Abstract
Eukaryotic ribosomes are assembled by a complex pathway that extends from the nucleolus to the cytoplasm and is powered by many energy-consuming enzymes 1-3. Nuclear export is a key, irreversible step in pre-ribosome maturation4-8, but mechanisms underlying the timely acquisition of export competence remain poorly understood. Here we show that a conserved GTPase Nug2/Nog2 (called NGP-1, Gnl2 or nucleostemin 2 in human9) plays a key role in the timing of export competence. Nug2 binds the inter-subunit face of maturing, nucleoplasmic pre-60S particles, and the location clashes with the position of Nmd3, a key pre-60S export adaptor10. Nug2 and Nmd3 are not present on the same pre-60S particles, with Nug2 binding prior to Nmd3. Depletion of Nug2 causes premature Nmd3 binding to the pre-60S particles, whereas mutations in the G-domain of Nug2 block Nmd3 recruitment, resulting in severe 60S export defects. Two pre-60S remodeling factors, the Rea1 ATPase and its co-substrate Rsa4, are present on Nug2-associated particles, and both show synthetic lethal interactions with nug2 mutants. Release of Nug2 from pre-60S particles requires both its K+-dependent GTPase activity and the remodeling ATPase activity of Rea1. We conclude that Nug2 is a regulatory GTPase that monitors pre-60S maturation, with release from its placeholder site linked to recruitment of the nuclear export machinery.
The conserved GTPase Nug2/Nog2 (Extended Data Fig. 1) is associated with a number of pre-60S particles located in the nucleoplasm, but was not detected on particles with a known cytoplasmic location (see also Extended Data Fig. 2). The bacterial homolog of Nug2, YlqF, binds directly to rRNA11, and we therefore used the CRAC UV cross-linking method to localize the binding site for yeast Nug2 within the pre-60S particle12. Direct contacts for Nug2 were identified only with the 25S rRNA, at sites in helices H38, H69, H71, H80, H81-83, H84-86, H89, H91-92 and H93 (Fig. 1a, c). Yeast 3-hybrid analyses confirmed interactions between Nug2 and these rRNA helices (Fig. 1b). Mapping the major rRNA crosslink sites of Nug2 onto the 60S subunit structure (Fig. 1d) showed a distinct cluster on the inter-subunit joining surface13. Nug2 binding sites overlap with regions occupied by the export factor Nmd3 in cryo-EM10. CRAC was therefore applied to Nmd3 to more precisely identify its binding sites, which were found to lie in H38, H69 and H89 of 25S rRNA (Fig. 1a, c, e). Strikingly, the Nug2 and Nmd3 binding sites overlapped in H38, H69 and H89 (Fig. 1c, f), suggesting that binding of these two proteins is mutually exclusive. To test this, pre-60S particles were purified with tagged Nug2 and shown to lack detectable Nmd3, and vice versa (Extended Data Fig. 2). Nmd3 is an essential nuclear export factor that recruits the export receptor Crm1 to the nascent 60S subunits14,15. These observations suggested that Nug2 acts as a “placeholder” to prevent premature recruitment of Nmd3 to earlier, export-incompetent pre-60S particles.
Figure 1. Nug2 binds to inter-subunit face of the pre-60S subunit clashing with export factor Nmd3.
(a) CRAC analyses of Nug2 and Nmd3 (performed twice; only sites were considered that were reproducibly found in both datasets). Total number of hits was plotted against the relative location along the rDNA. (b) Yeast 3-hybrid revealing interaction between Nug2 and identified 25S rRNA fragments. Negative control, empty vector and H25. (c) Nug2 (yellow) and Nmd3 binding sites (green) identified by CRAC and highlighted in the indicated 25S rRNA. (d, e) Mapping of CRAC Nug2 (yellow) and Nmd3 (green) binding sites on the 60S structure (PDB: 3O5H13). (f) Overlapping binding sites (red) of Nug2 (yellow) and Nmd3 (green).
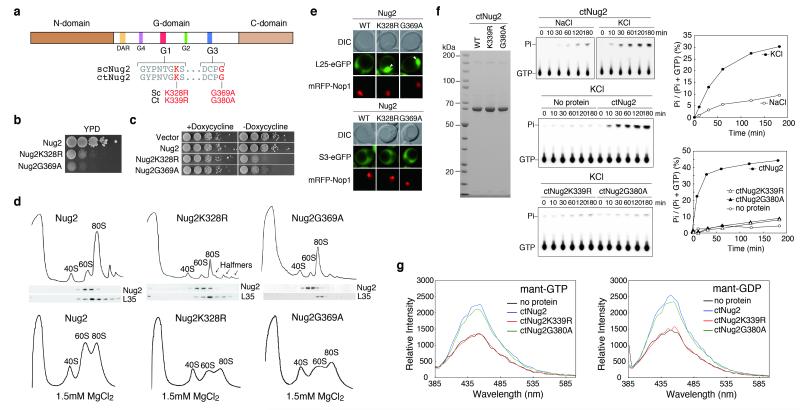
Like other GTP-binding proteins, Nug2 has characteristic G1, G3 and G4 motifs in its G-domain (Fig. 2a, Extended Data Fig. 1), suggesting that GTP-binding or hydrolysis16 might regulate dynamic interactions between Nug2 and the pre-ribosome. Dominant-negative mutations were previously described in two GTPases involved in ribosome biogenesis, the G1-motif of Lsg1 (K349N/R/T)17 and G3-motif of Nog1 (G224A)18. Orthologous G1- and G3-motif mutants, nug2K328R and nug2G369A, respectively (Fig. 2a, Extended Data Fig. 1), each showed severe growth defect phenotypes (Fig. 2b), and were also dominant-negative when overexpressed in the presence of chromosomal NUG2 (Fig. 2c). Pre-ribosome analysis by sucrose gradient centrifugation showed that the Nug2K328R and Nug2G369A proteins were efficiently assembled into pre-60S subunits, but induced a ‘half-mer’ polysome phenotype (in particular for Nug2K328R), characteristic of reduced 60S subunit synthesis (Fig. 2d). The reduced 60S levels were more apparent under low Mg2+ conditions that cause 80S ribosomes to dissociate into 60S and 40S subunits (Fig. 2d). The nug2K328R and nug2G369A strains showed nuclear accumulation of a RpL25-GFP reporter, but not RpS3-GFP, revealing a specific block in pre-60S nuclear export (Fig. 2e). We conclude that mutations in the GTPase domain of Nug2 allow recruitment to the pre-ribosomes, but block nuclear export.
Figure 2. K+-dependent GTPase activity of Nug2.
(a) Domain organization of Nug2. (b). Complementation of nug2Δ cells by NUG2, nug2K328R and nug2G369A on YPD plates. (c) Repression (+doxycycline) and overexpression (−doxycycline) of NUG2, nug2K328R and nug2G369A in NUG2 cells. (d) Polysomal (10mM MgCl2; upper panel) and ribosomal profiles (1.5mM MgCl2; lower panel) of NUG2, nug2K328R and nug2G369A cells analyzed by sucrose gradient centrifugation. Western analysis of gradient fractions using antibodies against Nug2 and RpL35 (upper panel) (e) Subcellular distribution of RpL25-GFP and RpS3-GFP in NUG2 and nug2 mutant cells analyzed by fluorescence microscopy. (f) GTPase activity of purified ctNug2 (SDS-PAGE; left panel) analyzed by thin-layer chromatography/autoradiography (middle panel). Ratio of hydrolyzed phosphate/total GTP plotted against time (right panel). (g) Binding of MANT-GTP (left panel) and MANT-GDP (right panel) to purified wild-type and mutant ctNug2. GTPase and binding assays were performed twice yielding highly reproducible datasets.
To determine the basis of the defects associated with Nug2K328R and Nug2G369A, we assayed in vitro guanine nucleotide-binding activity and GTP hydrolysis. Nug2 from Saccharomyces cerevisiae was unstable when expressed in E.coli (data not shown). In contrast, good yields were obtained for wild-type and mutant Nug2 from the eukaryotic thermophile Chaetomium thermophilum (ctNug2, ctNug2K339R and ctNug2G380A, respectively; Fig. 2f), whose thermostable proteins have superior biochemical properties19. ctNug2 is highly homologous to yeast Nug2 (74% identity; Extended Data Fig. 1), and can complement albeit not perfectly a yeast nug2Δ mutant (Extended Data Fig. 3). As Nug2 may act as a potassium-dependent GTPase20, we tested the cation requirement for GTP hydrolysis. The GTPase activity of ctNug2 was low in NaCl-containing buffer, but was substantially stimulated by KCl (Fig. 2f). In contrast, ctNug2K339R and ctNug2G380A exhibited only background GTPase activity (Fig. 2f). In binding assays, wild-type ctNug2 and ctNug2G380A readily bound fluorescent MANT-GTP or MANT-GDP, whereas ctNug2K339R did not (Fig. 2g). We conclude that ctNug2K339R is defective in GTP binding, whereas ctNug2G380A binds but cannot hydrolyse GTP. This K+-stimulated GTPase activity might regulate interaction of Nug2 with nascent 60S particles.
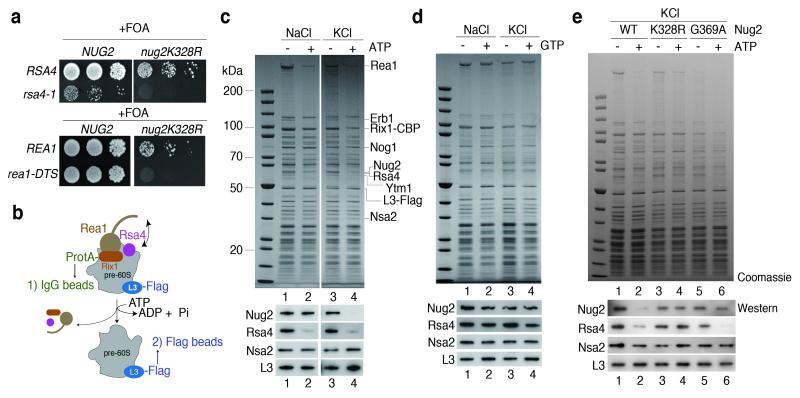
Nug2 is associated with nucleoplasmic pre-60S particles that also carry the Rix1-Ipi1-Ipi3 heterotrimer, the dynein-related AAA-ATPase Rea1 and its co-substrate Rsa4 (Extended Data Fig. 2; see also below and21). The enzymatic activity of Rea1 is required for the release of Ytm122 and Rsa421 and a genetic screen revealed synthetic lethality between the G1-motif mutant nug2K328R and the mutant alleles rea1-DTS and rsa4-121 (Fig. 3a). We therefore investigated whether ATP-dependent remodeling of the Rix1-particle by the AAA-ATPase activity of Rea121 is altered in particles containing Nug2K328R or Nug2G369A. Pre-60S particles carrying Flag-tagged RpL3 were affinity-purified with Rix1-TAP via IgG binding and TEV elution. The pre-60S particles were incubated in vitro to allow factor release, and then re-isolated on Flag-beads via RpL3-Flag (Fig. 3b). Consistent with previous data21, incubation of the pre-60S particles with ATP in Na+-containing buffer resulted in release of Rsa4 and Rea1, but not Nug2 (Fig. 3c). In contrast, incubation in K+-containing buffer caused the ATP-dependent release of Nug2, in addition to Rsa4 and Rea1 (Fig. 3c). Incubation with GTP in Na+ or K+-containing buffer did not induce the release of biogenesis factors (Fig. 3d). However, neither Nug2K328R nor Nug2G369A could be dissociated from pre-60S particles upon ATP treatment in K+ buffer (Fig. 3e). In the case of Nug2K328R (defective in GTP-binding), incubation with ATP in K+ buffer failed to release Rsa4, whereas pre-60S particles carrying Nug2G369A (defective in GTP-hydrolysis) still showed Rsa4 release upon ATP-treatment (Fig. 3e). Mutation of one of the six ATP-binding protomers of Rea1 (AAA2; rea1 K659A) inhibited remodeling, including Nug2 release (Extended Data Fig. 4). These findings indicate that the GTP-binding activity of Nug2 influences the remodeling activity of the Rea1 ATPase, whereas GTP hydrolysis is necessary for the final Nug2 release from the pre-60S subunit.
Figure 3. Nug2 release from pre-60S particles requires intrinsic K+-dependent GTPase and Rea1 ATPase activity.
(a) Synthetic lethality (sl) between alleles rsa4-121 or rea1-DTS21 and nug2K328R revealed by growth on 5-FOA. (b-e) ATP-dependent release of Rsa4 and Nug2 from purified pre-60S particles. Scheme of the release assay (b) and experimental analyses (c-e). Affinity-purified Rix1-particles carrying wild-type or mutant Nug2 were incubated with ATP or GTP in NaCl or KCl buffer, before matured pre-60S particles were re-isolated via RpL3-Flag affinity-purification. Final eluates were analyzed by SDS-PAGE and Coomassie staining (upper panel; indicated bands were identified by mass spectrometry) and Western blotting using the indicated antibodies (lower panel). All in vitro assays were performed at least twice with highly reproducible datasets.
In vitro, Rea1-dependent release of Rsa4 and Nug2 required only ATP and K+ without addition of GTP, whereas the mutational analyses suggested that GTPase activity is necessary for Nug2 release. These findings suggest that Nug2 on the Rix1-particle might have retained bound GTP during purification (which is possible due to its low intrinsic GTPase activity). Alternatively, ribosome-associated nucleotide diphosphate kinases can transfer the γ-phosphate from ATP to GDP to generate GTP-loaded GTPases23,24.
The pre-60S particles co-purified with Rix1 also contained small amounts of Ytm1 and Erb1 (Fig. 3c), which were previously described as nucleolar co-substrates for Rea122, and both were released by incubation with ATP in Na+ or K+ buffer (Fig. 3c).
To determine the step in 60S subunit biogenesis at which dissociation of Nug2 is disturbed in vivo, we affinity-purified different pre-60S particles from Nug2 wild-type and mutant cells using bait proteins that specifically enrich nucleolar, nucleoplasmic or cytoplasmic intermediates (Fig. 4a). The nug2K328R mutation did not markedly alter the biochemical composition of most pre-60S particles tested. The exception was Arx1-associated particles, which showed a marked depletion of the export adapter Nmd3 and the cytoplasmic factor Rei1 that stimulates recycling of Arx125 (Fig. 4a). Nmd3 was also largely absent from Arx1-particles purified from nug2G369A cells (Fig. 4b).
Figure 4. Nug2 release from the pre-60S subunit is linked to Nmd3 recruitment.
(a) Affinity-purification of the indicated TAP-tagged pre-60S factors from NUG2 or nug2K328R mutant cells. (*) position of Rei1 identified by mass spectrometry. (b) Affinity-purification of Arx1-TAP from NUG2, nug2K328R and nug2G369A cells. (c) Affinity-purification of Rix1-TAP from sAid-Nug2-sAid degron strain after time-dependent auxin treatment. (d) Affinity-purification of Nug2-TAP particles with (lane 1) or without (lane 2) subsequent Rix1-Flag immunodepletion. (a-d) SDS-PAGE and Coomassie staining (upper panel) and Western blotting using the indicated antibodies (lower panel). Protein bands indicated were identified by mass spectrometry. All affinity-purifications were performed at least twice, yielding highly reproducible datasets. (e) Model of pre-60S subunit maturation starting from the Rix1-particle with final Nmd3-Crm1-RanGTP recruitment.
To test the model that Nug2 depletion allows premature recruitment of Nmd3 we employed an auxin-inducible degron system28. Nug2 was expressed as a fusion protein (sAid-Nug2-sAid) with two copies of the sAid tag (small Auxin-inducible degron), which is targeted by the F-box E3 ubiquitin ligase TIR1 in the presence of auxin, inducing fast proteasomal degradation28 (Extended Data Fig. 5). Nmd3 is normally not detected on Rix1-associated particles, but was prematurely recruited to this pre-60S particle upon Nug2 depletion (Fig. 4c). Concomitant with Nmd3 association, the recovery of Rea1 and Rsa4 decreased during Nug2 depletion (Fig. 4c). We conclude that Nug2 promotes the stable association of Rsa4 and Rea1 with the Rix1-particles, while blocking premature recruitment of Nmd3.
To address the timing of Nug2 recruitment to pre-60S particles in comparison to Rea1, Rsa4 and the Rix1-Ipi1-Ipi3 complex, we employed a combination of affinity-purification and immunodepletion. Affinity-purification of Nug2-TAP yielded a mixture of different pre-60S particles including Rix1/Rea1-particles. Rix1-FLAG immunoprecipitation was used to deplete Rix1/Rea1-particles from this mixture, leaving Nug2 particles that contained Rsa4 and a number of intermediate pre-60S factors including Nog1, Arx1, Nug1, Nop53, Nsa3, Rpf2, Rlp7 and Nsa2, (Fig. 4d). However, this Nug2-particle lacked other (further upstream) pre-60S factors such as Ytm1, Erb1 and Has1, suggesting that it corresponds to the precursor particle to which Nug2 was recruited. These data complement previous findings that Nog2/Nug2 is the last “B-factor” to associate with pre-ribosomes after dissociation of Has129.
As outlined in Fig. 4e, we propose that a previously uncharacterized step in the reorganization of the evolving pre-60S subunit primes it for nuclear export. This involves a regulatory GTPase Nug2 that overlaps with the binding site for the essential nuclear export adaptor Nmd3. As long as intranuclear maturation is incomplete, the pre-60S subunit cannot be exported, since recruitment of this essential export factor is not possible. However, a late nucleoplasmic remodeling step, catalyzed by the AAA-ATPase Rea1 and its co-factor Rsa4, restructures the pre-60S particle, which could lead to both an rRNA and assembly factor rearrangement. This conformational change could also stimulate Nug2’s K+-dependent GTPase activity, thereby triggering its release from the matured pre-60S particles. We suggest that the Nug2 GTPase acts as molecular switch to proofread pre-ribosome maturation and regulate the acquisition of export competence. After this reorganization step, the binding site for Nmd3 becomes accessible on the pre-60S subunit, which further triggers Crm1 and RanGTP recruitment to generate nuclear export competence. Thus, our data indicate coordination between a remodeling AAA-ATPase and a conformation-sensing GTPase.
The human Nug2 orthologue Gnl2 is highly expressed in proliferating cells including cancer cells and involved in the control of cell cycle progression30. The discovery of the role of Nug2 during surveillance of ribosome biogenesis may help reveal the molecular mechanisms by which nucleostemin family members interconnect the elementary cellular processes of ribosome biogenesis and cell proliferation.
METHODS
Yeast strains and genetic methods
The S. cerevisiae strains used in this study are listed in Extended Data Table 1. Gene disruption and C-terminal tagging were performed as previously described31,32.
Plasmid constructs
All recombinant DNA techniques were performed according to standard procedures using Escherichia coli DH5α for cloning and plasmid propagation. Site-directed mutagenesis was performed by overlap-extension PCR. All cloned DNA fragments generated by PCR amplification were verified by sequencing. Plasmids used in this study are listed in Extended Data Table 2.
CRAC analysis
The CRAC experiments were performed as described12 using the Nug2- and Nmd3-HTP (His6-TEV-ProtA) strain. CRAC data were processed using pyCRAC (Webb, Hector, Kudla and Granneman, in preparation). Cells were UV-irradiated in the Megatron UV chamber1 at a dose of 1.6J/cm2 and processed as described12,33. The cDNAs from the Nug2 CRAC data were cloned into pCR4-TOPO (Invitrogen) and inserts were sequenced by Sanger sequencing. The cDNAs originating from Nmd3 CRAC experiments were sequenced on the Illumina MiSeq system (single-end 50b), according to manufacturers procedures. The MiSeq CRAC data were processed using the pyCRAC software suite (Webb, Hector, Kudla and Granneman, submitted; https://bitbucket.org/sgrann/pycrac). To remove potential PCR duplicates, the Nmd3 MiSeq data was collapsed using pyFastqDuplicateRemover. Reads subsequently mapped to the yeast genomic reference sequence (version 2008) using novoalign (www.novocraft.com). Plots of reads aligned to the 35S reference sequence were generated using pyPileup and GNUplot. Adapters using this experiment are listed in Extended Data Table 3.
Expression and purification of ctNug2
The gene encoding Chaetomium thermophilum Nug2 (accession number in the UniProtKB/TrEMBL protein data base: G0SBX1_CHATD) was cloned from cDNA by standard procedures as recently described19. Subsequently, the ctNug2 was inserted into yeast or E. coli expression plasmids (see below). Since the C-terminal extension of ctNug2 (511 - 627 aa) is not conserved (Extended Data Fig. 1), ctNug2 from 1 to 510 amino acids was cloned into pET21 vector for the in vitro experiments. ctNug2 was expressed by using pET-ctNug2-510-His6 plasmid in E. coli Rosetta-DE3 cells. Transformed cells were grown at 23°C in LB medium until reached OD of 0.6, IPTG was added to a final concentration of 0.1 mM. The cells were grown for additional 3 h and then harvested by centrifugation and stored frozen at −80°C. Frozen pellets were resuspended in buffer KCl200 (50 mM Tris, pH 8.0, 200 mM KCl, 5% glycerol, 0.01% NP-40, 2 mM ß-mercaptoethanol) with protease-inhibitor cocktail, and were broken by sonication (BANDELIN sonopuls 3200 with TITANTELLER TT13) on ice. Sonication was performed under these conditions: Amplitude: 50%, 3 seconds ON, 8 seconds OFF, processed for 10 minutes. The lysate was centrifuged at 18,000 rpm for 30 minutes at 4 °C. The supernatant fraction was applied to a SP-sepharose column, and it was washed with buffer KCl200. ctNug2-His6 was eluted by buffer KCl200 containing 300mM KCl. Next, the eluate fraction was applied to Ni2+-NTA column, and the column was washed with buffer KCl200. ctNug2-His6 was eluted by buffer KCl200 containing 250mM imidazole, before it was finally dialyzed against buffer KCl200.
Measurement of GTPase activity by single-turnover reactions
The GTPase activity experiments were performed as previously described34. 1μM ctNug2, ctNug2K339R or ctNug2G380A were incubated with a final concentration of 0.1 μM GTP containing 750 nCi of γ-P32-labeled GTP in buffer KCl300 (50 mM Tris, pH 8.0, 300 mM KCl, 10mM MgCl2, 1 mM DTT) or in buffer NaCl300 (50 mM Tris, pH 8.0, 300 mM NaCl, 10mM MgCl2, 1 mM DTT) for indicated time at 30°C. After the reaction, the hydrolyzed γ-phosphate was separated by thin-layer chromatography.
Guanine nucleotide binding experiments
1μM ctNug2, ctNug2K339R or ctNug2G380A were incubated with 0.1μM MANT-GTP or MANT-GDP in buffer KCl300 (50mM Tris, pH 8.0, 300mM KCl, 60mM MgCl2, 20mM EDTA, 1mM DTT). MANT-GTP or MANT-GDP are analogues of natural GTP or GDP, where either the ribose 2′-hydroxy or the 3′-hydroxy group has been esterified by the fluorescent methylisatoic acid with an Ex/Em = 355/448 nm. The fluorescence quantum yield of Mant fluorophore is very low in water and increases significantly in nonpolar solvents or upon binding to most proteins. This highly environmental sensitive fluorescence of Mant makes Mant-GTP/GDP useful for directly detecting the nucleotide-protein interactions. Accordingly, it was excited at 355 nm with a xenon lamp, and emission spectra were recorded between 385-600 nM with a 5 nm increment step using a Synergy 4 spectrophotometer (BioTek).
In vitro release assay.
Rix1-particle was affinity purified using IgG beads via Rix1-TAP from yeast strain (Rix1-TAP, RpL3-Flag) expressing NUG2, nug2K328R and nug2G369A followed by TEV protease cleavage at 4°C to release the Rix1-particle. The TEV-eluate (i.e. the released Rix1-particle) was incubated with 4mM ATP or 4mM GTP at 23 °C for 1 hour. After ATP or GTP treatment, the 60S particle was re-purified via affinity purification of L3-Flag using Flag beads. Buffer KCl100 (50 mM Tris, pH 8.0, 100 mM KCl, 10 mM MgCl2, 1 mM DTT) or buffer NaCl100 (50 mM Tris, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT) were used. Affinity purifications were performed as previously described35.
Immunodepletion of Rix1 by FLAG immunoprecipitation
Nug2-particle was affinity purified from yeast strain (Nug2-TAP, Rix1-Flag) via IgG beads. The TEV eluate was incubated twice with Flag beads at 4°C for 30min each to deplete the Rix1-associated Nug2 particle. The flow-through was used for the final Calmodulin purification step.
Miscellaneous
Additional methods used in this study and previously described were TAP-purifications of pre-60S particles36, sucrose gradient analysis to obtain ribosomal and polysomal profiles37, ribosomal export assays using the large subunit reporter Rpl25-GFP monitored by fluorescence microscopy38 and yeast 3-hybrid analysis39. Antibodies used for Western analysis in the following dilutions were anti-Nug240 1:10,000, anti-Rsa441 1:10,000, anti-Nmd315 1:5,000, anti-Mex67/Mtr242 1:10,000, anti-RpL3543 1:35,000, anti-RpL344 1:5000, anti-Nsa245 1:10,000, anti-RpL1046 1:1000, anti-Nog140 1:30,000, anti-Rei147 1:5,000, goat-anti-mouse 1:3,000 (Cat.-No. 170-6516) and mouse-anti-rabbit horse radish peroxidase conjugated antibodies 1:3,000 (Cat.-No. 170-6515, both BIORAD, Munich, Germany). Page Ruler Unstained Protein Ladder (Thermo Scientific, Rockford, Illinois, USA) was used as a protein marker, Brillant Blue G-Colloidal Concentrate Electrophoresis Reagent (Sigma-Aldrich, Munich, Germany) was used for Coomassie stain, and 4-12% NuPAGE Bis-Tris Gels (Novex, Darmstadt, Germany) together with NuPAGE MOPS SDS Running Buffer (Invitrogen, Darmstadt, Germany) were used for SDS-PAGE.
Extended Data
Acknowledgements
We thank Drs. M. Remacha, M. Fromont-Racine, A. W. Johnson, C. Dargemont, M. Seedorf, and J. Warner for antibodies. We thank the GenePool at the University of Edinburgh for performing the MiSeq sequencing, and Elisabeth Petfalski for performing the initial cross-linking test experiments. We thank Dr. Emma Thomson for careful reading the manuscript. This work was supported by a postdoctoral fellowship from Alexander von Humboldt Foundation to Y.M., and by the Welcome Trust to S.G. and D.T. (077248), and by grants from the Deutsche Forschungsgemeinschaft to E.H. (DFG Hu363/10-4).
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staley JP, Woolford JL., Jr. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Current opinion in cell biology. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Lafontaine DL. A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem Sci. 2010;35:267–277. doi: 10.1016/j.tibs.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Zemp I, Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581:2783–2793. doi: 10.1016/j.febslet.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai RY, Meng L. Nucleostemin: a latecomer with new tricks. The international journal of biochemistry & cell biology. 2009;41:2122–2124. doi: 10.1016/j.biocel.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sengupta J, et al. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol. 2010;189:1079–1086. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuo Y, et al. The GTP-binding protein YlqF participates in the late step of 50 S ribosomal subunit assembly in Bacillus subtilis. J Biol Chem. 2006;281:8110–8117. doi: 10.1074/jbc.M512556200. [DOI] [PubMed] [Google Scholar]
- 12.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci U S A. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 14.Gadal O, et al. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 17.Hedges J, West M, Johnson AW. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapik YR, Misra JM, Lau LF, Pestov DG. Restricting conformational flexibility of the switch II region creates a dominant-inhibitory phenotype in Obg GTPase Nog1. Mol Cell Biol. 2007;27:7735–7744. doi: 10.1128/MCB.01161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amlacher S, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 20.Ash MR, Maher MJ, Mitchell Guss J, Jormakka M. The cation-dependent G-proteins: in a class of their own. FEBS Lett. 2012;586:2218–2224. doi: 10.1016/j.febslet.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 21.Ulbrich C, et al. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 2009;138:911–922. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 22.Bassler J, et al. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell. 2010;38:712–721. doi: 10.1016/j.molcel.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikkawa S, et al. Conversion of GDP into GTP by nucleoside diphosphate kinase on the GTP-binding proteins. J Biol Chem. 1990;265:21536–21540. [PubMed] [Google Scholar]
- 24.Wertheimer AM, Kaulenas MS. GDP kinase activity associated with salt-washed ribosomes. Biochemical and biophysical research communications. 1977;78:565–571. doi: 10.1016/0006-291x(77)90216-9. [DOI] [PubMed] [Google Scholar]
- 25.Hung NJ, Johnson AW. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3718–3727. doi: 10.1128/MCB.26.10.3718-3727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao W, et al. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Bradatsch B, et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 29.Dembowski JA, Kuo B, Woolford JL., Jr. Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt545. doi:10.1093/nar/gkt545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chennupati V, et al. Signals and pathways regulating nucleolar retention of novel putative nucleolar GTPase NGP-1(GNL-2) Biochemistry. 2011;50:4521–4536. doi: 10.1021/bi200425b. [DOI] [PubMed] [Google Scholar]
- 31.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21 doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 32.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Granneman S, Petfalski E, Tollervey D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira-Cerca S, et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat Struct Mol Biol. 2012;19 doi: 10.1038/nsmb.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradatsch B, et al. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol. 2012;19:1234–1241. doi: 10.1038/nsmb.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nature biotechnology. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 37.Bassler J, et al. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 38.Hurt E, et al. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.SenGupta DJ, et al. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci U S A. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saveanu C, et al. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de la Cruz J, Sanz-Martinez E, Remacha M. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 2005;33:5728–5739. doi: 10.1093/nar/gki887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gwizdek C, et al. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci U S A. 2006;103:16376–16381. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frey S, Pool M, Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J Biol Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- 44.Vilardell J, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lebreton A, Saveanu C, Decourty L, Jacquier A, Fromont-Racine M. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J Biol Chem. 2006;281:27099–27108. doi: 10.1074/jbc.M602199200. [DOI] [PubMed] [Google Scholar]
- 46.Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J Cell Biol. 2012;197:747–759. doi: 10.1083/jcb.201112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebreton A, et al. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 49.Bassler J, Kallas M, Hurt E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J Biol Chem. 2006;281:24737–24744. doi: 10.1074/jbc.M604261200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.