Abstract
Background and Objectives
MUC1 is over-expressed and aberrantly glycosylated in >60% of human pancreatic cancer (PC). Development of novel approaches for detection and/or targeting of MUC1 are critically needed and should be able to detect MUC1 on PC cells (including cancer stem cells) and in serum.
Methods
The sensitivity and specificity of the anti-MUC1 antibody, TAB 004, was determined. CSCs were assessed for MUC1 expression using TAB 004-FITC on in vitro PC cell lines, and on lineage− cells from in vivo tumors and human samples. Serum was assessed for shed MUC1 via the TAB 004 EIA.
Results
In vitro and in vivo, TAB 004 detected MUC1 on >95% of CSCs. Approximately, 80% of CSCs in patients displayed MUC1 expression as detected by TAB 004. Shed MUC1 was detected serum in mice with HPAF-II (MUC1high) but not BxPC3 tumors (MUC1low). The TAB 004 EIA was able to accurately detect stage progression in PC patients.
Conclusions
The TAB 004 antibody may be explored as a therapeutic targeting agent for CSCs in PC. The TAB 004 EIA detected circulating MUC1 in a stage-dependent manner in patients with PC and thus may be explored as a PC stage diagnostic biomarker.
Keywords: pancreatic cancer, MUC1, cancer stem cells, TAB 004
INTRODUCTION
Pancreatic cancer (PC) has the worst prognosis of all cancers and is the fourth leading cause of cancer-related deaths in the United States [1]. Currently, therapies for PC are limited to surgical resection and adjuvant therapies, including chemotherapy and radiation therapy, but the median survival of patients diagnosed with PC is a dismal 4–6 months. Thus, PC remains a lethal diagnosis for the vast majority of patients due to high rate of recurrence and metastasis.
Cancer stem cells (CSCs) are defined as a subset of tumor cells that have the ability to self-renew and generate the diverse cells that comprised the original tumor. Although a debate still exists over PC stem cell markers, CD133+ [2], CD44+CD24+EpCAM+ [3], and aldehyde dehydrogenase (ALDH)-expressing pancreatic cells have been shown to possess “stem-like” properties [4]. Interestingly, Hermann et al. [2] identified a 14% overlap between CD44+CD24+EpCAM+ and CD133+ cells, suggesting that there may be more than one set of cell surface markers that may be used to enrich for pancreatic CSCs. Further a highly metastatic subpopulation of the CD133+ PC cells co-express CXCR4, the receptor for SDR-1 [2]. Clinically, the presence of CSCs is extremely important, as these cells need to be eradicated in order to provide long-term disease-free survival. Recent studies have shown that the CD133+ populations of PC cells are enriched after exposure to the chemotherapeutic agent, gemcitabine [2]. This supports the hypothesis that CSCs are resistant to conventional treatments and that these cells are the culprit behind cancer metastases and recurrence after clinical remission.
MUC1 (Mucin1) is a membrane tethered glycoprotein expressed on the apical surfaces of normal glandular epithelia but is over-expressed and aberrantly glycosylated in over 60% of human PDA [5]. In PC, tumor-associated MUC1 is a marker of an aggressive phenotype, as its expression is correlated with high metastases and poor prognosis [6][7]. MUC1 can be detected in the serum of patients with PC, as it is cleaved from the epithelial cells and released into circulation. High MUC1 serum levels are associated with progressive disease [8–10]. MUC1 synthesized by cancerous tissues displays an aberrant oligosaccharide profile giving rise to the expression of neomarkers such as sialyl-Lea (used in the CA19-9 test) [11–14], sialyl-Lex, and sialyl-Tn (TAG-72), as well as the cryptic epitopes such as Tn [15–17]. In addition, because of underglycosylation, the peptide core of the mucin becomes exposed such that epitopes within the core that are not accessible within normal tissue-derived MUC1 may serve as potential antigens [18,19]. Thus, differences between normal versus malignant MUC1 can provide for distinct epitopes that may show higher specificity for malignant tissues and can be explored as tumor-associated antigens in various anticancer applications.
To this end, we have assessed the applications of a novel antibody developed against tumor-associated MUC1, TAB 004. In this manuscript, we will evaluate the ability of TAB 004 to distinguish tumor-associated MUC1, to detect the presence of MUC1 on CSCs and to identify shed MUC1 in the serum of mice and patients with PC.
MATERIALS AND METHODS
Antibody Generation
TAB 004 (Patent # US-2011-0123442, PCT/US2011/037972) was generated by immunizing Balb/c mice with protein lysate from MUC1-expressing tumors that developed in a MUC1 transgenic mice that expressed human MUC1 [6]. Hybridomas were generated by fusion of spleen cells from immunized mice with myeloma cell line P3X63Ag8 to develop the monoclonal antibody TAB 004 which was identified by screening hybridomas. The antibody was then grown and isolated at the Immunology Core Facility (University of North Carolina at Chapel Hill).
Cell Generation and Cell Culture
Generation of BxPC3.Neo and BxPC3.MUC1 is summarized in [20]. These cells were maintained in complete RPMI (Invitrogen, Carlsbad, CA) containing 10% FCS, 1% penicillin/streptomycin, 1% glutaMAX™ (Invitrogen), and 3 μg/ml G418. The KCKO cell line was generated from a pancreatic tumor in Muc1 knockout mice [6][21]. The KCM cell line was isolated from a pancreatic tumor in a mouse that was transgenic for human MUC1 [6][21]. These cells were maintained in DMEM (Invitrogen) containing 10% FCS, 1% penicillin/streptomycin and 1% glutaMAX™. HPAF-II cells were maintained in MEM (Invitrogen) containing 10% FCS, 1% penicillin/streptomycin and 1% GlutaMAX™.
Western Blot
Briefly, cells were lysed in HEPES buffer (20 mmol/L HEPES, 150 mmol/L NaCl, 1% Triton X-100, 2 mmol/L EDTA) containing protease (Complete inhibitor cocktail; Roche, Indianapolis, IN) and phosphatase inhibitors (10 mmol/L sodium fluoride, 2 mmol/L sodium vanadate, 50 μmol/L ammonium molybdate). Equal quantities of lysate were loaded on SDS–PAGE gels [20]. TAB 004 and β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies were added at 1:1,000 dilution, incubated overnight at 4°C, followed by washes and detection with anti-mouse HRP at 1:2,500 dilution (Santa Cruz Biotechnology, Inc.).
Immunohistochemistry
Paraffin-embedded tissue from the nude mice and patients with PC were sectioned to 4 μm and placed on slides. Tissue was deparaffinized and hydrated via washes with 100% EtOH, 95% EtOH, 70% EtOH and then water. Antigen retrieval was performed for 30 min at 99°C followed by a 20-min cool down (RT) in Dako Target antigen retrieval (S1700, Dako, Carpinteria, CA). Sections were then blocked for 1 hr in 50% FBS in phosphate buffer saline (PBS), and then incubated over-night with TAB 004 (1:1,000 dilution). The next morning, sections were washed with 3 times with PBS and a detection antibody was added for 1 hr (goat anti-mouse HRP, 1:100, Dako). For all slides, 3,3″–Diaminobenzidine (Vector Laboratories, Burlington, CA) was used as the chromogen and hematoxylin was used as counterstain. Slides were then dehydrated, coverslipped, and viewed using light microscopy.
Human Sample Collection
Patients with suspected PC that were scheduled to undergo a Whipple procedure were consented for our IRB approved protocol (UNC Charlotte IRB protocol # 12-04-26 and CHS IRB file # 10-07-09B). Whole blood was collected during surgery and spun down at 2,000 rpm for 5 min. Serum was collected and frozen for later analysis. Resected tumors were collected from the Pathology Laboratory at CMC. The tumor sections were cut into small pieces and digested in 1 mg/ml Collagenase IV (Worthington Biochemical Corporation, Lakewood, NJ) in RPMI for 30 min at 37°C. The tumor was further digested mechanically through a 40 μm filter and spun down for lineage selection. Lineage negative cells were collected using the human Lineage Depletion kit with MACS columns (Miltenyi Biotec, Cambridge, MA). Cells were then subjected to flow cytometry analysis. Human sera and tissue sections were obtained separately from the NIH tissue repository for immunohistochemical (IHC), immunofluorescence (IF) and TAB 004 analysis.
Nude Mouse Studies
For BxPC3 tumors, 2-month-old, male athymic nude mice (Jackson Laboratory, Bar Harbor, ME) were injected with 5 × 106 BxPC3.Neo cells in 100 μl of PBS subcutaneously (s.c.) into the flank of the mice. Tumors were allowed to grow for 2 months. At sacrifice, serum was collected and tumors were fixed in 10% neutral-buffered formalin (pH 6.8–7.2) for a minimum of 24 hr.
For in vivo CSC assessment, 2-month-old, male athymic nude mice (Harlan Laboratories, Inc., Fredrick, MD) were subcutaneously injected with either 3 × 106 HPAF-II, 5 × 106 PANC-1 or 5 × 106 MiaPaCa-2 cells in PBS. Tumors grew for 25 days. At sacrifice, serum was collected, a section of the tumor was formalin-fixed, and another tumor section was digested as above and processed using the mouse Lineage Depletion Kit (Miltenyi Biotec).
Flow Cytometry
Cells were washed 2× with PBS/0.1% Sodium Azide/1% BSA buffer. Cells were then incubated with either FcBlock (BD Bioscience, San Jose, CA) or human IgG (Jackson ImmunoResearch Lab, Inc., West Grove, PA) for 10 min on ice. TAB 004 was conjugated to FITC using the Lightning-Link™ FITC conjugation kit (Innova Biosciences, Cambridge, UK). To assess MUC1 on Triple+ cells, the following antibody panel was used: TAB 004-FITC, EpCAM-PE (347198), CD24-PECy7 (311119), and CD44-APC (500890; BD Biosciences). Separate cells were also dually stained with TAB 004-FITC and CD133-APC (Clone AC133; Miltenyi Biotec). Data were collected using the BD LSRFortessa cell analyzer and analyzed using FlowJo software.
Immunofluorescence
Paraffin-embedded tissue from both nude mice and patients with PDA were sectioned to 4 μm and placed on slides. Tissue was deparaffinized and hydrated via washes with 100% EtOH, 95% EtOH, 70% EtOH and then water. Antigen retrieval was performed for 30 min at 99°C followed by a 20-min cool down at RT in Dako Target antigen retrieval (Dako, S1700). Sections were then blocked for 1 hr in 50%FBS/PBS, followed by 30 min in Image-iT™ FX Signal Enhancer (Molecular Probes, Grand Island, NY) all at RT. TAB 004-FITC (1:500 dilution) and CD133 (1:50 dilution; Santa Cruz Biotechnology Inc., sc-23797) were incubated overnight at 4°C. After washing with PBS, chicken anti-goat Alexa 647 was added for 1 hr at 1:100 dilution (Molecular Probes). Finally, sections were washed and treated with Prolong gold with DAPI (Molecular Probes); slides were coverslipped and left at 4°C until analysis. Immunofluorescence was analyzed using the Olympus FV1000.
TAB 004 Enzyme Immunoassay
Frozen serum collected from the nude mice with pancreatic tumors and from human patients was subjected to the TAB 004 enzyme immunoassay (EIA). High-binding 96-well plates (Corning Inc., Corning, NY) were coated with 2 μg/ml TAB 004 over night at RT. The next morning, the plate was blocked with 10% FBS in PBS for 1 hr at RT. Standards and samples (1:4 dilution for human sera and 1:2 dilution for mouse sera) were added to the wells and incubated for 2 hr at RT. The standard curve consisted of KCM cell lysate at a range from 0.98 to 250 μg/ml. The plate was then washed 3× with PBS-Tween. The detection antibody was then added at 4 μg/ml of Biotin-TAB 004 and incubated for 2 hr at RT. After a repeated wash, SA-HRP (Chemicon, Austrialia) at a 1:1,000 dilution was added for 2 hr at RT. After a final wash, the EIA was developed with 1Step Ultra TMB-ELISA solution (ThermoScientific, Waltham, MA) for approximately 30 min followed by the addition of stop solution. The plate was read at 450 nm using the μQuant plate reader (BioTek, Winooski, VT). For the samples collected from NIH, a similar procedure was followed, except MUC1 peptides were used for the standard and TAB 004 was conjugated directly to HRP for detection.
Statistics
Statistical analysis for the TAB 004 EIA using the serum from the nude mice was performed using a Mann–Whitney U-test. Data collected from patients using the TAB 004 EIA was logged to promote normality. A repeated-measures ANOVA was performed to test significance between groups. The P-values less than 0.5 were considered significant.
RESULTS
Generation and Characterization of TAB 004
TAB 004 is a monoclonal antibody that was generated by immunizing Balb/c mice with pancreatic tumor lysate that was isolated from a primary pancreatic ductal adenocarcinoma, which spontaneously developed a P48-KRASG12D mouse [22] that was transgenic for human MUC1 [23]. These mice are designated PDA.MUC1 mice or KCM mice and have been previously characterized [6][21]. Thus, the immunogen displayed human MUC1 in the context of a pancreatic tumor. Hybridomas were generated by fusion of spleen cells from immunized mice with myeloma cell line P3X63Ag8 to develop the monoclonal antibody TAB 004, which was identified by screening hybridomas. Epitope screening determined that the TAB 004 monoclonal antibody epitope was the sequence STAPPVHNV that is present within the MUC1 tandem repeats (TR) at amino acids 950–958. The complementary determinant region (CDR) sequence of the antibody was determined and found to be unique compared to other commercially available MUC1 antibodies (Supplementary Fig. 1). Recombinant antibody was tested for antigen binding using a MUC1 peptide. TAB 004 displayed antigen binding at 3 ng/ml (20 pM; Fig. 1A).
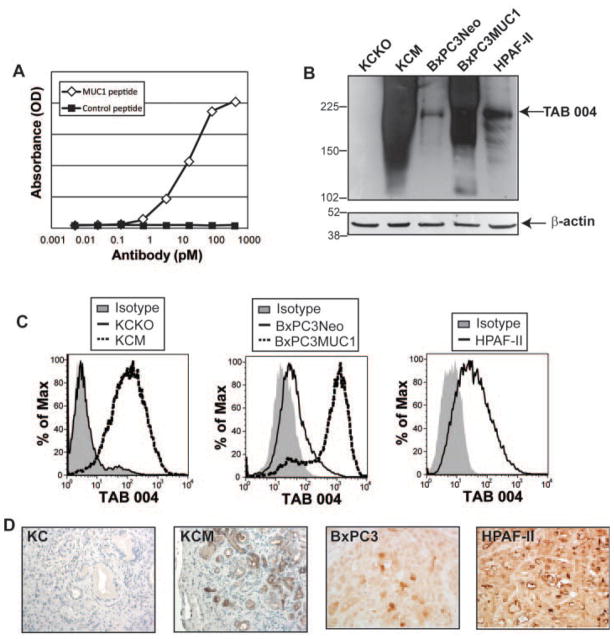
Fig. 1.
TAB 004 specifically detects human MUC1 in PC. A: Recombinant antibody was produced from CHO cells and antigen binding was determined against a MUC1 or control peptide via ELISA. Antibody concentration (as determined via an anti-human IgG ELISA) is plotted on the x-axis and absorbance for the ELISA is plotted on the y-axis. This assay was performed by LakePharma, San Francisco, CA. B,C: TAB 004 accurately detected MUC1 via Western blot (B) and flow cytometry (C) in KCM, BxPC3MUC1 and HPAF-II but not in KCKO and at low endogenous levels in BxPC3Neo. D: Immunohistochemistry for MUC1 using TAB 004 was performed on spontaneous pancreatic tumor in KC mice and KCM mice, which are transgenic for human MUC1, and on tumor sections from BxPC3 and HPAF-II tumors in nude mice. Representative images are displayed (n = 3 for KC, KCM and BxPC3 and n = 4 for HPAF-II).
Next, we assessed TAB 004 in vitro binding using the following PC cell lines: (1) KCKO—a murine PC cell line isolated from mice that are Muc1 null; (2) KCM—a murine PC cell line isolated from mice that are transgenic for human MUC1; (3) BxPC3.Neo—a human PC cell line that was transfected with the neomycin resistance empty vector; (4) BxPC3.MUC1—a human PC cell line that was transfected with full-length MUC1-expressing vector; and (5) HPAF-II—a human PC cell line known to have high endogenous MUC1. Using Western blotting and flow cytometry with the TAB 004 antibody, high levels of MUC1 were observed in KCM, BxPC3.MUC1 and HPAF-II. As expected, no MUC1 was detected in KCKO and low endogenous levels of MUC1 were detected in BxPC3.Neo cells (Fig. 1B,C). We also assessed TAB 004 specificity in vivo in transgenic mice and in nude mice with human tumors. Immunohistochemical staining using TAB 004 was performed on spontaneously forming pancreatic tumors taken from 34-week KC mice (P48-Cre with the LSL-KRASG12D mice) and KCM mice (KC mice that are transgenic for human MUC1) [21]. TAB 004 did not detect the murine MUC1 as no stain was observed in KC but strongly detected human MUC1 in the KCM mice (Fig. 1D). Additionally, nude mice were injected with BxPC3 (MUC1low) and HPAF-II (MUC1high) cells into the right flank. As was expected, low endogenous staining was observed in BxPC3 tumors and high staining was observed in the HPAF-II tumors (Fig. 1D) demonstrating the specificity of TAB 004.
TAB 004 Detects MUC1 on CSCs In Vitro
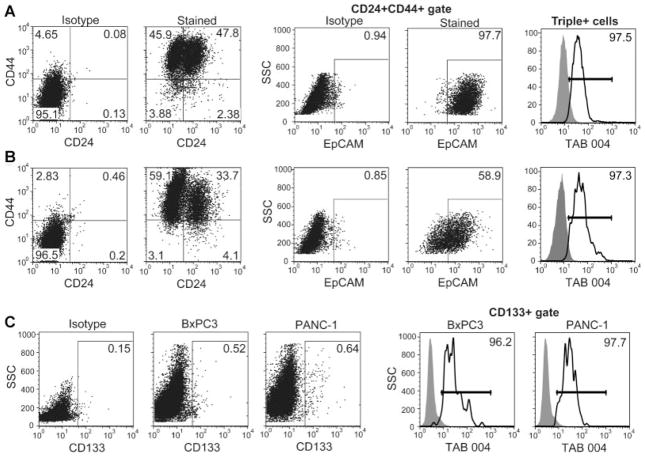
The expression of targetable markers on CSCs is vital to future therapy against these cancer-causing cells. Thus, we sought to determine the detectability of MUC1 on CSCs using the TAB 004 antibody. We began by performing flow cytometry to determine MUC1 levels on two populations of CSCs, (1) CD44+CD24+EpCAM+ (Triple+ cells) and (2) CD133+ cells. We observed surprisingly high levels of Triple+ cells in all cell lines tested (Fig. 2A—46.7% for BxPC3) (Fig. 2B—19.8% for PANC-1). TAB 004 detected MUC1 on over 97% of these Triple+ cells. The CSCs, detected using CD133, were observed at much lower levels compared to Triple+ cells (Fig. 2C—approximately 0.5%). This population again displayed high levels of MUC1, as recognized by the TAB 004 antibody.
Fig. 2.
Expression of MUC1 on PC CSCs in vitro. A,B: BxPC3 (A) and PANC-1 (B) cells were stained with CD24, CD44, EpCAM, and MUC1 using the TAB 004 antibody and subjected to flow cytometry. Triple+ CSCs were determined by gating for CD24+CD44+ cells and then gating for EpCAM expression. MUC1 levels were then assessed on these Triple+ cells. C: BxPC3 and PANC-1 cells were dually stained with CD133 and TAB 004 and subjected to flow cytometry to assess the level of MUC1 on CD133+ CSCs.
TAB 004 Detects MUC1 on CSCs In Vivo
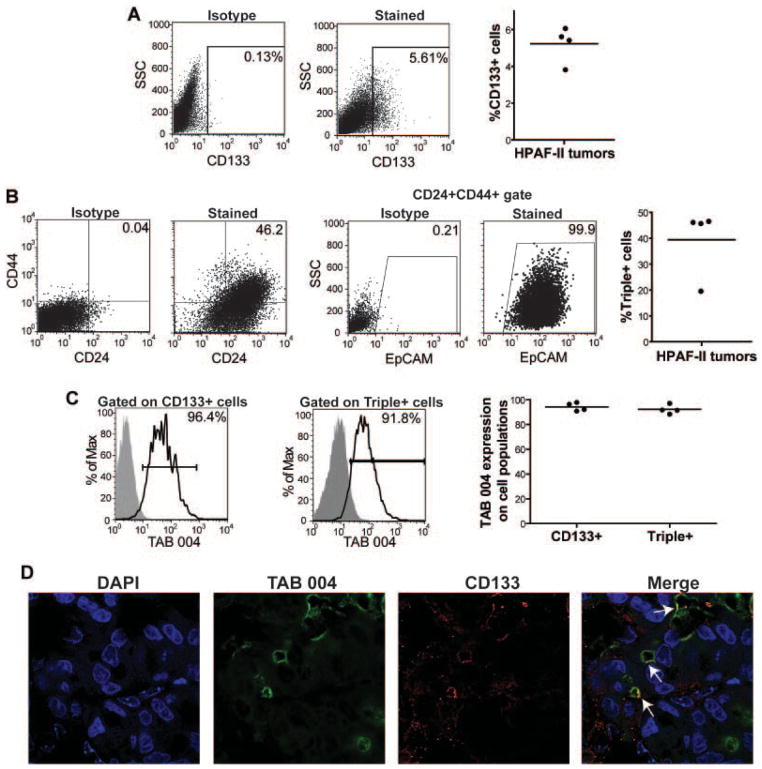
CSCs are known to be heavily influenced by their environment, and thus we evaluated this interaction in vivo. Nude mice were injected with the PC cell line HPAF-II. After 25 days, tumors were removed and assessed for CSC levels. Approximately 5% of the tumor was positive for CD133-expressing CSCs (Fig. 3A). We again observed unexpectedly high levels of Triple+ CSCs, which were about 40% of the tumor (Fig. 3B). On both populations of CSCs, TAB 004 detected MUC1 on over 95% of the cells (Fig. 3C). We observed similar levels of MUC1 on CSCs isolated from MiaPaCa-2 and PANC-1 tumors—average of 96.7% on MiaPaCa-2 Triple+ cells and 78.5% on CD133+ cells (n = 3) and average of 92.4% on PANC-1 Triple+ cells and 80.4% on CD133+ cells (n = 3; data not shown). Sections from the HPAF-II tumors were dually stained with the TAB 004 and CD133 antibodies to visualize the presence of these proteins within the tumors. While TAB 004 detected MUC1 throughout the tumor, CD133 expression was limited to a small population of tumor cells. TAB 004 staining was visually seen on these CD133+ cells, confirming our flow cytometric analysis (Fig. 3D).
Fig. 3.
Expression of MUC1 on PC CSCs in vivo. Male nude mice were injected with HPAF-II cells into the right flank. After 25 days, mice were sacrificed and tumor sections were digested to a single cell suspension. Lineage− cells were assessed for CSC and MUC1 levels via flow cytometry. A: A representative image of CD133 stain from the HPAF-II tumors is displayed with the isotype control. B: To assess Triple+ CSCs, CD24+CD44+ cells were first gated and then EpCAM expression on those cells was determined. Representative staining and isotype controls are displayed. C: Gated CD133+ and Triple+ CSCs were assessed for their MUC1 expression via TAB 004 staining. A representative sample is shown for each. D: A HPAF-II tumor section was assessed for TAB 004 and CD133 expression using immunoflourescence. TAB 004 staining was seen throughout the tumor and localized areas of CD133 were observed. Co-localization of TAB 004 and CD133 was observed (white arrows). For all experiments (n = 4 mice) and the experiment was repeated twice.
MUC1 Expression via TAB 004 Staining in Tumor Specimens Derived From Patients With Various Stages of PC
To this point, we have demonstrated the usage of TAB 004 in vitro and in vivo. Hence, we moved forward to assess the antibody functionality with human samples. We first assessed MUC1 expression detected by TAB 004 throughout the stage progression of PC. Immunohistochemical staining demonstrated that TAB 004 does not detect MUC1 in Stage 0 patients. However, TAB 004 detects MUC1 by Stage 2 disease and continues detection throughout Stages 3–4. We observed consistent levels of TAB 004 staining throughout stage progression, as confirmed by quantification of TAB 004 stain (Fig. 4 and data not shown).
Fig. 4.
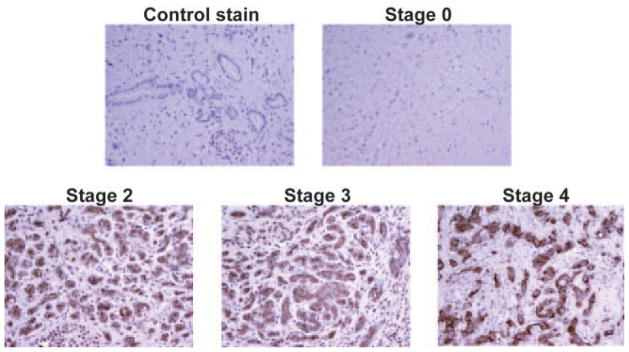
Human PC express MUC1 as detected by TAB 004 IHC. Sections from human PC samples were obtained from the NIH tissue repository. IHC was performed using the TAB 004 antibody. Secondary antibody alone served as the negative control. TAB 004 did not detect MUC1 in Stage 0 samples but was detected in all other stages. The image is a representative of n = 5.
TAB 004 Detected MUC1 Expression on CD133+ Cells in Tumor Specimens Derived From Patients With PC
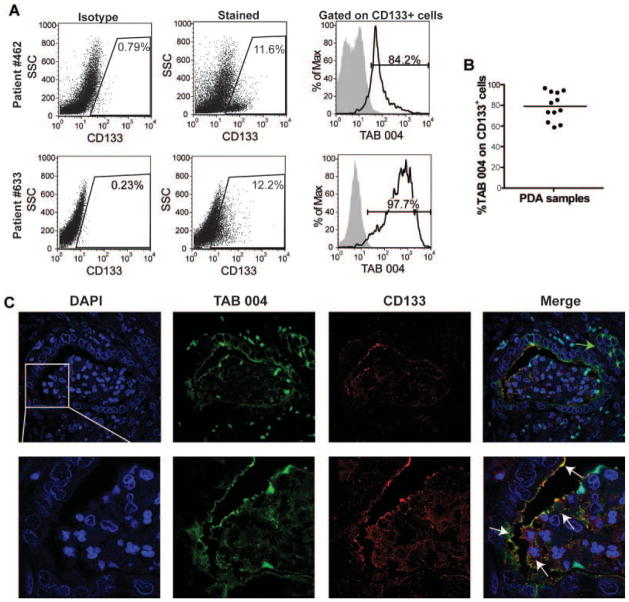
Patients with suspected or confirmed cases of PC that were undergoing a Whipple procedure were enrolled in our IRB-approved protocol, which allowed for the collection of tumor sections and blood. Patient demographics are displayed in Table I. Tumors were digested to single cell suspensions, and lineage-negative cells were assessed for MUC1 expression, as well as for a CSC marker, CD133. MUC1 expression detected by TAB 004 staining was observed in 15 of the 17 patients assessed. Levels of MUC1 positive cells and degree of MUC1 expression were highly variable, most likely due to the heterogeneous nature of the samples collected (Table I). We assessed CSC levels in patients via CD133 staining. Gating is displayed in Figure 5A and each sample was gated based on their individual isotype to control for staining variability. CD133+ cells were detected in 12 out of 17 patients. Patients had varying amounts of CD133 with an average of 8.9% (Fig. 5A and Table I). Using TAB 004, we assessed levels of MUC1 on CSCs in those patients with CD133+ cells. TAB 004 detected MUC1 on CD133+ CSCs on approximately 80% of the cells (Fig. 5B). Paraffin-embedded tissue samples were assessed for dual CD133 and MUC1 expression via immunofluorescence. Ductal adenomas stained positively for MUC1 via TAB 004 stain (green). Autofluorescent nuclear staining was also observed in the green channel, but this was easily distinguishable from the extracellular TAB 004 stain. Localized areas of intense CD133 stain (red) were also observed on ductal cells. The areas of CD133 positivity displayed TAB 004 staining as shown by the white arrows in the merged image (Fig. 5C). However, many areas of TAB 004 positivity do not also show CD133 expression (Fig. 5C, green arrow). This is to be expected as we would anticipate that not all MUC1 positive cells are CSCs, but that the majority of CD133+ CSCs express MUC1. This data confirms our in vitro and in vivo findings and emphasizes the use of TAB 004 to detect MUC1 on CSCs in patients with PC.
TABLE I.
Summary of Patient Demographics and Data
| Age | Gender | Race | Stage | MUC1 expression
|
CD133 | MUC1 on CD133 | Sera MUC1 | ||
|---|---|---|---|---|---|---|---|---|---|
| % positive | Geometric mean | ||||||||
| 64 | Male | Caucasian | 2a | 38.9 | 8.0 | ++ | 10.6 | 73.2 | 65.9 |
| 58 | Male | African American | 2b | 90.4 | 200.9 | +++ | 10.8 | 82.4 | 95.0 |
| 68 | Female | Caucasian | 2b | 69.9 | 56.6 | +++ | 0.6 | 93.2 | NA |
| 54 | Female | Caucasian | 2b | 75.1 | 105.0 | +++ | 6.4 | 85.1 | NA |
| 59 | Male | Caucasian | 2b | 46.3 | 27.6 | ++ | 8.3 | 75.1 | 61.8 |
| 69 | Male | Caucasian | 2b | 4.6 | 21.1 | + | 2.1 | 63.6 | NA |
| 53 | Female | Caucasian | 2a | −0.4 | 24.4 | − | −1.7 | — | 53.3 |
| 81 | Female | Caucasian | 2a | 9.7 | 2.0 | + | 3.4 | 92.3 | 70.5 |
| 34 | Male | Caucasian | 4 | * | * | * | * | * | 209.0 |
| 49 | Male | Caucasian | 2b | 85.8 | 165.2 | +++ | 12.0 | 96.6 | 49.9 |
| 78 | Male | Caucasian | 3 | 0.4 | −0.4 | − | −0.1 | — | 259.7 |
| 60 | Female | African American | 2a | 37.1 | 936.4 | ++ | −0.2 | — | 85.5 |
| 68 | Male | Caucasian | ** | 3.7 | 3.0 | + | 0.0 | — | NA |
| 67 | Male | African American | 2b | 73.3 | 136.3 | +++ | 46.1 | 94.3 | 62.4 |
| 75 | Male | Caucasian | 1b | 23.1 | 9.1 | ++ | 0.2 | 73.3 | 43.5 |
| 69 | Female | Caucasian | 2b | 16.8 | 8.8 | + | 0.0 | — | 32.9 |
| 80 | Male | Caucasian | 2b | 20.7 | 24.7 | ++ | 1.0 | 60.7 | 43.5 |
| 53 | Female | African American | 2b | 16.3 | 6.8 | + | 5.4 | 58.7 | 51.2 |
Patient age, gender, race, and stage of disease are displayed. Intratumoral MUC1 expression as detected by flow cytometry using TAB 004 is displayed as % positive cells and Geometric Mean. Percentage of cells positive for CD133 is displayed. TAB 004 detection of CD133+ cells is listed per patient (*not available because surgery not performed; **not available because patient expired during surgery). Serum MUC1 levels were detected using the TAB 004 EIA (NA—not available because serum samples not collected).
Fig. 5.
TAB 004 detects MUC1 on PC CSCs from human patients. Patients with suspected PC that were scheduled for a Whipple procedure were consented for our IRB approved protocol. A,B: Samples were collected immediately after surgery, digested to a single-cell suspension, selected for lineage− cells, dually stained with TAB 004-FITC and CD133-APC, and subjected to flow cytometry. Representative dot plots are displayed in A. CD133 levels for each patient were normalized by subjecting the isotype control levels to account for staining variability. MUC1 levels were assessed on gated CD133+ cells (n = 12). C: Sections from NIH tissue repository were assessed for TAB 004 and CD133 expression using immunoflourescence. TAB 004 detected MUC1 along ductal cells in the displayed lesion. Areas of TAB 004 positive cells that lacked CD133 expression were observed (green arrow). CD133 expression was also observed on the surface of ductal epithelial cells, although to a lesser extent than TAB 004. The merged image demonstrates areas that express CD133 and MUC1 as detected by TAB 004. The image is a representative of n = 6.
Detection of Shed MUC1 in the Serum of Mice and Patients With PC
It has been well established that MUC1 can be cleaved from tumor cells, thus releasing the protein into the circulation. Therefore, we tested the ability of the TAB 004 antibody to detect MUC1 in the circulation of mice and patients with PC using an EIA. The TAB 004 EIA detected circulating MUC1 in mice with HPAF-II tumors (MUC1high) but not in mice with MUC1low BxCP3 tumors (Fig. 6A). Serum collected from patients was subjected to the TAB 004 EIA. TAB 004 detected MUC1 in all patients assayed (Table I). In all but one case, the expression of MUC1 in the single cells isolated from the tumor specimens correlated with its expression in the circulation. However, in patient #11, we were unable to detect MUC1 in the tumor specimens but this patient had high levels of circulating MUC1. We are unable to determine the cause for this discrepancy other than technical error while staining the isolated tumor cells. Serum samples were also obtained from NIH to assess the ability of the TAB 004 EIA to detect stage progression in PDA. Stage 0 patients displayed an average of 15.7 Units/ml. TAB 004 detected significantly more MUC1 in circulation at each stage of progression (Stage 0 vs. Stage 2 and Stage 2 vs. Stage 3 P < 0.001; Stage 3 vs. Stage 4 P = 0.048; Fig. 6B). These data demonstrate the ability of the TAB 004 EIA to predict stage progression in PC.
Fig. 6.
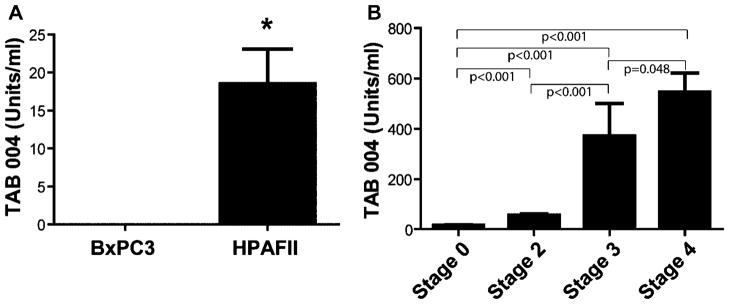
TAB 004 EIA detects MUC1 in the serum of mice and humans with PC. The TAB 004 EIA was performed on serum samples which uses TAB 004 as the capture and detection antibody in an immunoassay. A: Serum collected from nude mice with BxPC3 (MUC1low) and HPAF-II (MUChigh) tumors was assessed for shed MUC1 using the TAB 004 antibody. No MUC1 was detectable in the serum of mice with BxPC3 (n = 3), and MUC1 was present in the serum of all mice with HPAF-II tumors (n = 5; *P = 0.0325). B: Serum was collected from the NIH tissue repository to detect stage progression in PC patients. TAB 004 detected significantly more MUC1 in circulation at each stage of progression (n = 5; Stage 0 vs. Stage 2 and Stage 2 vs. Stage 3 P < 0.001; Stage 3 vs. Stage 4 P = 0.048).
DISCUSSION
In summary, we have demonstrated exciting applications for the novel MUC1 antibody, TAB 004. First we show the high sensitivity of TAB 004 for MUC1, with binding observed at 3–20 pM range. TAB 004 detected MUC1 with high specificity on PC cell lines and tumors. Further, we demonstrated MUC1 expression on CD44+CD24+EpCAM+ and CD133+ pancreatic CSCs using the TAB 004 antibody. Approximately, 95% of CSCs in vitro and in vivo were identified by the TAB 004 antibody. Expectedly, patient samples displayed more variability, but an average of 80% of CD133+ pancreatic CSCs were positive for MUC1 via TAB 004 staining. Confocal images show TAB 004 staining on CD133+ cells in murine and human tumors, confirming our results. Lastly, we developed an EIA using the TAB 004 antibody to detect circulating levels of tumor-associated MUC1 shed from pancreatic tumors. Detectable levels of MUC1 were observed in mice with HPAF-II tumors, which displayed high levels of intratumoral MUC1. However in tumors with low MUC1, BxPC3 tumors, tumor-associated MUC1 was undetectable in the serum, indicating the specificity of our EIA to MUC1. Importantly, TAB 004 was able to accurately detect shed tumor-associated MUC1 in the serum of patients with PC in a stage-specific manner. These data demonstrate the wide range of applications for the novel MUC1 antibody, TAB 004.
Much debate exists within the scientific community as to the appropriate markers for CSCs, which have been defined for each individual type of cancer. Pancreatic CSCs were initially identified by Simeone’s group, when they demonstrated the high tumorigenic potential of cells expressing EpCAM+CD44+CD24+ [3]. The observation was interesting as this definition differed from those CSCs originally identified in breast cancer as CD44+CD24−/low [24]. Thereafter, Hermann et al. [2] used CD133 as a marker to isolate PC cells with a significantly higher tumorigenic potential and demonstrated that this cell population was enriched in mice treated with chemotherapy. CD133 has also been identified in multiple reports as a marker of brain, colon, and lung CSCs [25]. Further, ALDH has also been used as a marker to identify PC stem cells, but ALDH+ and CD24+CD44+ cells showed very little overlap [4]. We assessed both levels of EpCAM+CD44+CD24+ and CD133+ cells in PC cell lines in vitro and in vivo. We observed a consistent expression MUC1 on both populations of cells as detected with the TAB 004 antibody. We choose to focus on CD133+ CSCs in the human samples for multiple reasons: (1) CD133 is a well-established marker for CSCs in multiple carcinomas including pancreatic adenocarcinoma and (2) the unexpectantly high levels of Triple+ CSCs (EpCAM+CD44+CD24+) that we observed in both in vitro and in vivo. It is well established that CSCs should comprise an inherently low population of the total cells within a tumor.
MUC1 is well known as a cell surface marker of epithelial cells, where it normally functions as a protective barrier. Therefore, MUC1 expression on CSCs is unexpected as these cells are mesenchymal in nature. However, one report investigated levels of MUC1 on H9 and H14 human embryonic stem cells. They found full-length MUC1 expression on newly differentiated human embryonic stem cells [26]. Further, another report investigated MUC1 levels on CSCs within the breast cancer cell line, MCF7. They found that approximately 77% of the CSCs, defined by the Side Population cells, were MUC1bright, corroborating our data [27]. Our recent study has demonstrated that MUC1 expression causes epithelial-to-mesenchymal transition (EMT) in PC cells [28]. Thus, we propose that a portion of the MUC1-expressing PC cells are undergoing EMT and retaining their MUC1 positivity, hence leading to MUC1 positive CSCs. The detectable expression of MUC1 on CSCs is extremely promising as a possible mechanism to target these cancer-initiating cells. In a recent study, mice that were vaccinated with PC cells expressing α-gal epitopes, displayed immune responses against CSCs, which we suggest is a result of the expression of MUC1 on the CSCs [29].
Since MUC1 is aberrantly expressed on tumor cells and is shed into the circulation it has long been proposed as a possible biomarker for various epithelial tumors. High MUC1 serum levels are associated with progressive disease and poor prognosis [8–10]. Currently, tests for several of these epitopes are available in commercial form for use in patient management, which include CA15-3 (Abbott Laboratories, Abbott Park, IL), CA27.29 (Bayer Diagnostics, Tarrytown, NY), and CA19-9 (Panomics, Inc., Redwood City, PA). Thus far, none have proven to be of significant diagnostic value due to low specificity as reported in the literature [30–33]. Thus, the American Society of Clinical Oncology does not recommend these tumor marker tests for recurrence screening. The TAB 004 EIA accurately detects shed MUC1 in the circulation of mice with MUC1high tumors but not in the circulation of MUC1low tumors (Fig. 6A), indicating the specificity of the test. Further using sera from patients with different stages of PC, we were able to predict patient stage based on the levels of shed MUC1 determined by the TAB 004 EIA. These results are extremely exciting and warrant further exploration of MUC1 (as detected by TAB 004) as a biomarker for PC.
The TAB 004 antibody differs from other commercially available antibodies in the manner in which it was generated and its unique CDR sequence. We have demonstrated its sensitivity and specificity to tumor-associated MUC1. Further, we have shown the novel expression of MUC1 on CSCs as recognized by TAB 004. This holds great promise as a possible target to deplete CSCs, which is necessary for elimination of the bulk tumor. TAB 004 can also detect shed MUC1 in the serum of patients with PC before they have progressed to later stage disease and may be explored as a marker of tumor stage. Early detection for PC is key to survival, as this cancer normally does not produce symptoms until later stage disease, is highly aggressive, and mostly fatal. Our lab is currently optimizing many uses for this antibody including targeted drug delivery and TAB 004 EIA for early detection of PC.
Supplementary Material
Acknowledgments
This work was supported by AACR Pancreatic Cancer Action Network Pilot Project (Mukherjee), NIH/NCI RO1 CA118944-01A1 (Mukherjee) and the UNCC Belk Endowment Funds (Mukherjee). We would like to acknowledge David Gray for his assistance with IHC and Confocal Microscopy at UNC-Charlotte. We are grateful to the Hepato–Pancreatico–Biliary Surgery research nurses at the Carolinas Medical Centers. Specifically, Whitney Rossman and Rondell Staten have been instrumental to these studies. We would like to acknowledge Tracy Walling in the Department of Surgery at Carolinas Medial Centers for her assistance with histology. We also thank the Vivarium staff at UNC-Charlotte.
Abbreviations
- PC
pancreatic cancer
- CSCs
cancer stem cells
Footnotes
Additional supporting information may be found in the online version of this article.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 4.Rasheed ZA, Yang J, Wang Q, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levi E, Klimstra DS, Andea A, et al. MUC1 and MUC2 in pancreatic neoplasia. J Clin Pathol. 2004;57:456–462. doi: 10.1136/jcp.2003.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tinder TL, Subramani DB, Basu GD, et al. MUC1 enhances tumor progression and contributes toward immunosuppression in a mouse model of spontaneous pancreatic adenocarcinoma. J Immunol. 2008;181:3116–3125. doi: 10.4049/jimmunol.181.5.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osako M, Yonezawa S, Siddiki B, et al. Immunohistochemical study of mucin carbohydrates and core proteins in human pancreatic tumors. Cancer. 1993;71:2191–2199. doi: 10.1002/1097-0142(19930401)71:7<2191::aid-cncr2820710705>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 8.Qu CF, Li Y, Song YJ, et al. MUC1 expression in primary and metastatic pancreatic cancer cells for in vitro treatment by (213)Bi-C595 radioimmunoconjugate. Br J Cancer. 2004;91:2086–2093. doi: 10.1038/sj.bjc.6602232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gronborg M, Bunkenborg J, Kristiansen TZ, et al. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 10.Hollingsworth MA, Strawhecker JM, Caffrey TC, et al. Expression of MUC1, MUC2, MUC3 and MUC4 mucin mRNAs in human pancreatic and intestinal tumor cell lines. Int J Cancer. 1994;57:198–203. doi: 10.1002/ijc.2910570212. [DOI] [PubMed] [Google Scholar]
- 11.Berberat P, Friess H, Kashiwagi M, et al. Diagnosis and staging of pancreatic cancer by positron emission tomography. World J Surg. 1999;23:882–887. doi: 10.1007/s002689900593. [DOI] [PubMed] [Google Scholar]
- 12.Jiang JT, Wu CP, Deng HF, et al. Serum level of TSGF, CA242 and CA19-9 in pancreatic cancer. World J Gastroenterol. 2004;10:1675–1677. doi: 10.3748/wjg.v10.i11.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zalatnai A. Pancreatic cancer—A continuing challenge in oncology. Pathol Oncol Res. 2003;9:252–263. doi: 10.1007/BF02893388. [DOI] [PubMed] [Google Scholar]
- 14.Shore S, Raraty MG, Ghaneh P, et al. Review article: Chemotherapy for pancreatic cancer. Aliment Pharmacol Ther. 2003;18:1049–1069. doi: 10.1111/j.1365-2036.2003.01781.x. [DOI] [PubMed] [Google Scholar]
- 15.Ichihara T, Nomoto S, Takeda S, et al. Clinical usefulness of the immunostaining of the tumor markers in pancreatic cancer. Hepatogastroenterology. 2001;48:939–943. [PubMed] [Google Scholar]
- 16.Frebourg T, Bercoff E, Manchon N, et al. The evaluation of CA19-9 antigen level in the early detection of pancreatic cancer. A prospective study of 866 patients. Cancer. 1988;62:2287–2290. doi: 10.1002/1097-0142(19881201)62:11<2287::aid-cncr2820621103>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 17.Ziske C, Schlie C, Gorschluter M, et al. Prognostic value of CA19-9 levels in patients with inoperable adenocarcinoma of the pancreas treated with gemcitabine. Br J Cancer. 2003;89:1413–1417. doi: 10.1038/sj.bjc.6601263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia. 2001;6:339–353. doi: 10.1023/a:1011379725811. [DOI] [PubMed] [Google Scholar]
- 19.Hollingsworth MA, Swanson BJ. Mucins in cancer: Protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 20.Sahraei M, Roy LD, Curry JM, et al. MUC1 regulates PDGFA expression during pancreatic cancer progression. Oncogene. 2012;31:4935–4945. doi: 10.1038/onc.2011.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besmer DM, Curry JM, Roy LD, et al. Pancreatic ductal adenocarcinoma (PDA) mice lacking Mucin 1 have a profound defect in tumor growth and metastasis. Cancer Res. 2011;71:4432–4442. doi: 10.1158/0008-5472.CAN-10-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 23.Rowse GJ, Tempero RM, VanLith ML, et al. Tolerance and immunity to MUC1 in a human MUC1 transgenic murine model. Cancer Res. 1998;58:315–321. [PubMed] [Google Scholar]
- 24.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 26.Hikita ST, Kosik KS, Clegg DO, et al. MUC1* mediates the growth of human pluripotent stem cells. PLoS ONE. 2008;3:e3312. doi: 10.1371/journal.pone.0003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelmann K, Shen H, Finn OJ. MCF7 side population cells with characteristics of cancer stem/progenitor cells express the tumor antigen MUC1. Cancer Res. 2008;68:2419–2426. doi: 10.1158/0008-5472.CAN-07-2249. [DOI] [PubMed] [Google Scholar]
- 28.Roy LD, Sahraei M, Subramani DB, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deguchi T, Tanemura M, Miyoshi E, et al. Increased immunogenicity of tumor-associated antigen, mucin 1, engineered to express alpha-gal epitopes: A novel approach to immunotherapy in pancreatic cancer. Cancer Res. 2010;70:5259–5269. doi: 10.1158/0008-5472.CAN-09-4313. [DOI] [PubMed] [Google Scholar]
- 30.Ugorski M, Laskowska A. Sialyl Lewis(a): A tumor-associated carbohydrate antigen involved in adhesion and metastatic potential of cancer cells. Acta Biochim Pol. 2002;49:303–311. [PubMed] [Google Scholar]
- 31.Magnani JL, Steplewski Z, Koprowski H, et al. Identification of the gastrointestinal and pancreatic cancer-associated antigen detected by monoclonal antibody 19-9 in the sera of patients as a mucin. Cancer Res. 1983;43:5489–5492. [PubMed] [Google Scholar]
- 32.Banfi G, Zerbi A, Pastori S, et al. Behavior of tumor markers CA19. 9, CA195, CAM43, CA242, and TPS in the diagnosis and follow-up of pancreatic cancer. Clin Chem. 1993;39:420–423. [PubMed] [Google Scholar]
- 33.Perkins GL, Slater ED, Sanders GK, et al. Serum tumor markers. Am Fam Physician. 2003;68:1075–1082. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.