Abstract
The bacterium, Vibrio parahaemolyticus was isolated from 776 patients at Hat Yai Hospital in Southern Thailand from 2006 to 2010. 51.3–73.6% of the isolates were tdh+ trh− and Group-specific PCR positive pandemic strains. A comparison of the number of V. parahaemolyticus isolates in this study and that from the same hospital in 2000–2005 indicates that this region of Thailandis endemic for V. parahaemolyticus.
Keywords: V. parahaemolyticus, pandemic strains, Thailand, GS-PCR
Introduction
Vibrio parahaemolyticus is a Gram-negative halophilic bacterium detected worldwide in the marine environment. It is a leading cause of gastroenteritis and generally associated with consumption of raw or improperly cooked seafood [1]. The infective dose of pathogenic V. parahaemolyticus has been estimated to be 105 to 107 pathogenic organisms [2]. The incubation period ranges from 4 to 96 h and the illness lasts for 2–3 days. Clinical symptoms include abdominal pain, watery diarrhea, nausea, vomiting and the disease is occasionally associated with headache, fever and chills [1]. However, not all strains of V. parahaemolyticus are pathogenic. Most clinical strains possess a major virulence factor, the thermostable direct hemolysin (TDH), and exhibit β-hemolysis on Wagatsuma agar [3]. Another virulence factor, the TDH-related hemolysin (TRH) has also been associated with some food-poisoning outbreaks [4]. TDH and TRH are encoded by the tdh and trh genes respectively. The percentage of tdh and trh genes detected in the environment varies and depends on both the location and the detection techniques. However, around 90% of clinical isolates possess either the tdh gene, the trh gene, or both [5–7].
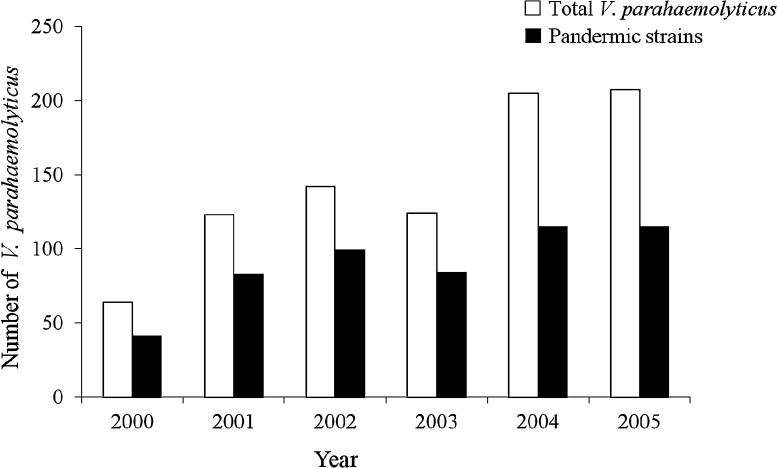
In Thailand, V. parahaemolyticus was first reported in 1970 [8]. Since then, infections have been reported from various communities [9, 10]. The incidence of infections in Thailand was unremarkable until 1996 when apandemic strain with the tdh virulence gene and O3:K6 serotype emerged [11, 12]. Group-specific PCR (GS-PCR) has been established to detect nucleotide variations unique to the pandemic clone within the 1,364 bp toxRS region [13]. The first report of the O3:K6 pandemic strain was obtained from patients at a Hat Yai Hospital in southern Thailand. One isolate of the O3:K6 pandemic strain was detected in shellfish purchased from the same area, indicating that people in this area were at risk of V. parahaemolyticus infection [12]. Thus, a surveillance program has been established at Hat Yai Hospital, the main public hospital in Songkhla province, as a public health warning. A total of 865 patients infected with V. parahaemolyticus were reported from 2000 to 2005 [7]. Approximately 64.1–69.7% of the isolates between 2000 and 2003 were pandemic strains, defined as GS-PCR positive tdh+ trh− (Fig. 1). However, in 2004 and 2005, the percentage of infections by these strains decreased to 56.1 and 55.5%, respectively. It has been postulated that people previously exposed to pandemic strains may have acquired immunity to the pandemic strains although they continue to be susceptible to non-pandemic strains. Hence, we report our continued surveillance of infection by V. parahaemolyticus in Hat Yai Hospital from 2006 to 2010.
Fig. 1.
Number of totaland pandemic strains of V. parahaemolyitcus isolated from patients in Hat Yai Hospital from 2000 to 2005.
Materials and Methods
Sample collection and bacteriology
Rectal swabs were obtained from patients with diarrhea in Hat Yai Hospital, Songkhla Province, Thailand, between 2006 and 2010. Each sample was plated on MacConkey, Salmonella-Shigella and Thiosulfate Citrate Bile-Salts Sucrose (TCBS) agars (Difco, Md., U.S.A.). After incubation at 37°C overnight, sucrose non-fermenter colonies on TCBS were selected and were identified as V. parahaemolyticus by standard biochemical tests. Confirmation of V. parahaemolyticus was performed by PCR targeted to the toxR gene.
ToxR investigation
The test isolate was grown in Luria-Bertani (LB) broth containing 1% NaCl with shaking at 160 rpm at 37°C overnight. One milliliter of the broth culture was centrifuged, and the bacterial cells were washed with sterile saline (0.85% NaCl) and then suspended in it. The cell suspension was boiled for 10 min and the supernatant was obtained by centrifugation, diluted 10-fold in distilled water, and used as the DNA template for PCR amplification. To investigate the toxR gene, PCR using primers T4 and T7 was performed, as described previously [14].
Detection of tdh and trh genes
To determine the presence of the tdh and trh genes, the DNA template was prepared as described above. PCR was carried out using the primers D3–D5 and R2–R6, respectively [15].
Serotyping
The serotype of V. parahaemolyticus was established by the slide-agglutination technique using anti-O and anti-K antibodies (Denka Seiken, Tokyo, Japan). The test isolate was grown in tryptic soy broth containing 3% NaCl at 37°C for 18 h, and the bacterial cells were suspended in saline (3% NaCl). For the determination of the K serotype, the bacterial cell suspension was subjected to agglutination with specific anti-K antibodies. For the O serotype, the bacterial cell suspension was autoclaved at 121°C for 1 h after which the autoclaved bacterial cells were agglutinated with specific anti-O antibodies.
GS-PCR
GS-PCR was carried out using the primers GS-VP1 and GS-VP2 and the reaction was carried out as previously described [13].
Statistical analysis
Student’s t test was used for comparison of the results.
Results
From 2006 to 2010, a total of 776 V. parahaemolyticus isolates were obtained from patients in Hat Yai Hospital (Table 1). The highest number was in 2006 and the lowest number was in 2009. The bacteria were classified into four groups: tdh+trh−, tdh+trh+, tdh−trh− and tdh−trh+ based on presence or absence of the tdh or trh toxin genes. Most of the clinical V. parahaemolyticus isolates were in the tdh+trh− group, which was divided into pandemic (GS-PCR positive) and non-pandemic (GS-PCR negative) strains. The predominant isolates of V. parahaemolyticus in each year were pandemic strains of which the highest number, 73.6%, and the lowest number, 51.3%, were from 2008 and 2010, respectively (Table 1). The numbers of isolates in the other three groups, tdh+trh+, tdh−trh− and tdh−trh+, were highest in 2007 (7.9%), 2010 (12.6%) and 2009 (2.7%) respectively. Two isolates of V. parahaemolyticus in the tdh−trh− group obtained in 2009 were GS-PCR positive.
Table 1.
Characteristics of V. parahaemolyticus isolates from Hat Yai Hospital from 2006 to 2010.
| Year | total | No. of isolates (%) |
||||
|---|---|---|---|---|---|---|
|
tdh+trh− |
tdh+trh+ | tdh−trh− | tdh−trh+ | |||
| GS-PCR positive | GS-PCR negative | |||||
| 2006 | 214 | 138 (64.5) | 52 (24.3) | 7 (3.3) | 16 (7.5) | 1 (0.4) |
| 2007 | 139 | 83 (59.7) | 26 (18.7) | 11 (7.9) | 17 (12.2) | 2 (1.5) |
| 2008 | 193 | 142 (73.6) | 32 (16.6) | 6 (3.1) | 11 (5.7) | 2 (1.0) |
| 2009 | 111 | 67 (60.4) | 28 (25.2) | 4 (3.6) | 9* (8.1) | 3 (2.7) |
| 2010 | 119 | 61 (51.3) | 37 (31.1) | 6 (5.0) | 15 (12.6) | 0 |
| total | 776 | 491 | 175 | 34 | 68 | 8 |
* Two isolates were GS-PCR positive
Every year the most prevalent serotype was O3:K6, which accounted for 46.8% of the total isolates (Table 2). Also, O3:K6 was the predominant serotype among the pandemic strains, followed by O4:K8 and O1:K25 (Table 3). The serotypes of isolates from non-pandemic strains were variable; the predominant serotypes were O1:K56, O4:K8, O1:KUT and O4:K9 (Table 2).
Table 2.
Serotypes of V. parahaemolyticus isolated between 2006 and 2010.
| Year (total isolate) | No. of isolates* | Serotype |
|---|---|---|
| 2006 (214) | 107 | O3:K6 |
| 31 | O4:K68 | |
| 19 | O1:K56 | |
| 13 | O4:K8 | |
| 5 | O1:KUT †, O4:K55 | |
| 4 | O11:KUT, O12:KUT | |
| 3 | O2:K3, O3:K29, O5:KUT, | |
| 2 | O1:K9, O4:K9, OUT:KUT | |
| 1 | O1:K25, O3:K5, O3:K29, O3:K53, O4:K34, O4:K49, O4:K11, O5:K19, O5:K61, O8:KUT, O11:K40 | |
| 2007 (139) | 73 | O3:K6 |
| 9 | O1:KUT | |
| 7 | O1:K25 | |
| 5 | O4:K8, O4:K68 | |
| 4 | O1:K56 | |
| 3 | O13:K5, O4:K11, O5:KUT, O8:KUT, O11:KUT | |
| 2 | O1:K69, O3:K5, OUT:KUT | |
| 1 | O1:K9, O1:K32, O1:K64, O2:K3, O2:K28, O3:K75, O3:KUT,O4:K13, O4:KUT, O9:KUT,O10:K19,O10:KUT, O12:KUT, O13:K75,OUT:K6 | |
| 2008 (193) | 106 | O3:K6 |
| 32 | O1:K25 | |
| 9 | O1:K56 | |
| 7 | O1:KUT | |
| 6 | O4:K8 | |
| 3 | O12:KUT | |
| 2 | O2:K3, O4:K13, O4:K53, O4:K55, O5:KUT, O8:K74, O9:K44 | |
| 1 | O1:K9, O1:K58, O1:K69, O3:K5, O3:K29, O3:K45, O3:K75, O4:K12, O4:K68, O5:K17, O5:K61, O8:K38, O8:K41, O8:K44, O9:K44, O11:K22 | |
| 2009 (111) | 49 | O3:K6 |
| 18 | O4:K68 | |
| 9 | O4:K9 | |
| 6 | O1:K56 | |
| 5 | O1:KUT | |
| 4 | O2:K3 | |
| 3 | O4:K8 | |
| 2 | O1:K25, O3:KUT, O8:K22, O11:K36 | |
| 1 | O1:K9, O3:K29, O4:K13, O4:K42, O4:K55, O5:K15, O8:K32, O9:K44, O10:KUT | |
| 2010 (119) | 47 | O3:K6 |
| 24 | O1:KUT | |
| 15 | O4:K9 | |
| 7 | O1:K56, O4:K8 | |
| 4 | O3:KUT | |
| 3 | O10:KUT, O11:KUT | |
| 2 | O4:K4, O4:KUT, O5:KUT | |
| 1 | O4:K3, O8:KUT, O8:K70 |
* No. of isolates per serotype.
† UT, Untypeable
Table 3.
Predominant serotypes of GS-PCR positive strains.
| Year | Serotypes |
|||
|---|---|---|---|---|
| O3:K6 | O4:K68 | O1:K25 | Others | |
| 2006 | 105 | 30 | 1 | 2 |
| 2007 | 70 | 5 | 7 | 1 |
| 2008 | 106 | 1 | 31 | 4 |
| 2009 | 47 | 16 | 2 | 2 |
| 2010 | 47 | 0 | 0 | 14 |
Discussion
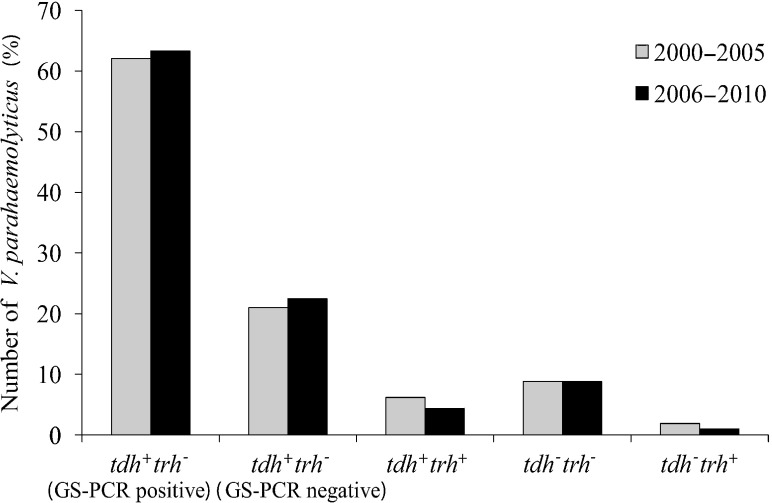
Infections caused by V. parahaemolyticus were investigated between 2006 and 2010. It was found that most of the isolates were pandemic strains and belonged to the O3:K6 serotype. This result is consistent with the findings since 2000 in Hat Yai Hospital, where O3:K6 pandemic strains were the main isolates [7]. This pandemic clone is a permanent part of the bacterial flora in this region. The reasons remains unknown, because it has been demonstrated that the effects of environmental factors such as temperature (either 4 or 50°C), acid stress (pH 4) and low-salinity stress on O3:K6 pandemic strains and other non-pandemic strains are not different [16]. In this study, the percentage of O3:K6 pandemic strains between 2006 and 2010 was alternatively high and low, but the total percentage median of pandemic strains detected in this study (63.3%) was not significantly different from that (62.1%) detected in 2000–2005 (Fig. 2). This indicates that the incidence of V. parahaemolyticus infection by the pandemic strains in this southern part of Thailand did not increase.
Fig. 2.
Comparison of the number of V. parahaemolyticus isolates in each of five groups of toxin gene profile from patients in Hat Yai Hospital between 2000–2005 and 2006–2010.
No significant difference was detected in any of the groups of V. parahaemolyticus (P > 0.05)
The clonal diversity of the pandemic strains from this study was similar to that observed in our previous investigation in 2000–2005 [7]. In 2010, however, serotype O4:K68 and O1:KUT were not detected, and four other serotypes (14 isolates) including O1:KUT (11), O4:K9 (2) and O3:KUT (1) were identified as pandemic strains (Table 3).
The number of isolates belonging to tdh−trh− group was higher than the number of isolates belonging to tdh+trh+ and tdh−trh+ groups and did not differ from the number previously detected in 2000–2005 (Fig. 2). Most clinical isolates of V. parahaemolyticus harbor either tdh or trh or both. These genes have been demonstrated to be the virulence genes involved in diarrhea pathogenicity [17, 18]. The mechanism of the disease caused by tdh−trh− V. parahaemolyticus is not clearly understood, but it has been demonstrated that a clinical V. parahaemolyticus strain lacking tdh gene resulted from the IS-mediated tdh gene deletion of a tdh+ strain [19, 20]. In this study, we also detected two isolates of tdh−trh− that belong to the O3:K6 serotype and GS-PCR positive pandemic group (Table 1). Therefore, it is possible that the tdh gene was deleted in those strains.
Mollusks are important sources of V. parahaemolyticus because they are filter feeders. In Thailand, consumption of mollusks, especially cockles contaminated with V. parahaemolyticus, is probably a major cause of acute gastroenteritis because the popular way to prepare this seafood is by semi-cooking. The average amount of V. parahaemolyticus reported from cockles was 5.1 × 103 cfu/g [21]. Infection by this bacterium is regularly encountered. An effective way to reduce V. parahaemolyticus contamination in cockles is needed. It has been reported that the number of V. parahaemolyticus in oysters decreased after they were stored at low temperatures of 0 to 5°C [22]. Therefore, simple refrigeration of cockles in the market may help to minimize the risk of infection.
In conclusion, investigation of V. parahaemolyticus isolates from Hat Yai Hospital from 2006 to 2010 revealed that the predominant isolates were pandemic O3:K6 strains. The number of isolates belonging to each toxin gene profile was not significantly different from the number from the same hospital in 2000–2005. This indicates that people in this area are constantly exposed to pathogenic V. parahaemolyticus at the same level. Therefore, this region of Thailand is probably endemic for V. parahaemolyticus. In the future it would be worthwhile to investigate the factors involved in this infection. It is also important to find methods to reduce the amount of V. parahaemolyticus in shellfish in the market.
Acknowledgements
This work was supported in part by funds from the Commission on Higher Education, Thailand, the Royal Golden Jubilee Ph.D. program (Grant No. PHD/0066/2550), National Science and Technology Development Agency, Thailand, and the Japan Society for the Promotion of Science, JSPS (KAKENHI 19101010).
References
- 1.Yeung PMS, Boor KJ. Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog Dis 2004; 1: 74–88 [DOI] [PubMed] [Google Scholar]
- 2.Sanyal SC, Sen PC. Human volunteer study on the pathogenicity of Vibrio parahaemolyticus. In Fujino T, Sakaguchi G, Sakazaki R, Takeda Y, eds. International Symposium on Vibrio parahaemolyticus Tokyo: Saikon Publishing Company; 1974
- 3.Nishibuchi M, Kaper JB. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect Immun 1995; 63: 2093–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda T, Ni Y, Miwatani T. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative V. parahaemolyticus and related to the thermostable direct hemolysin. Infect Immun 1988; 56: 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deepanjali A, Sanath Kumar H, Karunasagar I, Karunasagar I. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the Southwest Coast of India. Appl Environ Microbiol 2005; 71: 3575–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutierrez West CK, Klein SL, Lovell CR. High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a Pristine Estuary. Appl Environ Microbiol 2013; 79: 2247−2252 DOI: 10.1128/AEM.03792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wootipoom N, Bhoopong P, Pomwised R, Nishibuchi M, Ishibashi M, Vuddhakul V. A decrease in proportion of infections by pandemic Vibrio parahaemolyticus in Hat Yai Hospital, southern Thailand. J Med Microbiol 2007; 56: 1630–1638 [DOI] [PubMed] [Google Scholar]
- 8.Suthienkul O. Infectious Diseases. 2 ed. Bangkok: Holistic publishing; 1998.
- 9.Udompat P, Sukuonsakul M, Pokawattana L, Guharat S, Hanshaoworakul W. Food poisoning outbreak in a hotel, Bangkok. J Health Sci 2001; 13: 682–688 [Google Scholar]
- 10.Jatapai A, Moungthong B, Thunyaharn S, Huttayananont S, Rangsin R. An acute gastroenteritis outbreak of Vibrio parahaemolyticus O4:K55 in Nursing College, Thailand Trop Biomed 2010; 27: 265–274 [PubMed] [Google Scholar]
- 11.Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay AK, Garg S, Bhattacharya SK, Nair GB, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol 1997; 35: 3150–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuddhakul V, Chowdhury A, Laohaprertthisan V, Pungrasamee P, Patararungrong N, Thianmontri P, Ishibashi M, Matsumoto C, Nishibuchi M. Isolation of a pandemic O3:K6 clone of a Vibrio parahaemolyticus strain from environmental and clinical sources in Thailand. Appl Environ Microbiol 2000; 66: 2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, Wong HC, Depaola A, Kim YB, Albert MJ, Nishibuchi M. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol 2000; 38: 578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol 1999; 37: 1173–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, Takano J, Nishibuchi M, Takeda Y. Detection of thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes 1992; 6: 477–487 [DOI] [PubMed] [Google Scholar]
- 16.Wong HC, Liu SH, Wang TK, Lee CL, Chiou CS, Liu DP, Nishibuchi M, Lee BK. Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl Environ Microbiol 2000; 66: 3981–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda T, Iida T. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev Med Microbiol 1993; 4: 106–113 [Google Scholar]
- 18.Takahashi A, Sato Y, Shiomi Y, Cantarelli VV, Iida T, Lee M, Honda T. Mechanism of chloride secretion induced by thermostable direct hemolysin of Vibrio parahaemolyticus in human colonic tissue and intestinal epithelial cell line. J Med Microbiol 2000; 49: 801–810 [DOI] [PubMed] [Google Scholar]
- 19.Bhoopong P, Palittapongarnpim P, Pomwised P, Kiatkittipong A, Kamruzzaman M, Nakaguchi Y, Nishibuchi M, Ishibashi M, Vuddhakul V. Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J Clin Microbiol 2007; 45: 1544–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamruzzaman M, Bhoopong P, Vuddhakul V, Nishibuchi M. Insertion sequence-mediated deletion of thermostable direct hemolysin gene (tdh) in Vibrio parahaemolyticus. Gene 2008; 421: 67–73 [DOI] [PubMed] [Google Scholar]
- 21.Thongchan J, Bhoopong P, Yingkajorn M, Nishibuchi M, Vuddhakul V. Total number, virulence genes, and heterogeneity of Vibrio parahaemolyticus in a single shellfish. Science Asia 2013; 39: 230−235 [Google Scholar]
- 22.Shen X, Cai Y, Liu C, Liu W, Hui Y, Su YC. Effect of temperature on uptake and survival of Vibrio parahaemolyticus in oysters (Crassostrea plicatula). Int J Food Microbiol 2009; 136: 129–132 [DOI] [PubMed] [Google Scholar]