Abstract
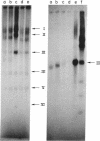
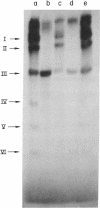
Transcription of T7 DNA by T7 RNA polymerase in vitro gives rise to six major size classes of RNAs comprising seven major T7 RNA species. These RNAs are all read from the r-strand of T7 DNA and are not derived from the early (leftmost on the conventional genetic map) region of the molecule. When artifically shortened T7 DNA templates are transcribed, four (I, II, IIIb, and VI) of the seven species are found to be truncated or deleted. This indicates that all are terminated near the right end of the T7 DNA molecule, probably at a common termination site near 98.5%. (Map positions are all given in terms of percentage of total length measured from the left end of the molecule.) Since the approximate lengths of the transcripts are known, the promotor sites for T7 RNA species I, II, IIIb, and VI are tentatively mapped at 56, 64, 83, and 97% on the T7 chromosome. Only a single major T3 RNA is transcribed by T7 RNA polymerase; analysis of transcripts directed by shortened T3 DNA templates indicates it is analogous to T7 RNA species IIIb. Hence the promotor and terminator sites for T3 species IIIb are tentatively mapped at 83 and 98.5%, respectively, on the T3 chromosome. The major transcripts read by T7 RNA polymerase from T3-T7 hybrid phage DNAs vary, depending on which regions of the T7 chromosome are present. This provides an alternative method of mapping the strong T7 promotor sites on the T7 chromosome.
Keywords: in vitro T7 RNAs, gel electrophoresis, exonuclease cleavage, T3-T7 hybrid phages
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973 Mar 25;248(6):2235–2244. [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs are generated by site-specific cleavages. Proc Natl Acad Sci U S A. 1973 May;70(5):1559–1563. doi: 10.1073/pnas.70.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez B., Lang D. Denaturation map of bacteriophage T7 DNA and direction of DNA transcription. J Mol Biol. 1972 Sep 28;70(2):239–251. doi: 10.1016/0022-2836(72)90536-0. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972 Sep 28;70(2):157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- Hyman R. W., Brunovskis I., Summers W. C. DNA base sequence homology between coliphages T7 and phiII and between T3 and phiII as determined by heteroduplex mapping in the electron microscope. J Mol Biol. 1973 Jun 25;77(2):189–196. doi: 10.1016/0022-2836(73)90330-6. [DOI] [PubMed] [Google Scholar]
- Hyman R. W. Physical mapping of T7 messenger RNA. J Mol Biol. 1971 Oct 28;61(2):369–376. doi: 10.1016/0022-2836(71)90386-x. [DOI] [PubMed] [Google Scholar]
- MacHattie L. A., Ritchie D. A., Thomas C. A., Jr, Richardson C. C. Terminal repetition in permuted T2 bacteriophage DNA molecules. J Mol Biol. 1967 Feb 14;23(3):355–363. doi: 10.1016/s0022-2836(67)80110-4. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., LEHMAN I. R., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. II. CHARACTERIZATION OF THE EXONUCLEASE ACTIVITY. J Biol Chem. 1964 Jan;239:251–258. [PubMed] [Google Scholar]
- Ritchie D. A., Thomas C. A., Jr, MacHattie L. A., Wensink P. C. Terminal repetition in non-permuted T3 and T7 bacteriophage DNA molecules. J Mol Biol. 1967 Feb 14;23(3):365–376. doi: 10.1016/s0022-2836(67)80111-6. [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Herrlich P., Schweiger M. Transcription of T3 and T7 early genes by T3 and T7 RNA polymerases. Mol Gen Genet. 1972;118(1):67–77. [PubMed] [Google Scholar]
- Siegel R. B., Summers W. C. The process of infection with coliphage T7. 3. Control of phage-specific RNA synthesis in vivo by an early phage gene. J Mol Biol. 1970 Apr 14;49(1):115–123. doi: 10.1016/0022-2836(70)90380-3. [DOI] [PubMed] [Google Scholar]
- Simon M. N., Studier F. W. Physical mapping of the early region of bacteriophage T7 DNA. J Mol Biol. 1973 Sep 15;79(2):249–265. doi: 10.1016/0022-2836(73)90004-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Bacteriophage T7. Science. 1972 Apr 28;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Summers W. C., Siegel R. B. Transcription of late phage RNA by T7 RNA polymerase. Nature. 1970 Dec 19;228(5277):1160–1162. doi: 10.1038/2281160a0. [DOI] [PubMed] [Google Scholar]
- Sutton W. D. A crude nuclease preparation suitable for use in DNA reassociation experiments. Biochim Biophys Acta. 1971 Jul 29;240(4):522–531. doi: 10.1016/0005-2787(71)90709-x. [DOI] [PubMed] [Google Scholar]
- de Wachter R., Fiers W. Preparative two-dimensional polyacrylamide gel electrophoresis of 32 P-labeled RNA. Anal Biochem. 1972 Sep;49(1):184–197. doi: 10.1016/0003-2697(72)90257-6. [DOI] [PubMed] [Google Scholar]