoyltransferase (CROT), hydroxyacyl-CoA-dehydrogenase (HADHB), and carnitine palmitoyltransferase 1A (CPT1A) (Table 1). HADHB encodes a subunit of the mitochondrial trifunctional protein that catalyzes the last three steps of fatty acid β-oxidation. CROT catalyses the transfer of short-chain fatty acids to carnitine, making them available for degradation by β-oxidation. CPT1A is not a β-oxidation gene per se but is required for the transport of long-chain fatty acids to mitochondria where β-oxidation takes place, and represents the rate limiting enzyme in regulation of fatty acid degradation (Jogl et al. 2004). Interestingly, the Drosophila homolog of CPT1A, CptI, contains a predicted binding site of the fly miR-33 homolog, which is embedded in an intron in the dSREBP gene (Gerin et al. 2010; Davalos et al. 2011). As insects are cholesterol auxotroph and do not appear to have an ABCA1 ortholog, this suggests that inhibition of fatty acid β-oxidation could represent a primordial function of miR-33, acting in concert with the fly SREBP host gene product to control fatty acid/lipid homeostasis (Table 1).
We show here that introduction of excess miR-33a/b precursors results in down-regulation of the protein levels of CROT, CPT1a and HADHB (Fig. 3a), whereas antisense-targeting of miR-33a/b causes accumulation of these proteins (Fig. 3b). These results indeed indicate regulation of β-oxidation genes by miR-33a/b. Similar results have been reported by others (Gerin et al. 2010; Davalos et al. 2011). In accord with miR-33 regulation of fatty acid β-oxidation genes, excess miR-33 resulted in increased intracellular triglyceride and fatty acid levels, as well as increased lipid droplet formation in human hepatoma cells and in the Drosophila fat body (Gerin et al. 2010; Davalos et al. 2011). Taken together, we and others find that SREBP-1 and miR-33b are indeed cooperating in a similar fashion to SREBP-2/miR-33a to control fatty acid/lipid homeostasis.
Control of glucose/energy homeostasis regulators AMPK, SIRT6, and IRS2 by miR-33
As discussed above, SREBPs integrate information about nutritional state such as insulin, glucose and AMP levels to coordinate appropriate lipid levels. It was therefore intriguing to also find AMPKα1, IRS-2 and SIRT6 on the list of predicted miR-33 targets. AMPKα1 is the catalytic subunit of AMPK, and directly inhibits proteins such as ACC and HMGCR, as well as SREBPs, as discussed above (Hardie 2011b). IRS-2, or insulin receptor substrate 2, is one of two such proteins that mediates the effect of insulin by acting as a molecular adaptor between the insulin receptor and downstream effectors (White 2003). Interestingly, depletion of IRS-2 (but not IRS-1) in mouse liver results in elevated SREBP-1c expression/activity and downstream lipogenic gene expression (Tobe et al. 2001; Matsumoto et al. 2002; Taniguchi et al. 2005; Kohjima et al. 2008), suggesting another potential functional link with the SREBP/miR-33 circuit. SIRT6 is an NAD+-dependent histone deacetylase with roles in metabolism and stress resistance (Kanfi et al. 2008; Kawahara et al. 2009; Lombard 2009; Michishita et al. 2009; Yang et al. 2009; Kaidi et al. 2010; Kanfi et al. 2010; Schwer et al. 2010; Tennen and Chua 2010; Xiao et al. 2010; Mao et al. 2011; Tennen et al. 2011), and has been shown to be a major regulator of glucose uptake (Xiao et al. 2010; Zhong et al. 2010). Importantly, recent studies of mice with liver-specific ablation of SIRT6 revealed upregulated hepatic glycolysis, lipogenesis and triglyceride synthesis, accompanied by steatosis (Kim et al. 2010). These data suggest that hepatic SIRT6 may also be functionally linked to the SREBP/miR-33 pathway. To investigate if these genes are indeed targeted by miR-33, we modulated miR-33a/b levels in HepG2 cells. As with the other targets, excess miR-33a/b increased the protein levels of IRS-2, AMPKα1 and SIRT6 (Fig. 3a), whereas depletion of miR-33a/b decreased their protein levels (Fig. 3b). As AMPK, IRS-2, and SIRT6 all appear to counteract hepatic SREBP-1c-dependent lipogenesis, these results indicate that miR-33b embedded within the SREBP-1c gene may cooperate with the SREBP-1c host gene product to promote hepatic lipid accumulation, similar to miR-33a cooperation with SREBP-2 in promoting elevated intracellular cholesterol levels.
Conclusions
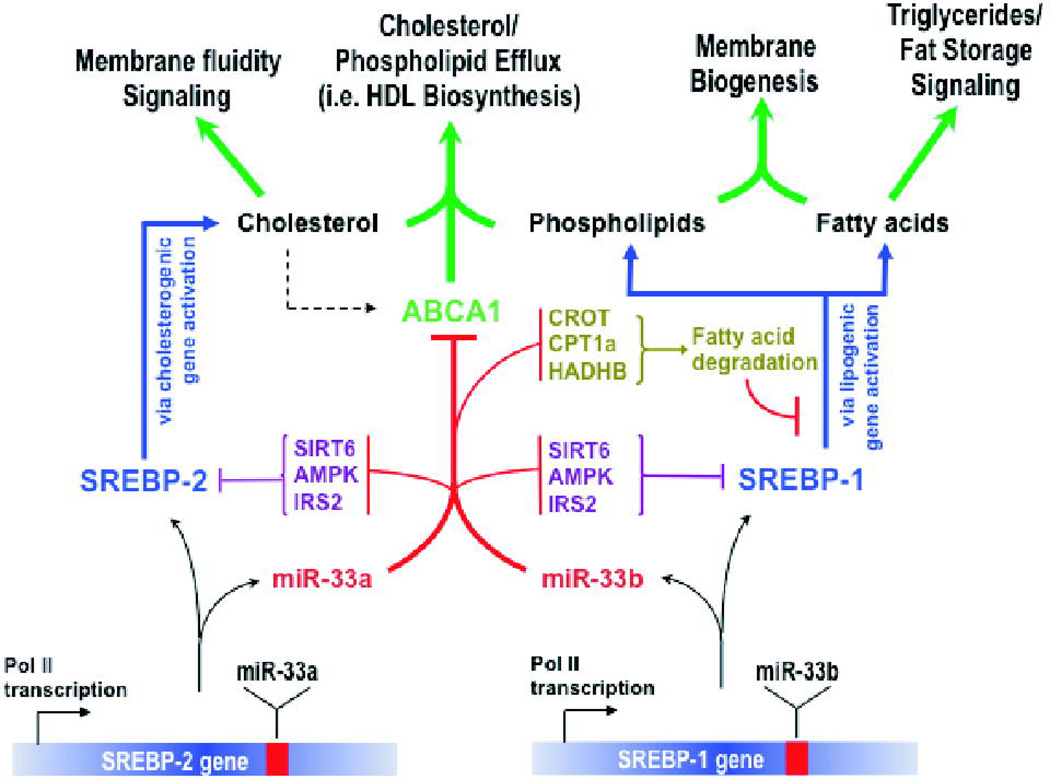
A number of independent studies have now shown the cooperation between SREBP-2 and its intronic microRNA miR-33a, adding a layer of regulation to the SREBP-2 control of cholesterol homeostasis (Fig. 4) (Gerin et al. 2010; Marquart et al. 2010; Najafi-Shoushtari et al. 2010; Rayner et al. 2010). The finding by us and others of a similar cooperation between SREBP-1 and miR-33b in regulation of lipid homeostasis suggests a more extensive and integrated network of functional interactions between SREBPs and their intronic miRNAs. These findings together indicate that an important function of miR-33 is inhibition of proteins counteracting SREBP-dependent activities. Indeed, miR-33 targeting of fatty acid β-oxidation genes would aid the SREBP-1 host gene product by preventing degradation of lipids produced by SREBP-1-regulated genes. Moreover, the finding that miR-33 controls levels of AMPKα1, a protein that counteracts the function of SREBP through the inhibition of downstream SREBP targets such as HMGCR and ACC as well as directly affecting SREBP itself, represents an excellent example of the notion that miR-33 acts as an “enforcer” of SREBP-dependent processes. The SIRT6 inhibition of miR-33 also follows this paradigm since it was shown that SIRT6 deacetylates histone H3 lysine 9 (H3K9) residues at the promoters of SREBP target genes such as FASN, ACC1 and SCD1 in the liver and directly inhibits their expression (Kim et al. 2010), thus effectively opposing SREBP-1 function. Finally, miR-33 inhibition of insulin receptor substrate 2 should promote SREBP-1 activity, as IRS-2 reduction has been shown to activate the other insulin receptor substrate, IRS-1, which in turn activates SREBP-1 (Kohjima et al. 2008). This regulatory network is schematically summarized in Fig. 4. This extensive cooperation of the SREBP transcription factors with miR-33a/b uncovers a novel concept of integrated and concerted regulation of metabolic networks by a transcription factor/microRNA circuits.
Figure 4.
The SREBP transcription factors act coordinately with their intronic microRNA miR-33a/b to regulate both fatty acid and cholesterol homeostasis. Whereas SREBPs activate genes involved in fatty acid and cholesterol biosynthesis and uptake such as FASN and HMGCR, miR-33a/b act to repress genes functioning in cholesterol efflux (ABCA1) and fatty acid β-oxidation (CROT, HADHB, CPT1A), as well as negative regulators of SREBPs such as AMPKα1, SIRT6 and IRS-2.
The cholesterol transporter ABCA1 was the first well-described miR-33 target in macrophages and hepatoma cells. ABCA1 mediates the initial and essential step in the formation of HDL. High LDL levels and low HDL levels are strongly associated with risk for cardiovascular disease. While statins are relatively successful in lowering LDL, treatments for elevating HDL levels such as niacin are only moderately efficacious. Hence, miR-33 may represent an attractive new target for raising HDL. Indeed, ABCA1 and HDL levels were elevated in mice carrying a homozygous deletion of miR-33a (Horie et al. 2010). More importantly, HDL levels were elevated in mice after miR-33a knockdown using an antisense approach (Najafi-Shoushtari et al. 2010; Rayner et al. 2010; Rayner et al. 2011), indicating that antisense targeting of miR-33 could indeed be clinically useful. Highlighting the therapeutic potential of miRNA-targeting antisense approaches, miR-122, a liver-specific miRNA that regulates hepatic cholesterol/lipid metabolism and hepatitis C virus replication (Girard et al. 2008), has been successfully targeted by LNA antisense oligonucleotides in nonhuman primates (Elmen et al. 2008; Lanford et al. 2010). miR-122-targeting LNAs (miravirsen) are now being evaluated in a Phase 2a clinical trial of chronic hepatitis C patients and preliminary results look promising. These LNA techniques, or approaches such as the so-called tiny LNAs targeting entire miRNA familes (Obad et al. 2011), represent exciting future directions for miR-33 antagonism for therapeutic purposes. With the expansion of the function of miR-33a/b into the regulation of fatty acid β-oxidation, insulin signaling, and energy homeostasis, therapeutic benefits may encompass not only ameliorated atherosclerosis, but also effects on the broader array of conditions associated with metabolic syndrome, including insulin resistance, hepatosteatosis, and circulating lipid abnormalities such as elevated triglycerides. In the clinically relevant conditions of metabolic syndrome, insulin levels are highly elevated which causes a strong transcriptional upregulation of SREBP-1c, and thus presumably of miR-33b, in the liver. This is in contrast to SREBP-2 and miR-33a, whose mRNA levels are only modestly changed in response to cholesterol levels, most likely because cholesterol regulation of SREBP-2 is chiefly acting at the post-transcriptional level (Brown et al. 2010). Thus, targeting both miR-33a/b in insulin resistance conditions that are associated with elevated VLDL and triglyceride levels and low HDL might prove beneficial. An important complication to studying this circuit in model organisms is the absence of miR-33b in rodents, making studies in non-human primates necessary to address the relevance and therapeutic potential of targeting miR-33b. Such studies should also allow addressing the broader impact of miR-33 antisense targeting on other possible/predicted downstream targets as well. Of interest could be predicted targets that would be involved in other functions of SREBP, including the regulation of cofactors for lipid production such as NADPH or S-adenosylmethionine. Of particular concern could be predicted targets that affect cancer biology such as CDK6, PIM-1 and p53. A recent study indicated indeed that miR-33a targets the proto-oncogene Pim-1, but not the predicted target CDK6, and suggested overexpression of miR-33a as an anti-cancer treatment (Thomas et al. 2011). However, another study described the regulation of p53 by miR-33 (Herrera-Merchan et al. 2010), suggesting a complex and possible context-dependent response to miR-33 manipulations. Final validation of miR-33 for therapeutic use in metabolic disorders thus awaits further detailed in vivo investigation in primate models and in humans.
Acknowledgments
This work was supported by funding from the following NIH grants: 1R21DK084459 and 1R01GM071449.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Goldstein JL. Medicine. HDL miR-ed down by SREBP introns. Science. 2010;328:1495–1496. doi: 10.1126/science.1192409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci U S A. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Fernandez-Hernando C, Suarez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011;22:86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, Ancellin N, Andreelli F, Saintillan Y, Grondin P, Kahn A, Thorens B, Vaulont S, Viollet B. Short-term overexpression of a constitutively active form of AMP-activated protein kinase in the liver leads to mild hypoglycemia and fatty liver. Diabetes. 2005;54:1331–1339. doi: 10.2337/diabetes.54.5.1331. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated protein kinase: a cellular energy sensor with a key role in metabolic disorders and in cancer. Biochem Soc Trans. 2011a;39:1–13. doi: 10.1042/BST0390001. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Sensing of energy and nutrients by AMP-activated protein kinase. Am J Clin Nutr. 2011b;93:891S–896S. doi: 10.3945/ajcn.110.001925. [DOI] [PubMed] [Google Scholar]
- Herrera-Merchan A, Cerrato C, Luengo G, Dominguez O, Piris MA, Serrano M, Gonzalez S. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle. 2010;9:3277–3285. doi: 10.4161/cc.9.16.12598. [DOI] [PubMed] [Google Scholar]
- Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogl G, Hsiao YS, Tong L. Structure and function of carnitine acyltransferases. Ann N Y Acad Sci. 2004;1033:17–29. doi: 10.1196/annals.1320.002. [DOI] [PubMed] [Google Scholar]
- Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, Kronfeld-Schor N, Cohen HY. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, et al. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohjima M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, Enjoji M, et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int J Mol Med. 2008;21:507–511. [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Griffiths B, Santos CR, Pende M, Schulze A. Regulation of the SREBP transcription factors by mTORC1. Biochem Soc Trans. 39:495–499. doi: 10.1042/BST0390495. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB. Sirtuins at the breaking point: SIRT6 in DNA repair. Aging (Albany NY) 2009;1:12–16. doi: 10.18632/aging.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Ogawa W, Teshigawara K, Inoue H, Miyake K, Sakaue H, Kasuga M. Role of the insulin receptor substrate 1 and phosphatidylinositol 3-kinase signaling pathway in insulin-induced expression of sterol regulatory element binding protein 1c and glucokinase genes in rat hepatocytes. Diabetes. 2002;51:1672–1680. doi: 10.2337/diabetes.51.6.1672. [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH. MicroRNAs in cardiometabolic disease. Curr Atheroscler Rep. 13:202–207. doi: 10.1007/s11883-011-0179-y. [DOI] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, Chiang CM, Veenstra TD, Kemper JK. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285:33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43(3):198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Schumacher B, Lombard DB, Xiao C, Kurtev MV, Gao J, Schneider JI, Chai H, Bronson RT, Tsai LH, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc Natl Acad Sci U S A. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 2008;7:365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Taniguchi CM, Ueki K, Kahn R. Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. J Clin Invest. 2005;115:718–727. doi: 10.1172/JCI23187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tennen RI, Bua DJ, Wright WE, Chua KF. SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun. 2011;2:433. doi: 10.1038/ncomms1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennen RI, Chua KF. Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem Sci. 2010;36:39–46. doi: 10.1016/j.tibs.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Lange-Grunweller K, Weirauch U, Gutsch D, Aigner A, Grunweller A, Hartmann RK. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 2011 doi: 10.1038/onc.2011.278. [DOI] [PubMed] [Google Scholar]
- Tobe K, Suzuki R, Aoyama M, Yamauchi T, Kamon J, Kubota N, Terauchi Y, Matsui J, Akanuma Y, Kimura S, et al. Increased expression of the sterol regulatory element-binding protein-1 gene in insulin receptor substrate-2(−/−) mouse liver. J Biol Chem. 2001;276:38337–38340. doi: 10.1074/jbc.C100160200. [DOI] [PubMed] [Google Scholar]
- Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, Finnegan DM, Shioda T, Hansen M, Yang F, Niebergall L, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011 doi: 10.1016/j.cell.2011.09.045. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF. Insulin signaling in health and disease. Science. 2003;302:1710–1711. doi: 10.1126/science.1092952. [DOI] [PubMed] [Google Scholar]
- Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, Jou W, Gius D, Deng CX. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem. 2010;285:36776–36784. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap F, Craddock L, Yang J. Mechanism of AMPK suppression of LXR-dependent Srebp-1c transcription. Int J Biol Sci. 2011;7:645–650. doi: 10.7150/ijbs.7.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellaturu CR, Deng X, Cagen LM, Wilcox HG, Mansbach CM, 2nd, Siddiqi SA, Park EA, Raghow R, Elam MB. Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J Biol Chem. 2009;284:7518–7532. doi: 10.1074/jbc.M805746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]