SUMMARY
pRb and p53 are two major tumor suppressors. Here, we found that p53 activates expression of Pirh2 and KPC1, two of the three ubiquitin ligases for p27. Loss of p53 in the absence of Skp2, the third ubiquitin ligase for p27, shrinks the cellular pool of p27 ubiquitin ligases to accumulate p27 protein. In the absence of pRb and p53, p27 was unable to inhibit DNA synthesis in spite of its abundance, but could inhibit division of cells that maintain DNA replication with re-replication. This mechanism blocked pRb and p53 doubly deficient pituitary and prostate tumorigenesis lastingly coexistent with BrdU-labeling neoplastic lesions, revealing an unconventional cancer cell vulnerability when pRb and p53 are inactivated.
INTRODUCTION
The prototype tumor suppressor retinoblastoma protein (pRb) is a transducer between the cells’ environment and gene expression machinery (Burkhart and Sage, 2008). Fully active pRb recruits chromatin-modifying proteins to the promoters of E2F target genes to repress genes for DNA replication, which can be sufficiently potent and permanent to induce cellular senescence (Chicas et al., 2010). Upstream, pRb is regulated by phosphorylation by cyclin-dependent kinases (CDKs). Various signaling pathways can activate expression of relevant CDKs and CKIs (cyclin dependent kinase inhibitors) to inactivate pRb (such as by cyclin D1/Cdk4 to induce tumorigenesis) or activate pRb (such as by p16Ink4A to induce senescence) (Sherr, 2012).
The other major tumor suppressor p53 is activated by oncogenic stress, such as the loss of pRb, directly or indirectly via Arf (Sherr, 2012). Activated p53 switches on its target genes to induce cell cycle arrest, senescence, and apoptosis to safeguard against tumorigenesis.
In experimental settings, most of the cells’ intrinsic antitumor mechanisms seem to function via p53, pRb, or both. Indeed, combined deletion of Rb1 (encoding pRb) and Trp53 (encoding p53) is very effective in inducing tumors in wide spectrum of tissues in mice. Clinically, inactivation of both pRb and p53 are frequent in various cancers and may explain, in large part, why cancers are difficult to treat. Recent studies discovered that deleting Skp2, a subunit of the SCF (CRL1) E3 ubiquitin ligase, can induce apoptosis to block pRb deficient pituitary tumorigenesis (Wang et al., 2010) or induce p53-independent senescence to block tumorigenesis in Arf−/− mice or Pten deficient prostate (Lin et al., 2010). These two findings might have conformed to the existing paradigm that p53 was activated to inhibit pRb deficient tumorigenesis and, vice versa, pRb activated to inhibit p53 deficient tumorigenesis when Skp2 is absent. Skp2 deletion however did not block tumorigenesis by ENU (N-ethyl-N-nitrosourea) (Wang et al., 2010), nor by Myc-driven lymphomagenesis (Old et al., 2010).
In this study, we asked whether combined loss of pRb and p53 would leave cells defenseless against tumorigenesis in the absence of Skp2.
RESULTS
Skp2 deletion blocks pRb and p53 doubly deficient pituitary tumorigenesis
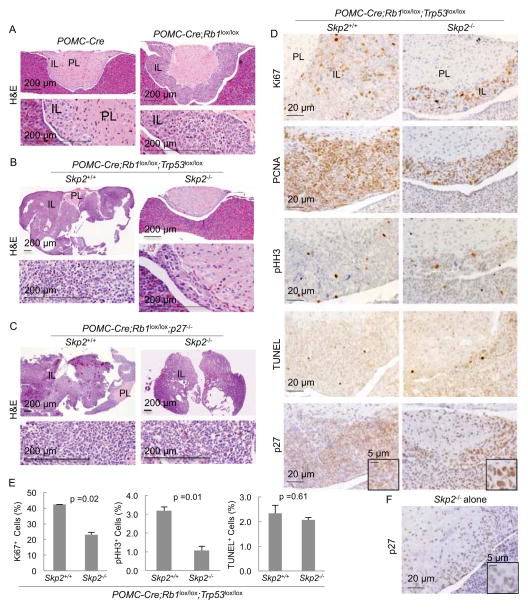
Deleting Rb1 using POMC-Cre is sufficient to induce melanotroph tumorigenesis across the entire intermediate lobe (IL) in the pituitary (Figure 1A) while deleting Trp53 did not do so (Figure S1A). Combined deletion of Rb1 and Trp53 greatly accelerated IL tumorigenesis (Figure 1B), demonstrating the safeguard role of p53 following loss of Rb1. Remarkably, Skp2 deletion still blocked this tumorigenesis (Figure 1B). In comparison, p27 knockout induced IL hyperplasia (Fero et al., 1996) (Kiyokawa et al., 1996) (Nakayama et al., 1996) (Figure S1B) and accelerated pRb deficient IL tumorigenesis to similar extent as deletion of Trp53; but Skp2 was dispensable in this context (Figure 1C).
Figure 1. Skp2 deletion blocks pRb and p53 doubly deficient, but not pRb and p27 doubly deficient, pituitary tumorigenesis.
(A–C) H&E sections of pituitary samples at 7 weeks of age. Higher magnification views of the IL are presented under lower magnification ones.
(D) Pituitary sections of indicated genotypes were stained as indicated. For the Skp2+/+;POMC-Cre;Rb1lox/lox;Trp53lox/lox genotype, we selected pituitaries that were not deformed by macroscopic tumors.
(E) Quantification of Ki67, pHH3, or TUNEL stain positive cells in (D).
(F) A representative image of Skp2−/− pituitaries stained for p27.
IL, intermediate lobe; PL, posterior lobe. Scales bars are as marked. Quantitative data are presented as average +/− SEM. Statistical analyses were by Student’s t test. See also Figure S1.
Surprisingly, although the Skp2 KO ILs did not develop tumors following co-deletion of Rb1 and Trp53 in them, they appeared to contain as much proliferation markers Ki67 and PCNA as the Skp2 WT, pRb and p53 doubly deficient ILs, which were undergoing rapid tumorigenesis (Figure 1D). Quantification of Ki67 positive cells on a percent (%) basis showed a reduction of about or less than two fold but it was statistically significant (Figure 1E). In the same samples, the mitotic marker pHH3 was reduced by three fold, suggesting a more significant inhibition in cell division (Figure 1D and 1E). Consistent with the substantial presence of proliferation markers, senescence-associated β-galactosidase (SA-β-gal) staining was negative in Skp2−/−;POMC-Cre;Rb1lox/lox;Trp53lox/lox IL (data not shown). Apoptosis was similarly infrequent in these two genotypes as measured by TUNEL staining (Figure 1D and 1E), demonstrating that, indeed, p53 functioned to induce apoptosis in Rb1 deficient pituitary tumorigenesis in absence of Skp2 (Wang et al., 2010). Thus, the complete block of the highly accelerated tumorigenesis co-existed, unexpectedly, with abundant proliferation markers (also see Figure S1C).
Another unexpected finding was that Skp2−/−;POMC-Cre;Rb1lox/lox;Trp53lox/lox melanotrophs contained more p27 protein than Skp2−/− melanotrophs (Figure 1D and 1F; Figure S1C). This finding was controlled with staining of p27−/− pituitary (Figure S1D). p27 accumulated in the nuclei, and p27 abundant nuclei were larger, suggesting accumulation of more DNA.
Altogether, these results provided indications that (1) in the absence of Skp2, codeletion of Rb1 and Trp53 could further increase p27 protein levels, reminiscent of a safeguard response, but, (2) this high-level p27 is unable to inhibit expression of proliferation markers and induce senescence in the absence of pRb and p53. Nevertheless, (3) the highly accelerated tumorigenesis was blocked.
Deleting Trp53 in Skp2KO MEFs activates a pRb safeguard to induce senescence
To further investigate the above unexpected findings, we generated mouse embryo fibroblasts (MEFs) of the same genotypes to study how Skp2 deletion blocked pRb and p53 doubly deficient tumorigenesis. We used Adeno-Cre to delete Trp53, Rb1, or both in MEFs controlled by parallel infection with Adeno-GFP. To simplify the text, we will call the various MEFs as p53KO, pRbp53DKO, or Skp2KO;pRbp53DKO, for examples.
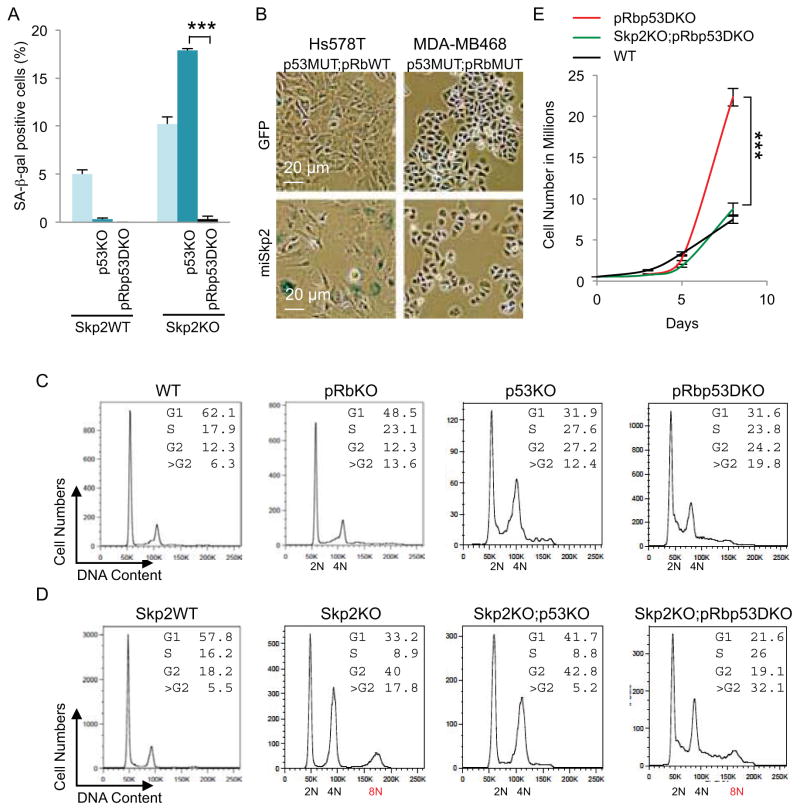
Inactivation of p53 can immortalize Skp2WT MEFs but, to the opposite, induced senescence in Skp2KO MEFs (Lin et al., 2010). In Figure 2A and Figure S2A and 2B, we show that Skp2WT;p53KO MEFs were prevented from senescence but Skp2KO;p53KO MEFs senesced more than Skp2KO MEFs, as measured by SA-β-gal staining. To study how p53KO paradoxically induced more senescence in Skp2KO MEFs, and to determine whether combined deletion of Rb1 would disable the senescence, we deleted Rb1 in Skp2KO;p53KO MEFs (resulting in Skp2KO;pRbp53DKO MEFs) and prevented senescence (Figure 2A). To model therapeutic inhibition of Skp2, we performed Skp2 knockdown in human breast cancer cell lines Hs578T and HCC1143, which contain functional pRb but are deficient for p53, and MDA-MB468 and BT549, which are deficient for both pRb and p53. Tandem two miR-30 based hairpins targeting Skp2 effectively reduced Skp2 protein and increased p27 protein in these cell lines (Figure S2C) (Sun et al., 2006). SA-β-gal staining revealed senescence in Hs578T, but not in MDA-MB468 cells, following Skp2 knockdown (Figure 2B and Figure S2D). Thus, inactivation of p53 and Skp2 together activated a pRb safeguard to induce senescence. With additional loss of pRb, this line of antitumor defense is disabled, consistent with the abundant presence of proliferation markers in Skp2−/−;POMC-Cre;Rb1lox/lox;Trp53lox/lox IL.
Figure 2. Skp2, Rb1 and Trp53 triply deficient MEFs show higher DNA replication and re-replication, accumulation at 8N, and proliferation speed of WT MEFs.
(A) Quantification of senescence marker SA-β-gal in MEFs of the indicated genotypes.
(B) Human breast cancer cell lines with known pRb and p53 status (as indicated) were transduced with lentiviruses expressing GFP or miRNA haipins targeting Skp2 (miSkp2). Five days after transduction, same numbers of cells were plated. SA-β-gal staining was performed three days later.
(C and D) DNA content FACS analysis of MEFs with indicated genotypes.
(E) Cell proliferation of the indicated MEFs is measured by counting cell numbers.
Quantitative data are presented as average +/− SEM. Statistical analyses were by Student’s t test. ***, p < 0.002. FACS results are representative of three experiments. See also Figure S2.
Deleting Trp53 and Rb1 induces DNA re-replication
Notably, although the Skp2KO;pRbp53DKO genotype disabled senescence in MEFs and induced proliferation markers in IL, it induced no tumorigenesis. We subjected MEFs of relevant genotypes to FACS analysis to study this unusual phenotype. As expected, deletion of Rb1 or Trp53 increased S phase cells (DNA content between 2N and 4N) and G2 phase cells (4N) (Figure 2C).
Unexpectedly, deletion of Rb1 or Trp53 also generated more >G2 cells (DNA content larger than 4N); combined deletion of Rb1 and Trp53 increased this population to 20% (Figure 2C). The continuous distribution of the larger DNA contents indicated DNA re-replication, which is defined as re-firing of certain DNA replication origins before the entire chromosomes were fully duplicated (Arias and Walter, 2007). These findings reveal that pRb inhibits DNA re-replication and p53 safeguards against it. Notably, while combined loss of pRb and p53 did not synergistically increase S phase cells, it did for DNA re-replication (Figure 2C). Thus, a significant portion of elevated DNA synthesis activity during tumorigenesis following loss of pRb and p53 might be in the form of re-replication.
Co-deleting Trp53 and Rb1 in Skp2KO MEFs induces accumulation of DNA re-replicating cells by mitotic entry delay
Consistent with previous reports that increased p27 in Skp2KO MEFs inhibited mitotic cyclin-Cdk (Nakayama et al., 2004), Skp2KO MEFs showed increased G2 population and an 8N peak. There was however no accumulation of cells between 4N and 8N (Figure 2D). This profile indicates no DNA re-replication (as defined above) but the fully duplicated chromosomes failed to segregate into daughter cells before they were fully duplicated again.
Deletion of Trp53 in Skp2KO MEFs, which induced senescence in them (Figure 2A), induced less DNA re-replication compared to p53KO MEFs (Figure 2C and 2D). Since pRb was activated in Skp2KO;p53KO MEFs to induce senescence (Figure 2A), a more active pRb might have inhibited DNA re-replication as well. Deletion of Trp53 also abolished the 8N peak, suggesting that a more active pRb also blocked chromosome re-duplication, likely due to the inhibition of DNA synthesis.
Skp2KO;pRbp53DKO MEFs, which were prevented from senescence, showed more S phase cells and DNA re-replication than Skp2KO;p53KO MEFs with return of the 8N peak, resulting in a total of 32% cells with DNA content larger than 4N (Figure 2D). This profile, together with the events leading to it, suggests that Skp2KO;pRbp53DKO allowed DNA re-replication to generate cells with >4N DNA content; and these cells gradually accumulated as an 8N peak. We further found that pHH3 positive cells were reduced from 1.13% in pRbp53DKO MEFs to 0.69% in Skp2KO;pRbp53DKO MEFs. These findings indicate the presence of a mitotic entry delay and are consistent with the significant decrease in pHH3 positive cells in Skp2−/−;POMC-Cre;Rb1lox/lox;Trp53lox/lox ILs (Figure 1D), while increased S phase and DNA re-replication explains the abundant presence of Ki67 positive and PCNA positive cells (Figure 1D and 1E).
Finally, we counted cells to determine actual proliferation. We found that Skp2KO;pRbp53DKO MEFs proliferated at the speed of WT MEFs, despite much larger S phase population. pRbp53DKO MEFs, despite similar S phase size as Skp2KO;pRbp53DKO MEFs, proliferated much faster than the latter, resulting in about three fold more cells at day 8 (Figure 2E). This difference in cell numbers resembled the tumorigenic potentials of these two genotypes in the pituitary. Similarly, human breast cancer cell lines MDA-MB468 and BT549 (mimicking pRbp53DKO) proliferated much faster than MDA-MB468-miSkp2 and BT549-miSkp2 (mimicking Skp2KO;pRbp53DKO) although the latter did not senesce (Figure 2B; Figure S2D and S2E).
These MEF studies modeled findings from pituitary and provide insights for the unusual phenotypes of the Skp2KO;pRbp53DKO genotype. Chromosome separation failure restricted actual cell proliferation, while DNA re-replication sustained presence of abundant proliferation markers.
p53 loss induces a p27 safeguard
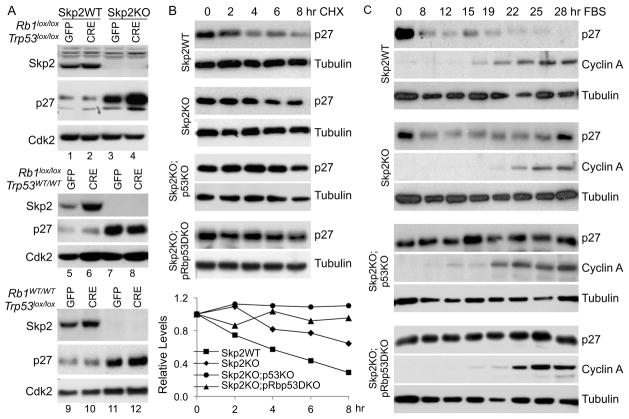
We next determined the molecular mechanisms for the phenotypes of Skp2KO;pRbp53DKO MEFs. Our finding that p27 protein levels were higher in Skp2−/−;POMC-Cre;Rb1lox/lox;Trp53lox/lox IL than in Skp2−/− IL (Figure 1D and 1F) suggested an elevation of a p27 safeguard by combined deletion of Rb1 and Trp53. We performed Western blots to determine whether p27 protein levels in Skp2KO;pRbp53DKO MEFs also mimicked those in pituitary. As expected, Skp2KO MEFs contained more p27 protein than Skp2WT MEFs (Figure 3A lanes 1 and 3) (Nakayama et al., 2000). Combined deletion of Rb1 and Trp53 did not change p27 protein levels in Skp2WT MEFs (Figure 3A lanes 1 and 2), but further increased p27 protein levels in Skp2KO MEFs (lanes 3 and 4). Thus, MEFs again mimicked IL.
Figure 3. Deletion of Trp53 triggers further stabilization of p27 protein in Skp2KO MEFs.
(A) Western blot analysis of indicated proteins in MEFs of indicated genotypes transduced in parallel by Adeno-GFP or Adeno-CRE.
(B) Cell extracts were prepared from MEFs of indicated genotypes at the indicated time points following the addition of cyclohexamide (CHX), and the amounts of p27 protein were measured by Western blot in comparison to tubulin. p27 protein level quantification is presented under the gels.
(C) Cell extracts were prepared from MEFs of indicated genotypes at indicated time points after being cultured in serum containing media following serum starvation for three days. The p27 protein level was determined by Western blot. The cyclin A protein level is shown to indicate cell cycle progression and the tubulin level is shown to indicate loading amounts.
See also Figure S3.
We investigated whether deleting Rb1 or Trp53 alone could further increase p27 proteins in Skp2KO MEFs. We found that Skp2KO;p53KO MEFs (Figure 3A lane 12), but not Skp2KO;pRbKO MEFs (lane 8) contained higher p27 proteins. Since p53 is gradually activated with passage of cultured MEFs (Figure S3A lanes 1 and 2), deletion of Trp53 in late passage Skp2KO MEFs induced higher amounts of p27 (Figure S3A lanes 6 and 7). Deletion of Rb1 activated p53 in early passage MEFs (Figure S3A lane 3), which might explain that combined deletion of Trp53 and Rb1 induced the most p27 protein in Skp2KO MEFs (Figure 3A).
These findings explained that deletion of Trp53 in Skp2KO MEFs induced higher levels of p27, which inhibits S phase cyclin-dependent kinases. Consequently, pRb is less phosphorylated and therefore more active. The activated pRb, in turn, inhibits DNA synthesis to induce senescence. This role of pRb as a transducer between inhibition of S phase cyclins and inhibition of DNA synthesis is essential since Skp2KO;pRbp53DKO MEFs resumed DNA synthesis and failed to senesce. In comparison, since Skp2KO;pRbp53DKO MEFs are delayed for mitotic entry, the inhibition of mitotic cyclins (by the high p27) can inhibit mitotic entry without an activated pRb.
To determine the functional significance of the high p27 in blocking mitotic entry, we knocked down p27 in Skp2KO;pRbp53DKO MEFs with two p27 targeting hairpins (Figure S3B). Hairpin 2 (sh2) reduced p27 protein in Skp2KO;pRbp53DKO MEFs to below that in Skp2WT;pRbp53DKO MEFs, abolished the 8N peak and reduced 4N-8N cells (Figure S3C), and increased proliferation of Skp2KO;pRbp53DKO MEFs to an extent faster than Skp2WT;pRbp53DKO MEFs (Figure S3D). sh1 knocked down p27 to a lesser degree and reduced 4N-8N cells and increased proliferation to a smaller extent. Thus, accumulation of 4N-8N DNA content and inhibition of proliferation of pRbp53DKO MEFs by Skp2 deletion was highly dependent on high p27 protein levels. When we used a CMV promoter to overexpress p27 in Skp2WT;pRbp53DKO MEFs (Figure S3E), p27 protein levels increased less than two fold (Figure S3F), cells did not accumulate 8N DNA content (data not shown), and proliferation rates were only slightly reduced (Figure S3G), suggesting that p27 protein degradation is the major regulation of p27 levels and functions in Skp2WT;pRbp53DKO MEFs.
The further increases in p27 protein levels were not associated with increases in p27 mRNA levels (Figure S3H), but with more stable p27 proteins (Figure 3B). This finding indicated that an Skp2-independent p27 degradation mechanism(s) was reduced by deletion of Trp53 in Skp2KO MEFs. That deletion of Trp53 did not induce higher p27 in Skp2WT MEFs suggested that Skp2 possesses sufficient p27 degradation activity to compensate for reduction in other p27 protein degradation mechanisms.
In addition to Skp2, p27 is ubiquitinated by KPC1 and Pirh2 ubiquitin ligases, which primarily function in early time points of cell cycle entry (Hattori et al., 2007; Kamura et al., 2004). We used serum starvation-release to determine whether Trp53 deletion affected p27 protein degradation in this time window. Figure 3C shows Skp2 KO did not prevent reduction of p27 protein levels in early time points (8, 12, 15 hr), but prevented its further reduction at 22 and 25 hr. Trp53 deletion or combined Trp53 and Rb1 deletion did not affect kinetics of p27 protein reduction in Skp2WT MEFs (data not shown), but prevented p27 protein reduction at 8, 12, 15 hr in Skp2KO MEFs (Figure 3C). This finding suggested that Trp53 deletion impaired the ability of KPC1 and/or Pirh2 to degrade p27.
Interestingly, Pirh2 was previously identified as a p53 target gene (Leng et al., 2003). We used Transcription Regulatory Element Database (Xuan et al., 2005) to study whether the KPC1 gene promoter contains a p53 binding site, which is a 10-nucleotide consensus sequence repeated once with a spacer of 0–13 nucleotides (El-Deiry et al., 1992) (Wei et al., 2006) (Figure S4A). The promoter of p21, a canonical p53 target gene, contains a p53 binding site with a matched 1st half and one-mismatch 2nd half (we call it a 0,1 site). The Pirh2 promoter contains a 0,1 site and the KPC1 promoter contains a 1,1 site (Figure S4B). Skp2 and p27 promoters contain a 1,2 site and a 2,1 site, respectively.
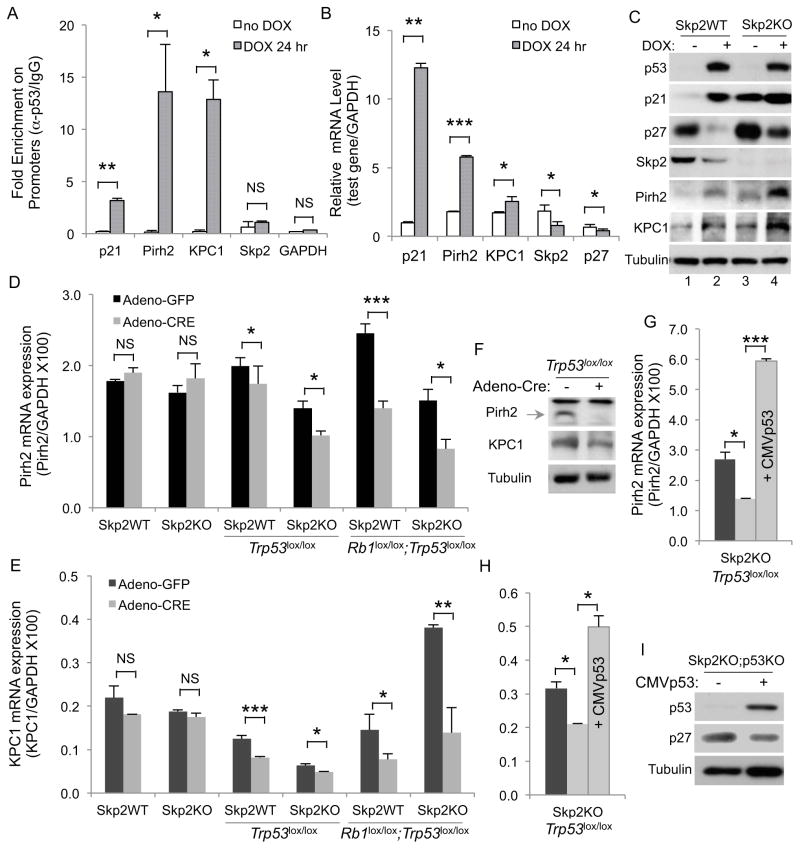
ChIP experiments in Figure 4A show that activation of p53 in WT MEFs by doxorubicin (DOX) stimulated recruitment of p53 to the promoters of p21, Pirh2 and KPC1, but not Skp2 and GAPDH. In agreement with the ChIP results, expressions of Pirh2 and KPC1 were stimulated by DOX, although to smaller degrees than the stimulation of p21 expression; but DOX did not stimulate expression of Skp2 or p27 (Figure 4B). Western blots show that p53, p21, Pirh2, and KPC1 proteins were increased, and Skp2 protein decreased, by DOX treatment (Figure 4C). We then predicted, and our results confirmed, that DOX counter-intuitively decreased p27 protein levels (lanes 1 and 2). Notably, DOX also reduced p27 protein levels in Skp2KO MEFs (lanes 3 and 4). Thus, strong p53 activation and acute Trp53 deletion have opposite effects on p27 protein levels. We explored the possibility that these changes in protein levels were due to changes in cell cycle profiles by DOX; we found that under the conditions used DOX did not dramatically change cell cycle profiles of WT or Skp2KO MEFs (Figure S4C).
Figure 4. p27 ubiquitin ligases Pirh2 and KPC1 are p53 target genes.
(A) ChIP with anti-p53 and control IgG following DOX treatment of WT MEFs. Fold enrichment (anti-p53/IgG) for the indicated promoters is presented.
(B) RT-qPCR of the indicated genes in WT MEFs following DOX treatment, normalized with GAPDH.
(C) Western blot of the indicated MEFs following treatment with DOX as in (B).
(D and E) Expression of Pirh2 (D) and KPC1 (E), in MEFs transduced in parallel by Adeno-GFP and Adeno-CRE, values were normalized with GAPDH.
(F) Western blots of Pirh2 and KPC1 before and after deletion of Trp53.
(G and H) Same as (D and E) with an additional sample in which p53 was ectopically expressed, as marked inside the bars.
(I) Western blots of Skp2KO;p53KO MEFs without or with ectopic p53 expression.
Quantitative data are presented as average +/− SEM. Student’s t test was used for statistical analysis. *, p <0.05; **, p < 0.01; ***, p < 0.002; NS, p >0.05. See also Figure S4.
We next determined effects of Trp53 deletion on expression of Pirh2 and KPC1. Figure 4D and 4E show that Trp53 deletion alone or with Rb1 reduced mRNA levels of Pirh2 and KPC1 in both Skp2WT and Skp2KO MEFs. The effects of Trp53 deletion were smaller than Trp53 and Rb1 co-deletion, likely because p53 was not highly activated in cultured MEFs before Rb1 was deleted (Figure 3A and Figure S3A). Western blots show that Pirh2 and KPC1 protein levels also decreased following deletion of Trp53 (Figure 4F). Skp2 expression was not reduced following Trp53 deletion or co-deletion with Rb1 (Figure S4D). When we re-expressed p53 in Skp2KO;p53KO MEFs, expression of Pirh2 and KPC1 increased (Figure 4G and 4H) and p27 protein levels reduced (Figure 4I). These results suggest that the combined reduction in Pirh2 and KPC1 expression shrinks the pool of the ubiquitin ligase for p27 further in Skp2KO;p53KO MEFs, therefore, increases p27 protein accumulation further.
Knockdown of Pirh2 or KPC1 can increase p27 protein in various human and mouse cells (Hattori et al., 2007; Kamura et al., 2004). For our study, we ectopically expressed Pirh2 and KPC1 in Skp2KO;pRbp53DKO MEFs to determine their negative effects on high p27 levels. While we succeeded in increasing Pirh2 levels by expression of CMV-huPirh2, expression of CMV-huKPC1 only slightly increased KPC1 protein levels in these MEFs (Figure S4E). Under these conditions, expression of Pirh2, KPC1, or both reproducibly decreased p27 protein levels, reduced 4N-8N cells, and increased proliferation (Figure S4E to S4G); but all these effects were smaller than those observed with p27 knockdown (Figure S3B to S3D). In MDA-MB468 and BT549 breast cancer cell lines with Skp2 knockdown (mimicking Skp2KO;pRbp53DKO MEFs), expression of CMV-huPirh2 and CMV-huKPC1 also increased proliferation, most effectively in BT549 cells (Figure S4H to S4K). These results confirmed the functions of Pirh2 and KPC1 in promoting p27 degradation and proliferation, but the extent of their overexpression and phenotypes are cell type dependent.
Skp2 deletion blocks pRb and p53 deficient prostate tumorigenesis
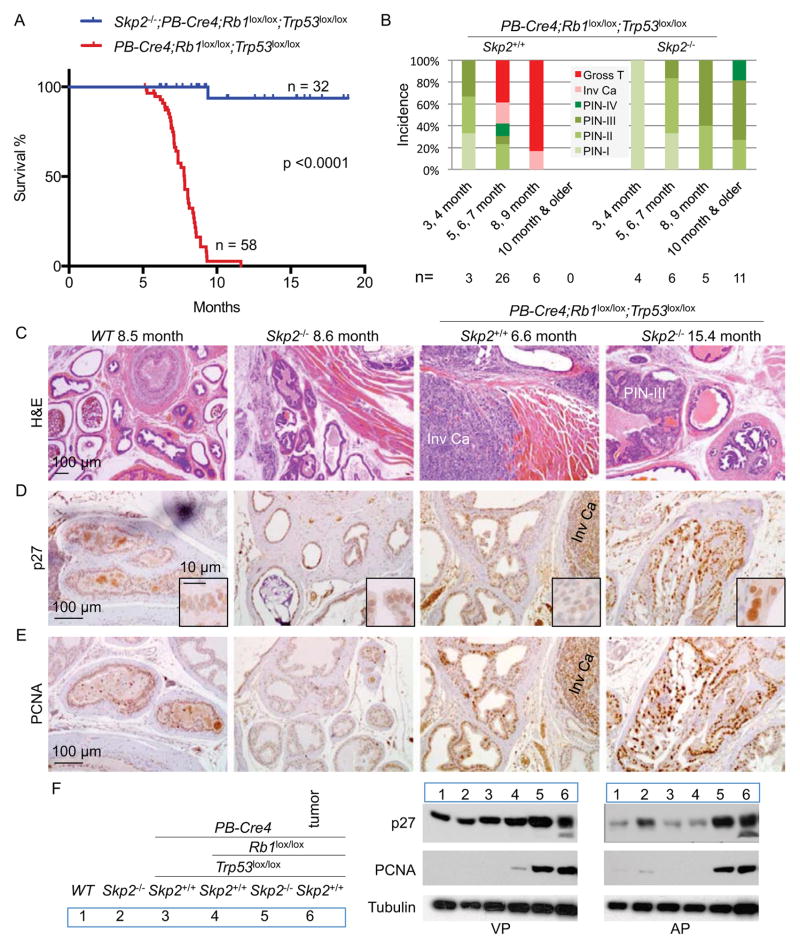
We determined whether our findings from pituitary and MEFs were applicable to another tissue. While Rb1 deletion is sufficient to induce pituitary tumorigenesis, it required combined deletion of Trp53 to induce tumorigenesis in the brain (Marino et al., 2000), lung (Meuwissen et al., 2003), bone (Walkley et al., 2008) (Berman et al., 2008), prostate (Zhou et al., 2006), ovary (Flesken-Nikitin et al., 2003), breast (Jiang et al., 2010), and liver (McClendon et al., 2011), suggesting that these tissues may mount a stronger p53 safeguard following Rb1 deletion. We studied the prostate. PB-Cre4;Rb1lox/lox;Trp53lox/lox mice developed rapid and invasive prostate cancer (Zhou et al., 2006). The prostate tumors became lethal from six months and killed all hosts (n=58) within one year (Figure 5A) (Zhou et al., 2006). In contrast, all (n=32) Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox mice survived like WT mice for the 19 months period (Figure 5A). Pathological diagnoses for these two cohorts are shown in Figure 5B. Prostatic intraepithelial neoplasias (PINs) were divided into four grades (Park et al., 2002). At 3–4 months, PINs developed in both cohorts (also see Figure S5A). PINs in PB-Cre4;Rb1lox/lox;Trp53lox/lox mice were of higher grades and quickly progressed to invasive carcinoma (Figure 5B and 5C; Figure S5B) and gross tumors during the 5–7 months period, when 20 out of 42 mice died. In the 8–9 months period, most tumors have become macroscopic and all mice died by the end of the 9th month. In contrast, PINs in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox mice never progressed beyond the PIN stage in a total of 26 mice examined, six of which were examined at 15–22 months.
Figure 5. Deletion of Skp2 blocks pRb and p53 doubly deficient prostate tumorigenesis inside PIN stages.
(A) Kaplan-Meier survival analysis of two cohorts of mice with indicated genotypes. One Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox mouse died from fighting at 9.4 months without prostate tumors.
(B) Pathological diagnoses of prostate lesions in mice of indicated genotypes in four age groups. Each prostate was serially sectioned and the most advanced lesions were the diagnoses.
(C–E) Sections of prostates from mice of four genotypes at the indicated ages stained with H&E (C), anti-p27 (D) and anti-PCNA (E). Invasive carcinoma (Inv Ca) and a PIN-III lesion are marked.
(F) Western blots of isolated ventral prostates (VP) and anterior prostates (AP) from 3–4 month old mice. A macroscopic tumor was included as marked.
See also Figure S5.
Figure 5D and 5F show that PIN lesions in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox mice contained higher levels of p27, as measured by IHC and Western blot of dissected ventral and anterior lobes, than in Skp2−/− glands. Nuclei expressing higher p27 protein were larger. PCNA expression was also high in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox PINs (Figure 5E).
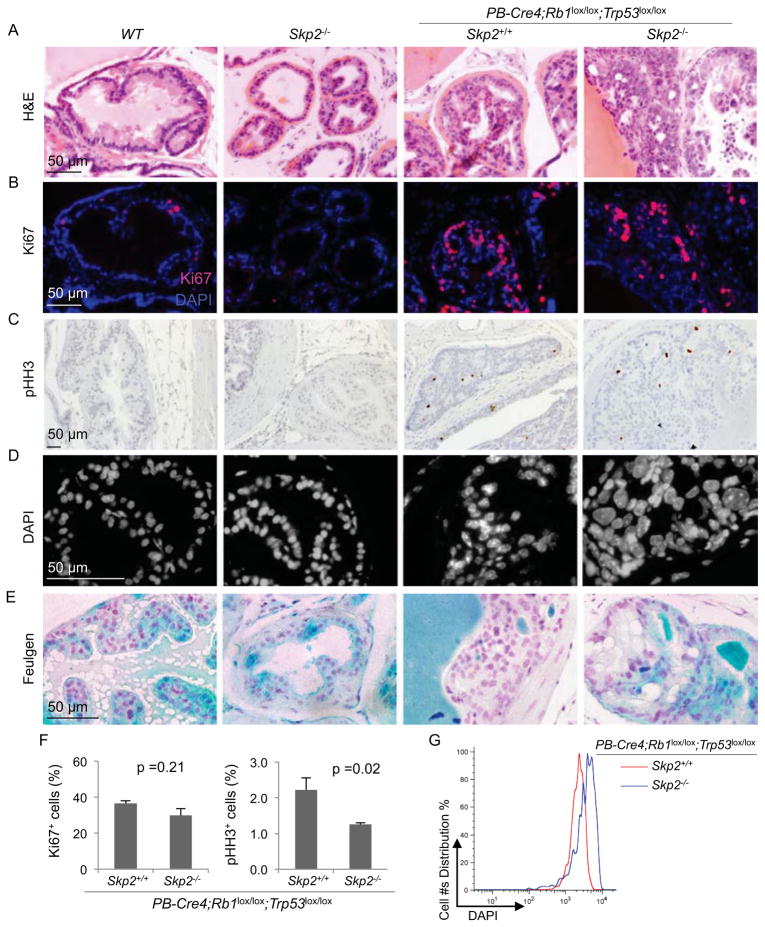
We used Ki67 staining to compare the proliferation status of PINs. Normal glands of WT and Skp2−/− mice contained few Ki67 positive cells; PINs contained more (Figure 6A and 6B). Remarkably, abundance of Ki67 positive cells was similar between PINs in PB-Cre4;Rb1lox/lox;Trp53lox/lox mice and PINs in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox mice. Presence of pHH3 on the other hand was significantly reduced from 2.23% to 1.26% (Figure 6C and 6F). If PINs in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox mice were undergoing DNA re-replication with mitotic entry block as demonstrated by the MEF model of this genotype (Figure 2D), their nuclei should accumulate larger amounts of DNA. We used DAPI and Feulgen stains to study this and found that nuclei in PINs of Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox genotype contained significantly more DAPI and Feulgen staining than the other three genotypes (Figure 6D, 6E, and 6G).
Figure 6. Comparisons of proliferation markers and nuclear sizes in prostate of the indicated genotypes.
(A–E) Sections of prostates from mice with indicated genotypes at same ages as those shown in Figure 5C were stained with H&E (A), anti-Ki67 (B), anti-pHH3 (C), DAPI (D), and Feulgen (E).
(F) Quantification of Ki67 positive cells and pHH3 positive cells from (B) and (C), respectively.
(G) DAPI stained slides were quantified by iCys® Research Imaging Cytometer and iCys® Cytometric Analysis Software.
Quantitative data are presented as average +/− SEM. Student’s t test was used for statistical analysis.
Thus, the ability of Skp2 deletion to block pRb and p53 doubly deficient tumorigenesis and the mechanisms underlining this block were essentially the same in prostate and pituitary, and as modeled in MEFs.
Life long block in BrdU-labeling PINs
We next addressed the possibility that S phase Skp2KO;pRbp53DKO MEFs (as determined by DNA content) and proliferating Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox PINs (as determined by Ki67 and PCNA expression) might not be actually synthesizing DNA, as reasons for slow proliferation for MEFs and non-tumorigenic for PINs. Cells containing >4N DNA content due to re-replication were found to cease DNA synthesis by activating p53-dependent checkpoints (Zhu et al., 2004). If cells of the Skp2KO;pRbp53DKO genotype had ceased DNA synthesis, it would implicate a mechanism to inhibit DNA synthesis (and therefore tumorigenesis) following re-replication in the absence of pRb and p53.
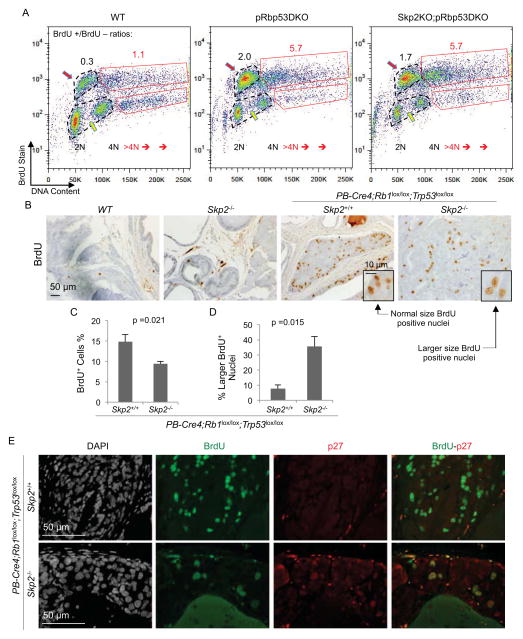
We first addressed this issue by analyzing BrdU labeled (30 minutes) Skp2KO;pRbp53DKO MEFs in parallel with WT and pRbp53DKO MEFs. For these three genotypes, cells defined as in S phase by DNA content on x-axis (positions pointed by yellow arrows) were up shifted to become BrdU positive on y-axis (positions pointed by red arrows), indicating that they were actively synthesizing DNA during the labeling period (Figure 7A). The ratios of BrdU positive cells vs negative cells were similar between pRbp53DKO MEFs and Skp2KO;pRbp53DKO MEFs (2.0 and 1.7, respectively) but much higher than that of WT MEFs (0.3), consistent with results in Figure 2C and 2D. Cells containing >4N DNA content were also largely BrdU positive. The ratios of BrdU positive >4N cells to BrdU negative >4N cells were 5.7 for both pRbp53DKO and Skp2KO;pRbp53DKO MEFs, but was 1.1 for WT MEFs. Thus, there was no indication for a pRb and p53 independent inhibitory checkpoint effect on DNA synthesis in response to DNA re-replication.
Figure 7. Sustained active DNA synthesis in Skp2KO;pRbp53DKO MEFs and in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox PINs.
(A) Representative (from three experiments) FACS plots of DNA content by PI and BrdU content by anti-BrdU. Numbers of BrdU positive and negative cells in regular cell cycle were determined within the dotted black circles, and the ratios indicated in black numbers above the circles. Numbers of cells with >4N DNA contents were similarly analyzed with red boxes and red numbers.
(B and C) Prostate sections from BrdU injected mice with indicated genotypes were stained with BrdU (B), quantification of BrdU positive cells was shown in (C).
(D) Quantification of normal sized and larger sized BrdU positive nuclei as portions of the entire BrdU positive cell populations.
(E) Prostates sections from BrdU injected mice with indicated genotypes were stained with DAPI, anti-BrdU and anti-p27. The Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox sample was from a 21.7 months old mouse, showing persistence of PIN cells containing enlarged nuclei with high p27 and BrdU incorporation.
Quantitative data in (C and D) are presented as average +/− SEM. Student’s t test was used for statistical analysis. See also Figure S6.
We labeled mice with BrdU (2 hours) to determine whether prostate PINs were actively synthesizing DNA. We found numerous BrdU positive cells in PINs of both genotypes although labeling frequency was reduced from 14.85% to 9.39% by the deletion of Skp2 (Figure 7B and 7C). Although this was a statistically significant reduction, it was less than two fold. We noticed that BrdU positive nuclei in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox PINs were larger than those in PB-Cre4;Rb1lox/lox;Trp53lox/lox PINs (compare the two inserts in Figure 7B). Figure 7D shows that 35.7% of BrdU positive cells contained larger nuclei in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox PINs, compared to 7.6% in PB-Cre4;Rb1lox/lox;Trp53lox/lox PINs. These findings suggest that DNA synthesis activities in Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox PINs were largely from DNA re-replication, explaining the presence of larger nuclei in them. Furthermore, BrdU positive and larger nuclei also contained higher p27 as shown by double staining for BrdU and p27 (Figure 7E).
Results in this section solidify the salient feature of the Skp2 deletion-mediated block of pRb and p53 doubly deficient tumorigenesis. In the absence of pRb and p53, deregulation of DNA replication in the form of re-replication persists in neoplastic lesions that are apparently blocked for life by higher levels of p27. Figure S6 shows more examples of BrdU-labeling PINs in four Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox mice with ages ranging from 11 to 22 months.
Blocked Skp2KO;pRbp53DKO PIN cells succumb to apoptosis
Although pRb and p53 doubly deficient prostate tumorigenesis did not progress beyond PINs in Skp2−/− mice as old as 22 months, the PIN lesions persisted coexistent with active DNA synthesis. Although entry to mitosis was blocked as indicated by significant reduction in pHH3 positive cells (Figure 6C and 6F; Figure S6), more than half of the pHH3 positive cells were in metaphase. Mitotic figures were also present in H&E stained sections of blocked PIN lesions. These features appeared inadequate to explain the life long block of pRb and p53 doubly deficient prostate tumorigenesis in Skp2−/− mice and would be compatible for selection of resistant cells.
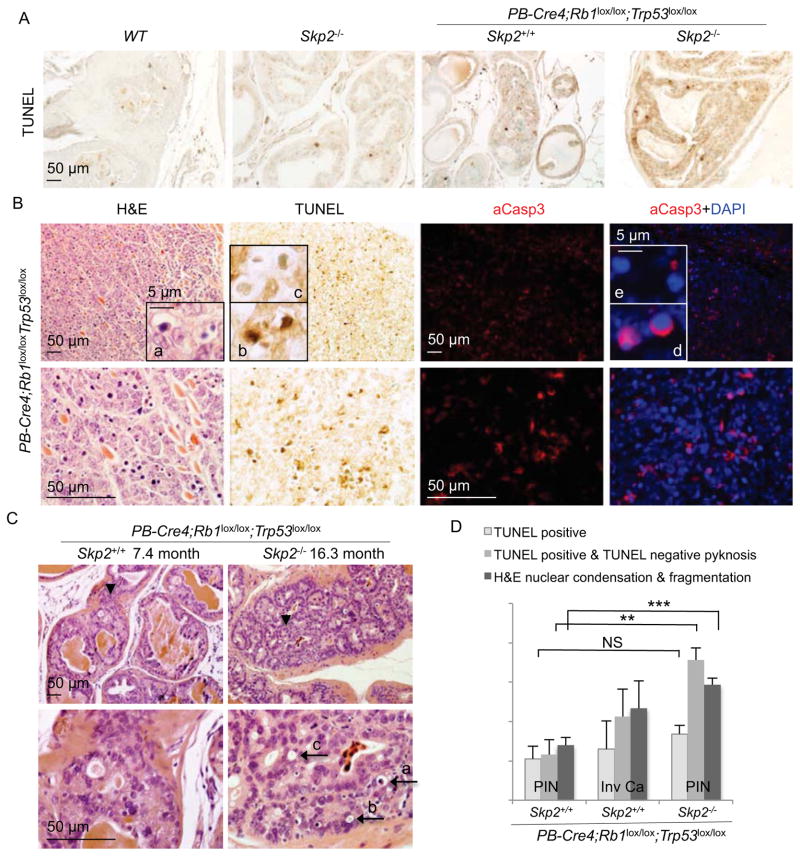
To determine whether this block to tumorigenesis also involved cell elimination, we measured the rates of apoptosis in these PINs. TUNEL stain positive cells were about 5.3% in pRbp53DKO PINs and increased to 8.4% in Skp2KO;pRbp53DKO PINs, which is not statistically significant (Figure 8A and 8D). Invasive carcinomas showed similar rates of apoptosis as measured by TUNEL. Detection of activated caspase 3 (aCasp3) yielded similar results. We next used nuclear condensation and fragmentation morphology (also called apoptotic bodies or pyknosis) to detect apoptosis (Figure 8B insert a). We unexpectedly found that morphologically apoptotic cells were TUNEL positive (insert b) as well as TUNEL negative (insert c). Similarly, cells with condensed nuclei (by DAPI stain) were positive or negative for aCasp3 (inserts d and e). After we confirmed the abilities of our TUNEL and aCasp3 staining to detect apoptosis by examining intestine villi (Figure S7), we re-evaluated apoptosis combining TUNEL stain with morphological assessment. The results (Figure 8C and 8D) show that apoptosis rates remained similar with determination by TUNEL alone for DKO PINs, but increased by two fold in invasive cancer and TKO PINs. Areas with as much as 20% apoptotic cells were frequently observed in TKO PINs. Black arrows a, b, and c in Figure 8C point to a typical step-wise process of late apoptosis leading to the elimination of the cell. The block of pRb and p53 doubly deficient prostate tumorigenesis in Skp2−/− mice therefore was not solely due to cytostatic mechanisms. Apoptotic elimination of blocked Skp2KO;pRbp53DKO PIN cells could contribute to the life-long block of tumorigenesis by reducing the possibility for emergence of resistant cells by, for example, epigenetically silencing p27 expression.
Figure 8. Blocked Skp2−/−;PB-Cre4;Rb1lox/lox;Trp53lox/lox PIN cells succumb to apoptosis.
(A) TUNEL staining of prostate sections of the indicated genotypes.
(B) Comparison of apoptosis detection by nuclear morphology (insert a), TUNEL stain (inserts b and c), and aCasp3 and DAPI costains (inserts d and e) in a microscopic tumor.
(C) Representative H&E stained prostate PIN section of the indicated genotypes. Lower panels show higher magnification views of areas pointed by black arrowheads in the corresponding upper panels. Black arrows point to typical apoptotic morphologies, as described in text.
(D) Quantification of apoptosis rates in the indicated lesions by three different methods as marked. Quantitative data are presented as average +/− SEM. Student’s t test was used for statistical analysis. **, p < 0.01; ***, p < 0.002; NS, p >0.05.
See also Figure S7.
Discussion
Tumorigenesis is a multistep process. Because of the cells’ intrinsic antitumor safeguards, first oncogenic events seldom succeed. pRb and p53 tumor suppressors are major effectors of antitumor safeguards and are therefore frequently inactivated in cancer. To learn how to treat cancers after all the antitumor mechanisms exemplified by pRb and p53 have been exhausted, we conducted the current study. To completely and irreversibly inactivate pRb and p53, we deleted the genes encoding them.
In this context, we discovered that deletion of Skp2 unmasked a Trp53 deletion induced safeguard to elevate p27 protein levels. In the absence of Skp2, loss of activated p53 shrinks the pool of p27 ubiquitin ligases further by reducing expression of p27 E3s Pirh2 and KPC1 and, therefore, accumulates p27 protein further. This rise of p27 in Skp2KO;p53KO cells effectively inhibited S phase to the extreme of cellular senescence. Critically, additional deletion of Rb1 disabled this senescence antitumor mechanism. Not only DNA replication but also re-replication became uninhibitable even in the presence of high levels of p27. These findings add a dramatic piece of evidence for the importance of keeping at least one of these two major tumor suppressors for the prevention and treatment of cancer.
Unexpectedly however, our study consistently show that pRb and p53 doubly deficient prostate tumorigenesis in the absence of Skp2 was blocked lastingly. This unique co-existence of BrdU-labeling neoplastic lesions and a tumor-free life generated a counterintuitive preclinical model of complete block to tumorigenesis. In addition to revealing a vulnerability of pRb and p53 doubly deficient tumors, this model suggests an unconventional concept for diagnosis, treatment, and management for a worst type of cancer. At surface, detection of abundant proliferation markers would immediately lead to the diagnosis of a more aggressive and poorer prognostic cancer, while in fact they could be lastingly blocked neoplastic lesions.
Up to now, tumor block is associated with dramatic reduction of proliferation markers down to the extremes of cellular senescence. While ostensibly elevated to block tumorigenesis, cellular senescence, the most studied safeguard, could also secrete cytokines and growth factors to promote tumorigenesis (Coppe et al., 2010; Krtolica et al., 2001; Kuilman and Peeper, 2009). Recent evidence showed that oncogenic Nras induced senescence in hepatocytes to induce liver cancer when the senescence cells were not cleared (Kang et al., 2011); and doxorubicin treated Wnt1-induced mammary tumors more effectively in the absence of p53 induced cellular senescence than in its presence (Jackson et al., 2012).
Persistent DNA re-replication could provide different ways to overcome tumor block and promote tumorigenesis. DNA re-replication is logically a source for gene amplifications and genomic instability. On an on-going basis, these changes can generate new mutations that inhibit p27 expression or promote p27 protein degradation and/or nuclear export to disable the block to pRb and p53 doubly deficient tumorigenesis. This scenario is expected to lead to rapid tumorigenesis since pRb and p53 are already inactivated. The fact that this scenario had not materialized up to the old age of 22 months of mouse life is surprising. It is however possible that this blocking mechanism for pRb and p53 doubly deficient tumorigenesis will only delay tumor progression to end stage within the longer timeframes of typical human cancers. On the other hand, our findings may nevertheless suggest that Skp2 inactivation also stabilized the genome via mechanisms yet to be identified. Finally, further studies are required to determine the effects of inhibiting Skp2 on established tumors that are caused by inactivation of pRb and p53 but likely have incurred additional mutations during tumorigenesis.
Targeting Skp2 to treat cancer has already been actively pursued. The finding that Skp2 deletion can induce p53-independent senescence (Lin et al., 2010) broadens its application to p53 deficient cancers and a recent report has provided a Skp2 inhibitor with this ability pre-clinically (Chan et al., 2013). Our study now broadens its application further to include pRb and p53 doubly deficient cancers and sheds more light on the design of Skp2 inhibitors. Protein degradation in the ubiquitin proteasome system (Ciechanover, 2005) proceeds via a set of hierarchic steps starting with target-specific polyubiquitination and ending in degradation of polyubiquitinated proteins in the proteasomes. For Skp2 mediated p27 ubiquitination and degradation, the most target specific step is the interaction between SCF(CRL1)Skp2/Cks1 with T187 phosphorylated p27 and an inhibitor blocking this interface has recently been identified (Wu et al., 2012). Inhibitors targeting the interaction between Skp2 and Skp1 in SCF E3 (Chen et al., 2008) (Chan et al., 2013) blocks ubiquitination of p27 together with other substrates of SCFSkp2, but might spare substrates of SCF E3s employing other F-box proteins. Further upstream for broader substrate spectrum are inhibitors targeting the cullin subunit in all CRL E3s (Soucy et al., 2009). Inhibitors for ubiquitin conjugating enzymes (E2s) have also been identified. Targeting CDC34, which functions with CRLs, inhibited p27 ubiquitination but also a larger group of CRL substrates (Ceccarelli et al., 2011). While these above inhibitors, and their designs, all can inhibit Skp2-mediated p27 ubiquitination, with varying degrees of selectivity, they do not inhibit Pirh2 and KPC1 since they are not CRL type E3s and do not depend on CDC34 as E2 for the ubiquitination of p27.
Proteasome inhibitors, by definition, do not target specific proteins or specific groups of proteins. Impressively however, the proteasome inhibitor bortezomib was approved by FDA and has demonstrated remarkable efficacy towards multiple myeloma and mantle cell lymphoma. Stabilization of p27 might be an important mechanism for its therapeutic efficacy, although other effects caused by non-specific accumulation of proteins might also be responsible, especially for its side effects. In this respect, the proteasome inhibitor argyrin A (Nickeleit et al., 2008) exhibited antitumor activities in a p27-dependent manner. Since argyrin A is a more specific proteasome inhibitor than bortezomib (Nickeleit et al., 2008), its intriguing effects could suggest that truly specific inhibition of proteasomes could indeed effectively treat cancer “specifically” through p27 stabilization. It will be important to determine whether this is because inhibiting proteasomes disabled the whole cellular pool of p27 E3s including Skp2, Pirh2, KPC1, and likely others (Cao et al., 2011). Our study provides a basis for why pan-inhibition of p27 degradation mechanisms would be more effective than specific inhibition of Skp2 for cancer treatment.
Experimental Procedures
Mice
POMC-Cre mice (Balthasar et al., 2004), PB-Cre4 mice (Wu et al., 2001), Rb1lox/lox mice (Sage et al., 2003), Trp53lox/lox mice (Jonkers et al., 2001), Skp2+/− mice (Nakayama et al., 2000) and p27−/− mice (Fero et al., 1996) have been described. POMC-Cre mice (from B. Lowell Lab) were obtained on FVB background, Rb1lox/lox mice (from T. Jacks Lab) on C57BL6Jx129Sv background, Trp53lox/lox mice (from NCI mouse repository) on FVBx129 background, Skp2−/− mice on C57BL6Jx129Sv background, p27−/− mice (from The Jackson Laboratory) on C57BL6J background, and PB-Cre4 mice (from NCI mouse repository) on C57BL6JxCg background. Subsequent breedings did not select for or against any particular strain background. Testing genotypes were studied with control genotypes from littermates. Genotyping details are in Supplemental Information. Animals were housed in the Albert Einstein College of Medicine animal facility. Mouse procedures were reviewed and approved by Einstein Animal Care Committee, conforming to accepted standards of humane animal care. Mouse pathology studies were conducted together with Einstein Cancer Center Mouse Pathology Core.
MEFs, human breast cancer cell lines, and treatments
MEFs were prepared from E13.5 embryos and cultured in DMEM containing 10% FBS. Parallel infections with Adeno-Cre or Adeno-GFP were used to delete or mock-delete Rb1 or/and Trp53 in MEFs containing floxed Rb1 or/and floxed Trp53. Controls also included infection with Adeno-Cre of MEFs without floxed Rb1 or Trp53. Knockdown or overexpression was by lentiviral transduction and drug selection with puromycin (#BP2956-100, Fisher Scientific) or blasticidin (#ant-bl-1, InvivoGen). DNA damage response was generated by treatment with 1 μM doxorubicin (#BP2516-1, Fisher Scientific). Human breast cancer cell lines MDA-MB468, BT549, Hs578T and HCC1143 (from ATCC) were cultured in RPMI-1640 medium containing 10% FBS, 1% Penicillin/Streptomycin, and 1% Glutamine. Knockdown of Skp2 by microRNA was achieved by lentiviral transuction at near 100% efficiency. See Supplemental Information for details.
SA-β-gal stain for senescence, FACS for DNA content and BrdU labeling, and cell number counting for proliferation determination
Standard protocols were used; details are in Supplemental Information.
Western blots, RT-qPCR, and ChIP
Antibodies used for Western blots are Skp2 (H435), cyclin A (C-19), p21 (C-19), Pirh2 (FL261), PCNA (PC10) and Cdk2 (C-19) from Santa Cruz Biotechnology. Mouse anti-p27 was from BD Bioscience (#610242), mouse anti-p53 from Cell signaling Technology (1C12) and Santa Cruz Biotechnology (SC-126). Antibody to α-Tubulin was from Sigma-Aldrich (T6074). Antibody to KPC1 was described previously (Kamura et al., 2004). See Supplemental Information for experimental details and sequences of primers.
Cycloheximide (CHX) chase and serum starvation-release
For protein stability analysis, MEFs were plated into 60 mm dishes at 70% 80% confluence. CHX (#239764, Calbiochem) was added at 50 μg/ml. At the indicated time points, cell extracts were prepared and Western blotted. Quantification of protein levels was performed by Image J software. Serum starvation of MEFs was in 100 mm dishes at 80–90% confluence in DMEM (1% Pen-Strep, 1% Glutamine) with 0.2% FBS. After 72 hr, cells were aliquoted into 60 mm dishes and restimulated with DMEM (1% Penicillin/Streptomycin, 1% Glutamine) containing 10% FBS.
IHC, IF, TUNEL, BrdU labeling
Antibodies included: PCNA (PC10, Santa Cruz Biotechnology), BrdU (Ab-2, Calbiochem), phospho-Histone H3 (Cell Signaling Biotechnology), Ki67 (SP6, Vector), activated caspase 3 (#9664S, Cell Signaling), mouse anti-p27 (#610242, BD Bioscience) and rabbit anti-p27 (ab92741, Abcam). TUNEL staining was performed with an Apoptosis Detection Kit (Millipore, S7100). For BrdU labeling, mice were injected with 0.4% BrdU (B5002, Sigma-Aldrich) at 25 μl/gram of body weight 2 hr before sacrifice. See Supplemental Information for experimental details.
Statistical analysis
In the survival analysis, difference in Kaplan-Meier survival curves was analyzed by log-rank test (Prism 6 Software). Differences in Ki67, phospho-histone H3, BrdU, and TUNEL positive cells between indicated samples were analyzed by Student’s t-test. p<0.05 is considered as statistically significant.
Supplementary Material
Significance.
Since pRb and p53 together are responsible for most antitumor mechanisms, their combined loss is frequent in cancers and explains in large part why cancers are difficult to treat. We discovered that deleting Skp2 in mice allowed a p27 safeguard to be activated by the loss of p53 and pRb to block the otherwise aggressive tumorigenesis. This block to tumorigenesis lasted well into mouse old age. A counterintuitive feature of this block is that DNA synthesis persisted in the blocked neoplastic cells before they were eliminated by apoptosis. Thus, pRb and p53 doubly deficient tumors might be effectively treated by Skp2 inhibitors but inhibition of DNA synthesis might not be a useful diagnostic criterion as traditionally practiced.
Highlights.
p53 transactivates two of the three p27 ubiquitin ligases
In the absence of Skp2, deletion of p53 triggers a p27 safeguard
Abundant p27 inhibits cell division, not DNA synthesis, in the absence of p53 and pRb
BrdU-labeling pRb and p53 deficient lesions are prevented from tumorigenesis
Acknowledgments
This work was supported by NIH grants RO1CA127901 and RO1CA131421 (LZ). Albert Einstein Comprehensive Cancer Research Center (5P30CA13330) and Liver Research Center (5P30DK061153) provided core facility support. We thank Dr. Sarah Schweber of the Oncology Division for suggestions of human breast cancer cell lines, and Dr. Jinghang Zhang of Einstein FACS facility for assistance in using iCys® Research Imaging Cytometer and iCys® Cytometric Analysis Software. HZ is a recipient of DOD PCRP Postdoctoral Fellowship (PC121837), and LZ was a recipient of the Irma T. Hirschl Career Scientist Award.
Footnotes
Author contributions: HZ and LZ conceived and designed the research; HZ, FB, HF, ZL and JC performed the research; KN and KN provided Skp2 mutant mice and KPC1 antibody; HZ, JL and LZ analyzed the data; HZ and LZ wrote the manuscript; and all authors reviewed the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Balthasar N, Coppari R, McMinn J, Liu SM, Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC, Jr, Elmquist JK, et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–891. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein ML, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008;105:11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nature reviews Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Xue L, Han L, Ma L, Chen T, Tong T. WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) delays cellular senescence by promoting p27(Kip1) degradation in human diploid fibroblasts. J Biol Chem. 2011;286:33447–33456. doi: 10.1074/jbc.M111.225565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, et al. Pharmacological Inactivation of Skp2 SCF Ubiquitin Ligase Restricts Cancer Stem Cell Traits and Cancer Progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–387. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annual review of pathology. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nature Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorignesis, and female sterility in p27Kip1-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- Flesken-Nikitin A, Choi KC, Eng JP, Shmidt EN, Nikitin AY. Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res. 2003;63:3459–3463. [PubMed] [Google Scholar]
- Hattori T, Isobe T, Abe K, Kikuchi H, Kitagawa K, Oda T, Uchida C, Kitagawa M. Pirh2 promotes ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. Cancer Res. 2007;67:10789–10795. doi: 10.1158/0008-5472.CAN-07-2033. [DOI] [PubMed] [Google Scholar]
- Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, Tavana O, Yang P, Manshouri T, Li Y, et al. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Deng T, Jones R, Li H, Herschkowitz JI, Liu JC, Weigman VJ, Tsao MS, Lane TF, Perou CM, et al. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120:3296–3309. doi: 10.1172/JCI41490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004;6:1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- Kiyokawa H, Kineman RD, Manova-Todorova K, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27Kip1. Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nature reviews Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- McClendon AK, Dean JL, Ertel A, Fu Z, Rivadeneira DB, Reed CA, Bourgo RJ, Witkiewicz A, Addya S, Mayhew CN, et al. RB and p53 cooperate to prevent liver tumorigenesis in response to tissue damage. Gastroenterology. 2011;141:1439–1450. doi: 10.1053/j.gastro.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K-i. Mice lacking p27Kip1 display incresased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama TI, shida N, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T, Nakayama KI. Skp2-Mediated Degradation of p27 Regulates Progression into Mitosis. Dev Cell. 2004;6:661–672. doi: 10.1016/s1534-5807(04)00131-5. [DOI] [PubMed] [Google Scholar]
- Nickeleit I, Zender S, Sasse F, Geffers R, Brandes G, Sorensen I, Steinmetz H, Kubicka S, Carlomagno T, Menche D, et al. Argyrin a reveals a critical role for the tumor suppressor protein p27(kip1) in mediating antitumor activities in response to proteasome inhibition. Cancer Cell. 2008;14:23–35. doi: 10.1016/j.ccr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Old JB, Kratzat S, Hoellein A, Graf S, Nilsson JA, Nilsson L, Nakayama KI, Peschel C, Cleveland JL, Keller UB. Skp2 directs Myc-mediated suppression of p27Kip1 yet has modest effects on Myc-driven lymphomagenesis. Mol Cancer Res. 2010;8:353–362. doi: 10.1158/1541-7786.MCR-09-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Walls JE, Galvez JJ, Kim M, Abate-Shen C, Shen MM, Cardiff RD. Prostatic intraepithelial neoplasia in genetically engineered mice. The American journal of pathology. 2002;161:727–735. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Ink4-Arf Locus in Cancer and Aging. Wiley interdisciplinary reviews. Developmental biology. 2012;1:731–741. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Sun D, Melegari M, Sridhar S, Rogler CE, Zhu L. A multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. BioTechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, Sellers RS, Nakayama K, Nakayama KI, Cobrinik D, et al. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/− mice. Nat Genet. 2010;42:83–88. doi: 10.1038/ng.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chemistry & biology. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, Sangiorgi FO, Maxson RE, Sucov HM, Roy-Burman P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mechanisms of development. 2001;101:61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- Xuan Z, Zhao F, Wang J, Chen G, Zhang MQ. Genome-wide promoter extraction and analysis in human, mouse, and rat. Genome Biol. 2005;6:R72. doi: 10.1186/gb-2005-6-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, Nikitin AY. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006;66:7889–7898. doi: 10.1158/0008-5472.CAN-06-0486. [DOI] [PubMed] [Google Scholar]
- Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.