Abstract
Recombinant elastin-like protein (ELP) polymers display several favorable characteristics for tissue repair and replacement as well as drug delivery applications. However, these materials are derived from peptide sequences that do not lend themselves to cell adhesion, migration, or proliferation. This report describes the chemoselective ligation of peptide linkers bearing the bioactive RGD sequence to the surface of ELP hydrogels. Initially, cystamine is conjugated to ELP, followed by the temperature-driven formation of elastomeric ELP hydrogels. Cystamine reduction produces reactive thiols that are coupled to the RGD peptide linker via a terminal maleimide group. Investigations into the behavior of endothelial cells and mesenchymal stem cells on the RGD-modified ELP hydrogel surface reveal significantly enhanced attachment, spreading, migration and proliferation. Attached endothelial cells display a quiescent phenotype.
Keywords: Elastin-like polypeptide, Tissue engineering, Viscoelastic, Protein polymer, Biomimetic
1. Introduction
Experimental small-diameter vascular grafts underperform due to thrombogenicity of the lumenal surface and neointimal hyperplasia attributed to a mechanical mismatch between synthetic materials and arterial tissue [1,2]. Recombinant elastin-like protein (ELP) materials have emerged in principle as attractive biomaterials for vascular applications because they are based on amino acid sequences from elastin, a key structural protein of the native vasculature [3]. We have recently described triblock ELP polymers with hydrophilic, elastomeric midblock sequences flanked by self-associating, hydrophobic endblocks in an ABA triblock format [4–6]. Notably, triblock ELPs are highly soluble under cool, aqueous conditions, but form elastomeric gels when solutions are raised above an inverse transition temperature. Under these conditions the more hydrophobic endblocks coacervate to form physical crosslinks while the midblocks remain elastomeric and solvated. As an example, LysB10, a 209 kDa triblock ELP, has an inverse transition temperature of 13 °C [6]. Due to the inclusion of lysine residues at the block interfaces, LysB10 can be crosslinked by chemical strategies that reinforce physical crosslinking. Our studies have shown that triblock ELPs are non-thrombogenic [7], can be highly biostable [8], permit controlled drug release [9] and can be used in protein-based composites that mimic native artery mechanics [10]. Despite these advantages, the peptide sequences typically employed in ELP design do not support cell adhesion.
Poor patency rates of synthetic polymers have motivated strategies to promote luminal endothelialization [11–17], often through covalent tethering of biomolecules [18]. Because these reactions are typically between the amino acid side chains and activated surface functional groups, uncontrolled or non-specific covalent binding often results. Herein, we describe maleimide–thiol chemistry for hydrogel surface biofunctionalization. The high degree of specificity and reactivity of sulfhydryl groups with maleimide moieties to form stable thioether bonds has been exploited extensively for the construction of immobilized antibodies, enzymes and peptide-conjugated haptens [19–22]. Maleimide reacts approximately 1000 times faster with thiols than with amines at neutral pH and below, and this reaction is widely used for conjugation of cysteine-containing peptide and proteins [23–25].
The RGD sequence was explored as a model peptide conjugate, given its ubiquitous nature in extracellular matrix (ECM) proteins, including fibronectin, vitronectin, fibrinogen, von Willebrand factor, as well as collagen, and its ability to bind numerous integrins, notably α5β1 and αvβ3 [26]. Several biomaterials have been functionalized with RGD, including polymers such as poly(ethylene glycol) (PEG) hydrogels [27,28], polyacrylamide [29,30], poly(2-hydroxyethyl methacrylate) [31,32], poly(lactic acid-co-lysine) [34,35], poly(propylene fumarate) [33,34] and polyurethanes [35,36]; and biopolymers, including collagen [37,38], fibrin [14,39], hyaluronic acid [40,41], alginate [42,43], dextran [44,45], ELPs [46–48] and silk-like proteins [49].
2. Materials and methods
2.1. Reagents, antibodies and cells
All solvents and reagents were purchased from commercial sources and were used as received, unless otherwise noted. The peptide sequences GRGDSP and GRGESP were synthesized by Ana-Spec (Fremont, CA). Porcine mesenchymal stem cells (pMSCs) were a kind gift from Dr. Steven Stice (University of Georgia). The biosynthetic strategy for the expression and purification of the recombinant ELP triblock polymer, LysB10, has been described previously [6].
2.2. Cystamine modification of LysB10
LysB10 was chemically modified utilizing aqueous carbodiimide chemistry (Scheme 1) [24]. Cystamine (Sigma Aldrich) was added to the solution at 20-fold molar excess to a cooled solution of LysB10 (10 mg ml−1, 4 °C PBS), followed by N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC) at 5-fold molar excess relative to cystamine. After stirring (72 h, 4 °C) cystamine-modified LysB10 polymer was purified by dialysis and lyophilization (81% yield).
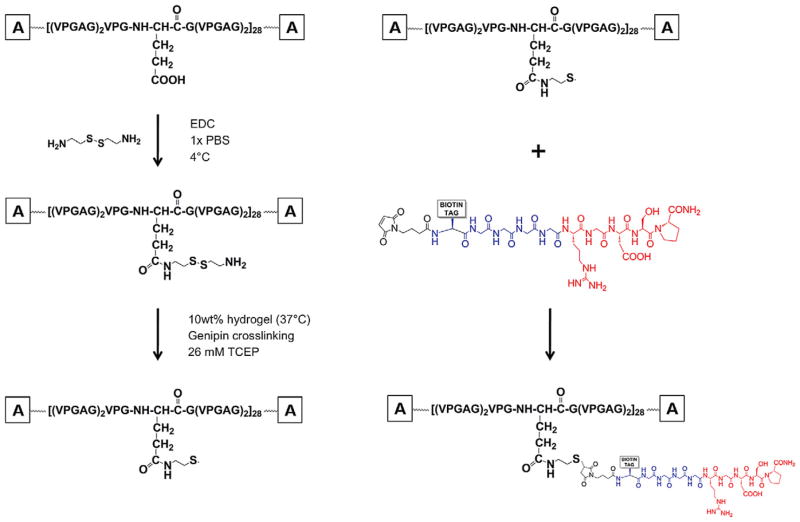
Scheme 1.
Reaction scheme of LysB10 modification and peptide coupling. Amide bond formation was mediated by the carbodiimide through the carboxylic group of the amino acid and the amine of cystamine, resulting in thiolated LysB10. The plastic domains of LysB10 are represented as “A” endblocks. Hydrogel formation was achieved by placing 10 wt.% thiol–LysB10 solution at 37 °C, well above the transition temperature of the protein polymer. Lysine residues of the protein polymer were crosslinked with a 6mgml−1 genipin solution for 24 h, followed by stringent PBS rinsing to remove all genipin. The thiol groups were reduced with the addition of 26 mM Tris(2-carboxyethyl)phosphine (TCEP) to form free sulfhydryls. After rinsing the gels with three 20 min PBS washes, thiol-reactive peptide linker was incubated for 2 h at room temperature to form a thioether bond with the protein polymer. Peptides were generated via solid-phase synthesis, with key features incorporated in the design. The N-terminus of the molecule contains the thiol-reactive maleimide linker (black). Four glycine residues (blue) act as a spacer between the cell-binding RGD domain (red) and the remaining sequence to facilitate ligand–integrin presentation. A biotinyl–PEG3 tag was incorporated into the peptide for detection of the molecule.
2.3. Solid-phase peptide synthesis
The RGDSP (Arg–Gly–Asp–Ser–Pro) peptide was synthesized manually on a Rink amide resin using the standard Fmoc amino acid coupling strategy [50]. Briefly, Fmoc-Pro-Rink amide resin (1.0 g, 0.45 mmol g−1) was loaded into a fritted column equipped with a plastic cap. The resin was swelled by stirring gently with 20 ml dichloromethane (DCM) for 10 min and filtered. This procedure was repeated with 4 × 20 ml portions of DCM and followed by 4 × 20 ml portions of dimethylformamide (DMF). Deprotection of Fmoc was performed with 20% piperidine in DMF (2 × 10 ml) for 20 min. The protected amino acids used in the coupling sequences are Fmoc–Ser–OH, Fmoc–Asp(tBu)–OH, Fmoc–Arg(Pbf), Fmoc–Glu(biotinyl–PEG)–OH and 3-maleimido propinoic acid. Except for the Fmoc–Glu(biotinyl–PEG)–OH coupling reaction, every successive coupling was performed with four equivalents of Fmoc–amino acid preactivated for 5 min with HBTU/HOBt in 10 ml DMF. The Fmoc–Glu(biotinyl–PEG)–OH reaction was performed with PyBOP/HOBt as the coupling reagent. The progress of the deprotection/coupling was followed at every cycle by performing the ninhydrin test using the Kaiser kit. Cleavage/deprotection of the peptide from the resin was accomplished by gently stirring in 20 ml of a 1/2/2/95 of water/ethanedithiol/triethylsilane/trifluoroacetic acid for 2 h and filtering. The peptide was obtained as a yellow syrup, which was precipitated as a white solid by adding the cold ether (50 ml). The precipitated peptide was collected and purified by RP-HPLC using Varian C18 columns (150 × 10.0 mm) with a gradient elution of 95/5 to 50/50 of water/acetonitrile in 30 min. The fraction eluted at 11.5 min was collected and lyophilized to obtain desired peptide as a white powder (453 mg, 68% yield). ESI-HRMS calculated for C60H96N19O22S1 [M+H]+ = 1466.66980, found 1466.49657.
2.4. Genipin crosslinking and thiol modification of cystamine–LysB10 hydrogels
Protein polymer gels were formed by pipetting 40 μl of cystamine–LysB10 solution (100 mg ml−1) uniformly into wells of a polystyrene 96-well plate at 4 °C, followed by incubation well above the LysB10 inverse transition temperature (37 °C, 1 h). Hydrogels were crosslinked with genipin solution (6 mg ml−1) in PBS for 24 h, followed by stringent PBS rinsing. Thiol groups were reduced with the addition of 26 mM Tris(2-carboxyethyl)phosphine (TCEP) for 6 h. After rinsing with three 20 min PBS washes, thiolated hydrogels were either conjugated to the RGD peptide or assayed for extent of thiol modification. Unmodified LysB10 hydrogels were generated similarly, with the exclusion of the reducing (TCEP) step. The extent of thiol modification was determined by incubation of Ellman’s reagent in phosphate buffer (4 mg ml−1 Ellman’s reagent, 0.1 M sodium phosphate, 1 mM EDTA, pH 8.0) for 15 min at room temperature. Absorbance at 412 nm was measured and concentration values were obtained from comparison of measurements to a standard curve generated from cysteine dilutions in phosphate buffer.
2.5. Conjugation of the peptide linker to LysB10
In pilot experiments, a maleimide–PEG2–biotin linker (5, 10, 25, 50, 150 or 1000 μgml−1) was incubated for 2 h at room temperature with unmodified or thiol-modified LysB10 hydrogels, followed by incubation in PBS with daily buffer changes for 5 days to minimize nonspecific binding. Biotin conjugation was determined using the fluoreporter biotin quantification assay kit (Molecular Probes). Identical conjugation was performed with RGD–maleimide linker with consistency of results verified by biotin assay (Scheme 1).
2.6. Cell culture, adhesion and proliferation
Human umbilical vein endothelial cells (HUVEC, Clonetics) were maintained in endothelial growth medium-2 (EGM-2, 2% serum, Clonetics). pMSCs were cultured in α-MEM basal medium supplemented with 10% fetal bovine serum, 50 U ml−1 penicillin, 50 μgml−1 streptomycin and 2 mM L-glutamine. Cells were incubated (5% CO2, 37 °C) and passed every 2 days. HUVECs and pMSCS between passages 3 and 9 were used in all experiments.
Cell adhesion was studied on thiol-modified or unmodified LysB10, incubated with various concentrations of RGD linker and rinsed, as described above. Positive controls consisted of fibronectin (50 μgml−1) absorbed to polystyrene wells. Cells were harvested with Cell Dissociation Solution (Sigma) and centrifuged (220g, 5 min). HUVEC and pMSCs were resuspended at 200,000 cells ml−1 in basal medium containing 0.5% bovine serum albumin (BSA) or in low-serum media (1% serum), respectively. Cell suspension (100 μl) was plated onto LysB10 surfaces, incubated at 37 °C for 2 h, and washed thereafter three times with PBS. Adherent cells were determined using the CyQuant Cell Proliferation Assay Kit (Molecular Probes) and the results normalized to fibronectin-coated control surfaces.
Specificity of cell adhesion was studied on unmodified or thiol-modified LysB10 gels exposed to either a RGD or a PEG2–biotin linker at 50 μgml−1. Cells were harvested and seeded as above, although some groups were treated with soluble GRGDSP (2 mM) or soluble GRGESP peptide (2 mM) for 30 min prior to plating. Cells were counted as described above and all data normalized to fibronectin-coated control surfaces.
Cell proliferation was evaluated using the CyQuant Cell Proliferation Assay Kit. Cells were harvested and seeded onto various LysB10 gels at a density of 5000 cells per well. After an initial 2 h incubation period, non-adherent cells were removed with media washes and substrate-bound cells were cultured for another 48 h. Proliferation was characterized by comparison of cell counts at 2 and 48 h.
2.7. Radial migration assay
Cell migration on LysB10 hydrogels and fibronectin-coated wells (n = 4) was defined using the Oris cell migration assay FLEX kit (Platypus Technologies) in which cells were seeded into 96-well plates consisting of opaque wells and transparent bottoms. A cell-seeding stopper (d = 2 mm) was positioned on the hydrogel surface, creating a central detection zone initially free from cell adhesion. Cells were harvested as above and resuspended at 400,000 cells ml−1 in serum-free basal media (HUVECs) or low-serum media (pMSCs). Cell suspensions were treated with mitomycin C (10 μgml−1, Sigma Aldrich) to arrest proliferation. A total of 100 μl of cell suspension was seeded onto the outer annular region of the protein gel surfaces (30 mm2), and allowed to adhere for 6 h at 37 °C, at which point the stoppers were removed to permit migration into the central detection zone (3.14 mm2). In reference wells, stoppers remained in place until wells were read, as pre-migration controls. Following removal of unbound cells by rinsing with complete media, wells were filled with 150 μl of complete media, and incubated at 37 °C for 36 h. Following three PBS washes, migration was quantified by Calcein AM staining after incubation with 2 μM Calcein AM for 1 h. Migrated cells in the central detection zone were analyzed with a fluorescent plate reader. The Oris detection mask prevented signal detection of the outer annular region, such that the detected fluorescent signal was isolated from cells that had migrated only into the central detection zone. Readings from pre-migration control wells were subtracted from the post-migration wells to eliminate background. Data was normalized to the fibronectin-coated wells as a positive control.
2.8. Immunofluorescence characterization of cell morpohology and HUVEC activation
Cell morphology was observed by fluorescently labeling actin. HUVEC activation and quiescence was studied by immunofluorescent staining of ICAM-1 and E-selectin. Experiments were performed in polystyrene 8-well chamber slides (Nalge Nunc, International). Fibronectin controls were formulated by adsorbing 50 μgml−1 solutions overnight at 4 °C. Suspensions of HUVECs (15000 cells per well) were seeded onto the slides and cultured for a period of 2 h (F-actin staining) or 4 h (ICAM-1 and E-selectin staining) in serum-free medium. Similarly prepared pMSCs were cultured in low-serum (1% FBS) media. To activate HUVECs, 100 ng ml−1 tumor necrosis factor alpha (TNFα) was added to cells cultured on fibronectin-coated slides for 4 h prior to immunostaining. Subsequently, specimens were fixed in 4% paraformaldehyde (10 min), permeabilized with PBS containing 0.5% Triton X-100 (10 min), rinsed with 100 mM glycine (10 min) and incubated with block buffer (PBS+/+, 0.2% Triton X-100, 6% goat serum, room temperature, 1 h). For F-actin staining, cells were incubated with Alexa Fluor 568-conjugated phalloidin for 30 min. To evaluate HUVEC activation/quiescence, 10 μg ml−1 solutions of E-selectin and ICAM-1 monoclonal antibodies were incubated for 1 h. Primary antibody incubation was followed by 45 min incubation with 2.5 μgml−1 biotinylated goat anti-mouse IgG secondary and 30 min incubation with 2.5 μgml−1 streptavidin-AlexaFluor 488 tertiary. Nuclei were counterstained with Prolong Gold mounting medium containing DAPI, and specimens were evaluated with confocal microscopy.
2.9. Statistical analysis
Comparisons were performed by ANOVA or paired, two-tailed Student’s t-test, with P < 0.05 significance. Results are presented as mean ± standard deviation. Data represent characteristic results from a particular experimental run (n = 4 for all groups) from at least three independent runs.
3. Results
3.1. Extent of thiol and RGD linker modification
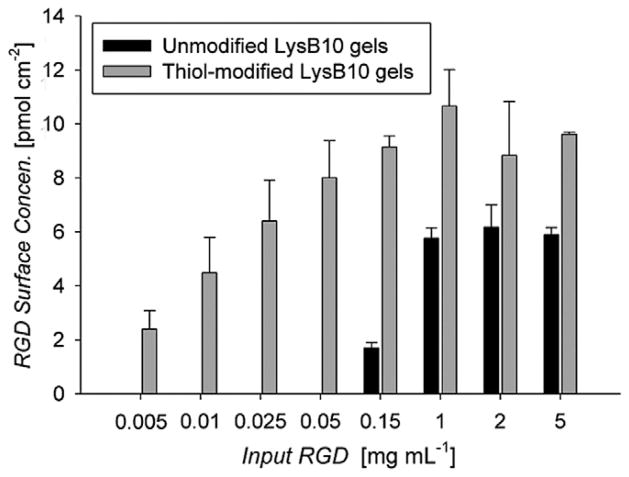
Following cystamine modification and TCEP reduction of LysB10, Ellman’s reaction was performed to determine the extent of thiol modification. Modification was 54.7 ± 1.9% and 48.6 ± 2.5% in solution and as hydrogels, respectively, which corresponds to modifying 13–15 of the 28 available carboxylic groups per molecule. In the absence of thiol reduction no modification was observed. By monitoring biotin concentration, conjugation of the RGD linker was explored at linker concentrations ranging from 5 to 5000 μg ml−1, with bound peptide steadily increasing until a plateau was observed at 1 mg ml−1 (Fig. 1). Passive adsorption onto unmodified hydrogels was not observable at concentrations below 150 μgml−1, and then increased with increasing linker concentration, despite the 5 day PBS rinsing step. Linker surface densities were calculated assuming a 50 nmpenetration of reactants [42,51]. The grafting efficiency of a 50 μgml−1 RGD linker solution was 37%, which corresponds to five RGD peptides per LysB10 molecule.
Fig. 1.
Coupled RGD peptide as a function of the amount of input peptide in surface-modified LysB10. Data represent one of three similar experiments, with each condition run in quadruplicate. Peptide conjugation was assessed with the use of the biotin tag. In order to determine the moles of biotin conjugated to the LysB10 hydrogel surface, the fluoreporter biotin quantitation assay kit was utilized.
3.2. Cell attachment
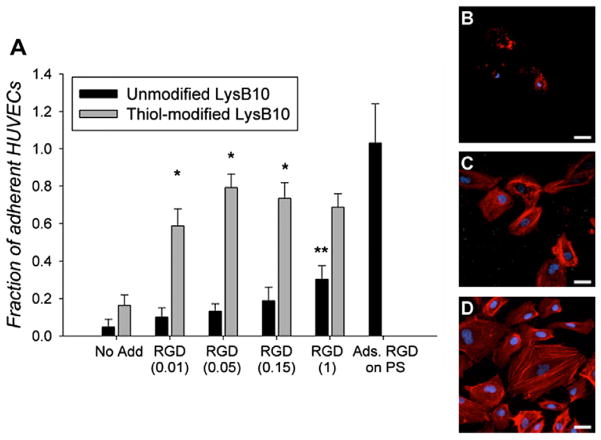
The conjugation of RGD linkers to LysB10 gels supported HUVEC and pMSC adhesion. HUVEC attachment and spreading was observed within 2 h when cells were seeded onto protein gels modified with 10, 50, 150 and 1000 μgml−1 of RGD linker (Fig. 2). Minimal HUVEC adhesion was observed on hydrogels in the absence of RGD treatment. Cell adhesion on thiol-modified LysB10 treated with 10 or 50 μgml−1 of RGD linker was approximately 6-fold higher than that observed on unmodified LysB10. However, further increases in RGD peptide concentration from 50 to 1000 μgml−1 did not increase HUVEC adhesion. The maximum level of cell adhesion onto RGD modified protein gels was ~80% of that noted on fibronectin-coated polystyrene surfaces. pMSC adhesion was also enhanced on LysB10 surfaces by the presence of the linker. Although some pMSC attachment was observed on thiol-modified gels in the absence of RGD linker, pMSC adhesion increased nearly 5-fold when thiol-modified gels were treated with RGD-linker (Fig. 3A). Furthermore, cell adhesion on RGD-conjugated LysB10 gels was equivalent to that observed on fibronectin-coated polystyrene surfaces. For both cell types, spreading was greatly enhanced on RGD-conjugated LysB10, with well-developed actin fiber networks apparent. However, cells assumed a rounded morphology, with actin molecules distributed in a nonspecific manner, when plated onto unmodified LysB10 gels surfaces (Figs. 2B–D and 3B–D).
Fig. 2.
(A) HUVEC adhesion to varying LysB10 hydrogel surfaces after 2 h. RGD peptide concentrations ranging from 0.01 to 1 mg ml−1 were added to unmodified and thiol-modified LysB10 surfaces. 50 μg ml−1 fibronectin adsorbed to polystyrene served as a positive control, and all data was normalized to this control. Data represent one of three similar experiments, with each condition run in quadruplicate. *P < 0.01 compared to unmodified LysB10–RGD at the same concentration. **P < 0.05 compared to unmodified LysB10–no add control. Representative confocal images of HUVECs cultured on LysB10 gels are shown, with white bars representing 20 μm. Ten weight per cent unmodified LysB10 with adsorbed 50 μgml−1 RGD linker (B), modified LysB10 with conjugated 50 μg ml−1 RGD linker (C), and 50 μgml−1 fibronectin coating (D). Fluorescently labeled actin is visualized in red.
Fig. 3.
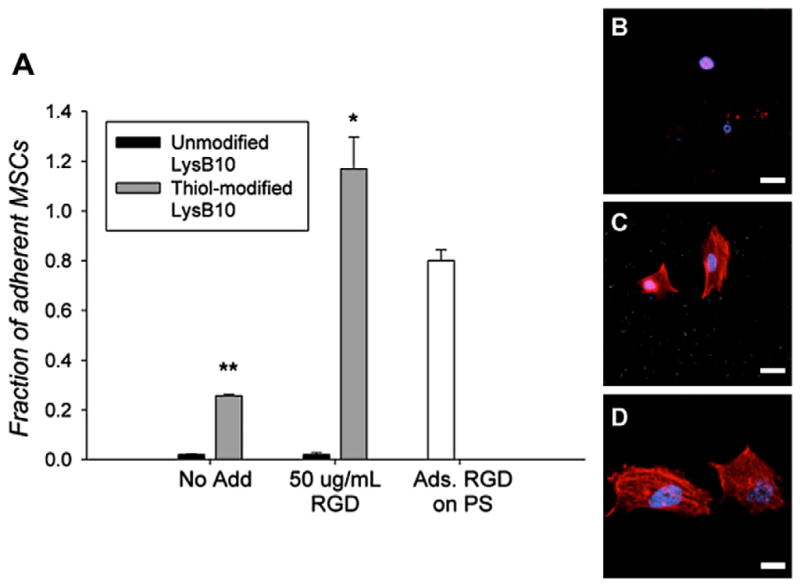
(A) MSC adhesion to various LysB10 hydrogel surfaces after 2 h assay. RGD peptide was added to the gels at a concentration of 50 μg ml−1. Fibronectin adsorbed (50 μg ml−1) to polystyrene served as a positive control, and all data was normalized to this control. Data represent one of three similar experiments, with each condition run in quadruplicate. *P < 0.05 compared to thiol-modified LysB10–no add. **P < 0.05 compared to unmodified LysB10–no add control. Representative confocal images of MSCs cultured on LysB10 gels are shown, with white bars representing 20 μm. Ten weight per cent unmodified LysB10 with adsorbed 50 μg ml−1 RGD linker (B), modified LysB10 with conjugated 50 μgml−1 RGD linker (C), and 50 μgml−1 fibronectin coating (D). Fluorescently labeled actin is shown in red.
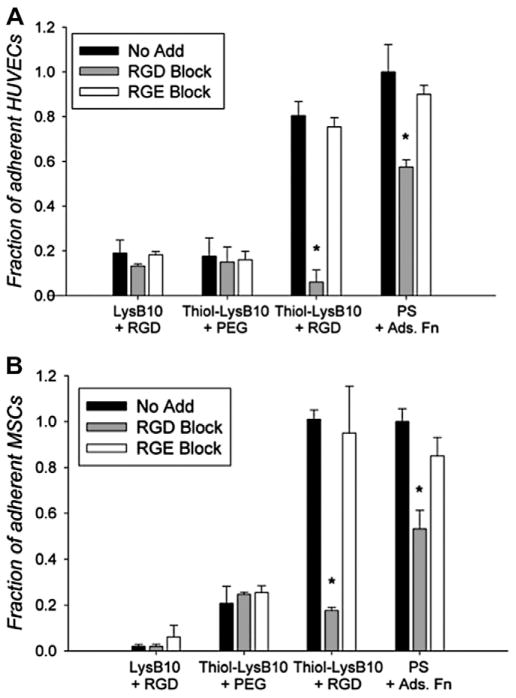
3.3. Cell specificity
Upon addition of soluble GRGDSP to the seeding medium, adhesion of HUVECs and pMSCs onto RGD-LysB10 gel surfaces decreased 8-fold and 5-fold, respectively (Fig. 4). Cell attachment was not affected by the soluble sequence GRGESP, confirming integrin-specific interactions. As expected, minimal non-specific cell adhesion was observed on unmodified LysB10 gels or those treated with a PEG linker that did not bear a terminal RGD peptide sequence.
Fig. 4.
HUVEC (A) and pMSC (B) adhesion and specificity to treated LysB10 hydrogel surfaces. Hydrogels were treated with either 50 μg ml−1 RGD linker or 50 μg ml−1 PEG linker (without RGD). Cells were treated with soluble GRGDSP (2 mM) and soluble GRGESP peptide (2 mM) for 30 min prior to plating. All data was normalized to the fibronectin, no add control. Fibronectin (50 μg ml−1) was adsorbed onto polystyrene. Data represent one of three similar experiments, with each condition run in quadruplicate. *P < 0.05 compared to no-add treatment group.
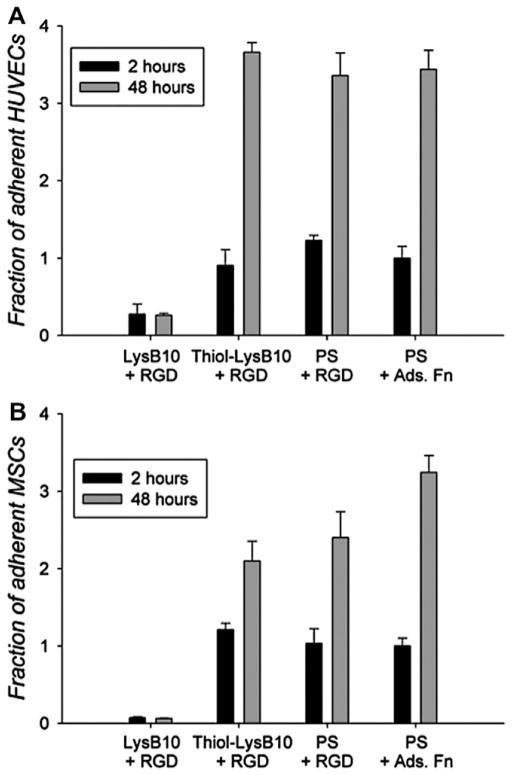
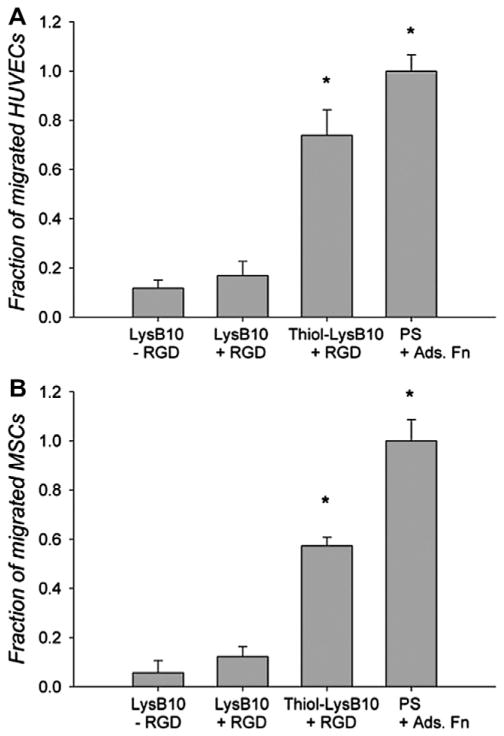
3.4. Cell proliferation, migration and activation
RGD–LysB10 promoted a 3.5-fold increase in HUVEC cell number (Fig. 5A), closely matching the proliferation rate of cells seeded onto fibronectin-coated polystyrene. The rate of pMSC proliferation was somewhat lower on RGD-grafted LysB10, as compared to fibronectin-coated surfaces (Fig. 5B). The radial migration assay, which tracked cell migration over 36 h period from a seeded, outer annulus area into a central region, demonstrated enhanced migration of HUVECs and pMSCs on RGD–LysB10 surfaces (Fig. 6). HUVEC migration was significantly greater on RGD–LysB10 than on unmodified LysB10 (6-fold increase, P < 0.05). Likewise, pMSC migration was 10-fold higher on RGD–LysB10 surfaces (P < 0.05). As expected, cell spreading and migration was not observed on unmodified LysB10 gels. HUVECs grown on RGD–LysB10 surfaces revealed limited ICAM-1 and E-selectin staining consistent with a non-proinflammatory, quiescent phenotype (Fig. 7).
Fig. 5.
Proliferation rate of (A) HUVECs and (B) MSCs over a 48 h period. Cells were seeded onto various LysB10 gels at a density of 5,000 cells per well for 2 h. Unbound cells were removed with media washes and substrate-bound cells were maintained in culture for another 48 h period. All cell counts were normalized to the 2 h adhesion value on fibronectin-coated polystyrene. Cell counts at 48 h were compared to those at 2 h for each treatment group.
Fig. 6.
Radial migration assay of (A) HUVECs and (B) MSCs on modified surfaces. Cells were seeded onto an outer annulus area and monitored for motility into an inner radial zone over a 36 h period. Quantification was achieved with fluorescent measurement of the number of migrated cells into the detectable inner zone, which was normalized against the number of migrated cells on fibronectin-coated polystyrene. *P < 0.05 compared to non-RGD treated, unmodified LysB10.
Fig. 7.
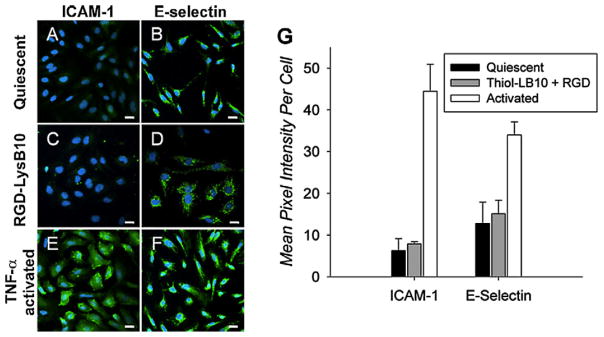
Representative confocal images of HUVECs cultured on various substrates. Cells that were cultured on fibronectin-coated slides without TNF-α stimulation (A, B) maintained a quiescent phenotype. Activation was achieved with the addition of TNF-α to the culture medium (E, F). HUVEC activation or quiescence was compared to that on RGD-conjugated LysB10 films (C, D). Markers of HUVEC activation were ICAM-1 (A, C, E) and E-selectin (B, D, F). Quantification demonstrated that RGD–LysB10 was similar to quiescent controls (G). White bars indicate 20 μm.
4. Discussion
Recombinant ELPs, based upon repeats and variants of the pentapeptide Val–Pro–Gly–[X]–Gly, have shown potential in applications such as drug delivery, tissue engineering, and as coatings for implanted medical devices [4,7,52–57]. However, ELP sequences do not support cell adhesion. As a case in point, cell seeding on unmodified LysB10 surfaces was associated with minimal cell adhesion or migration, limited cell spreading and no measureable proliferation. To extend the use of these materials to tissue engineering applications, investigators have inserted various bio-adhesive peptide sequences into ELPs, including RGD [58], the CS5 sequence from fibronectin [59], laminin-derived sequences or the collagen-binding domain [60]. Compared to the linker conjugation strategy employed here, these approaches have the advantage of circumventing the chemical steps required to synthesize and conjugate the peptide linker. However, modifying the parent DNA sequence for each ELP variant through genetic cloning and recombinant protein expression are time-intensive and expensive processes, particularly given the need to optimize the number, distribution and presentation of incorporated cell-binding sequences. The synthesis of short peptides with functionalized end-groups may present a more efficient approach for screening multiple sequences and related ELP variants and provide the opportunity to incorporate alternative ligands with non-canonical amino acids or chemical groups that cannot be processed via genetic engineering. Additionally, although further studies would need to be performed, conducting chemical conjugation after gel formation may allow the sequestration of ligands at the biomaterial surface.
Modification of the triblock protein polymer, LysB10, proceeded through carbodiimide-catalyzed incorporation of free sulfhydryls under aqueous conditions. Nearly 50% of the accessible carboxylic acid groups (~14) per LysB10 molecule were converted to thiol groups. Concentrated solutions of modified LysB10 gelled above the inverse transition temperature by coacervation or physical crosslinking of the hydrophobic endblock domains. The presence of lysine units at the interface of hydrophobic and hydrophilic blocks afforded an opportunity for subsequent chemical crosslinking of gels with genipin, a cytocompatible crosslinker, in order to further stabilize physical crosslinks [61–63].
A RGD–maleimide linker was designed and conjugated to the gel surface to optimize ligand presentation to cell surface integrin receptors (Scheme 1). Enhanced cell adhesion has been shown for the sequence GRGDSP due to the contributions that the flanking residues make toward the overall peptide conformation and stability. Since the RGD sequence is typically presented in native matrix proteins as an exposed loop that interacts with the binding site of the integrin receptor, the peptide sequence was designed with a short spacer consisting of four glycine residues. An N-terminal maleimide facilitated covalent linkage to the thiol-modified surface, and detection of the immobilized peptides was afforded by the addition of a biotin moiety. Commercially available maleimide–PEG2–biotin was chosen for model studies due to its similarity to the RGD peptide linker and was utilized to examine whether maleimide linkage to free sulfhydryls to form a thioether bond would affect cell adhesion in the absence of a peptide ligand.
Functionalization of ELP protein polymer surfaces with the RGD linker had a profound effect on cell adhesion, proliferation and migration. Increasing linker concentration resulted in an increase in peptide surface density to a maximum level of 11 pmol cm−2, or 8 RGD moieties per LysB10 molecule, above a linker concentration of 1 mg ml−1. This apparent limit in grafting efficiency may be due to steric hindrance associated with the binding of multiple ligands onto a LysB10 backbone. Non-specific adsorption of RGD linker was observed upon exposure of unmodified LysB10 to linker concentrations exceeding 150 μg ml−1. HUVEC adhesion studies demonstrated that a linker concentration of 50 μgml−1, leading to a peptide density of 8 pmol cm−2 (5 RGD moieties per LysB10 molecule), was sufficient to promote robust cellular adhesion, with minimal non-specific binding of the linker. Diminished cell adhesion after exposure to soluble GRGDSP peptide confirmed that cell binding was due to the presence of the RGD peptide. Likewise, proliferation and migration studies with both HUVECs and pMSCs were consistent with these findings. Given the association of an activated endothelial cell state with a prothrombotic phenotype, we further demonstrated that HUVECs grown on RGD functionalized protein polymers displayed a quiescent phenotype.
Integrin-mediated cellular functions occur via diverse mechanisms. Previous studies have shown that the density of ECM proteins regulates cell adhesion, spreading and migration speed. However, differences in experimental protocols and surface chemistry and roughness have resulted in a range of reported values for the minimal surface concentration required for cell adhesion and spreading. For example, Massia and Hubbell have reported that a minimal RGD peptide density of 10 fmol cm−2 is required for fibroblast cell spreading, focal contact and stress fiber formation on modified glass surfaces [64]. In contrast, Patel et al. have demonstrated a higher RGD peptide density ranging from 0.2 to 3 pmol cm−2 on an interpenetrating polymer network coating for robust endothelial cell adhesion and spreading [29]. In the present study, RGD–LysB10 protein polymers displayed favorable attachment and proliferation at grafting densities ranging from 4 to 12 pmol cm−2.
Our results may also be viewed in the context of other studies on modified ELPs. Kaufmann et al. noted large increases in murine osteoblast plating efficiency after recombinant modification of ELP gels with RGD sequences, although migration and proliferation were not investigated [48]. They reported high plating efficiency with the insertion of two RGD sequences in each 21.5 kDa ELP molecule. Using a genetic engineering strategy, Liu and colleagues inserted RGD and CS5 cell-binding sequences into an elastin-mimetic protein and observed increased spreading and adhesion of HUVECs [54,58]. Cell migration speed did not depend upon the concentration of RGD sequences. Likewise, Nakamura et al. observed that the genetic insertion of RGD sequences into ELPs led to HUVEC attachment and migration that was equivalent to that on fibronectin and laminin [46]. However, in contrast to the triblock protein polymer used in our studies, high background levels of cell attachment were observed (~40% of that on fibronectin and laminin) on these unmodified elastin-like protein polymers.
Integrin specificity is critical in directing cell fates such as migration, proliferation and differentiation, as different integrins trigger specific signaling pathways. An acknowledged limitation of the RGD sequence, which binds multiple integrins, is an inability to elicit responses based on closely defined intracellular pathways. In the future, greater specificity may be achieved with alternative linker sequences or sequence combinations. For example, in order to promote α5β1-mediated adhesion, both the RGD sequence in the 10th type III repeat of fibronectin, as well as its synergy site, the PHSRN sequence in the 9th type III repeat domain, could be present in combination [65]. Another peptide with increased integrin affinity and selectivity is the sequence REDV, which binds integrin α4β1 and is capable of interaction with endothelial cells, but not platelets [66].
The results of this study demonstrate that chemical conjugation of a bioactive ligand via maleimide–thiol chemistry is a viable means of functionalizing surfaces of elastin-like protein polymers. HUVEC and MSC attachment, actin fiber formation, proliferation, radial migration and HUVEC activation states were characterized in order to evaluate the efficacy of RGD-functionalized LysB10, and in all cases linker addition was found to significantly improve cell–material interactions.
Supplementary Material
Appendix A. Figures with essential colour discrimination
Certain figures in this article, particularly Figs. 2, 3, and 7 and Scheme 1 are difficult to interpret in black and white. The full colour images can be found in the on-line version, at doi:10.1016/j.actbio.2011.10.027.
References
- 1.Abbott WM, Megerman J, Hasson JE, L’Italien G, Warnock DF. Effect of compliance mismatch on vascular graft patency. J Vasc Surg. 1987;5:376–82. [PubMed] [Google Scholar]
- 2.Conte MS. The ideal small arterial substitute: a search for the Holy Grail? FASEB J. 1998;12:43–5. doi: 10.1096/fasebj.12.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Patel A, Fine B, Sandig M, Mequanint K. Elastin biosynthesis: the missing link in tissue-engineered blood vessels. Cardiovasc Res. 2006;71:40–9. doi: 10.1016/j.cardiores.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Wright ER, Conticello VP. Self-assembly of block copolymers derived from elastin-mimetic polypeptide sequences. Adv Drug Deliv Rev. 2002;54:1057–73. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 5.Nagapudi K, Brinkman WT, Thomas BS, Park JO, Srinivasarao M, Wright E, et al. Viscoelastic and mechanical behavior of recombinant protein elastomers. Biomaterials. 2005;26:4695–706. doi: 10.1016/j.biomaterials.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Sallach RE, Cui W, Wen J, Martinez A, Conticello VP, Chaikof EL. Elastin-mimetic protein polymers capable of physical and chemical crosslinking. Biomaterials. 2009;30:409–22. doi: 10.1016/j.biomaterials.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan SW, Haller CA, Sallach RE, Apkarian RP, Hanson SR, Chaikof EL. The effect of a recombinant elastin-mimetic coating of an ePTFE prosthesis on acute thrombogenicity in a baboon arteriovenous shunt. Biomaterials. 2007;28:1191–7. doi: 10.1016/j.biomaterials.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 8.Sallach RE, Cui W, Balderrama F, Martinez AW, Wen J, Haller CA, et al. Long-term biostability of self-assembling protein polymers in the absence of covalent crosslinking. Biomaterials. 2009;31:779–91. doi: 10.1016/j.biomaterials.2009.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagapudi K, Brinkman W, Leisen J, Thomas B, Wright E, Haller C, et al. Protein-based thermoplastic elastomers. Macromolecules. 2005;38:345–54. [Google Scholar]
- 10.Caves JM, Kumar VA, Martinez AW, Kim J, Ripberger CM, Haller CA, et al. The use of microfiber composites of elastin-like protein matrix reinforced with synthetic collagen in the design of vascular grafts. Biomaterials. 2010;31:7175–82. doi: 10.1016/j.biomaterials.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosselin C, Vorp DA, Warty V, Severyn DA, Dick EK, Borovetz HS, et al. EPTFE coating with fibrin glue, FGF-1, and heparin: effect on retention of seeded endothelial cells. J Surg Res. 1996;60:327–32. doi: 10.1006/jsre.1996.0052. [DOI] [PubMed] [Google Scholar]
- 12.Hubbell JA, Massia SP, Desai NP, Drumheller PD. Endothelial cell-selective materials for tissue engineering in the vascular graft via a new receptor. Biotechnology. 1991;9:568–72. doi: 10.1038/nbt0691-568. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Hill A, Imran M. In vitro and in vivo studies of ePTFE vascular grafts treated with P15 peptide. J Biomater Sci Polym Ed. 2005;16:875–91. doi: 10.1163/1568562054255754. [DOI] [PubMed] [Google Scholar]
- 14.Meinhart JG, Schense JC, Schima H, Gorlitzer M, Hubbell JA, Deutsch M, et al. Enhanced endothelial cell retention on shear-stressed synthetic vascular grafts precoated with RGD-cross-linked fibrin. Tissue Eng. 2005;11:887–95. doi: 10.1089/ten.2005.11.887. [DOI] [PubMed] [Google Scholar]
- 15.Nishibe T, O’Donnel S, Pikoulis E, Rich N, Okuda Y, Kumada T, et al. Effects of fibronectin bonding on healing of high porosity expanded polytetrafluoroethylene grafts in pigs. J Cardiovasc Surg. 2001;42:667–73. [PubMed] [Google Scholar]
- 16.Randone B, Cavallaro G, Polistena A, Cucina A, Coluccia P, Graziano P, et al. Dual role of VEGF in pretreated experimental ePTFE arterial grafts. J Surg Res. 2005;127:70–9. doi: 10.1016/j.jss.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Seifalian AM, Tiwari A, Hamilton G, Salacinski HJ. Improving the clinical patency of prosthetic vascular and coronary bypass grafts: the role of seeding and tissue engineering. Artif Organs. 2002;26:307–20. doi: 10.1046/j.1525-1594.2002.06841.x. [DOI] [PubMed] [Google Scholar]
- 18.Michael KE, Vernekar VN, Keselowsky BG, Meredith JC, Latour RA, García AJ. Adsorption-induced conformational changes in fibronectin due to interactions with well-defined surface chemistries. Langmuir. 2003;19:8033–40. [Google Scholar]
- 19.Ghosh SS, Kao PM, McCue AW, Chappelle HL. Use of maleimide–thiol coupling chemistry for efficient syntheses of oligonucleotide-enzyme conjugate hybridization probes. Bioconjug Chem. 1990;1:71–6. doi: 10.1021/bc00001a009. [DOI] [PubMed] [Google Scholar]
- 20.King HD, Dubowchik GM, Walker MA. Facile synthesis of maleimide bifunctional linkers. Tetrahedron Lett. 2002;43:1987–90. [Google Scholar]
- 21.Dubowchik GM, Walker MA. Receptor-mediated and enzyme-dependent targeting of cytotoxic anticancer drugs. Pharmacol Ther. 1999;83:67–123. doi: 10.1016/s0163-7258(99)00018-2. [DOI] [PubMed] [Google Scholar]
- 22.Lattuada L, Gabellini M. Straightforward synthesis of a novel maleimide–DTPA bifunctional chelating agent. Synthetic Comm. 2005;35:2409–13. [Google Scholar]
- 23.Ji S, Zhu Z, Hoye TR, Macosko CW. Maleimide functionalized poly(-caprolactone)-block-poly(ethylene glycol) (PCL-PEG-MAL): synthesis, nanoparticle formation, and thiol conjugation. Macromol ChemPhys. 2009;210:823–31. doi: 10.1002/macp.200900025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermanson GT. Bioconjuate Techniques. 2. Amsterdam: Elsevier; 2008. [Google Scholar]
- 25.Lateef SS, Boateng S, Hartman TJ, Crot CA, Russell B, Hanley L. GRGDSP peptide-bound silicone membranes withstand mechanical flexing in vitro and display enhanced fibroblast adhesion. Biomaterials. 2002;23:3159–68. doi: 10.1016/s0142-9612(02)00062-5. [DOI] [PubMed] [Google Scholar]
- 26.Pfaff M. Recognition sites of RGD-dependent integrins. In: Eble JAK, Klaus, editors. Integrin–Ligand Interaction. Austin: R.G. Landes Company; 1997. pp. 101–21. [Google Scholar]
- 27.Villanueva I, Weigel CA, Bryant SJ. Cell-matrix interactions and dynamic mechanical loading influence chondrocyte gene expression and bioactivity in PEG–RGD hydrogels. Acta Biomater. 2009;5:2832–46. doi: 10.1016/j.actbio.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salinas CN, Anseth KS. The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials. 2008;29:2370–7. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel S, Tsang J, Harbers GM, Healy KE, Li S. Regulation of endothelial cell function by GRGDSP peptide grafted on interpenetrating polymers. J Biomed Mater Res Part A. 2007;83A:423–33. doi: 10.1002/jbm.a.31320. [DOI] [PubMed] [Google Scholar]
- 30.Stile RA, Healy KE. Thermo-responsive peptide-modified hydrogels for tissue regeneration. Biomacromolecules. 2001;2:185–94. doi: 10.1021/bm0000945. [DOI] [PubMed] [Google Scholar]
- 31.Tugulu S, Silacci P, Stergiopulos N, Klok H-A. RGD-functionalized polymer brushes as substrates for the integrin specific adhesion of human umbilical vein endothelial cells. Biomaterials. 2007;28:2536–46. doi: 10.1016/j.biomaterials.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Jacob JT, Rochefort JR, Bi J, Gebhardt BM. Corneal epithelial cell growth over tethered-protein/peptide surface-modified hydrogels. J Biomed Mater Res B. 2005;72B:198–205. doi: 10.1002/jbm.b.30131. [DOI] [PubMed] [Google Scholar]
- 33.He X, Ma J, Jabbari E. Effect of grafting RGD and BMP-2 protein-derived peptides to a hydrogel substrate on osteogenic differentiation of marrow stromal cells. Langmuir. 2008;24:12508–16. doi: 10.1021/la802447v. [DOI] [PubMed] [Google Scholar]
- 34.Shin H, Jo S, Mikos AG. Modulation of marrow stromal osteoblast adhesion on biomimetic oligo[poly(ethylene glycol) fumarate] hydrogels modified with Arg-Gly-Asp peptides and a poly(ethylene glycol) spacer. J Biomed Mater Res. 2002;61:169–79. doi: 10.1002/jbm.10193. [DOI] [PubMed] [Google Scholar]
- 35.Lin YS, Wang SS, Chung TW, Wang YH, Chiou SH, Hsu JJ, et al. Artif Organs. 2001;25:617. doi: 10.1046/j.1525-1594.2001.025008617.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang DA, Ji J, Sun YH, Shen JC, Feng LX, Elisseeff JH. In situ immobilization of proteins and RGD peptide on polyurethane surfaces via poly(ethylene oxide) coupling polymers for human endothelial cell growth. Biomacromolecules. 2002;3:1286. doi: 10.1021/bm0255950. [DOI] [PubMed] [Google Scholar]
- 37.Burgess BT, Myles JL, Dickinson RB. Quantitative analysis of adhesion-mediated cell migration in three-dimensional gels of RGD-grafted collagen. Ann Biomed Eng. 2000;28:110–8. doi: 10.1114/1.259. [DOI] [PubMed] [Google Scholar]
- 38.Grzesiak JJ, Pierschbacher MD, Amodeo MF, Malaney TI, Glass JR. Enhancement of cell interactions with collagen/glycosaminoglycan matrices by RGD derivatization. Biomaterials. 1997;18:1625–32. doi: 10.1016/s0142-9612(97)00103-8. [DOI] [PubMed] [Google Scholar]
- 39.Schense JC, Bloch J, Aebischer P, Hubbell JA. Enzymatic incorporation of bioactive peptides into fibrin matrices enhances neurite extension. Nat Biotech. 2000;18:415–9. doi: 10.1038/74473. [DOI] [PubMed] [Google Scholar]
- 40.Kim J, Park Y, Tae G, Lee KB, Hwang CM, Hwang SJ, et al. Characterization of low-molecular-weight hyaluronic acid-based hydrogel and differential stem cell responses in the hydrogel microenvironments. J Biomed Mater Res A. 2009;88A:967–75. doi: 10.1002/jbm.a.31947. [DOI] [PubMed] [Google Scholar]
- 41.Cui F, Tian W, Hou S, Xu Q, Lee IS. Hyaluronic acid hydrogel immobilized with RGD peptides for brain tissue engineering. J Mater Sci Mater Med. 2006;17:1393–401. doi: 10.1007/s10856-006-0615-7. [DOI] [PubMed] [Google Scholar]
- 42.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Du KT, Fang Q, Gu Y, Mihardja SS, Sievers RE, et al. The use of human mesenchymal stem cells encapsulated in RGD modified alginate microspheres in the repair of myocardial infarction in the rat. Biomaterials. 2010;31(27):7012–20. doi: 10.1016/j.biomaterials.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 44.Ferreira LS, Gerecht S, Fuller J, Shieh HF, Vunjak-Novakovic G, Langer R. Bioactive hydrogel scaffolds for controllable vascular differentiation of human embryonic stem cells. Biomaterials. 2007;28:2706–17. doi: 10.1016/j.biomaterials.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massia SP, Stark J. Immobilized RGD peptides on surface-grafted dextran promote biospecific cell attachment. J Biomed Mater Res. 2001;56:390–9. doi: 10.1002/1097-4636(20010905)56:3<390::aid-jbm1108>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura M, Mie M, Mihara H, Nakamura M, Kobatake E. Construction of multi-functional extracellular matrix proteins that promote tube formation of endothelial cells. Biomaterials. 2008;29:2977–86. doi: 10.1016/j.biomaterials.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Monteiro GA, Fernandes AV, Sundararaghavan HG, Shreiber DI. Positively and negatively modulating cell adhesion to type I collagen via peptide grafting. Tissue Eng A. 2011;17(13–14):1663–73. doi: 10.1089/ten.tea.2008.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaufmann D, Fiedler A, Junger A, Auernheimer J, Kessler H, Weberskirch R. Chemical conjugation of linear and cyclic RGD moieties to a recombinant elastin-mimetic polypeptide – a versatile approach towards bioactive protein hydrogels. Macromol Biosci. 2008;8:577–88. doi: 10.1002/mabi.200700234. [DOI] [PubMed] [Google Scholar]
- 49.Meinel L, Hofmann S, Karageorgiou V, Kirker-Head C, McCool J, Gronowicz G, et al. The inflammatory responses to silk films in vitro and in vivo. Biomaterials. 2005;26:147–55. doi: 10.1016/j.biomaterials.2004.02.047. [DOI] [PubMed] [Google Scholar]
- 50.Bodanszky M, Bodanszky A. The Practice of Peptide Synthesis. 2. Heidelberg: Springer-Verlag; 1994. [Google Scholar]
- 51.Cook AD, Hrkach JS, Gao NN, Johnson IM, Pajvani UB, Cannizzaro SM, et al. Characterization and development of RGD-peptide-modified poly(lactic acid-co-lysine) as an interactive, resorbable biomaterial. J Biomed Mater Res. 1997;35:513–23. doi: 10.1002/(sici)1097-4636(19970615)35:4<513::aid-jbm11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 52.Herrero-Vanrell R, Rincon AC, Alonso M, Reboto V, Molina-Martinez IT, Rodriguez-Cabello JC. Self-assembled particles of an elastin-like polymer as vehicles for controlled drug release. J Control Release. 2005;102:113–22. doi: 10.1016/j.jconrel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Dreher MR, Raucher D, Balu N, Michael Colvin O, Ludeman SM, Chilkoti A. Evaluation of an elastin-like polypeptide-doxorubicin conjugate for cancer therapy. J Control Release. 2003;91:31–43. doi: 10.1016/s0168-3659(03)00216-5. [DOI] [PubMed] [Google Scholar]
- 54.Liu JC, Heilshorn SC, Tirrell DA. Comparative cell response to artificial extracellular matrix proteins containing the RGD and CS5 cell-binding domains. Biomacromolecules. 2004;5:497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 55.Richman GP, Tirrell DA, Asthagiri AR. Quantitatively distinct requirements for signaling-competent cell spreading on engineered versus natural adhesion ligands. J Control Release. 2005;101:3–12. doi: 10.1016/j.jconrel.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 56.Woodhouse KA, Klement P, Chen V, Gorbet MB, Keeley FW, Stahl R, et al. Investigation of recombinant human elastin polypeptides as non-thrombogenic coatings. Biomaterials. 2004;25:4543–53. doi: 10.1016/j.biomaterials.2003.11.043. [DOI] [PubMed] [Google Scholar]
- 57.Sallach RE, Cui W, Balderrama F, Martinez AW, Wen J, Haller CA, et al. Long-term biostability of self-assembling protein polymers in the absence of covalent crosslinking. Biomaterials. 2009;31:779–91. doi: 10.1016/j.biomaterials.2009.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu JC, Tirrell DA. Cell response to RGD density in cross-linked artificial extracellular matrix protein films. Biomacromolecules. 2008;9:2984–8. doi: 10.1021/bm800469j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welsh ER, Tirrell DA. Engineering the extracellular matrix: a novel approach to polymeric biomaterials. I. Control of the physical properties of artificial protein matrices designed to support adhesion of vascular endothelial cells. Biomacromolecules. 2000;1:23–30. doi: 10.1021/bm0002914. [DOI] [PubMed] [Google Scholar]
- 60.Nakamura M, Mie M, Mihara H, Kobatake E. Construction of multi-functional extracellular matrix proteins that promote tube formation of endothelial cells. Biomaterials. 2008;29:2977–86. doi: 10.1016/j.biomaterials.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 61.Chang WH, Chang Y, Lai PH, Sung HW. A genipin-crosslinked gelatin membrane as wound-dressing material: in vitro and in vivo studies. J Biomater Sci Polym Ed. 2003;14:481–95. doi: 10.1163/156856203766652084. [DOI] [PubMed] [Google Scholar]
- 62.Sung H-W, Chang W-H, Ma C-Y, Lee M-H. Crosslinking of biological tissues using genipin and/or carbodiimide. J Biomed Mater Res. 2003;64A:427–38. doi: 10.1002/jbm.a.10346. [DOI] [PubMed] [Google Scholar]
- 63.Touyama IK, Takeda Y, Yatsuzuka M, Ikumoto T, Moritome N, Shingu T, et al. Studies on the blue pigments produced from genipin and methylamine. II. On the formation mechanisms of brownish-red intermediates leading to the blue pigment formation. Chem Pharm Bull. 1994;42:1571–8. [Google Scholar]
- 64.Massia SP, Hubbell JA. An RGD spacing of 440 nm is sufficient for integrin alpha V beta 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. J Cell Biol. 1991;114:1089–100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cutler SM, García AJ. Engineering cell adhesive surfaces that direct integrin alpha 5 beta 1 binding using a recombinant fragment of fibronectin. Biomaterials. 2003;24:1759–70. doi: 10.1016/s0142-9612(02)00570-7. [DOI] [PubMed] [Google Scholar]
- 66.Drumheller PD, Hubbell JA. Polymer networks with grafted cell adhesion peptides for highly biospecific cell adhesive substrates. Anal Biochem. 1994;222:380–8. doi: 10.1006/abio.1994.1506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.