Abstract
Steroidogenic factor 1 (SF1) is a nuclear receptor encoded by the NR5A1 gene. SF1 affects both sexual and adrenal development through the regulation of target gene expression. Genotypic male and female SF1 knockout mice have adrenal and gonadal agenesis with persistent Müllerian structures and early lethality. There have been several reports of NR5A1 mutations in individuals with 46,XY complete gonadal dysgenesis (CGD) or other disorders of sex development (DSD) with or without an adrenal phenotype. To date microdeletions involving NR5A1 have been reported in only two patients with DSDs. We report a novel microdeletion encompassing NR5A1 in a patient with 46,XY DSD and developmental delay. The phenotypically female patient initially presented with mild developmental delay and dysmorphisms. Chromosome analysis revealed a 46,XY karyotype. A 1.54 Mb microdeletion of chromosome 9q33.3 including NR5A1 was detected by array CGH and confirmed by FISH. Normal maternal FISH results indicated that this was most likely a de novo event. Since most NR5A1 mutations have been ascertained through gonadal or adrenal abnormalities, the additional findings of developmental delay and minor facial dysmorphisms are possibly related to haploinsufficiency of other genes within the 1.54 Mb deleted region. This report further confirms the role of NR5A1 deletions in 46,XY DSD and reinforces the utility of aCGH in the work up of DSDs of unclear etiology.
Keywords: NR5A1; SF1; 46,XY disorder of sex development; 46,XY DSD; 9q33 microdeletion
1. Introduction
Steroidogenic factor-1 (SF1) is a nuclear receptor that affects both sexual development and adrenal development through the regulation of target gene expression [1]. Genotypic male and female SF1 knockout mice have gonadal and adrenal agenesis, persistent Müllerian structures, and a reduced lifespan, probably due to adrenocortical insufficiency [2, 3]. SF1 is encoded by the NR5A1 gene, and the first three published human mutations in this gene were all missense mutations [4–6]. The first patient had a heterozygous mutation causing 46,XY complete gonadal dysgenesis (CGD) and adrenal failure [4]. The second was a heterozygous mutation in a 46,XX prepubertal girl presenting with adrenal failure only [6]. Both papers excluded a dominant negative mechanism of action for the reported mutation. Despite the second report, a screen of patients with idiopathic adrenal failure demonstrated that NR5A1 mutations are unlikely to be common causes of adrenal failure without a disorder of sex development (DSD) [7]. The third patient presented similarly to the first but was the product of a consanguineous marriage and was homozygous for a presumably milder NR5A1 missense mutation [5].
Since these initial discoveries, there have been other reports of various types of NR5A1 mutations (including missense, nonsense, and frameshift) in individuals with different forms of 46,XY DSD without an adrenal phenotype [8–11]. For example, Lin et al. found missense mutations in four 46,XY individuals. Two had female genitalia, one with inguinal testes and remnant Müllerian structures and the other with labial testes and no Müllerian structures. A third patient had clitoral enlargement, labioscrotal folds, labioscrotal testes, and remnant Müllerian structures. The last patient had a small phallus, hypospadias, high scrotal testes, and absent Müllerian structures [8]. Recently, there was a report of a small (3.1 to 4.8 kb) deletion encompassing two exons of NR5A1 in a patient with 46,XY DSD lacking an adrenal phenotype [12]. In addition, missense changes, in-frame deletions, frameshift, and nonsense mutations in NR5A1 were found in 46,XX females with isolated ovarian insufficiency [11, 13]. The DSD phenotypes can be quite variable even within the same family demonstrating that a single NR5A1 allele can have varied expressivity [11, 14]. Furthermore, heterozygous missense mutations were discovered in males with moderate to severe spermatogenic failure [15]. The missense mutations associated with male infertility were in or near the HINGE domain of the SF1 protein. Only two microdeletions detected by array comparative genomic hybridization (aCGH) involving NR5A1 have been reported. The first is a 3 Mb deletion encompassing several genes including NR5A1 and LIMX1B (the gene responsible for nail-patella syndrome) in a 46,XY female described to have genitopatellar syndrome and developmental delay. She was reported to have clitoromegaly, inguinal ovotestes with attached Müllerian and Wolffian duct remnants, and dysmorphic facies [16]. The second is an approximately 970 kb microdeletion including the NR5A1 gene in a patient with isolated 46,XY DSD including clitoromegaly, no fusion of the labia majora, a shallow vaginal entrance, and gonads palpable in the labium [17].
Here we present a novel 1.54 Mb microdeletion including NR5A1 in a 6 ½ year-old patient with 46,XY DSD, mild developmental delay, minor dysmorphisms, and normal adrenal function. This aberration was identified by aCGH and confirmed by fluorescence in situ hybridization (FISH). Maternal FISH analysis indicated that the microdeletion most likely occurred de novo. This report expands upon the range of mutations associated with NR5A1, and illustrates the value of aCGH studies in patients presenting with unusual or syndromic disorders of sexual development.
2. Clinical report
2.1 Patient
A 6 ½ year-old 46,XY female with mild developmental delay and minor dysmorphisms presented for reevaluation due to a new twin pregnancy in her mother. The patient’s antenatal course was uncomplicated and she met her early developmental milestones on time. At 2 ½ years she lost communication skills and previously acquired language. Subsequent developmental evaluation resulted in the diagnosis of Pervasive Developmental Disorder, Not Otherwise Specified (PDD-NOS). Karyotype at age 3, done for developmental delays, was reported to be 46,XY. On further evaluation, pelvic sonogram did not identify a uterus or gonads. A second study a year later also did not identify uterus, ovaries or gonadal tissue in the pelvis or inguinal regions. Gonadal testosterone showed poor response to hCG stimulation but normal T to DHT ratio. AntiMüllerian hormone, FSH, LH, and inhibin B were in the prepubertal range. Adrenal hormone production after ACTH stimulation was normal. Sequencing of the AR, SRD5A2, SRY, NR5A1 and WT1 genes did not detect any pathogenic mutations. Detailed discussion with the mother included recommendations for explorative surgery and removal of gonadal tissue if found. The mother decided to defer the surgery until the child was older. Both parents were healthy, non-consanguineous, and there were no fertility problems or developmental delays in the family. At age 6 ½ years, the patient was evaluated again. She was receiving physical, speech and occupational therapies and had made developmental progress. Patient’s facial features included prominent forehead, long palpebral fissures, slightly cupped ears, malar flattening, full mouth, and 3 to 4 small café-au-lait macules over the trunk. She was phenotypically female with normal height (−0.064 SDS for females) and normal prepubertal female external genitalia at Tanner stage I. The mother’s present twin pregnancy was spontaneously conceived, with normal 46,XY karyotype on both twins. At this visit, aCGH testing was ordered with informed consent as part of the clinical evaluation of the patient.
2.2 Array CGH
Patient aCGH was performed on a previously described custom microarray according to the manufacturer's instructions (Agilent Human CGH custom 4 × 44k; Agilent Technologies, Santa Clara, CA, USA) [18]. This array has enriched subtelomere coverage with an average resolution of 5 kb in the subtelomeres and a 125 kb resolution throughout the remaining genome. The data were analyzed with DNA Analytics 5.0.14 software (Agilent Technologies). Aberrations were identified using the DNA Analytics software via the Aberration Detection Method-1 algorithm with a sensitivity threshold of 6.0 and a data filter that rejected aberrations that did not include at least 4 probes with a log2 ratio ± 0.25. All quality control metrics passed.
2.3 FISH
FISH using the 9q subtelomere probe (9q TelVysion, Spectrum Orange, Abbott Molecular, Des Plaines, IL, USA) and the BAC RP11-412C6 probe (Spectrum Green, Empire Genomics, Buffalo, NY, USA) was performed according to manufacturer’s instructions. Twenty-five interphase nuclei and ten metaphases were scored for the presence or absence of two signals per probe and a normal control was examined in addition to the patient, prenatal, and maternal samples. All FISH analyses were performed using an ImagePoint cooled CCD video camera (Photometrics, Tucson, AZ, USA) with a Nikon, Labophot-2A fluorescence microscope (Nikon, Melville, NY, USA). Digital image analyses were performed using a CytoVision Probe system and FISH software (Applied Imaging), and all preparations were counterstained with DAPI (Vector Labs, Burlingame, CA, USA).
2.4 Genomic rearrangement
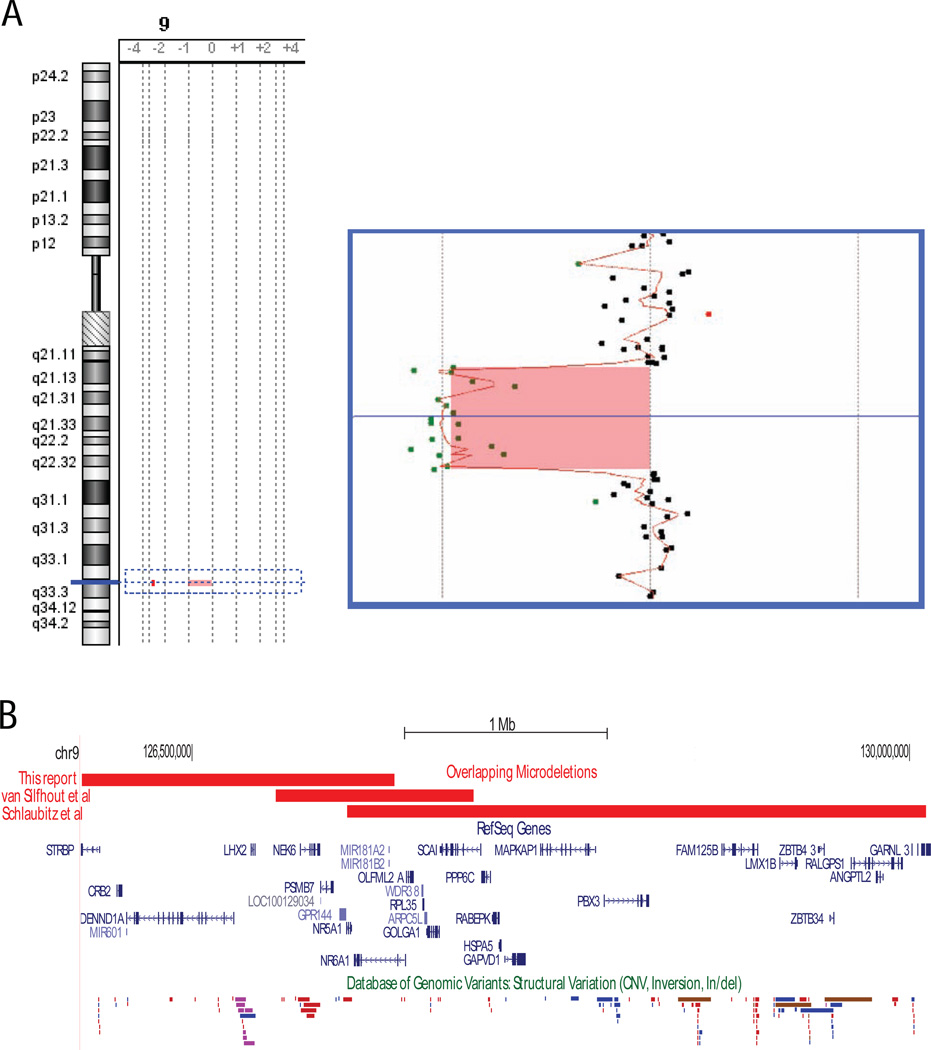
Array CGH revealed a novel 1.54 Mb interstitial 9q33.3 microdeletion (Figure 1). The deleted region encompassed 10 genes including NR5A1. The minimally deleted region was from 125,944,826 to 127,483,435 (1.54 Mb) and the maximally deleted region was from 125,887,272 to 127,542,311 (1.66 Mb) (hg19). This deletion partially overlaps with two previously reported deletions that also include NR5A1 [16, 17] (Figure 1). While the Database of Genomic Variants (DGV) database contains CNVs in this region detected in healthy controls, none are as large as the pathogenic 9q33.3 deletions [19]. One study recorded in the DGV reports NR5A1 deletions in four healthy children encompassing six interrogated single nucleotide polymorphisms (SNPs) on a SNP-based microarray (44.7 kb) [20]. However, the gender and age of these individuals are not available and without detailed clinical information, the significance is unclear.
Figure 1.
Array CGH detection of a novel 9q33.3 microdeletion in the patient. (A) On the left is an ideogram of chromosome 9. The area outlined by the dotted box is enlarged on the right. The Y-axis is genomic position and the X-axis is the log2 ratio of the signal of the patient relative to the control. Black filled circles represent probes with a log2 ratio between −0.25 and +0.25. Red filled circles are probes with a log2 ratio greater than 0.25 and green filled circles are probes with a log2 ratio less than −0.25. The red line is the moving average and the shaded region is the area called as an aberration by the Agilent software. (B) The pertinent region is depicted on the hg19 assembly as adapted from the UCSC genome browser (http://genome.ucsc.edu/) [25]. The 9q33.3 deletion detected in this report is compared to the other deletions reported in the literature. The color coding of the Database of Genomic Variants (DGV) is as follows: blue indicates gains, red indicates losses, brown indicates both gains and losses have been detected, and purple represents inversions [19].
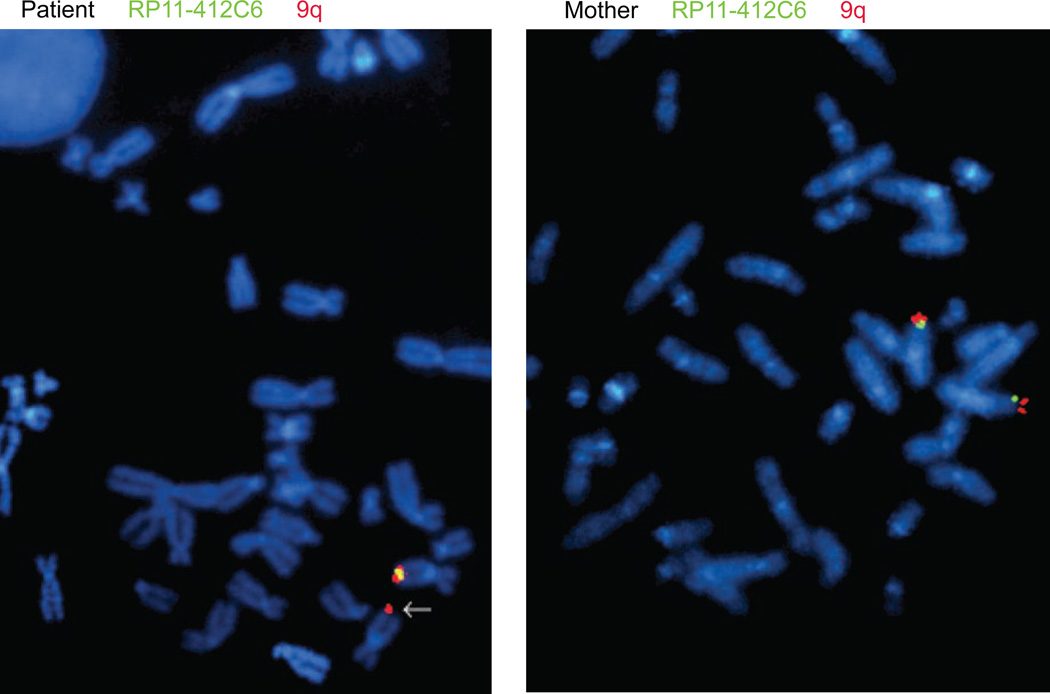
FISH with the BAC probe RP11-412C6 confirmed the presence of the deletion in the patient and its absence in the mother as well as the cultured chorionic villi from the mother’s ongoing twin pregnancy (both fetuses 46,XY) (Figure 2 and data not shown). The father was unavailable for testing. The RP11-412C6 probe hybridized to a 151 kb region containing GPR144, NR5A1, and NR6A1. The 9q subtelomere probe was used as an internal control. Only a single RP11-412C6 signal was observed in the nuclei and on metaphases of the affected individual.
Figure 2.
FISH confirms the 9q33.3 microdeletion in the patient but not in the mother. Representative metaphase spreads of the patient (left) and mother (right) are depicted. The arrow head points to the chromosome 9 with a deleted RP11-412C6 signal.
3. Discussion
We report the detection of a novel 9q33.3 microdeletion in a patient with 46,XY DSD, minor dysmorphisms, mild developmental delay, and normal adrenal function. The microdeletion includes NR5A1 (the gene encoding SF1). Mutations in this gene cause 46,XY CGD with or without adrenal failure, other 46,XY DSDs, 46,XX ovarian insufficiency, 46,XY infertility, and isolated adrenal failure in one case.
FISH analysis indicated that this aberration was not inherited from the mother. The father was unavailable for testing. It is unlikely that the father carries this microdeletion because missense mutations in specific domains of the SF1 protein have been associated with male infertility and to our knowledge, 46,XY males with dominant-acting NR5A1 mutations are rarely fertile. Those reported belong to families segregating missense mutations causing more mild DSD phenotypes than observed in our patient [14, 21]. In one of these reports, a NR5A1 mutation was transmitted by an unaffected father who seemed to be mosaic for the mutation [21]. It is noteworthy that the mother was carrying twins with an XY karyotype, and knowledge that the precise abnormality for the XY DSD was a de novo deletion provided much needed reassurance for the family.
The gonadal phenotypic spectrum of mutations associated with NR5A1 mutations ranges from isolated male infertility or female ovarian insufficiency to 46,XY complete gonadal dysgenesis. Including this present report, the three microdeletions encompassing NR5A1 all have relatively severe gonadal phenotypes, but none have adrenal insufficiency [16, 17]. In contrast, reported mutations within the same gene resulted in a range of findings on the gonadal phenotypic spectrum and a few have had adrenal insufficiency [2, 4, 5, 8, 9, 13, 15]. Of note, the sequencing of the NR5A1 gene done during the patient’s initial work-up, was normal. This illustrates the fact that other technologies, such as aCGH or Multiplex Ligation-dependent Probe Amplification, are needed to detect deletions or duplications of genes.
This 1.54 Mb microdeletion and the 3 Mb microdeletion reported by Schlaubitz et al. had additional phenotypic abnormalities that are likely to be due to the deletion of genes neighboring NR5A1, such as LMX1B [16] which appeared to be responsible for the patellar abnormalities in Schlaubitz et al’s patient. Our patient has developmental delay and minor facial dysmorphism in addition to DSD. Since NR5A1 mutations have not been reported to be associated with developmental delay or facial dysmorphism, one or more of the other ten genes in the 1.54 Mb deleted region is likely to be responsible. While none of these other genes have been well-characterized in humans, studies in model organisms have implicated two of them in brain development: STRBP and LHX2 [22, 23].
In a study of 116 patients with DSDs of unknown etiology, Tannour-Louet et al. identified clinically relevant submicroscopic deletions and duplications in 21.5% of patients presenting with gonadal dysgenesis, ambiguous genitalia, or genitourinary defects. Recurrently occurring imbalances were found in 5p15.3, 9p24.3, 22q12.1 and Xq28 leading to identification of novel candidate genes for DSDs [24]. These and other genes, such as NR5A1, are likely to influence the complex sex determination and differentiation pathways by dosage sensitive effects or other mechanisms. Array CGH will therefore significantly increase the yield of diagnosis in this group of disorders. Knowledge of the precise defect will help in providing prognosis and genetic counseling.
Acknowledgements
TB is the recipient of a Ruth L. Kirschstein National Research Service Award (NIH 5T32GM082773-05).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENT: The authors have nothing to disclose.
References
- 1.Ferraz-de-Souza B, Lin L, Achermann JC. Steroidogenic factor-1 (SF-1, NR5A1) and human disease. Mol Cell Endocrinol. 2011;336:198–205. doi: 10.1016/j.mce.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 3.Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, Simburger K, Milbrandt J. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci U S A. 1995;92:10939–10943. doi: 10.1073/pnas.92.24.10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Achermann JC, Ito M, Hindmarsh PC, Jameson JL. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet. 1999;22:125–126. doi: 10.1038/9629. [DOI] [PubMed] [Google Scholar]
- 5.Achermann JC, Ozisik G, Ito M, Orun UA, Harmanci K, Gurakan B, Jameson JL. Gonadal determination and adrenal development are regulated by the orphan nuclear receptor steroidogenic factor-1, in a dose-dependent manner. J Clin Endocrinol Metab. 2002;87:1829–1833. doi: 10.1210/jcem.87.4.8376. [DOI] [PubMed] [Google Scholar]
- 6.Biason-Lauber A, Schoenle EJ. Apparently normal ovarian differentiation in a prepubertal girl with transcriptionally inactive steroidogenic factor 1 (NR5A1/SF-1) and adrenocortical insufficiency. Am J Hum Genet. 2000;67:1563–1568. doi: 10.1086/316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin L, Gu WX, Ozisik G, To WS, Owen CJ, Jameson JL, Achermann JC. Analysis of DAX1 (NR0B1) and steroidogenic factor-1 (NR5A1) in children and adults with primary adrenal failure: ten years' experience. J Clin Endocrinol Metab. 2006;91:3048–3054. doi: 10.1210/jc.2006-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin L, Philibert P, Ferraz-de-Souza B, Kelberman D, Homfray T, Albanese A, Molini V, Sebire NJ, Einaudi S, Conway GS, Hughes IA, Jameson JL, Sultan C, Dattani MT, Achermann JC. Heterozygous missense mutations in steroidogenic factor 1 (SF1/Ad4BP, NR5A1) are associated with 46,XY disorders of sex development with normal adrenal function. J Clin Endocrinol Metab. 2007;92:991–999. doi: 10.1210/jc.2006-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallet D, Bretones P, Michel-Calemard L, Dijoud F, David M, Morel Y. Gonadal dysgenesis without adrenal insufficiency in a 46, XY patient heterozygous for the nonsense C16X mutation: a case of SF1 haploinsufficiency. J Clin Endocrinol Metab. 2004;89:4829–4832. doi: 10.1210/jc.2004-0670. [DOI] [PubMed] [Google Scholar]
- 10.Correa RV, Domenice S, Bingham NC, Billerbeck AE, Rainey WE, Parker KL, Mendonca BB. A microdeletion in the ligand binding domain of human steroidogenic factor 1 causes XY sex reversal without adrenal insufficiency. J Clin Endocrinol Metab. 2004;89:1767–1772. doi: 10.1210/jc.2003-031240. [DOI] [PubMed] [Google Scholar]
- 11.Warman DM, Costanzo M, Marino R, Berensztein E, Galeano J, Ramirez PC, Saraco N, Baquedano MS, Ciaccio M, Guercio G, Chaler E, Maceiras M, Lazzatti JM, Bailez M, Rivarola MA, Belgorosky A. Three new SF-1 (NR5A1) gene mutations in two unrelated families with multiple affected members: within-family variability in 46,XY subjects and low ovarian reserve in fertile 46,XX subjects. Horm Res Paediatr. 2011;75:70–77. doi: 10.1159/000320029. [DOI] [PubMed] [Google Scholar]
- 12.Barbaro M, Cools M, Looijenga LH, Drop SL, Wedell A. Partial deletion of the NR5A1 (SF1) gene detected by synthetic probe MLPA in a patient with XY gonadal disorder of sex development. Sexual Development. 2011;5:181–187. doi: 10.1159/000328821. [DOI] [PubMed] [Google Scholar]
- 13.Lourenco D, Brauner R, Lin L, De Perdigo A, Weryha G, Muresan M, Boudjenah R, Guerra-Junior G, Maciel-Guerra AT, Achermann JC, McElreavey K, Bashamboo A. Mutations in NR5A1 associated with ovarian insufficiency. N Engl J Med. 2009;360:1200–1210. doi: 10.1056/NEJMoa0806228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciaccio M, Costanzo M, Guercio G, De Dona V, Marino R, Ramirez PC, Galeano J, Warman DM, Berensztein E, Saraco N, Baquedano MS, Chaler E, Maceiras M, Lazzatti JM, Rivarola MA, Belgorosky A. Preserved Fertility in a Patient with a 46,XY Disorder of Sex Development due to a New Heterozygous Mutation in the NR5A1/SF-1 Gene: Evidence of 46,XY and 46,XX Gonadal Dysgenesis Phenotype Variability in Multiple Members of an Affected Kindred. Horm Res Paediatr. 2012 doi: 10.1159/000338346. [DOI] [PubMed] [Google Scholar]
- 15.Bashamboo A, Ferraz-de-Souza B, Lourenco D, Lin L, Sebire NJ, Montjean D, Bignon-Topalovic J, Mandelbaum J, Siffroi JP, Christin-Maitre S, Radhakrishna U, Rouba H, Ravel C, Seeler J, Achermann JC, McElreavey K. Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am J Hum Genet. 2010;87:505–512. doi: 10.1016/j.ajhg.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlaubitz S, Yatsenko SA, Smith LD, Keller KL, Vissers LE, Scott DA, Cai WW, Reardon W, Abdul-Rahman OA, Lammer EJ, Lifchez CA, Magenis E, Veltman JA, Stankiewicz P, Zabel BU, Lee B. Ovotestes and XY sex reversal in a female with an interstitial 9q33.3-q34.1 deletion encompassing NR5A1 and LMX1B causing features of Genitopatellar syndrome. Am J Med Genet A. 2007;143A:1071–1081. doi: 10.1002/ajmg.a.31685. [DOI] [PubMed] [Google Scholar]
- 17.van Silfhout A, Boot AM, Dijkhuizen T, Hoek A, Nijman R, Sikkema-Raddatz B, van Ravenswaaij-Arts CM. A unique 970kb microdeletion in 9q33.3, including the NR5A1 gene in a 46,XY female. Eur J Med Genet. 2009;52:157–160. doi: 10.1016/j.ejmg.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Scott SA, Cohen N, Brandt T, Toruner G, Desnick RJ, Edelmann L. Detection of low-level mosaicism and placental mosaicism by oligonucleotide array comparative genomic hybridization. Genet Med. 2010;12:85–92. doi: 10.1097/GIM.0b013e3181cc75d0. [DOI] [PubMed] [Google Scholar]
- 19.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 20.Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O'Hara R, Casalunovo T, Conlin LK, D'Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philibert P, Polak M, Colmenares A, Lortat-Jacob S, Audran F, Poulat F, Sultan C. Predominant Sertoli cell deficiency in a 46,XY disorders of sex development patient with a new NR5A1/SF-1 mutation transmitted by his unaffected father. Fertil Steril. 2011;95:1788, e1785–e1789. doi: 10.1016/j.fertnstert.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 22.Pires-daSilva A, Nayernia K, Engel W, Torres M, Stoykova A, Chowdhury K, Gruss P. Mice deficient for spermatid perinuclear RNA-binding protein show neurologic, spermatogenic, and sperm morphological abnormalities. Dev Biol. 2001;233:319–328. doi: 10.1006/dbio.2001.0169. [DOI] [PubMed] [Google Scholar]
- 23.Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 24.Tannour-Louet M, Han S, Corbett ST, Louet JF, Yatsenko S, Meyers L, Shaw CA, Kang SH, Cheung SW, Lamb DJ. Identification of de novo copy number variants associated with human disorders of sexual development. PLoS One. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]