Abstract
Leukemia cells are protected by various components of its microenvironment, including marrow stromal cells (MSCs). To understand the molecular mechanisms underlying this protection, we cultured acute lymphoblastic leukemia (ALL) cells with MSCs and studied the effect of the latter on the molecular profiling of ALL cells at the mRNA and protein levels. Our results indicated that activated Wnt signaling in ALL cells is involved in MSC-mediated drug resistance. Blocking the Wnt pathway sensitized the leukemia cells to chemotherapy and improved overall survival in a mouse model. Targeting the Wnt pathway may be an innovative approach to the treatment of ALL.
Keywords: Acute lymphoblastic leukemia, bone marrow stromal cells, Wnt pathway, Wnt pathway inhibitor, chemotherapy resistance
1. Introduction
Acute lymphoblastic leukemia (ALL), one of the most aggressive hematological malignancies, affects patients of all ages [1]. Recent advances in our understanding of this disease and the development of new therapies have greatly improved outcomes in ALL patients. Nevertheless, a substantial number of ALL patients experience disease relapse and treatment resistance, leading to death from the disease. Although large prospective clinical trials have reported complete remission rates of 78% to 93% in adults, only about 40% of patients experience long-term disease-free survival [2]. Minimal residual disease, which is strongly associated with relapse, is a major challenge in treating ALL [3].
A growing amount of evidence suggests that disease relapse and treatment resistance in ALL patients are largely due to the protection of leukemia cells by various components of the bone marrow microenvironment, especially marrow stromal cells (MSCs) [4]. It was shown that primary ALL cells cultured in vitro underwent spontaneous apoptosis, whereas ALL cells cultured with MSCs in a serum-free environment underwent less spontaneous apoptosis [5]. Indirect communication through extracellularly secreted growth factors [6] and direct contact between leukemia cells and MSCs appear to be essential to ALL cell survival and chemoresistance [7]. The interaction between ALL cells and MSCs may trigger molecular changes that lead ALL cells to chemotherapy resistance.
Multiple pathways and factors are involved in the protection of leukemia cells by MSCs. For example, CXCR4, which is expressed on leukemia blasts and interacts with CXCL12 (which is secreted by MSCs), was found to be necessary for leukemia cell survival and proliferation [6], and MSC-induced activation of phosphoinositide 3-kinase (PI3K)/Akt signaling plays a critical role in the chemoresistance of acute myeloid leukemia [8]. Aberrant activation of the Wnt pathway has been observed in hematological malignancies [9; 10]. However, the role of Wnt pathway in the cross-talk between ALL cells and MSCs is uncertain and the effect of Wnt pathway inhibition on ALL development and progression in vivo needs to be assessed.
In the present study, we determined the effects of human and murine MSCs on the survival and drug resistance of human ALL cell lines and primary ALL cells. Our results indicated that MSCs induce activation of the Wnt pathway in ALL cells and that this activation contributes to the survival of ALL cells. An important finding was that blocking the Wnt pathway with a pharmacologic inhibitor partially overcame MSC protection of ALL cells both in vitro and in vivo.
2. Materials and methods
2.1. Cell culture
This study was approved by the institutional review board of The University of Texas MD Anderson Cancer Center. Residual peripheral blood samples of 9 patients with ALL were obtained from an institutional tumor bank with informed consent. Peripheral blood mononuclear cells were isolated using Ficoll-HyPaque (Sigma, St. Louis, MO, USA). All experiments were carried out using freshly isolated cells.
The human leukemia cell lines Reh [11], RS4;11 [12], and SEMK2 [13] were provided by Dr. Patrick Zweidler-McKay at the MD Anderson Cancer Center. The human MSC line HS was derived from bone marrow and immortalized using human telomerase reverse transcriptase (a gift from Dr. Jianguo Wen at Methodist Hospital, Houston, TX), the human MSC line HS-5 [14] was obtained from the American Type Culture Collection (Manassas, VA, USA), and the murine MSC line M2-10B4 [15] was provided by Dr. Jan Burger at MD Anderson. All cells were cultured in RPMI 1640 medium supplemented with 2.05 mM L-glutamine (HyClone, Logan, UT, USA), 10% fetal bovine serum (Abnova, Walnut, CA, USA), and penicillin-streptomycin (Lonza, Walkersville, MD, USA) at 37°C in 5% CO2 in a humidified incubator. For co-culture with HS-5 cells, 1% sodium pyruvate was added.
2.2. Co-culture of ALL cells with MSCs
MSCs were seeded on 12-well plates at a concentration of 5×104 cells/ml/well. The next day, ALL cells were added to the confluent MSC layer at a ratio of 10:1. After the ALL cells adhered to the MSC layer, cytarabine (Ara-C; Sigma) was added, with or without the β-catenin inhibitor XAV939 (Sigma). At the indicated times ALL cells were collected, leaving the adherent stromal layer intact, and washed with phosphate-buffered saline (PBS) for subsequent analyses.
2.3. Rehluc and Reh7TGC cell generation
Luciferase cDNA was generated from pGL4.10 (Promega, Madison, WI, USA) using polymerase chain reaction (PCR) and subcloned into the green fluorescent protein (GFP)-tagged plasmid pMIG (Addgene, Cambridge, MA, USA). Recombinant pMIG-GFP/luciferase, pUMVC, and pMD2.G (generously provided by Dr. Mani Sendurai at MD Anderson) were co-transfected into 293T packaging cells using Fugene 6 (Roche, Indianapolis, IN, USA) to generate GFP/luciferase-expressing retrovirus. Reh cells were transduced using retroviral supernatants, and 48 hours later GFP-expressing Rehluc cells were sorted using a FACSAria flow cytometer (BD Biosciences, San Diego, CA, USA). For determining Wnt/β-catenin signaling activity, 7xTcf-eGFP/SV40-mCherry reporter lentivirus was prepared by co-transfecting 293T cells with 7TGC (from Dr. Roel Nusse at Stanford University) vector along with packaging plasmids, pPAX2 and pMD2.G. Viral supernatants were harvested 72 hours after the transfection and used to transduce Reh cells for generating Reh7TGC cells.
2.4. Proliferation analysis
MSCs were seeded onto 96-well plates at a concentration of 2×103 cells/well. The next day, Rehluc cells were cultured at a density of 2×104 cells/well, with or without a confluent MSC layer. At the indicated times, 150 µg of D-luciferin (Caliper LifeSciences, Hopkinton, MA, USA) was added to each well. Luminescence was determined using a MicroBeta Jet microplate scintillation and luminescence counter (PerkinElmer, Shelton, CT, USA).
2.5. Apoptosis analysis
ALL cells were stained with fluorescein isothiocyanate-annexin V and propidium iodide (PI) (both from BD Biosciences) according to the manufacturer’s instructions. Flow cytometric analysis was performed using a FACSCalibur flow cytometer (Beckman Coulter, Miami, FL, USA) and the FlowJo software program (TreeStar, Ashland, OR, USA). Cells that were annexin V- and PI-positive were considered apoptotic.
2.6. Cell cycle analysis
ALL cells were fixed in 70% ethanol overnight at 4°C and then stained with PBS containing 50 µg/ml PI, 0.2% Tween 20, and 1 mg/ml RNase at 4°C overnight. Samples were analyzed using a FACSCanto flow cytometer (BD Biosciences) and the FlowJo software program. Cells in the sub-G1 phase were considered apoptotic, and cells in the S+G2M phase were considered proliferative.
2.7. Western blot analysis
ALL cells were lysed on ice for 30 minutes in lysis buffer containing 150 mM NaCl, 1% Triton X-100, 5 mM ethylenediaminetetraacetic acid, 10 mM Tris-Cl (pH 7.4), 5 mM dithiothreitol, 0.1 mM phenylmethanesulfonyl flouride, and the complete protease inhibitor cocktail (Roche). Cell lysates were centrifuged at 13,000 g for 15 minutes at 4°C, and the supernatant was stored at −80°C until use. Thirty micrograms of protein was separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred onto polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked in PBS-Tween containing 5% nonfat dried milk, incubated with primary antibodies overnight, and then incubated with a secondary antibody (diluted 1:3000) conjugated with species-specific horseradish peroxidase for 2 hours. Antibodies against GSK3β, phosphorylated GSK3β, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), cyclin A, cyclin D1, cyclin E, Cdk2, and Cdk6 were from Santa Cruz Biotechnology (Santa Cruz, CA, USA), β-catenin from BD (NJ, USA), and all others were from Cell Signaling Technology (Danvers, MA, USA). Immunoreactive bands were visualized using an enhanced chemoluminescence detection system (Pierce Biotechnology, Rockford, IL, USA). Signal intensity was quantified with Quantity One software (Bio-Rad).
2.8. RNA isolation and microarray hybridization and analysis
Reh cells cultured with or without HS or HS-5 MSCs and treated with Ara-C for 48 hours were collected. Independent triplicate samples were used in the experiments. Total RNA was prepared using TRI reagent (Ambion, Austin, TX, USA). Biotin-labeled complementary RNA was synthesized using the Illumina TotalPrep RNA amplification kit (Ambion). Complementary RNA was then hybridized to the HumanHT-12 v3 Expression BeadChip (Illumina, San Diego, CA, USA) according to the manufacturer's instructions, and the bead chips were scanned with a BeadArray Reader (Illumina). Microarray data were normalized using the quantile normalization method in the Linear Models for Microarray Data package in the R language, and the expression values of each gene were log2-transformed for further analysis. BRB-ArrayTools was used for statistical analysis of gene expression data. The t-test was applied to identify the differentially expressed genes between 2 groups. Cluster analysis was performed using the Cluster software program, and a heatmap was generated using the Treeview software program. Microarray data analysis for identification of pathways was performed using IPA software version 6.3 (Ingenuity Systems, Redwood, CA, USA). Primary microarray data are available through the GeneExpression Omnibus of the National Center for Biotechnology Information at http://www.ncbi.nlm.nih.gov/geo/ (Accession number: GSE36372).
2.9. Reverse-phase protein array (RPPA)
Reh cells were lysed in lysis buffer (1% Triton X-100, 150 nmol/L NaCl, 50 nmol/L HEPES [pH 7.4], 1 mmol/L ethylene glycol tetraacetic acid, 1.5 nmol/L MgCl2, 100 nmol/L NaF, 10 nmol/L NaPPi, 10% glycerol, 1 nmol/L phenylmethylsulfonyl fluoride, 10 µg/mL aprotinin, and 1 nmol/L Na3VO4) for 30 minutes, with frequent vortexing on ice, and centrifuged for 15 minutes at 14,000 rpm. The cell lysate supernatants were collected and stored at −80°C until use. RPPA experiments were performed by the RPPA laboratory in the Department of Molecular Therapeutics at MD Anderson [16].
2.10. Quantitative reverse transcription PCR (qRT-PCR)
Total cellular mRNA was extracted using Trizol reagent (Takara, Liaoning, China). Reverse transcription was performed using a PrimeScript RT reagent kit with gDNA Eraser kit (Takara). Quantitative PCR was performed according to the instructions of SYBR Premix Ex Taq II (Takara) with the ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA, USA). Primer sets used for these analyses are listed in Supplementary Table 1.
2.11. ALL cell transplantation and treatment in vivo
The use of mice in this study was approved by the Institutional Animal Care and Use Committee of MD Anderson. NOD/SCID mice were purchased from The Jackson Laboratory (Sacramento, CA, USA) and bred and maintained in a specific pathogen-free barrier facility at MD Anderson. Reh cells (4×106) were transplanted via the tail vein into 6- to 8-week-old mice that had been sublethally irradiated (250 cGy) 12 to 24 hours prior to transplantation. Rehluc cell expansion in vivo was monitored by bioluminescent imaging (BLI) using the IVIS100 imaging system (Caliper LifeSciences). Images were acquired after intraperitoneal injection of 150 mg/kg D-luciferin 5 days after transplantation and then every 3 days. Chemotherapy was initiated when clear Rehluc signals were detected. Cohorts of mice were randomly assigned to 1 of 4 groups: vehicle control (n = 5), Ara-C (n = 5), XAV939 (n = 6), or Ara-C plus XAV939 (n = 6). Ara-C (60 mg/kg) and XAV939 (20 mg/kg) were administered intraperitoneally for 3 consecutive days. Treatment effects were monitored by imaging, peripheral blood smear analysis, and flow cytometry analysis. Mice were euthanized with CO2 at the terminal stage of disease (moribund appearance).
2.12. Statistical analysis
Statistical analyses were performed using GraphPad Prism for Windows version 5.00 (GraphPad Software, La Jolla, CA, USA). Statistical differences between mean values were evaluated using the 2-tailed Student’s t-test (paired or unpaired, as appropriate). A p value < .05 was considered statistically significant.
3. Results
3.1. MSCs protected ALL cells from spontaneous and Ara-C-induced apoptosis
To establish an MSC-ALL cell co-culture system, we cultured 3 leukemia cell lines (Reh, RS4;11, and SEMK2) with or without MSCs in the presence or absence of 1 µM Ara-C. At 24, 48, and 72 hours, the mean apoptotic rates of Reh cells treated with Ara-C in medium alone were 16.6±5.1%, 42.7±7.9%, and 58.1±2.1%, respectively; those of Reh cells treated with Ara-C in the presence of the murine MSC line M2-10B4 were 8.9±2.1%, 13.2±2.7%, and 33.1±5.1%, respectively; and those of Reh cells treated with Ara-C in the presence of the human MSC line HS were 4.8±0.5%, 9.9±4.1%, and 16.2±4.3%, respectively (Fig. 1A). Similar protection by MSCs was observed in RS4;11 and SEMK2 ALL cells. ALL cells cultured with or without MSCs in the absence of Ara-C showed minimal apoptosis.
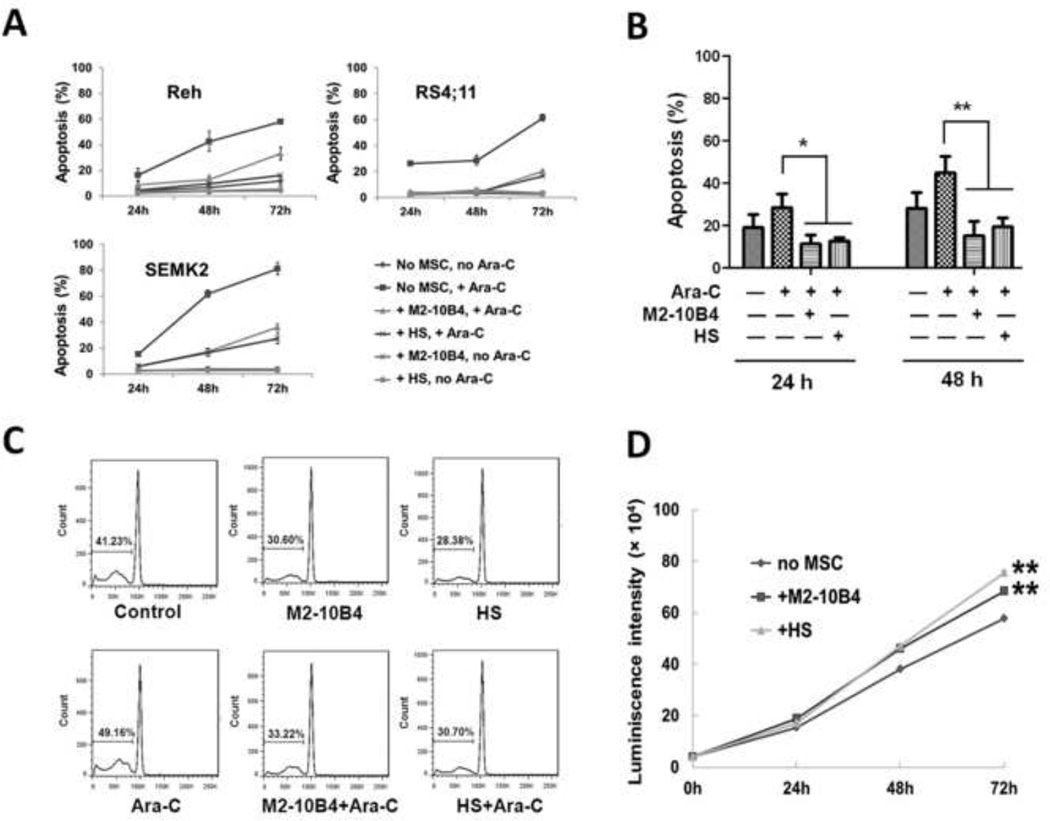
Fig. 1. Effect of MSCs on apoptosis and proliferation of ALL cells.
(A) MSCs decreased apoptosis in human ALL cell lines. Apoptosis was measured using flow cytometry after annexin V staining at 24, 48, and 72 hours. The concentration of Ara-C was 1 µM. ALL cell lines Reh, RS4;11, and SEMK2 were cultured with or without MSCs (M2-10B4 or HS cells). (B) MSCs decreased apoptosis in patient ALL cells ex vivo. Apoptosis was measured using flow cytometry after annexin V staining at 24 and 48 hours. Concentration of Ara-C was 1 µM. Data were obtained by testing blood samples from patients 1–6. *p < .05; **p < .01. (C) Cell cycle analysis of patient ALL cells cultured with or without MSCs for 48 hours and treated with or without 1 µM Ara-C. Cell cycle distribution was determined using flow cytometry after PI staining. Percentages of cells in the sub-G1 phase are indicated. Few primary ALL cells were in proliferative phases (S and G2/M). The same patterns were observed with samples from patients 7 and 8; data from patient 7 are presented. (D) MSCs promoted the proliferation of Rehluc cells. Rehluc cells were cultured with or without MSCs. Luminescence intensity, which was proportional to the number of viable Rehluc cells, was determined at 0, 24, 48, and 72 hours. **p < .01.
The level of protection from Ara-C (1 µM)-induced apoptosis provided by murine MSCs (M2-10B4 cells) was generally lower than those provided by human MSCs (HS cells). Ara-C did not induce substantial apoptosis in the MSCs. Thus, the MSCs treated with Ara-C remained as viable as the MSCs not treated with Ara-C (data not shown). We also treated Reh cells with daunorubicin, and similar to what we observed with cells treated with Ara-C, MSCs protected the leukemia cells from daunorubicin-induced apoptosis (data not shown).
Next, we determined the effect of MSCs on Ara-C-induced apoptosis in primary leukemia cells from patients with ALL. Peripheral blood mononuclear cells, of which more than 95% were ALL cells, from 6 patients (patients 1 to 6) (Supplementary Table 2) were cultured, and apoptosis was determined by flow cytometry using annexin V and PI staining. The rates of Ara-C-induced apoptosis in ALL cells cultured without MSCs were 28.5±15.4% and 44.7±19.2% after 24 and 48 hours, respectively (Fig. 1B). In leukemia cells cultured with M2-10B4 cells the corresponding rates were 11.4±9.7% and 15.2±16.3% (p < .05 and p < .01), and in leukemia cells cultured with HS cells the rates were 12.6±4.2% and 19.3±10.5% (p < .05 and p < .01). Cell cycle analysis of primary ALL cells also revealed that the presence of MSCs led to a decrease in apoptosis. The percentage of sub-G1 ALL cells decreased from 41.2% at 48 hours in the control group to 30.6% in the presence of M2-10B4 cells and 28.4% in the presence of HS cells (Fig. 1C). The same pattern emerged with the addition of Ara-C (49.2%, 33.2%, and 30.7, respectively). These data suggest that MSCs provide ALL cells with substantial protection from spontaneous and Ara-C-induced apoptosis, confirming the annexin V data (see Fig. 1B).
To determine the effect of MSCs on ALL cell proliferation, we introduced luciferase into Reh cells (Rehluc), cultured the Rehluc cells with MSCs, and measured luminescence intensity. Rehluc cell proliferation was significantly increased after culture with HS cells (p < .01) or M2-10B4 cells (p < .01) for 72 hours (Fig. 1D). Increased cell proliferation in the presence of MSCs was confirmed with cell cycle analysis. At 24, 48, and 72 hours, the percentage of proliferative S+G2M ALL cells was higher when ALL cells were cultured with M2-10B4 cells or HS cells (Table 1).
Table 1.
Effect of MSCs on percentage of proliferative Reh cells
| Co-culture | Time (hours) | ||
|---|---|---|---|
| 24 | 48 | 72 | |
| None | 31.9 ± 1.1 | 34.7 ± 1.4 | 39.6 ± 1.6 |
| M2-10B4 | 32.5 ± 0.7 | 38.8 ± 1.0 | 46.2 ± 2.1 |
| HS | 34.5 ± 1.5 | 38.7 ± 0.9 | 46.2 ± 1.4 |
Data are percentage of Reh cells in S+G2M phase of cell cycle following culture with or without MSCs for indicated number of hours. Triplicate tests were performed with specimen from patient 9
3.2. MSC protection of ALL cells from apoptosis was associated with decreased cleavage of apoptotic proteins and a modified cell cycle response
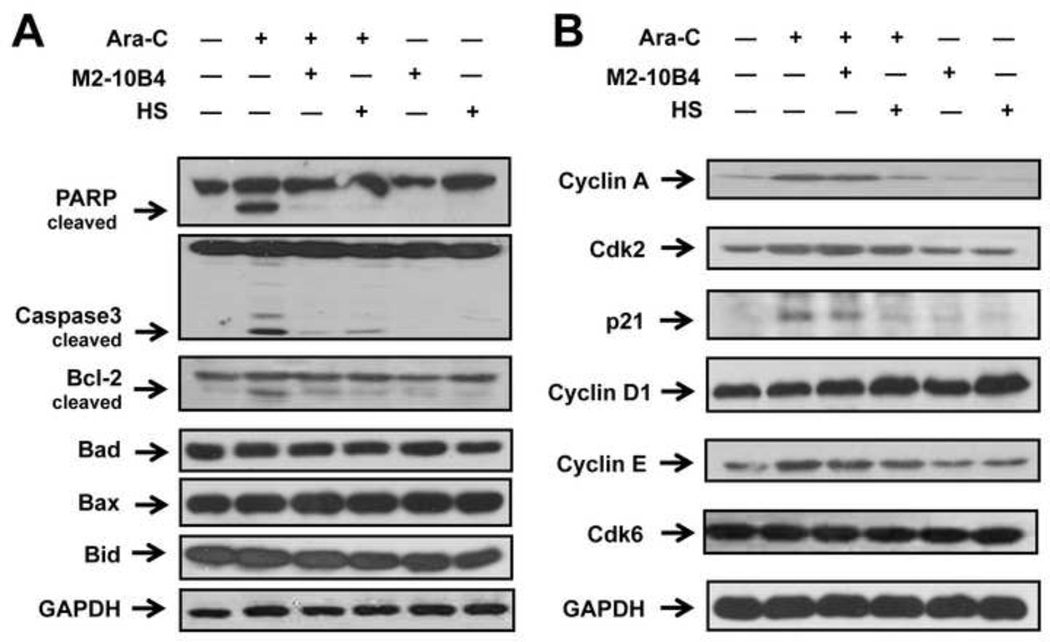
Because poly(RDP-ribose) polymerase (PARP) and caspase-3 are involved in apoptosis, we determined their cleavage status in Reh cells cultured with or without MSCs and treated with Ara-C. Western blot analysis showed that treatment of Reh cells with Ara-C alone resulted in cleavage of PARP (89 kDa) and caspase-3 (17 kDa) (Fig. 2A). When Reh cells were cultured with MSCs, cleavage diminished by 92% and 84%, respectively, in the presence of human M2-10B4 cells and by 97% and 68%, respectively, in the presence of murine HS cells. Similar results were observed for Bcl-2 (more than 80% reduction in the presence of M2-10B4 or HS cells) but not other Bcl-2 family members (Bad, Bax, and Bid; Fig. 2A). We confirmed these findings using RS4;11 ALL cells (data not shown). These results suggest that diminished cleavage of caspase-3, PARP, and Bcl-2 by MSCs contributes to the protection by MSCs of ALL cells from Ara-C-induced apoptosis.
Fig. 2. Expression of apoptosis and cell cycle-related proteins in ALL cells cultured with MSCs.
Reh cells were cultured with or without MSCs and treated with 1 µM Ara-C for 48 hours. (A) Western blot analysis of PARP, caspase-3, Bcl-2, Bad, Bax, and Bid expression in Reh cells. (B) Western blot analysis of cyclin A, Cdk2, p21, cyclin D1, cyclin E, and Cdk6 expression in Reh cells. GAPDH was used as the loading control. Results are representative of 3 independent experiments.
Because alterations in cell cycle checkpoints can modify the chemosensitivity of malignant cells [17], we hypothesized that the presence of MSCs would modify cell cycle-related protein expression in ALL cells in response to Ara-C treatment. Western blot analysis showed that cyclin A was upregulated in Reh cells treated with Ara-C but was downregulated in Reh cells cultured with M2-10B4 cells (by 14%) or HS cells (by 73%) (Fig. 2B). Likewise, p21 protein expression was upregulated in Reh cells treated with Ara-C but was decreased in Reh cells cultured with M2-10B4 cells (by 38%) or HS cells (by 66%). Moreover, MSCs increased cyclin D1 expression (by 8% with M2-10B4 cells and 35% with HS cells). MSCs did not notably change Cdk2, cyclin E, or Cdk6 expression.
3.3. Protection of ALL cells by MSCs was associated with specific modulation in gene expression
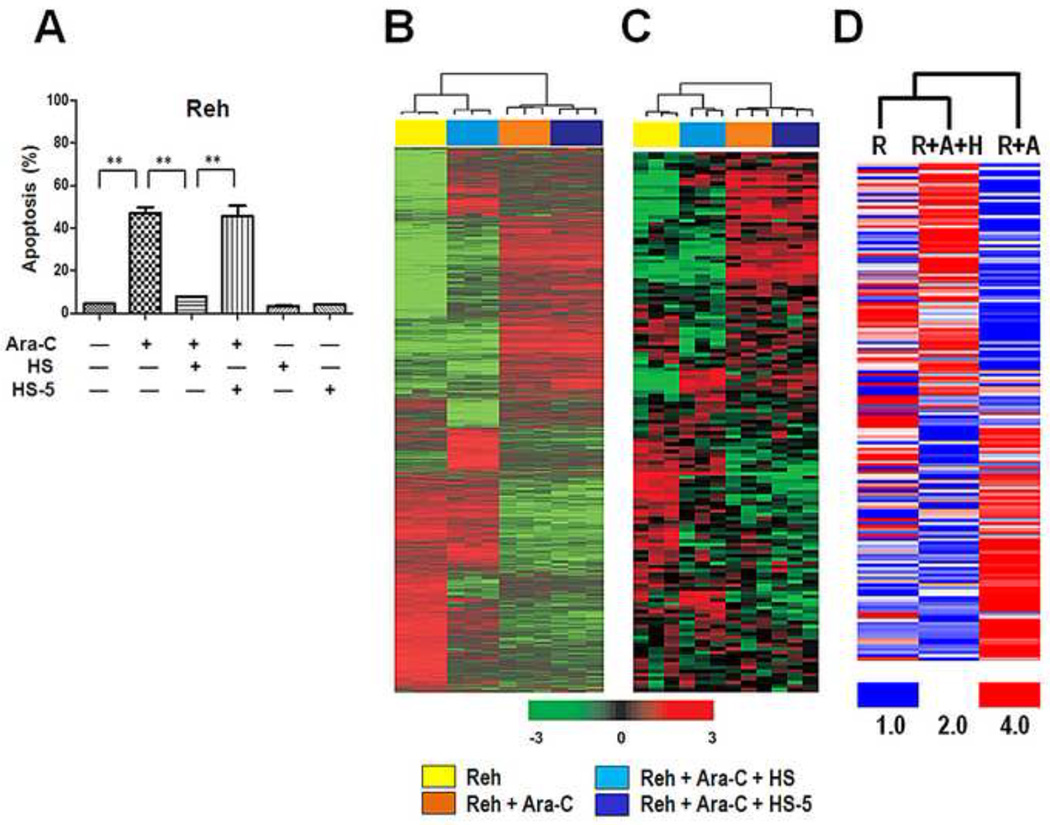
To understand the molecular alterations in ALL cells associated with drug resistance in the presence of MSCs, we performed gene expression microarray profiling and compared results from untreated Reh ALL cells, Reh cells treated with 1 µM Ara-C (Reh + Ara-C), and Reh cells treated with 1 µM Ara-C in the presence of HS cells (Reh + Ara-C + HS). Because Ara-C in the presence of human stromal cell line HS-5, in contrast to cell line HS, failed to provide any significant protection to ALL cells (Fig. 3A), we included Reh + Ara-C + HS-5 for comparison to identify genes involved in the protection of Reh cells by HS cells. Using the 2-sample t-test, this comparison revealed that 4305 genes were significantly expressed. A total of 904 genes were upregulated and 857 genes were downregulated in the Reh + Ara-C + HS group, for a fold change of ≥1.5 (data not shown). Using ingenuity pathway analysis, we found these genes to be mostly involved in cell death, the cell cycle, and cell proliferation. The microarray data were validated using semi-qRT-PCR on selected genes (data not shown).
Fig. 3. Hierarchical clustering analysis of gene expression microarray data.
(A) HS MSCs but not HS-5 MSCs decreased apoptosis in Reh cells. Apoptosis was measured using flow cytometry after annexin V staining at 48 hours. Concentration of Ara-C was 1 µM. **p < .01. (B) Gene expression microarray profiling was performed on Reh cells left untreated or treated with 1 µM Ara-C for 48 hours, with or without culture with MSCs. Three independent mRNA samples were tested for each of the 4 groups (Reh, Reh + Ara-C, Reh + Ara-C + HS, and Reh + Ara-C + HS-5). Data set that represents 5563 genes after low-variance filtering (SD > 0.25) was centralized by subtracting median expression level across samples for clustering analysis. Data are presented in matrix format in which rows represent individual gene and columns represent Reh cells at different co-culture conditions. Each cell in the matrix represents the expression level of a gene. Red and green colors reflect relative high and low expression levels, respectively, as indicated in the scale bar (log2-transformed scale). (C) Clustering analysis of 145 genes from the 254 genes that distinguished between poor and good treatment response (Table S3 in reference [18]). (D) Clustering analysis of reverse-phase protein array data. R, Reh cells; A, Ara-C; H, HS cells.
To determine the overall similarity in the gene expression profiles among the 12 sets of microarray data (3 sets for each group), we performed a hierarchical clustering analysis. As expected, initially, the 3 sets in each group were clustered. At the next level, the profiles of the Reh group were clustered with those of the Reh + Ara-C + HS group and the profiles of the Reh + Ara-C group were clustered with those of the Reh + Ara-C + HS-5 group (Fig. 3B), suggesting that HS cells diminished the pro-apoptotic effect of Ara-C and at least partly restored the original gene expression profile, whereas the HS-5 cells had minimal effect on the gene expression profile of the Reh cells in the presence of Ara-C.
We hypothesized that Reh cells cultured with the protective HS cells mimic ALL cells in patients with a high risk of relapse, whereas Reh cells cultured with the non-protective HS-5 cells would correspond to ALL cells in patients with a low risk of ALL relapse. Cario et al. [18] identified 254 genes that were distinguishable between these 2 ALL patient groups. We performed clustering analysis of 145 of the 254 genes (the other 109 genes were unidentifiable in our microarray by mapping gene symbol) and found the same clustering pattern. That is, the Reh group clustered with the Reh + Ara-C + HS group and the Reh + Ara-C group clustered with the Reh + Ara-C + HS-5 group (Fig. 3C). These clustering data indicated that our in vitro culture system with the protective and non-protective MSCs significantly correlated with clinically high risk and low risk, respectively, of disease relapse in vivo in the overall gene expression of ALL cells.
We also performed RPPA and determined the expression levels of 154 proteins that are known to be involved in apoptosis, the cell cycle, and several signaling pathways. RPPA results revealed that, in agreement with the gene expression microarray data, the overall expression pattern of Reh + Ara-C + HS cells was more similar to that of Reh cells than to that of Reh + Ara-C cells (Fig. 3D). These findings also suggest that MSCs antagonized the apoptotic effect of Ara-C and protected the ALL cells.
We thought it interesting that our microarray data showed that the expression of several members of the Wnt signaling pathway, including Lef1 (+2.53), c-Myc (+1.65), and CCNDBP1 (+1.47), was increased in Reh cells cultured with MSCs compared with in Reh cells cultured without MSCs (data not shown). This information suggests that the Wnt signaling pathway is involved in the protection by MSCs of ALL cells. RPPA analysis also revealed a slight increase in the expression of C-Myc (+1.6) in Reh cells cultured with MSCs (data not shown). These findings allowed us to further characterize the role of the Wnt pathway in the protection of ALL cells by MSCs.
3.4. MSCs activated the Wnt pathway in ALL cells
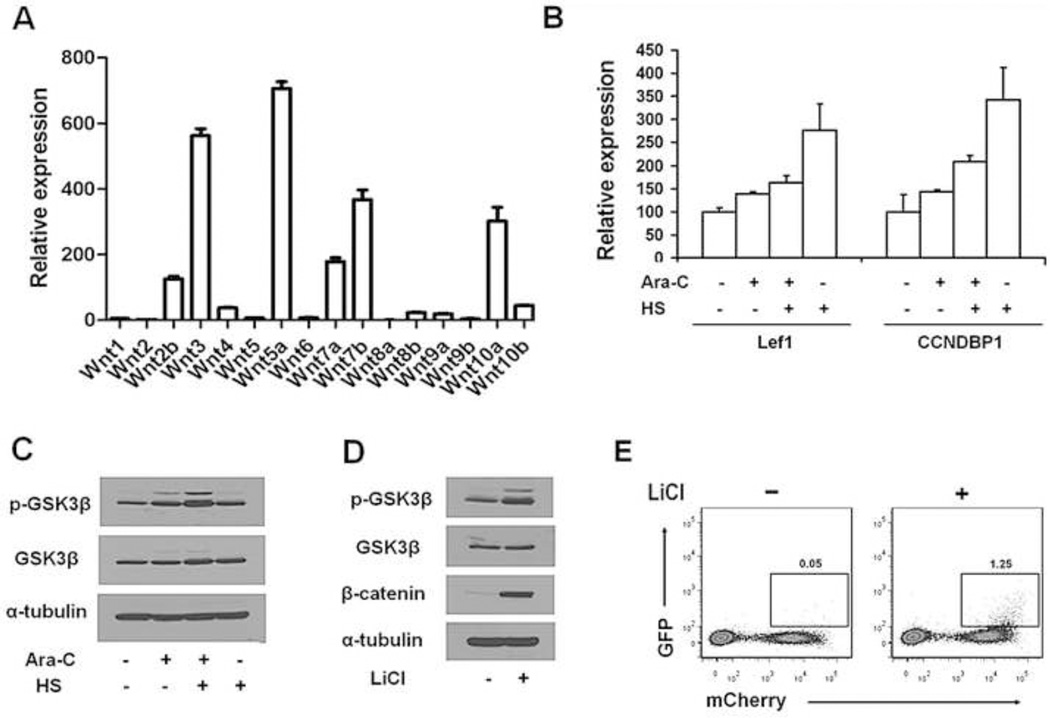
To activate the Wnt pathway in ALL cells, MSCs should express Wnt ligands. qRT-PCR detected the mRNA expression of all 16 Wnt ligands tested (Fig. 4A). The expression levels were markedly variable, with Wnt5a and Wnt3 at the highest levels and Wnt8a and Wnt2 the lowest. qRT-PCR also showed increased expression of the Wnt pathway members Lef1 and CCNDBP1 in Reh cells cultured with HS cells (Fig. 4B). Compared with untreated Reh cells in medium alone, the Wnt pathway suppressor GSK3β showed a more than 100% increase in phosphorylation in Reh cells cultured with HS cells and treated with Ara-C (Fig. 4C), an indication of GSK3β inactivation. We then demonstrated that treatment of Reh7TGC cells with the GSK3β inhibitor lithium chloride increased GSK3β phosphorylation by 130% (Fig. 4D), which was accompanied by a more than 900-fold increase in the level of β-catenin (Fig. 4D) and a 24-fold increase in Wnt signaling activity (Fig. 4E). These data suggest that GSK3β inactivation plays an important role in the activation of Wnt signaling in Reh cells.
Fig. 4. Activation of Wnt signaling in ALL cells by MSCs.
(A) Relative mRNA expression levels of Wnt pathway ligands in HS cells as determined by qRT-PCR. (B) Expression of Lef1 and CCNDBP1 mRNA in Reh cells as determined by qRT-PCR. Culture conditions and Ara-C treatment were the same as described in Figure 1A. (C) Phosphorylated (p-)GSK3β and total GSK3β in Reh cells as determined by Western blot analysis. Culture conditions and Ara-C treatment were the same as described in Figure 1A. (D) Inhibition of GSK3β with lithium chloride (LiCl) increased levels of p-GSK3β and β-catenin in Reh7TGC cells. Reh7TGC cells were cultured in the presence or absence of 10 mM LiCl for 48 hours. (E) Increased Wnt signaling activity in Reh7TGC cells in the presence of LiCl. Reh cells transduced with Wnt signaling reporter 7xTcf-eGFP/SV40-mCherry that carried mCherry as a marker were cultured for 48 hours in the presence or absence of 10 mM LiCl and analyzed by flow cytometry. Numbers indicate percentage of GFP positive cells analyzed.
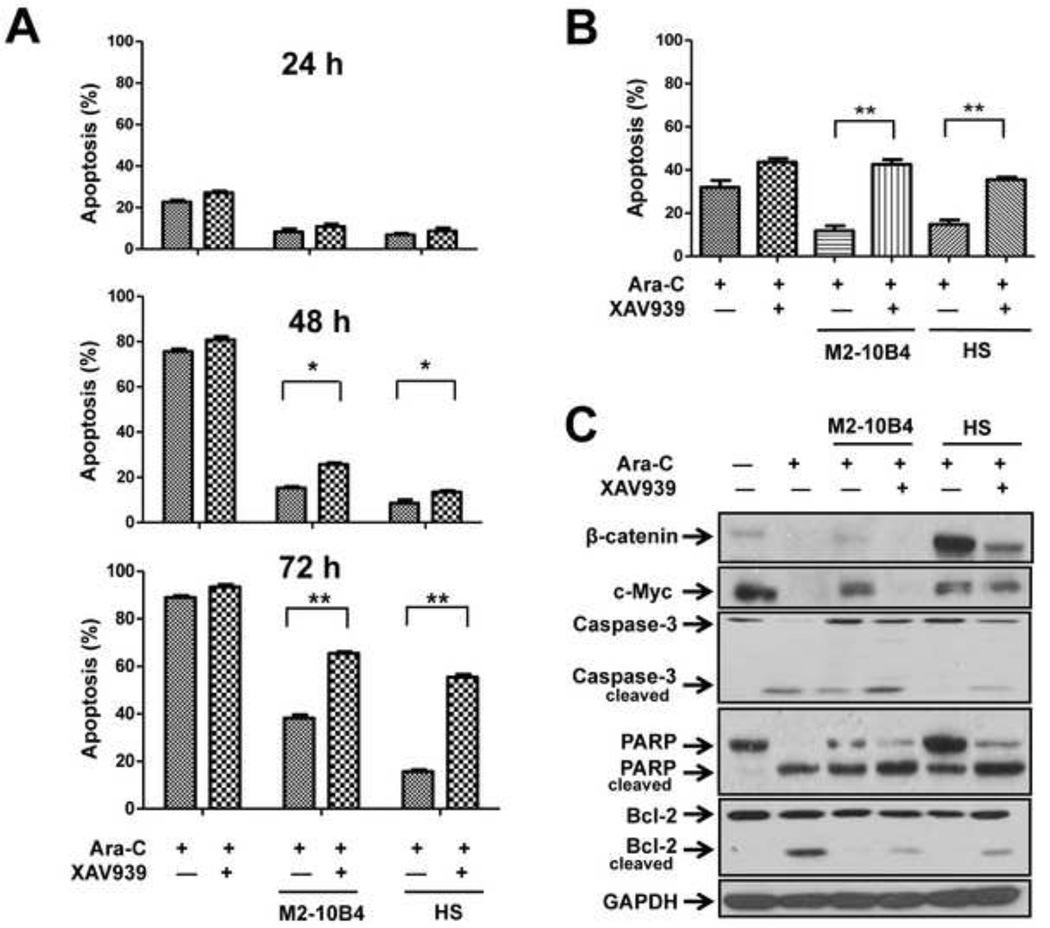
To further understand the role of β-catenin in leukemia cell survival, we determined the effect of the specific β-catenin inhibitor XAV939 on Reh cell apoptosis and found that XAV939 enhanced Ara-C-induced apoptosis in a time-dependent manner. At 72 hours after culture with M2-10B4 cells, 66.4±1.0% and 35.6±2.4% of Reh cells had undergone apoptosis in the presence and absence of XAV939, respectively (p < .01) (Fig. 5A). The corresponding values at 72 hours after culture with HS cells were 56.97±2.1% and 16.96±1.2% (p < .01). Similarly, MSCs protected human primary ALL cells from spontaneous and Ara-C-induced apoptosis (Fig. 5B). Such protection, however, was significantly diminished by the addition of the β-catenin inhibitor XAV939 (p < .01 for both M2-10B4 cells and HS cells).
Fig. 5. Effect of Wnt signaling inhibitor XAV939 on leukemia cell apoptosis.
ALL cells were cultured with or without MSCs and treated with Ara-C (1 µM) or a combination of Ara-C and XAV939 (40 µM). (A) Apoptotic rate in Reh cells treated for 24, 48, or 72 hours. *p < .05; **p < .01. (B) Apoptotic rate in patient primary ALL cells treated for 48 hours. **p < .01. Samples from patients 7–9 were tested. (C) Western blot analysis of β-catenin, c-Myc, caspase-3, PARP, and Bcl-2 expression in Reh cells treated for 72 hours.
Cell cycle analysis of primary ALL cells revealed that the percentage of cells in the sub-G1 phase was lower in the ALL cells cultured with M2-10B4 or HS cells than in ALL cells cultured in medium alone (Supplementary Table 3). Treatment with Ara-C induced a higher level of apoptosis in ALL cells cultured in medium alone than in ALL cells cultured with M2-10B4 or HS cells, suggesting that MSCs partially protected ALL cells from apoptosis. However, the addition of XAV939 increased the level of apoptosis in ALL cells cultured with M2-10B4 or HS cells. These data indicate that activation of Wnt signaling is critical to the protection of ALL cells by MSCs.
To confirm that XAV939 diminishes the protection provided by MSCs by inhibiting β-catenin, we determined the protein expression levels of β-catenin and other members of the Wnt signaling pathway in Reh cells. Western blot analysis revealed that the level of β-catenin was reduced by 72% when the cells were treated with Ara-C, was essentially restored in the presence of M2-10B4 cells and increased by 3.6-fold in the presence of HS cells, and then was diminished again when XAV939 was added (Fig. 5C). The same trend was observed for the level of c-Myc. In contrast, cleavage of caspase-3, PARP, and Bcl-2 was increased in Reh cells treated with XAV939. Similar results were observed for RS4;11 ALL cells (data not shown). These findings suggest that culture with MSCs activates the Wnt signaling pathway in ALL cells, contributing to the protection of ALL cells by MSCs. Conversely, blocking Wnt signaling with a β-catenin inhibitor could overcome this protection.
3.5. XAV939 enhanced the efficacy of Ara-C in vivo
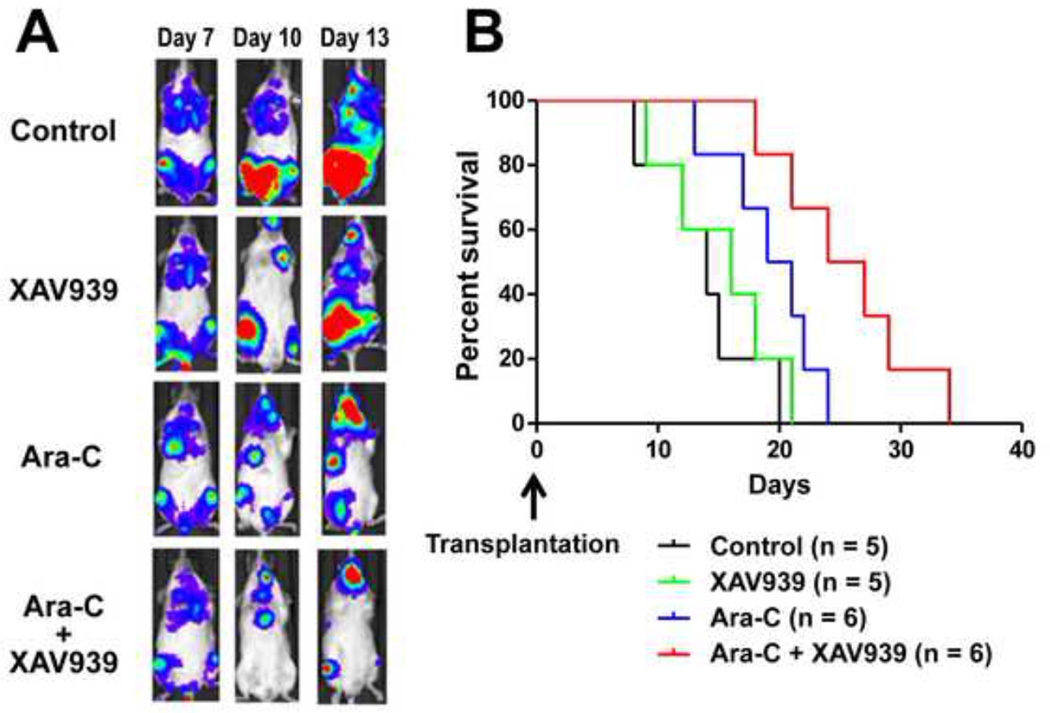
To determine the efficacy of XAV939 in vivo, we established leukemia models by xenografting Rehluc cells into NOD/SCID mice. We found that the Rehluc cells initially homed to the bone marrow and then gradually disseminated into the peripheral blood, in a manner similar to that of leukemia progression in humans. Seven days after injection, leukemia cells were clearly detectable by BLI. The mice were left untreated (control mice) or were treated with XAV939, Ara-C, or both. Mice treated with XAV939 alone had slightly lower BLI signals than untreated mice did (Fig. 6A) and a slightly longer median survival duration (16 versus 14 days) (Fig. 6B).
Fig. 6. Sensitization of Rehluc ALL cells by XAV939 to Ara-C treatment in vivo.
(A) BLI of mice transplanted with Rehluc cells. Mice were treated with 40 µM XAV939, 1 µM Ara-C, or both on day 7, when Rehluc cells were detectable on BLI, or were left untreated. The results from 1 representative animal over time from each group are shown. (B) Kaplan-Meier survival curve of transplanted mice. Median overall survival duration of mice treated with both XAV939 and Ara-C was longer than that of mice treated with Ara-C alone (p < .05).
Treatment with Ara-C had a higher inhibitory effect on xenograft growth and resulted in more prolonged survival (20 days) than did the treatment with XAV939. The combination of Ara-C and XAV939 resulted in the greatest therapeutic efficacy and the longest survival duration (25.5 days) of the 4 groups. Kaplan-Meier survival curves revealed that the overall survival of mice treated with Ara-C plus XAV939 was significantly better than that of mice treated with Ara-C alone (p < .05). These data indicate that the β-catenin inhibitor sensitizes leukemia cells to Ara-C and eventually leads to improved treatment outcome.
4. Discussion
The bone marrow microenvironment plays an important role in the initiation and progression of leukemia as well as in the persistence of minimal residual disease and disease relapse [19]. Various components of this microenvironment, particularly MSCs, regulate ALL survival, proliferation, and drug resistance by producing growth factors, cytokines, and intracellular signals by direct contact with the cells [6; 20]. Disrupting the cross-talk between leukemia cells and their milieu is of ultimate importance in eradicating the disease. In this study, we found that MSCs promoted ALL cell proliferation and inhibited apoptosis through Wnt pathway activation and that inhibition of β-catenin sensitized leukemia cells to Ara-C treatment both in vitro and in vivo.
Our extensive gene expression microarray analyses indicated that the co-culture system we used in this study is valid for studying the role of MSCs in the protection of ALL cells. As expected, treatment of ALL cells with Ara-C caused significant changes in the gene expression profiles in ALL cells. These changes was reversed in ALL cells cultured with the protective HS MSCs, but not in ALL cells cultured with the non-protective HS-5 MSCs. Whether the HS-5 cells originated in vivo from an MSC that did not possess the capability to protect ALL cells or had undergone a genetic or epigenetic alteration that resulted in the loss of this capability remains unknown. Nevertheless, HS-5 cells could serve as an excellent control in determining the ways in which M2-10B4 and HS cells protect ALL cells. Our gene expression profiles of ALL cells co-cultured with protective or non-protective MSCs correlated well with gene signatures that distinguish between clinical high risk and low risk of ALL relapse, indicating that our profiles reflect the functionality of MSCs in the protection of ALL cells and that our in vitro system is a valuable model of the in vivo scenario in patients.
We found that both human MSCs and murine MSCs significantly reduced apoptosis in ALL cells. Apparently, an exact species match between ALL cells and MSCs is not required for this effect. Our data indicated that MSCs reduced cleavage of caspase-3, PARP, and Bcl-2 and decreased p21 protein expression levels, suggesting that MSCs modulate the response of leukemia cells to chemotherapy through molecules involved in apoptosis and cell cycle regulation. Although it is commonly accepted that MSCs promote leukemia cell survival, it is unclear whether they promote cell proliferation. For example, Gaundar et al. [21] found that MSCs increased the proliferation of ALL cells by producing platelet-derived growth factor and vascular endothelial growth factor. In contrast, Zhu et al. reported that MSCs inhibited the proliferation of K562 cells and primary human leukemia cells [22]. Our results demonstrated that MSCs increase Reh cell proliferation. The use of different cell lines might account for the controversial results of the effect of MSCs on leukemia cell proliferation. Further studies are needed to reach a definitive conclusion.
Multiple signaling pathways, which may synergistically cooperate in ALL cells, are related to chemotherapy resistance. Notably, the PI3K/Akt pathway has been reported to protect leukemia cells by upregulating Bcl-2 [23]. Consistent with this finding, we found that MSCs stimulated the phosphorylation of Akt in Reh cells and that this effect was partially blocked by the Akt inhibitor LY294002 (data not shown). The CXCR4/CXCL12 axis participates in cell homing and migration and is considered essential to the development of chemoresistance in childhood ALL [24]. CXCR4 inhibitors have been shown to overcome the protection of acute myeloid leukemia cells by MSCs [24]. However, we tested the CXCR4 inhibitor AMD3100 in the present study and found no significant increase in Ara-C-induced apoptosis in ALL cells (data not shown). It is likely that different types of leukemia cells have different active signaling pathways in response to external stress.
Wnt signaling is highly conserved evolutionarily and is involved in embryogenesis and the maintenance of homeostasis in tissues by regulating cellular processes such as proliferation, differentiation, survival, and angiogenesis [25]. Wnt signaling activity is dependent on the interplay between pathway activators such as Lef1, C-Myc, CCNDBP1, and β-catenin and pathway suppressors such as GSK3β. In the absence of Wnt ligand stimulation, GSK3β phosphorylates β-catenin and causes β-catenin degradation. However, stimulation from Wnt ligands increases the phosphorylation of GSK3β which lead to β-catenin accumulation. Translocation of β-catenin into the nucleus induces the expression of target genes. Aberrant Wnt signaling is associated with several diseases, including cancer [26; 27]. The role of Wnt signaling in solid tumors, such as colorectal cancer, has been relatively well studied [28]; however, its role in hematological malignancies remains to be explored. Wnt family genes have been found to be expressed in patient ALL cells and to mediate cell proliferation and survival [29]. Activation of Wnt signaling has been shown to be epigenetically regulated by aberrant methylation of Wnt antagonists in acute myeloid leukemia and to be predictive of poor prognosis [30]. Expression of β-catenin has been demonstrated in most multiple myeloma cells but not in normal plasma cells, and it has been found to enhance myeloma cell proliferation [9].
Our microarray analysis of the gene expression profile indicated that the MSCs activated the Wnt pathway in ALL cells. Noticeable changes were detected in Wnt pathway members, including MYC, LEF1, CCNDBP1, and GSK3β. Although microarray analysis did not reveal an overt change in the mRNA expression of β-catenin, the expression level of its protein was significantly higher in ALL cells cultured with MSCs than in ALL cells cultured in medium alone. To elucidate the underlying mechanisms, we investigated the role of GSK3β in this process and found increased GSK3β phosphorylation in co-cultured ALL cells. Similarly, inhibition of GSK3β with lithium chloride increased its phosphorylation, which was accompanied by markedly increased β-catenin and Wnt signaling activity. These results suggested that MSCs, which expressed various Wnt ligands, activated the Wnt signaling pathway at least in part through phosphorylation (inactivation) of GSK3β. On the basis of these findings, we hypothesized that targeting the Wnt signaling pathway could be a useful therapeutic strategy for attenuating the protection by MSCs of ALL cells from chemotherapy.
XAV939 is a new β-catenin inhibitor that promotes β-catenin degradation by inhibiting tankyrase [31]. In this study, we treated ALL cells with XAV939 and assessed the effect of Wnt signaling inhibition on cell survival. XAV939 significantly reversed MSC-mediated resistance to Ara-C in human primary ALL cells and cell lines. Interestingly, XAV939 alone inhibited the proliferation of leukemia cells and MSCs but did not significantly induce apoptosis in these cells (data not shown). Reduced β-catenin and c-Myc protein expression levels in ALL cells in the presence of XAV939 suggested that the enhancing effect of XAV939 on Ara-C-induced apoptosis of ALL cells was mediated by blockade of Wnt signaling activation. We confirmed the effect of XAV939 in a mouse model: Ara-C and XAV939 together slowed leukemia progression in NOD/SCID mice more so than Ara-C alone did, and this combination prolonged the median survival duration.
In summary, we showed that MSCs provide effective protection for primary ALL cells and ALL cell lines from chemotherapy-induced apoptosis. Such protection is in part mediated by MSC activation of the Wnt pathway in ALL cells. Targeting Wnt signaling is a potential novel therapeutic strategy for disrupting the microenvironmental support of ALL cells.
Supplementary Material
Acknowledgements
We thank Dr. J. G. Wen at Methodist Hospital, Houston, Texas, USA, for the human bone marrow MSC cell line HS; Dr. Patrick Zweidler-McKay at MD Anderson for the human leukemia cell lines Reh, RS4;11, and SEMK2, Dr. Jan Burger at MD Anderson for the murine MSC line M2-10B4, and Dr. Roel Nusse at Stanford University for 7TGC reporter vector (obtained from Addgene; plasmid-24304). This work was supported in part by Research Scholar Grant 119645-RSG-10-131-01-DDC to X.S. from the American Cancer Society, a Physician Scientist Program Award to X.S. from MD Anderson, a scholarship award and National Natural Science Foundation of China (no. 81200385) to Y.Y. from the China Scholarship Council, an award to Z.C. from the High Level Health Technology Innovation of Elite Culture Program of Zhejiang Province, and the MD Anderson Support Grant CA016672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The online version of this article contains supporting information.
Authorship and Disclosures
Y.Y., Y.G., Z.C., and X.S. initiated the study, designed the experiments, and wrote the paper. Y.Y., S.M., B.S., J.Z., S.K., and J.S.L. performed the experiments and statistical analyses. The authors have no conflicts of interest to disclose.
References
- 1.Pui CH, Evans WE. Acute lymphoblastic leukemia. N. Engl. J. Med. 1998;339:605–615. doi: 10.1056/NEJM199808273390907. [DOI] [PubMed] [Google Scholar]
- 2.Fielding AK, Richards SM, Chopra R, Lazarus HM, Litzow MR, Buck G, Durrant IJ, Luger SM, Marks DI, Franklin IM, McMillan AK, Tallman MS, Rowe JM, Goldstone AH. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood. 2007;109:944–950. doi: 10.1182/blood-2006-05-018192. [DOI] [PubMed] [Google Scholar]
- 3.Kern W, Danhauser-Riedl S, Ratei R, Schnittger S, Schoch C, Kolb HJ, Ludwig WD, Hiddemann W, Haferlach T. Detection of minimal residual disease in unselected patients with acute myeloid leukemia using multiparameter flow cytometry for definition of leukemia-associated immunophenotypes and determination of their frequencies in normal bone marrow. Haematologica. 2003;88:646–653. [PubMed] [Google Scholar]
- 4.Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322:1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- 5.Manabe A, Coustan-Smith E, Behm FG, Raimondi SC, Campana D. Bone marrow-derived stromal cells prevent apoptotic cell death in B-lineage acute lymphoblastic leukemia. Blood. 1992;79:2370–2377. [PubMed] [Google Scholar]
- 6.Juarez J, Dela Pena A, Baraz R, Hewson J, Khoo M, Cisterne A, Fricker S, Fujii N, Bradstock KF, Bendall LJ. CXCR4 antagonists mobilize childhood acute lymphoblastic leukemia cells into the peripheral blood and inhibit engraftment. Leukemia. 2007;21:1249–1257. doi: 10.1038/sj.leu.2404684. [DOI] [PubMed] [Google Scholar]
- 7.Mudry RE, Fortney JE, York T, Hall BM, Gibson LF. Stromal cells regulate survival of B-lineage leukemic cells during chemotherapy. Blood. 2000;96:1926–1932. [PubMed] [Google Scholar]
- 8.Tabe Y, Jin L, Tsutsumi-Ishii Y, Xu Y, McQueen T, Priebe W, Mills GB, Ohsaka A, Nagaoka I, Andreeff M, Konopleva M. Activation of integrin-linked kinase is a critical prosurvival pathway induced in leukemic cells by bone marrow-derived stromal cells. Cancer Res. 2007;67:684–694. doi: 10.1158/0008-5472.CAN-06-3166. [DOI] [PubMed] [Google Scholar]
- 9.Dutta-Simmons J, Zhang Y, Gorgun G, Gatt M, Mani M, Hideshima T, Takada K, Carlson NE, Carrasco DE, Tai YT, Raje N, Letai AG, Anderson KC, Carrasco DR. Aurora kinase A is a target of Wnt/beta-catenin involved in multiple myeloma disease progression. Blood. 2009;114:2699–2708. doi: 10.1182/blood-2008-12-194290. [DOI] [PubMed] [Google Scholar]
- 10.Gelebart P, Anand M, Armanious H, Peters AC, Dien Bard J, Amin HM, Lai R. Constitutive activation of the Wnt canonical pathway in mantle cell lymphoma. Blood. 2008;112:5171–5179. doi: 10.1182/blood-2008-02-139212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenfeld C, Goutner A, Choquet C, Venuat AM, Kayibanda B, Pico JL, Greaves MF. Phenotypic characterisation of a unique non-T, non-B acute lymphoblastic leukaemia cell line. Nature. 1977;267:841–843. doi: 10.1038/267841a0. [DOI] [PubMed] [Google Scholar]
- 12.Stong RC, Korsmeyer SJ, Parkin JL, Arthur DC, Kersey JH. Human acute leukemia cell line with the t(4;11) chromosomal rearrangement exhibits B lineage and monocytic characteristics. Blood. 1985;65:21–31. [PubMed] [Google Scholar]
- 13.Pocock CF, Malone M, Booth M, Evans M, Morgan G, Greil J, Cotter FE. BCL-2 expression by leukaemic blasts in a SCID mouse model of biphenotypic leukaemia associated with the t(4;11)(q21;q23) translocation. Br. J. Haematol. 1995;90:855–867. doi: 10.1111/j.1365-2141.1995.tb05207.x. [DOI] [PubMed] [Google Scholar]
- 14.Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995;85:997–1005. [PubMed] [Google Scholar]
- 15.Lemoine FM, Humphries RK, Abraham SD, Krystal G, Eaves CJ. Partial characterization of a novel stromal cell-derived pre-B-cell growth factor active on normal and immortalized pre-B cells. Exp. Hematol. 1988;16:718–726. [PubMed] [Google Scholar]
- 16.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer. Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 17.Banker DE, Groudine M, Willman CL, Norwood T, Appelbaum FR. Cell cycle perturbations in acute myeloid leukemia samples following in vitro exposures to therapeutic agents. Leuk. Res. 1998;22:221–239. doi: 10.1016/s0145-2126(97)00174-4. [DOI] [PubMed] [Google Scholar]
- 18.Cario G, Stanulla M, Fine BM, Teuffel O, Neuhoff NV, Schrauder A, Flohr T, Schafer BW, Bartram CR, Welte K, Schlegelberger B, Schrappe M. Distinct gene expression profiles determine molecular treatment response in childhood acute lymphoblastic leukemia. Blood. 2005;105:821–826. doi: 10.1182/blood-2004-04-1552. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Jacamo R, Shi YX, Wang RY, Battula VL, Konoplev S, Strunk D, Hofmann NA, Reinisch A, Konopleva M, Andreeff M. Human extramedullary bone marrow in mice: a novel in vivo model of genetically controlled hematopoietic microenvironment. Blood. 2012 doi: 10.1182/blood-2011-11-389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishii K, Katayama N, Miwa H, Shikami M, Masuya M, Shiku H, Kita K. Survival of human leukaemic B-cell precursors is supported by stromal cells and cytokines: association with the expression of bcl-2 protein. Br. J. Haematol. 1999;105:701–710. doi: 10.1046/j.1365-2141.1999.01380.x. [DOI] [PubMed] [Google Scholar]
- 21.Gaundar SS, Bradstock KF, Bendall LJ. p38MAPK inhibitors attenuate cytokine production by bone marrow stromal cells and reduce stroma-mediated proliferation of acute lymphoblastic leukemia cells. Cell Cycle. 2009;8:2975–2983. [PubMed] [Google Scholar]
- 22.Zhu Y, Sun Z, Han Q, Liao L, Wang J, Bian C, Li J, Yan X, Liu Y, Shao C, Zhao RC. Human mesenchymal stem cells inhibit cancer cell proliferation by secreting DKK-1. Leukemia. 2009;23:925–933. doi: 10.1038/leu.2008.384. [DOI] [PubMed] [Google Scholar]
- 23.Lee BH, Ruoslahti E. alpha5beta1 integrin stimulates Bcl-2 expression and cell survival through Akt, focal adhesion kinase, and Ca2+/calmodulin-dependent protein kinase IV. J.Cell. Biochem. 2005;95:1214–1223. doi: 10.1002/jcb.20488. [DOI] [PubMed] [Google Scholar]
- 24.Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling X, Frolova O, Levis M, Rubin JB, Negrin RR, Estey EH, Konoplev S, Andreeff M, Konopleva M. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood. 2009;113:6215–6224. doi: 10.1182/blood-2008-05-158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akiyama T. Wnt/beta-catenin signaling. Cytokine Growth Factor Rev. 2000;11:273–282. doi: 10.1016/s1359-6101(00)00011-3. [DOI] [PubMed] [Google Scholar]
- 26.You A, Fokas E, Wang LF, He H, Kleb B, Niederacher D, Engenhart-Cabillic R, An HX. Expression of the Wnt antagonist DKK3 is frequently suppressed in sporadic epithelial ovarian cancer. J. Cancer Res. Clin. Oncol. 2011;137:621–627. doi: 10.1007/s00432-010-0916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CL, Liu D, Ishikawa S, Nakashima T, Nakashima N, Yokomise H, Kadota K, Ueno M. Wnt1 overexpression promotes tumour progression in non-small cell lung cancer. Eur. J. Cancer. 2008;44:2680–2688. doi: 10.1016/j.ejca.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, Garcia JM, Munoz A, Esteller M, Gonzalez-Sancho JM. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116–4121. doi: 10.1038/sj.onc.1209439. [DOI] [PubMed] [Google Scholar]
- 29.Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br. J. Haematol. 2007;138:338–348. doi: 10.1111/j.1365-2141.2007.06667.x. [DOI] [PubMed] [Google Scholar]
- 30.Valencia A, Roman-Gomez J, Cervera J, Such E, Barragan E, Bolufer P, Moscardo F, Sanz GF, Sanz MA. Wnt signaling pathway is epigenetically regulated by methylation of Wnt antagonists in acute myeloid leukemia. Leukemia. 2009;23:1658–1666. doi: 10.1038/leu.2009.86. [DOI] [PubMed] [Google Scholar]
- 31.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.