Abstract
Human immunodeficiency virus type 1 (HIV-1) circulating recombinant form (CRF) 07_BC has caused serious HIV-1 epidemics among injecting drug users (IDUs) in East Asia. Little is known about the characteristics of the virus and its impact on disease progression among the infected individuals. In this study, we compared immunological progression between 423 IDUs infected with CRF07_BC and 194 men who have sex with men (MSM) with primary subtype B infection, and a representative full-length CRF07_BC molecular clone, pCRF07_BC, was constructed to characterize the virus. We found that IDUs infected with CRF07_BC had significantly slower immunological progression in the Cox proportional hazards model (hazard ratio: 0.30; 95% confidence interval: 0.13–0.69; P=0.004). The constructed recombinant CRF07_BC viruses had a reduced processing of the Gag/Gag-Pol polyproteins, a decreased incorporation of Vpr in the virus particle, tethering of virus particles on the plasma membrane and decreased virus growth kinetics. These phenotypes are related to the unique 7-amino acid deletion in the p6 of CRF07_BC, since complementation of the 7-amino acid in pCRF07_BC could improve the defective phenotypes. In summary, compared with MSM infected with HIV-1 subtype B, IDUs infected with CRF07_BC had slower immunological progression, which is likely correlated with interference of virus particle maturation by the 7-amino acid deletion in p6.
Keywords: disease progression, growth kinetics, HIV subtype, injecting drug use, men who have sex with men, primary HIV infection
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) is characterized by its extensive genetic diversity, which can cause biological changes and affect virus pathogenicity, fitness and transmissibility.1 Several prospective, observational studies have demonstrated that various subtypes or circulating recombinant forms (CRFs) of HIV-1 result in different rates of disease progression.2,3,4,5,6,7 Nevertheless, only a few investigations were conducted in the modern era of HIV care to compare the disease progression between patients infected with subtype B and those infected with non-B subtypes.3,8
In Taiwan, male homosexual contact is the most common transmission route for HIV infection and these patients are mainly infected with subtype B.9 In 2003–2007, CRF07_BC, the most common strains among injecting drug users (IDUs) in China, was introduced to Taiwan along the drug trafficking routes and has caused an outbreak of HIV-1 infections among IDUs.10,11 The outbreak that reached its peak in 2005 and decreased after 2006 with the implementation of harm reduction program has changed the landscape of HIV epidemics in Taiwan, with injecting drug use having emerged as the second most common transmission route for HIV. IDUs who were infected with HIV during this outbreak shares about 97% nucleotide homology of HIV sequences and 98% of them were co-infected with hepatitis C virus (HCV).12 Despite the fact that CRF07_BC infections have posed a serious public health threat in East Asia, the natural history of CRF07_BC infections has never been well studied.
In CRF07_BC isolates, a 1–13-amino acid deletion is frequently observed downstream of the highly conserved Pro-Thr-Ala-Pro (PTAP) motif of p6gag.13 The PTAP motif is critical for the late stage of viral assembly, and mutations introduced to alter the motif may lead to accumulation of virus particles on the plasma membrane.14,15 In a previous study, HIV-1 strains isolated from three out of eight long-term non-progressors were shown to harbor amino acid deletion at R16/F17, F17/G18 and E34/L35, respectively.16 These short deletions are all located between the PTAP and LXXLF motifs of p6gag. Although subsequent studies, using sequential short deletion mutants between E13 and T21 of p6gag, as well as a long fragment deletion mutant (S14–I31 of p6gag), suggested that these mutations had relatively tolerable effects on Gag processing and in vitro viral infectivity,17,18 the influences of deletion between P30–Y36 of p6gag, as observed in CRF07_BC in Taiwan, were not addressed.
In this study, we aimed to compare the immunological progression between two HIV-infected, antiretroviral therapy-naive cohorts: IDUs infected with CRF07_BC during the outbreak and patients who presented with sexually transmitted primary HIV infection with subtype B; and to investigate the influence of 7-amino acid deletion within p6 on viral replication by constructing a full-length CRF07_BC molecular clone.
MATERIALS AND METHODS
Study patients and setting
An IDU is defined as a person who had injected drugs before HIV seroconversion.19 In Taiwan, injecting drug use is a criminal offense, and HIV screening is mandatory for all prison inmates and persons entering correctional facilities. All IDUs reported as new cases of HIV infection from 2003 to 2007, with follow-up at the National Taiwan University Hospital and Yun-Lin Branch, were enrolled. Most HIV-infected IDUs received their first diagnosis of HIV infection when they were incarcerated, and the earliest date of the report to the health authorities was used as the date of infection. The non-IDU cohort included patients with primary HIV-1 infection, whose transmission routes were sexual contact, from January 1997, at the National Taiwan University Hospital and Far Eastern Memorial Hospital. The date of infection was estimated as described previously.20 Both of the cohorts received free-of-charge HIV care, with CD4 cell count and plasma HIV RNA load (plasma viral load (PVL)) measurements every 3–6 months according to the national treatment guidelines. All patients were antiretroviral therapy-naive, and had at least two CD4 counts during follow-up. The study was approved by the Research Ethics Committee of the participating hospitals (200905047R) and all subjects who agreed to participate in this study gave written informed consent.
Outcome assessment
Immunological progression was used as the primary endpoint, since IDUs usually have higher non-AIDS-related mortality and fewer Kaposi's sarcomas than men who have sex with men (MSM), which may confound the analysis of clinical outcomes (death or development of AIDS).19 Immunological progression was defined as consecutive CD4 cell count <350 cells/mm3 more than three months after HIV infection was diagnosed.20 The timing of immunological progression was estimated as the mid-point between the last date of CD4 cell count ≥350 cells/mm3 and the first date of CD4 cell count <350 cells/mm3.2 Participants were followed until 29 February 2012 and were censored if they died, were lost to follow-up, or initiated antiretroviral therapy, whichever occurred first.
Western blot analysis
Western blot was performed as described previously.21 The antibodies used were anti-p24gag (ab9071; Abcam Plc, MA, USA), anti-protease (ab8327; Abcam Plc, Cambridge, ENG, UK) and anti-Vpr (vN-20) (sc-17493; Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibodies. The secondary antibodies used were goat anti-mouse antibody (81–6520; Invitrogen, Carisbad, CA, USA) and donkey anti-goat antibody (sc-2020; Santa Cruz Biotechnology) conjugated with horseradish peroxidase. Hybridization signal was determined using the Fuji LAS-4000 luminescent image analyzer (Fujifilm Life Science, MA, USA).
Ultrathin transmission electron microscopy
pCRF07_BC, pCRF07_BC+p6, or HXB2 plasmids were respectively transfected into 293T cells using lipofectamine (Invitrogen). After 48 h, cells were trypsinized and resuspended in ice-cold PBS, pH 7.4. After centrifugation at 9000 rpm. for 1 min at 4 °C (Allegra 64R centrifuge; Beckman Coulter, Pasadena, CA, USA), PBS, pH 7.4 was removed and cell pellets were fixed by 2% glutaldehyde (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at 4 °C. Cell pellets were picked up and then dehydrated with graded ethanol solution (Merck, Whitehouse, NJ, USA). Finally, cells were embedded in Spurr's resin (Sigma-Aldrich). Embedded cell blocks were cut in ultrathin sections, collected on copper grids and stained with 1% uranyl acetate and 1% lead citrate (Sigma-Aldrich). The grids were then observed by the JEOL JEM-1400 electron microscope (JEOL, Tokyo, Japan).
Statistical analysis
Data were analyzed using SPSS (IBM Corporation, Armonk, NY, USA) and SAS (SAS Institute, NC, USA). Categorical data were analyzed using Chi-square or Fisher's exact tests, as appropriate, and continuous variables were compared using the Wilcoxon test. Time to immunological progression, the interval between the estimated dates of infection and immunological progression, was assessed with Kaplan–Meier plots. Cox proportional hazards regression analysis was used to estimate hazard ratios of immunological progression. Variables with a P value of <0.1 and with biological plausibility in univariate analyses were later selected for multivariate models. All tests were two-tailed and a P value of <0.05 was considered significant.
RESULTS
Baseline characteristics
The demographic and clinical characteristics of 423 IDUs and 194 non-IDU patients with HIV-1 infection in this study are shown in Table 1. These two groups differed significantly in several aspects. In general, the IDUs were older, were more likely to be female, had higher CD4 cell counts and lower PVL at baseline, and had higher prevalence of chronic hepatitis B or C viruses (HBV or HCV) co-infection. As reported previously,12 virtually all the IDUs that were diagnosed during the outbreak (from 2003 to 2007) were co-infected with HCV (98.1%, 363/370). Among the IDUs whose blood samples were available for genotyping, 93.4% (199/213) of them were infected with CRF07_BC, followed by CRF01_AE in 9 (4.2%) patients. IDUs with subtype data had higher PVL at baseline than those without subtype data (mean PVL, 4.18 vs. 3.88 log10 copies/mL; P<0.01), but did not differ with respect to other demographic characteristics (data not shown). The non-IDU group was mainly MSM (96.9%, 188/194) and infected with subtype B (91.7%, 144/157), and 62.3% (121/194) of them acquired HIV-1 infection between 2008 and 2011. A relatively lower percentage of non-IDU patients were infected with CCR5-tropic viruses as compared with that in the IDUs (86.6% vs. 94.5%, P=0.03).
Table 1. Comparisons of baseline characteristics between the IDUs and the non-IDU group.
| Characteristica | IDUs (n=423) | Non-IDU (n=194) | P |
|---|---|---|---|
| Male, N (%) | 372 (87.9%) | 194 (100%) | <0.01 |
| Age (year) | 32.2 (26.7–38.3) | 28.5 (23.9–33.0) | <0.01 |
| CD4 cell counts (cells/mm3) | 481 (388–629) | 404 (317–557) | <0.01 |
| PVL (log10 copies/mL) | 3.99 (3.47–4.39) | 5.17 (4.52–5.88) | <0.01 |
| Subtype, N (%) | <0.01 | ||
| B | 2/213 (0.9%) | 144/157 (91.7%) | |
| CRF07_BC | 199/213 (93.4%) | 0/157 (0) | |
| CRF01_AE | 9/213 (4.2%) | 4/157 (2.5%) | |
| Others | 3/213 (1.4%) | 9/157 (5.7%) | |
| Coreceptor tropismb, N (%) | |||
| CCR5-tropic only | 121/128 (94.5%) | 123/142 (86.6%) | 0.03 |
| Risk factor, N (%) | <0.01 | ||
| MSM/bisexual | 0 (0) | 188 (96.9%) | |
| Heterosexual | 0 (0) | 6 (3.1%) | |
| IDU | 423 (100%) | 0 (0) | |
| HBsAg (+), N (%) | 76/368 (20.7%) | 21/183 (11.5%) | 0.01 |
| Anti-HCV (+), N (%) | 363/370 (98.1%) | 6/184 (3.3%) | <0.01 |
| Year of diagnosis | <0.01 | ||
| 1997–2003 | 2 (0.5%) | 20 (10.3%) | |
| 2004–2007 | 403 (95.3%) | 53 (27.3%) | |
| 2008–2011 | 18 (4.2%) | 121 (62.3%) | |
| Number of CD4 cell measurements per patient | 4 (3–6) | 4 (3–8) | 0.64 |
Abbreviations: HBsAg, hepatitis B surface antigen; HCV, hepatitis C virus; IDU, injecting drug users; MSM, men who have sex with men; PVL, plasma HIV RNA load.
Data are median (interquartile range) for continuous variables.
Genotypic tropism analysis was conducted to determine the coreceptor usage using the protocol developed by Dr PR Harrigan of the BC Centre for Excellence in HIV/AIDS laboratory.22 A 10% false positive rate (FPR) was used in the G2P algorithm. The accession numbers of submitted sequences are KC787218–KC787345.
Immunological progression
The median follow-up duration was 37.7 months (interquartile range: 11.6–59.8 months) and there was no difference in the numbers of CD4 cell measurements per patient between IDU and non-IDU groups. To avoid misclassification, patients were excluded from the outcome analyses if they were infected with strains other than CRF_07BC in the IDU group and other than subtype B in the non-IDU group. Overall, 119 (35.7%) IDUs and 86 (53.4%) non-IDU patients developed immunological progression (P<0.001). Two IDUs and one non-IDU patient had opportunistic infections during the follow-up.
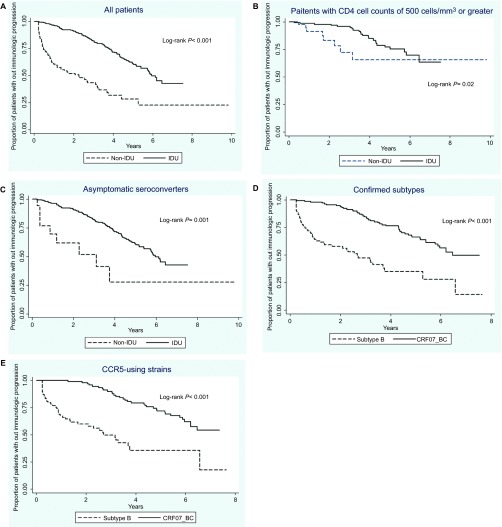
Kaplan–Meier plots showed a significantly shorter time to immunological progression in the non-IDU group than the IDU group (log-rank, P<0.001) (Figure 1A). Since non-IDU patients had lower baseline CD4 cell counts and many of them presented with acute retroviral syndrome, the symptomatic seroconverters and patients with baseline CD4 cell counts <500 cells/mm3 were excluded in the sensitivity analyses to eliminate their influence on immunological progression. The differences remained statistically significant (Figures 1B and 1C). An analysis restricted to patients with confirmed subtype yielded similar results. Overall, 50 (48.1%) subtype B-infected patients and 39 (36.8%) CRF07_BC-infected patients developed immunological progression. The median interval between the estimated date of infection and the onset of immunological progression was 30.7 months for patients infected with subtype B and 74.3 months for those infected with CRF07_BC (log-rank, P<0.001) (Figure 1D). Finally, when the analysis was confined to CCR5-using strains, CRF07_BC was still associated with significantly slower immunological progression (Figure 1E).
Figure 1.
Kaplan–Meier estimates of time to immunological progression in (A) all patients; (B) patients with initial CD4 cell count ≥500 cells/mm3; (C) asymptomatic seroconverters; (D) patients with confirmed infection with subtype B or CRF07_BC; and (E) patients with CCR5-using viruses, either subtype B or CRF07_BC.
In the multivariate Cox model after controlling for age, gender, baseline CD4 cell counts, PVL and symptoms of seroconversion, we found that patients infected with CRF07_BC had significantly slower immunological progression than those infected with subtype B (adjusted hazard ratio: 0.30; 95% confidence interval: 0.13–0.69; P=0.004) (Table 2).
Table 2. Prediction of covariates contributing to time to immunological progression among patients with confirmed infection with subtype B or CRF07_BC using the Cox model.
| Multivariate | |||
|---|---|---|---|
| HR | 95% CI | P | |
| CD4 cell counts (per 50 cells/mm3 increase) | 0.80 | 0.73–0.87 | <0.001 |
| Baseline PVL (per 1 log10 copies/mL increase) | 0.96 | 0.69–1.35 | 0.83 |
| Age (per 10-year increase) | 0.99 | 0.70–1.41 | 0.97 |
| Gender | |||
| Female | 1 | ||
| Male | 1.06 | 0.38–2.99 | 0.91 |
| Subtype | |||
| B | 1 | ||
| CRF07_BC | 0.30 | 0.13–0.69 | 0.004 |
| Coreceptor tropism | |||
| CCR5 | 1 | ||
| CXCR4 | 2.05 | 0.89–4.69 | 0.09 |
| Seroconversion | |||
| Asymptomatic | 1 | ||
| Symptomatic | 1.19 | 0.51–2.74 | 0.69 |
Abbreviations: CI, confidence interval; HR, hazard ratio; PVL, plasma HIV RNA load.
Characteristics of CRF07_BC molecular clone
Because a slow immunological progression was observed in CRF07_BC-infected IDUs as compared to subtype B-infected MSM, and both populations have the same ethnic background, the role of CRF07_BC in immunological progression was further investigated. A full-length, infectious CRF07_BC molecular clone, pCRF07_BC, was constructed (Supplementary Figure S1A). Phylogenetic analysis (Supplementary Figure S1B) and bootscanning analysis (Supplementary Figure S1C) indicated that pCRF07_BC was indeed a CRF07_BC subtype as described previously.23 The pCRF07_BC clone was representative of circulating CRF07_BC among the HIV-infected IDUs in Taiwan, since it exhibited very high sequence similarity to clinical CRF07_BC strains retrieved from IDU patients (pol median similarity: 99% range: 91%–100%) (env median similarity: 95% range: 81%–100%) (Supplementary Figures S2A and S2B). From the evolution tree analysis, pCRF07_BC and other CRF07_BC sequences retrieved from Taiwanese IDUs were closely related, but distinct from the CRF07_BC isolated from China (Supplementary Figure S2C), suggesting that the CRF07_BC outbreak in Taiwanese IDUs was originated from a single or very closely related CRF07_BC viruses.
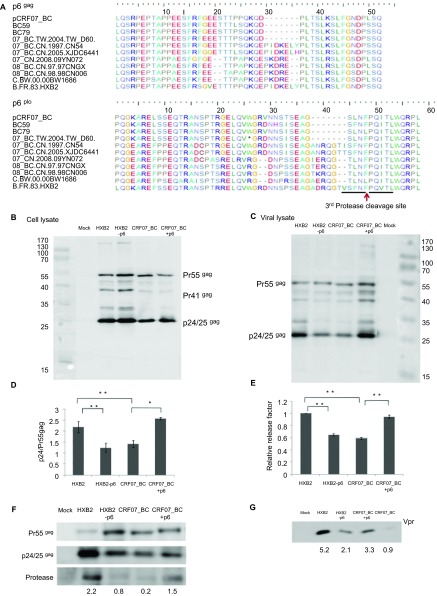
Reduced Pr55gag processing of CRF07_BC
A characteristic feature of pCRF07_BC and CRF07_BC isolated from HIV-infected IDUs in Taiwan was a 7-amino acid deletion in p6 (Figure 2A). In the p6pol, the 7-amino acid deletion was located immediately upstream of the C-terminal SNSF motif, whose deletion was shown to abrogate the regulation of protease activation by p6pol.25 As shown in Figures 2B and 2C, the amounts of p24gag, the final products of Gag processing, were reduced in both cell lysates and viral lysates from 293T cells transfected with pCRF07_BC. The ratio of p24gag to Pr55gag decreased significantly in pCRF07_BC-transfected cells as compared to that in subtype B HXB2-transfected cells (Figure 2D) and the viral release factor was significantly reduced in CRF07_BC viral lysates (Figure 2E). The reduced Pr55gag/Pr160Gag-Pol processing also resulted in decreased levels of protease and Vpr in the viral lysates of CRF07_BC (Figures 2F and 2G). The abrogated processing of Pr55gag/Pr160Gag-Pol and incorporation of Vpr proteins were likely correlated with the 7-amino acid deletion in p6, since the phenotypes were improved in the mutant CRF07_BC molecular clone with complementation of the 7-amino acid, pCRF07_BC+p6, and the reverse phenotypes were observed when the corresponding 7-amino acid was removed in HXB2-p6 (Figure 2).
Figure 2.
Gag and Gag-Pol polyprotein processing of CRF07_BC viruses. (A) Alignment of p6gag and p6pol amino acid sequences. The p6gag and p6pol amino acid sequences of pCRF07_BC were aligned with the prototype subtype B′ RL, CRF07_BC and CRF08_BC from China and subtype C from India. These sequences are aligned using MEGA5.0 and output data by BioEdit 7.1.9.24 Dashes indicate deletion of amino acids in comparison with the reference prototype B′ RL. The third protease cleavage site at the N-terminus of Pol was indicated. (B) Expression of Pr55gag; p41gag and p24/p25gag viral proteins in cell lysates of 293T transfectants. 293T cells were transfected with pCRF07_BC, pCRF07_BC+p6, HXB2 or HXB2-p6 plasmids. Forty-eight hours after transfection, the cell pellets were dissolved in lysis buffer and electrophoresed on a 10% SDS-PAGE. Gag proteins were detected by anti-p24gag antibody. For B and C, at least three independent experiments were conducted and the representative results were shown. (C) Expression of Pr55gag and p24/p25gag viral proteins in the viral lysates. The cell-free supernatants from 293T transfectants were centrifuged at 20 000 rpm. for 2 h through a 20% (w/v) sucrose cushion. The virus pellets were dissolved in lysis buffer and electrophoresed on a 10% SDS-PAGE. Gag protein was detected by anti-p24gag antibody. (D) Ratios of cellular p24gag/Pr55gag. The amounts of p24gag and Pr55gag in the Western blot were quantified using ImageJ software (http://rsb.info.nih.gov/ij/). The ratios represent the mean of three independent experiments with its respective standard deviation indicated. (E) Relative release factor. Relative release factor was calculated as the ratio of p24gag in the viral lysates/(Pr55gag+p24gag) in the cellular lysates. The data shown here represent the mean of three independent experiments with its respective standard deviation indicated. Incorporation of (F) protease and (G) Vpr in virus particle. The same amounts of viral lysates, as determined by the Bradford assay, were used in the F and G. The amount of protease and Vpr in the viral lysates was detected by anti-protease and anti-Vpr antibody, respectively. The signal strength of protease was quantified by ImageJ (http://rsb.info.nih.gov/ij/) and the values below the gel image represent the mean of three independent experiments. *P≤0.05; **P≤0.01.
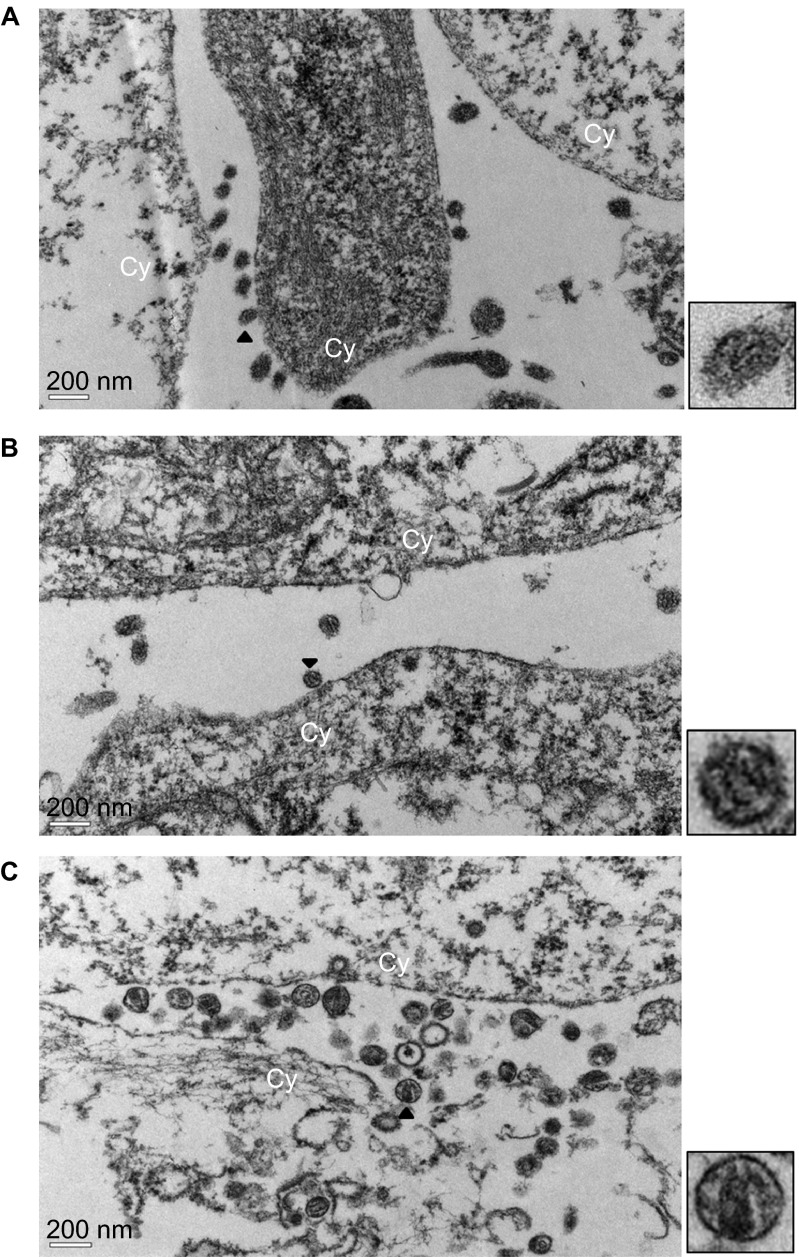
Electron microscopy analysis of virus particles
The maturation of HIV-1 into infectious virus particles requires the processing of Pr55gag and Pr160Gag-Pol polyproteins by protease,26 and the deletion of p6 was shown to significantly disrupt virus particle production and result in accumulation of tethered, immature virions on the plasma membrane.14,27 The influence of reduced Pr55gag and Pr160Gag-Pol processing on virus morphology was examined using the thin-section electron microscopy analysis. As shown in Figure 3A, viruses were tethered on the plasma membrane of 293T cells transfected with pCRF07_BC and the viruses did not have the characteristic cone-shaped core structures, as those observed in cells transfected with subtype B plasmid (Figure 3C). The virus release and formation of the cone-shaped core structure were improved in cells transfected with pCRF07_BC+p6 (Figure 3B).
Figure 3.
TEM analysis of HIV-1 viral particles. Thin sections of 293T cells transfected with (A) pCRF07_BC, (B) pCRF07_BC+p6 or (C) HXB2RU3 plasmids were analyzed by TEM. The zoomed images of virus particles indicated by arrow heads were shown at the right-hand side of the respective figures. Cy indicated the location of cytoplasm membrane. TEM, transmission electron microscopy.
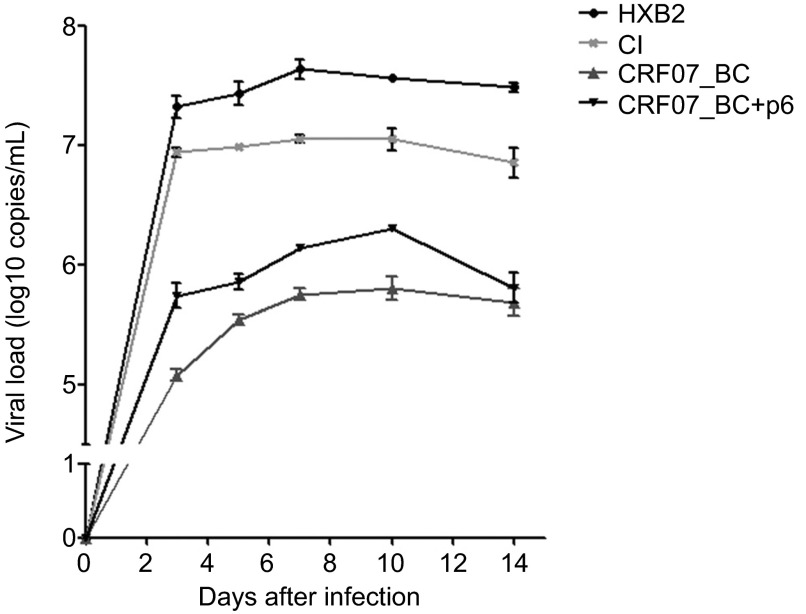
Growth kinetics of CRF07_BC clone
The growth kinetics of CRF07_BC in peripheral blood mononucleated cell was determined since reduced Pr55gag/Pr160Gag-Pol processing and incorporation of Vpr in virus particles were both shown to affect viral infectivity. As shown in Figure 4, CRF07_BC exhibited a reduced replication capability as compared to both subtype B reference strains, HXB2 (CXCR4-tropism) and CI (CCR5-tropism). The complementation of the 7 amino acids back to pCRF07_BC, pCRF07_BC+p6, was shown to have a significant improvement of the replication capability of CRF07_BC.
Figure 4.
Growth kinetics of CRF07_BC and subtype B in PBMCs. PBMCs were infected with equal amounts of CRF07_BC, CRF07_BC+p6, and two subtype B viruses, HXB2RU3 (CXCR4-tropism) and CI (CCR5 tropism). The culture supernatants were harvested at days 3, 5, 7, 10 and 14, and the virus titers in the supernatants were determined by real-time PCR with the condition described previously.21 The values represent the mean of three independent experiments and the error bars represent the standard deviation. PBMC, peripheral blood mononucleated cell; PCR, polymerase chain reaction.
DISCUSSION
In this study, we found that IDUs who were infected with CRF07_BC during the HIV outbreak in Taiwan had a significantly lower risk of immunological progression, compared with the sexually transmitted group, mainly MSM who were recently infected with subtype B. The differences remained statistically significant in multiple sensitivity analyses. To our knowledge, this is the first study on the natural course of CRF07_BC infection. The rate of immunological progression in our non-IDU group was similar to that observed in the recent European cohort infected with subtype B.28 The estimated time from seroconversion to CD4 cell counts <350 cells/mm3 was 4.25 years for 30- to 35-year-old IDUs, from Concerted Action on Seroconversion to AIDS and Death in Europe data, in which subtype B was the predominant strain (90%);29 yet the median time to immunological progression was longer for our IDUs infected with CRF07_BC (6.19 years).
HIV-1 subtypes are usually segregated among people with different risk factors and people living in different regions.30 As a result, comparisons of virulence between different subtypes are often hindered by potential confounders, such as race, socioeconomic status, access to health care and transmission modes.31 Inevitably, the two groups (MSM and IDUs) in our study, which represent the two leading HIV-infected populations in Taiwan, had unparalleled epidemiological backgrounds. However, many confounding factors that were difficult to control could not explain the slower immunological progression in IDUs. The older age and higher rates of co-infections with HBV and HCV in IDUs would unlikely lead to better outcomes.32,33 Because HIV-infected women have similar disease progression to men, the effect of higher proportion of women in the IDUs might be neglected.34,35 The impact of transmission routes on disease progression has been debated for a long time. However, after adjustments made for age, non-AIDS mortality and Kaposi's sarcoma, no notable effect on disease progression was found between different risk groups.19,35 In addition, rates of CD4 decline are also comparable between MSM and IDUs.29,36 While the IDUs had lower PVL and higher CD4 cell counts at baseline in our study, they remained in slower progression than non-IDU group after adjustments for these two strong predictors in the Cox model. Finally, after we confined our analysis to CCR5-using strains, infection with CRF07_BC was still significantly associated with slower immunological progression (Figure 1E). Hence, we hypothesize that viral diversity may play a major role in the difference of immunological progression.
Previously, deletion of the first 36 amino acids of p6 was shown to significantly reduce Pr55gag/Pr160Gag-Pol polyprotein processing and virus budding.14 Later, a 2-amino acid in-frame deletion in the p6gag from three out of eight long-term non-progressors has been reported16 and these short deletions were all located between the highly conserved PTAP and LXXLF motifs of p6gag. While the PTAP motif is critical for the late stage of viral assembly and mutations introduced to alter this motif may lead to accumulation of virus particles on the plasma membrane,14,15 the LXXLF motif is participating in the virus budding through interaction with the cellular factor, AIP1/ALIX.37 Although controversial results were reported in subsequent studies,17,18 the influences of deletion between P30-Y36 of p6gag, as observed in CRF07_BC in Taiwan, were not addressed in any of these studies. In the present study, we demonstrated that the naturally occurring 7-amino acid deletion in the p6 of CRF07_BC correlated with reduced Pr55gag/Pr160Gag-Pol processing, tethering of virus particles on plasma membrane and reduced incorporation of Vpr in virus particles, which could contribute to reduced replication of CRF07_BC viruses.
Besides the 7-amino acid deletion in p6gag, several characteristic genetic variations were also observed in pCRF07_BC and most CRF07_BC sequences, and their roles in slow immunological progression cannot be neglected. First, a S46F variation was observed in the highly conserved 41KGLGISY47 motif of CRF07_BC Tat, which resulted in a significantly lower transactivation activity on either authentic long terminal repeat or subtype B long terminal repeat compared to subtype B Tat (Supplementary Figure S3). Whether the reduced Tat transactivation activity is correlated with the relatively reduced viral protein expression of CRF07_BC (Figure 2B) requires further clarification. Second, an R50N variation was observed in Rev, which is known to export unspliced or partially spliced HIV-1 transcripts from nucleus.38 Because R50N is located within the RNA binding/nuclear localization domain (35RQARRNRRRRWRERQR50), it might affect Rev function by disrupting the binding of Rev to rev response element. Third, several variations were observed in the accessory genes vif, vpr and vpu. Vif is known to promote HIV-1 infectivity through inducing the degradation of the intrinsic host defense protein, APOBEC3G, by recruitment of the Cullin 5-Elongin B-Elongin C E3 ubiquitin ligase.39 In CRF07_BC, a 1-amino acid insertion, Asn, was observed within the motif of Cullin 5 binding domain. While whether this insertion has steric hindrance effect on Cullin 5 binding remains to be determined, a correlation of a 2-amino acid insertion (between D61 and A62) in Vif with a 20-fold reduction of virus replication in long-term non-progressors was reported by Alexander et al.40 Vpr is involved in nuclear localization of viral pre-integration complexes and induction of G2/M cell cycle arrest by blocking formation of the p34cdc2 cyclin B complex.41,42 Lum and his colleagues43 first reported that the frequency of R77Q mutant was higher among long-term non-progressors and this mutation impaired the induction of T-cell apoptosis by Vpr. Such observation was also reported by Mologni et al.44 from Italy. Another Vpr variation F72L was shown to present in long-term non-progressors, and was correlated with impaired nuclear import and viral incorporation of Vpr.45 In our study, both R77Q and F72Y were observed uniquely in CRF07_BC and a reduced incorporation of Vpr was observed in CRF07_BC viral lysates (Figure 2G). Whether these variations are correlated with reduced CD4+ T-cell depletion observed in CRF07_BC-infected IDUs is currently under investigation.
Our study has several limitations. First, although the numbers of CD4 and PVL measurements were similar between the two groups of patients, follow-up of the IDUs tended to be less regular after they were released from the prisons, which may preclude us from exactly determining the onset of immunological progression. Second, we do not have IDUs infected with CRF07_BC that exhibits full-length p6 as controls to demonstrate the significance of 7-amino acid deletion in p6. In China, the appearance of CRF07_BC with 7-amino acid deletion was around 2003, which was about the same time of CRF07_BC outbreak in Taiwan. Controversial results about the effect of 7-amino acid deletion on HIV-1 viral load and CD4 counts have been reported.13,46 A higher CD4 count and lower HIV viral loads in patients infected with CRF07_BC with a deletion were reported by Song et al.,13 and, limited by the smaller sample size, the difference was not statistically significant. Another report by Meng et al.46 described a more rapid increase of HIV viral loads in patients infected with CRF07_BC with 7-amino acid deletion than those infected with CRF07_BC without the 7-amino-acid deletion. Third, data on the clinical outcomes of our cohorts, such as mortality and progression to AIDS, are limited. During the follow-up, only three patients developed HIV-related opportunistic infections, mainly because all our patients had been provided with free-of-charge access to HIV care, including combination antiretroviral therapy and monitoring of CD4 count and plasma HIV RNA load, and had relative high CD4 counts (≥350 cells/mm3). With respect to mortality, a prior nationwide study from Taiwan Centers for Disease Control have clearly demonstrated the differences of clinical characteristics and outcomes between the IDU and non-IDU groups.47 The mortality rates in MSM and IDUs were 2.9 and 2.5 deaths per 100 person-years, respectively. However, AIDS-related deaths accounted for 64.2% of the total deaths in the MSM group, but only 16.9% in the IDU group (P<0.0001). Deaths from other medical diseases or from accident deaths were both higher in the IDU group than in sexual transmission groups. Only 1.6% of the IDUs developed AIDS within 3 months after HIV diagnosis, compared to 19.4% in the MSM group.47 These clinical data further support our findings of slow disease progression in the IDU group. Finally, we did not investigate the factors such as nutrition and socioeconomic status, human leukocyte antigen (HLA) alleles and access to health care, all of which can be potential confounders of disease progression. For example, HLA-B7 is associated with poor cytotoxic T lymphocyte responses and increased viremia in subtype B-infected individuals,48,49 and HLAB57 is associated with slow progression in subtypes B- and C-infected persons.48 Nevertheless, based on our previous study, the prevalence of these protective HLA is relatively rare in Taiwanese population and would be less likely to account for the observed slower immunological progression among IDUs.50
In conclusion, IDUs who were infected with the outbreak strain, CRF07_BC, had significantly slower immunological progression, compared with MSM who were infected with subtype B. The heterogeneity in immunological progression could be attributed to subtype differences, especially the naturally occurring 7-amino acid deletion observed in p6 of CRF07_BC.
Acknowledgments
The authors would like to thank Dr Tun-Hou Lee, Department of Immunology and Infectious Diseases, Harvard School of Public Health, for the critical review of the paper, and the staff of the Statistical Analysis Laboratory, Department of Medical Research, Kaohsiung Medical University Hospital and Kaohsiung Medical University for the assistance in statistical analysis. This work was supported by grants from the National Science Council, Taiwan (98-2314-B-002-043-MY2) and from National Taiwan University, Taipei, Taiwan (100R71806 and 101R7806).
Footnotes
Note: Supplementary Information for this article can be found on Emerging Microbes and Infections' website (http://www.nature.com/EMI/).
Supplementary Information
References
- Arien KK, Vanham G, Arts EJ. Is HIV-1 evolving to a less virulent form in humans. Nat Rev Microbiol. 2007;5:141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwanuka N, Laeyendecker O, Robb M, et al. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis. 2008;197:707–713. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- Keller M, Lu Y, Lalonde RG, Klein MB. Impact of HIV-1 viral subtype on CD4+ T-cell decline and clinical outcomes in antiretroviral naive patients receiving universal healthcare. AIDS. 2009;23:731–737. doi: 10.1097/QAD.0b013e328326f77f. [DOI] [PubMed] [Google Scholar]
- Vasan A, Renjifo B, Hertzmark E, et al. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis. 2006;42:843–852. doi: 10.1086/499952. [DOI] [PubMed] [Google Scholar]
- Kaleebu P, French N, Mahe C, et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002;185:1244–1250. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- Kanki PJ, Hamel DJ, Sankale JL, et al. Human immunodeficiency virus type 1 subtypes differ in disease progression. J Infect Dis. 1999;179:68–73. doi: 10.1086/314557. [DOI] [PubMed] [Google Scholar]
- Kiwanuka N, Robb M, Laeyendecker O, et al. HIV-1 viral subtype differences in the rate of CD4+ T-cell decline among HIV seroincident antiretroviral naive persons in Rakai district, Uganda. J Acquir Immune Defic Syndr. 2010;54:180–184. doi: 10.1097/QAI.0b013e3181c98fc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng OT, Lin L, Laeyendecker O, et al. Increased rate of CD4+ T-cell decline and faster time to antiretroviral therapy in HIV-1 subtype CRF01_AE infected seroconverters in Singapore. PLoS ONE. 2011;6:e15738. doi: 10.1371/journal.pone.0015738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SY, Chen MY, Lee CN, et al. Trends of antiretroviral drug resistance in treatment-naive patients with human immunodeficiency virus type 1 infection in Taiwan. J Antimicrob Chemother. 2008;61:689–693. doi: 10.1093/jac/dkn002. [DOI] [PubMed] [Google Scholar]
- Chang SY, Sheng WH, Lee CN, et al. Molecular epidemiology of HIV type 1 subtypes in Taiwan: outbreak of HIV type 1 CRF07_BC infection in intravenous drug users. AIDS Res Hum Retroviruses. 2006;22:1055–1066. doi: 10.1089/aid.2006.22.1055. [DOI] [PubMed] [Google Scholar]
- Lin HH, Shih YL, Liu YC, et al. An epidemic of HIV type I CRF07_BC infection among injection drug users in Taiwan. J Acquir Immune Defic Syndr. 2006;42:248–255. doi: 10.1097/01.qai.0000214818.80539.da. [DOI] [PubMed] [Google Scholar]
- Liu JY, Lin HH, Liu YC, et al. Extremely high prevalence and genetic diversity of hepatitis C virus infection among HIV-infected injection drug users in Taiwan. Clin Infect Dis. 2008;46:1761–1768. doi: 10.1086/587992. [DOI] [PubMed] [Google Scholar]
- Song YH, Meng ZF, Xing H, et al. Analysis of HIV-1 CRF07_BC gag p6 sequences indicating novel deletions in the central region of p6. Arch Virol. 2007;152:1553–1558. doi: 10.1007/s00705-007-0973-6. [DOI] [PubMed] [Google Scholar]
- Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L, Weiskopf E, Greenough TC, et al. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J Virol. 2000;74:4361–4376. doi: 10.1128/jvi.74.9.4361-4376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleiber G, Peters S, Martinez R, Cmarko D, Meylan P, Telenti A. The central region of human immunodeficiency virus type 1 p6 protein (Gag residues S14–I31) is dispensable for the virus in vitro. J Gen Virol. 2004;85:921–927. doi: 10.1099/vir.0.19576-0. [DOI] [PubMed] [Google Scholar]
- Pikora CA, Wittish C, Desrosiers RC. p6gag of human and simian immunodeficiency viruses is tolerant to small in-frame deletions downstream of the late domain. Virology. 2006;346:479–489. doi: 10.1016/j.virol.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Prins M, Veugelers PJ. Comparison of progression and non-progression in injecting drug users and homosexual men with documented dates of HIV-1 seroconversion. European Seroconverter Study and the Tricontinental Seroconverter Study. AIDS. 1997;11:621–631. doi: 10.1097/00002030-199705000-00010. [DOI] [PubMed] [Google Scholar]
- Goujard C, Bonarek M, Meyer L, et al. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis. 2006;42:709–715. doi: 10.1086/500213. [DOI] [PubMed] [Google Scholar]
- Lin PH, Ke YY, Su CT, et al. Inhibition of HIV-1 Tat-mediated transcription by a coumarin derivative, BPRHIV001, through the Akt pathway. J Virol. 2011;85:9114–9126. doi: 10.1128/JVI.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson LC, Moores A, Low AJ, et al. Improved detection of CXCR4-using HIV by V3 genotyping: application of population-based and “deep” sequencing to plasma RNA and proviral DNA. J Acquir Immune Defic Syndr. 2010;54:506–510. doi: 10.1097/QAI.0b013e3181d0558f. [DOI] [PubMed] [Google Scholar]
- Piyasirisilp S, McCutchan FE, Carr JK, et al. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J Virol. 2000;74:11286–11295. doi: 10.1128/jvi.74.23.11286-11295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Partin K, Zybarth G, Ehrlich L, DeCrombrugghe M, Wimmer E, Carter C. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1991;88:4776–4780. doi: 10.1073/pnas.88.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausslich HG, Traenckner AM, Rippmann F. Expression and characterization of genetically linked homo- and hetero-dimers of HIV proteinase. Adv Exp Med Biol. 1991;306:417–428. doi: 10.1007/978-1-4684-6012-4_54. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76:105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen M, Pasternak AO, Grijsen ML, et al. HIV-1 dual infection is associated with faster CD4+ T-cell decline in a cohort of men with primary HIV infection. Clin Infect Dis. 2012;54:539–547. doi: 10.1093/cid/cir849. [DOI] [PubMed] [Google Scholar]
- Lodi S, Phillips A, Touloumi G, et al. Time from human immunodeficiency virus seroconversion to reaching CD4+ cell count thresholds <200, <350, and <500 Cells/mm3: assessment of need following changes in treatment guidelines. Clin Infect Dis. 2011;53:817–825. doi: 10.1093/cid/cir494. [DOI] [PubMed] [Google Scholar]
- Spira S, Wainberg MA, Loemba H, Turner D, Brenner BG. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J Antimicrob Chemother. 2003;51:229–240. doi: 10.1093/jac/dkg079. [DOI] [PubMed] [Google Scholar]
- Kuritzkes DR. HIV-1 subtype as a determinant of disease progression. J Infect Dis. 2008;197:638–639. doi: 10.1086/527417. [DOI] [PubMed] [Google Scholar]
- Sulkowski MS, Moore RD, Mehta SH, Chaisson RE, Thomas DL. Hepatitis C and progression of HIV disease. JAMA. 2002;288:199–206. doi: 10.1001/jama.288.2.199. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos GK, Paraskevis D, Hatzitheodorou E, et al. Impact of hepatitis B virus infection on the progression of AIDS and mortality in HIV-infected individuals: a cohort study and meta-analysis. Clin Infect Dis. 2009;48:1763–1771. doi: 10.1086/599110. [DOI] [PubMed] [Google Scholar]
- Langford SE, Ananworanich J, Cooper DA. Predictors of disease progression in HIV infection: a review. AIDS Res Ther. 2007;4:11. doi: 10.1186/1742-6405-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Time from HIV-1 seroconversion to AIDS and death before widespread use of highly-active antiretroviral therapy: a collaborative reanalysis. Collaborative Group on AIDS Incubation and HIV Survival including the CASCADE EU Concerted Action Concerted Action on SeroConversion to AIDS and Death in Europe. Lancet. 2000;355:1131–1137. [PubMed] [Google Scholar]
- CASCADE Collaboration. Differences in CD4 cell counts at seroconversion and decline among 5739 HIV-1-infected individuals with well-estimated dates of seroconversion. J Acquir Immune Defic Syndr. 2003;34:76–83. doi: 10.1097/00126334-200309010-00012. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Mehle A, Thomas ER, Rajendran KS, Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J Biol Chem. 2006;281:17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- Alexander L, Aquino-DeJesus MJ, Chan M, Andiman WA. Inhibition of human immunodeficiency virus type 1 (HIV-1) replication by a two-amino-acid insertion in HIV-1 Vif from a nonprogressing mother and child. J Virol. 2002;76:10533–10539. doi: 10.1128/JVI.76.20.10533-10539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Rexach M, Zybarth G, et al. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Cohen OJ, Nie Z, et al. Vpr R77Q is associated with long-term nonprogressive HIV infection and impaired induction of apoptosis. J Clin Invest. 2003;111:1547–1554. doi: 10.1172/JCI16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mologni D, Citterio P, Menzaghi B, et al. Vpr and HIV-1 disease progression: R77Q mutation is associated with long-term control of HIV-1 infection in different groups of patients. AIDS. 2006;20:567–574. doi: 10.1097/01.aids.0000210611.60459.0e. [DOI] [PubMed] [Google Scholar]
- Caly L, Saksena NK, Piller SC, Jans DA. Impaired nuclear import and viral incorporation of Vpr derived from a HIV long-term non-progressor. Retrovirology. 2008;5:67. doi: 10.1186/1742-4690-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng ZF, Hu HL, Qiu C, et al. Transmission of new CRF07_BC strains with 7 amino acid deletion in Gag p6. Virol J. 2011;8:60. doi: 10.1186/1743-422X-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Huang YF, Hsiao CF, et al. Trends of mortality and causes of death among HIV-infected patients in Taiwan, 1984–2005. HIV Med. 2008;9:535–543. doi: 10.1111/j.1468-1293.2008.00600.x. [DOI] [PubMed] [Google Scholar]
- Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- Tang J, Tang S, Lobashevsky E, et al. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76:8276–8284. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HY, Hung CC, Lin PH, et al. Incidence of abacavir hypersensitivity and its relationship with HLA-B*5701 in HIV-infected patients in Taiwan. J Antimicrob Chemother. 2007;60:599–604. doi: 10.1093/jac/dkm243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.