Abstract
The treatment of patients with lung cancer is increasingly individualised. Rather than treating lung cancer as a single disease, clinicians are often called upon to consider the precise histology and molecular biology of each tumour in addition to the individual characteristics of each patient. Paralleling advances in lung cancer management, advances in the detection of lung cancer are changing practice. Lung cancer screening promises to find disease at a curable stage; however, the high false positive rate in screening trials has clinical and fiscal ramifications which demand attention. Biomarkers able to stratify for the risk of cancer, prognosticate the course of disease, or predict the response to treatment are in increasing demand. This paper summarizes some of the clinical problems faced by those treating lung cancer patients, and examines how knowledge about the role of microRNAs in lung cancer biology may change patient management.
Keywords: biomarker, microRNA, lung cancer, thoracic oncology, screening, early detection, tumor biology, prognostic marker, predictive marker.
Background
Lung cancer causes more deaths than any other cancer. A predicted 186 970 men and 82 640 women will die of lung cancer in the European Union in 2013 1. Although the majority of lung cancer patients are men, since 2009 there has been a 7% increase in the incidence of lung cancer in European women, likely in part due to changes in smoking behaviour. The global impact of lung cancer is also likely to increase: in 2010 respiratory cancers caused 1.5 million deaths worldwide, and accounted for 19% of all deaths from cancer 2. Although smoking is the predominant cause of most lung cancers, other environmental factors such as radon and asbestos are relevant in some patients 3. Genetic and epigenetic factors which may predispose individuals to the disease are also under investigation 4. Unfortunately, most lung cancer patients are diagnosed at an advanced stage of disease, and, although new drugs offer small subsets of these patients improved overall survival and reasonable quality of life, the majority of patients can only be treated with palliative chemotherapy. Overall survival remains poor, and many patients die within a few months of diagnosis.
Lung Cancer: One Name for Many Diseases
The term lung cancer is often used as if the term referred to a single disease. However, lung cancer is not one, but rather a group of diseases. Physicians have long recognised that small cell lung cancers (SCLCs) generally behave differently than non-small cell lung cancers (NSCLC). Because SCLC metastasizes earlier and is initially more chemosensitive than NSCLC, the two types of lung cancer are treated differently. Over the past two decades it has become clear that NSCLC itself is a clinically and biologically heterogeneous group of lung cancers, and should not be treated as a single disease entity.
Diversity of histologies
The two main subgroups of NSCLC are adenocarcinoma and squamous cell carcinoma. The appearance of these tumours at light microscopy differs substantially, suggesting that their aetiology and biology differ as well. However, it was not until the publication of clinical trials with pemetrexed that it became clear that the histological subgroups of NSCLC respond differently to some chemotherapeutic substances 5; 6. Data showing a higher risk of severe haemoptysis in patients with squamous cell lung cancer treated with bevacizumab compared with other histological subgroups added to the awareness that distinct treatment algorithms are needed for squamous cell lung cancer as compared to the so-called nonsquamous types of NSCLC, namely adenocarcinoma and large cell carcinoma 7.
Diversity of aetiologies
The causal association of lung cancer with cigarette smoking has been clear since the 1950s. However, the strong carcinogenic effect of cigarette smoking and the high proportion of smokers in many countries have often overshadowed the fact that lung cancer is not always caused by smoking. We now recognise that at least 10 % of lung cancer patients are never smokers; and that tumours in never smokers are biologically distinct 8. In addition, other carcinogens have been shown to cause lung cancer, including arsenic, which seems to be associated with a specific type of squamous cell lung cancer 9.
Diversity of response to therapy
The response to chemotherapy varies considerably not only between patient subgroups, but also between individuals within subgroups. A randomized trial in which patients with NSCLC were treated with first line pemetrexed and cisplatin or gemcitabine and cisplatin demonstrated that the subgroup of patients with adenocarcinoma benefitted from pemetrexed more than from gemcitabine. The contrary was true for nonsquamous histologies 5. A retrospective analysis of a second line trial of pemetrexed vs. docetaxel showed the same correlation between histology and treatment efficacy for pemetrexed but not for docetaxel 6. The relevance of histology for the treatment with pemetrexed was also confirmed in a trial of maintenance therapy 10. Such histological subgroup differences in treatment efficacy likely exist for other chemotherapeutic substances as well. A metaanalysis of the efficacy of cisplatin in subgroups demonstrated that cisplatin is more effective in nonsquamous tumours 11. Side effects of some therapies also appear to vary between subgroups. For instance, the antiangiogenic agent bevacizumab is not given to patients with squamous cell tumours due to an increased incidence of fatal bleeding in this group 7.
Diversity of molecular biologies
The detection of activating EGFR mutations in a subgroup of NSCLC patients generated a surge of interest in the genetic changes in lung cancer 12. Over the past decade there has been an explosion of biological knowledge in this field, stemming in part from genome-wide association studies, and fuelling the search for drugable targets and so-called driver mutations 13. Mutations associated with a specific treatment can be found in more than 50% of adenocarcinomas. An increasing number of such drugable mutations have also been identified in squamous cell lung cancer 14. Table 1 provides a summary of some genetic changes currently under investigation as treatment targets in NSCLC.
Table 1.
Drugable driver mutations in non small cell lung cancer (modified from Kris, MG ea. ASCO 2011 and Hammerman P ea. WCLC 2011).
| Adenocarcinoma | |
| K-Ras | 22 % |
| EGFR | 17 % |
| EML4-ALK | 7 % |
| B-Raf | 2 % |
| Other | 6% |
| No driver mutation detected | 46 % |
| Squamous cell lung cancer | |
| FGFR1 amplification | 20 - 25 % |
| FGFR2 mutation | 5 % |
| PIK3CA mutation | 9 % |
| PTEN mutation/deletion | 18 % |
| DDR2 mutation | 4 % |
The Clinician's Dilemma: Which treatment is right for which patient?
Treatment for both early stage and advanced lung cancer is often associated with significant morbidity and even mortality. Clinicians must therefore weigh the chances of benefit against the risks of treatment when advising patients. Localized NSCLC treated with surgery or radiotherapy often recurs, resulting in a limited prognosis despite initially localised disease. Additional systemic therapy reduces the chance of recurrence for a small number of patients; however, many of those treated do not benefit either because their tumour would not have recurred even without additional therapy, or because their tumour recurs despite therapy. Both of these groups continue to suffer the toxicity of additional therapy unnecessarily because it is not yet possible to accurately predict which patients at risk of recurrence will benefit.
Patients with metastases are generally treated with platin doublet chemotherapy. These agents act nonspecifically and result in only slight improvements in patient survival. The efficacy in the individual patient and tumour can only be judged during or after treatment. Although studies are ongoing, predictive biomarkers for the efficacy of chemotherapy are not yet available for routine clinical use.
Biomarkers which can predict the individual prognosis - especially after potentially curative treatment for local disease - would help determine the need for additional systemic treatment. Similarly, for those patients with advanced disease, predictive markers for the efficacy of various systemic agents would reduce the rate of primary progression under therapy and thus reduce patient exposure to ineffective and often toxic treatments.
Prognostic vs. Predictive Biomarkers
Biomarkers can be either prognostic, predictive or both. Prognostic biomarkers correlate with the clinical course of disease, often quantified as overall survival, independent of which type of therapy is given. In contrast, predictive biomarkers correlate with the response to a specific treatment, and can thus be used to select a treatment likely to be effective against a particular tumour.
Prognostic markers
There are a large number of prognostic markers available to clinicians caring for lung cancer partients. Currently the most clinically relevant prognostic factors are patient characteristics such as performance status, generally described using the ECOG or Karnofsky scales, and stage of disease.
Prognostic biomarkers are of scientific value because they suggest a strong link, and may imply a causal relationship, between a characteristic of tumour growth, for instance invasiveness or the potential to metastasize, and a specific molecule. For many molecules investigated primarily in in vitro models of disease, this represents an important first step from the bench to the bedside. Although prognostic markers are not able to directly predict the response to specific therapies, they may be taken into account when deciding how, or how often, to monitor patients for signs of disease progression, or when weighing the risks and benefits of a treatment.
Many clinical attributes and biomarkers have been examined for their prognostic value in lung cancer. The stage of disease, the performance status of the patient and the histology (in particular the distinction between small cell and non-small cell lung cancer) are widely accepted prognostic markers 15. The prognostic relevance of the subgroups of NSCLC is under investigation. Especially in early lung cancer the histological subtype of adenocarcinoma seems to be relevant to prognosis 16. With the advent of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in advanced NSCLC it has become clear that the presence of an activating EGFR mutation is also of prognostic and also predictive relevance 17.
Predictive markers in lung cancer therapy
Several predictive markers are relevant for the choice of systemic therapy in advanced NSCLC. Histology, in this case the distinction between squamous cell and non-squamous tumours, is predictive of response to pemetrexed. While patients with non-squamous NSCLC profit from pemetrexed, those with squamous cell cancer do not 18. Histology is also predictive of the risk of serious side effects during treatment with bevacizumab. An activating EGFR mutation predicts response and progression-free survival in patients treated with EGFR-TKIs such as erlotinib and gefitinib 19, 20; and EML4-ALK fusion 21 is predictive of response to ALK-TKIs such as crizotinib. Potential predictive biomarkers for the efficacy of specific chemotherapeutic agents have been described, for example thymidilate synthase for pemetrexed and ERCC1 for cisplatin, but these markers are not yet universally accepted or validated 22; 23. ERCC1, which is related to the DNA-repair mechanisms, demonstrated predictive value in the Bio-IALT trial 23. However, this finding was not reproducible 24, and ERCC1 has not yet found a place in routine clinical decision making. The predictive biomarkers currently used to individualise treatment in advanced NSCLC are summarized in table 2.
Table 2.
The Individualised Treatment of Advanced Lung Cancer: Predictive Markers Currently in Clinical Use.
| Systemic Treatment | Predictive Marker |
|---|---|
| First-Line Treatment Options | |
| cisplatin or carboplatin paired with a second chemotherapeutic agent such as gemcitabine, docetaxel, paclitaxel or vinorelbine | none currently established for widespread clinical use |
| cisplatin or carboplatin paired with pemetrexed | non-squamous histology |
| epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) such a erlotinib or gefitinib | Activating EGFR mutation, in particular in exon 19 or 21 |
| bevacizumab in combination with platin-based doublet chemotherapy | non-squamous histology (due to increased risk of serious complications in patients with squamous cell histology) |
| Second- and Further-Line Treatment Options | |
| monotherapy with docetaxel | none currently established for widespread clinical use |
| monotherapy with pemetrexed | non-squamous histology |
| epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) such a erlotinib or gefitinib | Activating EGFR mutation, in particular in exon 19 or 21 (note: erlotinib is also effective as a second line agent in patients without EGFR mutation) |
| anaplastic lymphoma kinase tyrosine kinase inhibitor (ALK-TKI) such as crizotinib | EML4-ALK translocation |
Biomaterial sampling in Lung Cancer: when tissue is an issue
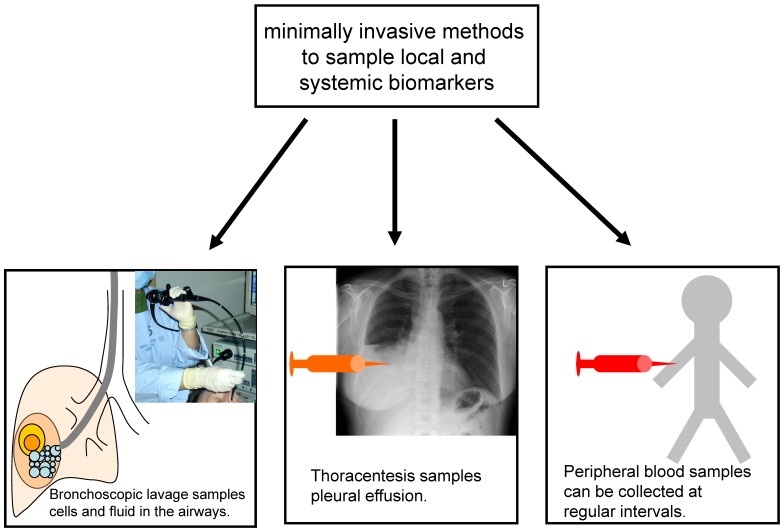
A detailed pathological and molecular workup of a tumour specimen, including analysis of possible prognostic and predictive markers, requires a sufficient number of tumour cells. While resected specimens are almost always large enough for these analyses, bronchoscopic biopsies and fine needle aspirations are often much smaller, in some cases capturing only a few tumour cells, and therefore present a challenge. Techniques to optimise the yield of small biopsies and aspirates, such as imaging with endobronchial ultrasound or CT, the use of several passes with the aspiration needle, and the cell block technique for processing cytological material, are of clinical importance. The difficulty in collecting enough cells to make a definitive diagnosis is particularly relevant in the setting of CT-screening for lung cancer. Because 97% of CT-detected nodules are non-malignant 25 and many of the patients screened are heavy smokers with pulmonary comorbidities, the risks and benefits of invasive diagnostic procedures have to be weighed carefully. It is neither feasible nor clinically sensible to biopsy every lesion detected by CT. The measurement of biomarkers in minimally invasive samples such as blood, lavage and sputum may help assess the risk of cancer in patients with lesions detected by screening, or may even help pre-select patients at particularly high risk of cancer for screening. Some of the minimally invasive methods currently used to collect samples for biomarker analysis are illustrated in figure 1.
Fig 1.
While histology and cytology remain critical for establishing the diagnosis of lung cancer, minimally invasive biomarker sampling may allow for the risk-stratification of individuals being considered for lung cancer screening, and may improve monitoring during and after lung cancer therapy.
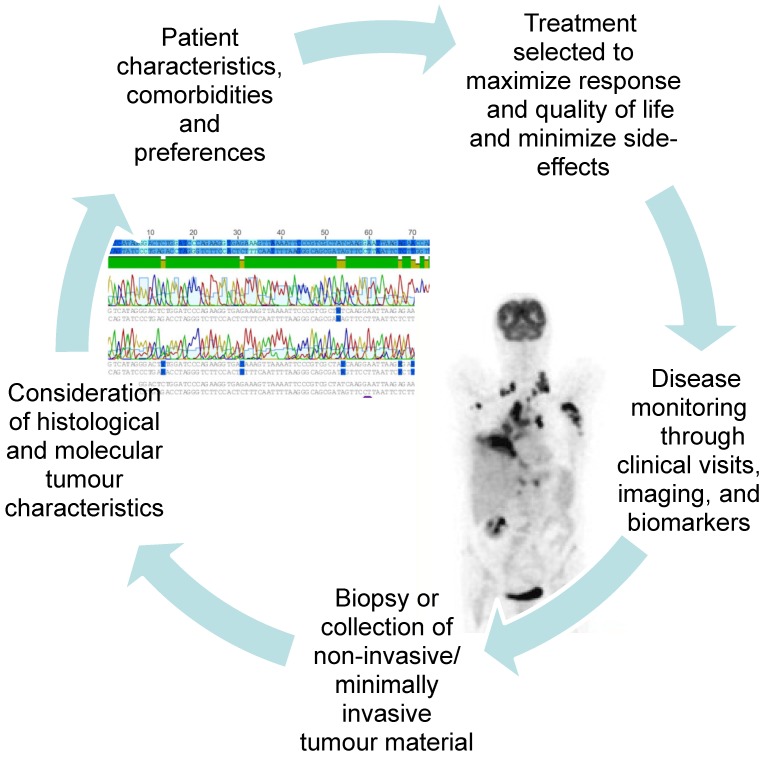
It is often difficult to get enough tissue to reach an exact histological diagnosis and perform the molecular analyses needed to select an individualised treatment at the time of first diagnosis. However, getting enough tissue to perform molecular analyses often becomes even more challenging as the tumour develops resistance to treatment and progresses. The advent of new molecular targets has taught us to look for and treat driver mutations, and when the tumour progresses, to look for new changes in the tumour's biology which may suggest a specific second line treatment. In addition to the tumour biopsy required at the time of first diagnosis, clinicians now often consider re-biopsy during the course of disease or at the time of progression. Figure 2 illustrates the increasing integration of the repeated collection of tumour biopsies and minimally invasive samples into patient care. This shift in our approach to lung cancer therapy demands that minimally invasive sampling and alternative biomaterials such as serum, BAL, and pleural fluid be developed to their fullest potential.
Fig 2.
As lung cancer therapies become increasingly tailored to the individual patient and tumour, re-sampling of biomaterials during the course of treatment and at progression are gaining relevance.
MicroRNAs
MicroRNAs (miRNAs) are short non-coding nucleotide sequences which interact with complementary sequences on target mRNAs. Through these interactions miRNAs are able to inhibit the translation of mRNAs and thus downregulate the expression of specific proteins. Hundreds of miRNAs have been identified, and, as each may interact with multiple mRNAs, the potential for interaction is enormous. A standardised nomenclature for miRNAs has been suggested, and newly discovered miRNAs and the genes which encode them are assigned sequential numbers 26, 27. Early data from C. elegans and Drosophila showed that miRNAs are involved in a multitude of critical developmental processes, including cell proliferation and apoptosis 28.
The altered expression of miRNAs in human disease states may provide insights into the pathomechanisms of disease, and offer novel means of diagnosing, monitoring and perhaps even treating patients. Clinically, the use of miRNAs as biomarkers is particularly appealing because of their apparent stability in both archived patient material, such as formalin fixed paraffin embedded (FFPE) biopsies 29 and samples of easily accessible bodily fluids such as serum, sputum, and pulmonary lavage 30; 31. The age and comorbidities of many lung cancer patients often complicate the use of invasive diagnostic biopsies, and so minimally invasive techniques are particularly appealing to clinicians caring for these patients.
Oncogenic miRNAs
The upregulation or downregulation of a number of miRNAs has been found in association with a variety of human cancers. Over ten years ago, Calin and colleagues reported that miR15 and miR16 are located at the chromosome 13q14 region which is often deleted in B cell chronic lymphocytic leukemia (B-CLL) 32. The authors hypothesised that these miRNAs regulate genes involved in normal B cell differentiation, and that their downregulation or deletion may be causally linked to the development of CLL. Since then a multitude of associations between miRNA levels and cancer, including lung cancer, have been reported. These associations are summarized in several detailed review articles 33 34.
miRNA in Lung Cancer: Interactions with known Lung Cancer Pathways
MiRNAs interact with complementary mRNA sequences to regulate protein expression. Studies in other tumour entities have identified a large number of interactions between miRNAs and proteins in pathways known to play a role in cancer. For instance, miR15 and miR16 may induce cell apoptosis by targeting B cell lymphoma 2 (BCL2) 35. Thus, a downregulation of miR15 and miR16 downregulates cell apoptosis and promotes carcinogenesis. There is growing evidence that the regulation of proteins involved pathways crucial to the pathogenesis of lung cancer is also regulated by miRNAs 36.
Rat Sarcoma (RAS)
Mutations in RAS-genes, in particular in KRAS, are of clinical importance in human lung cancer 37, 38. There is evidence that miRNAs from the let-7 family regulate RAS expression in human cells. In 2005 Johnson and colleagues reported that let-7 targets the C.elegans let-60 gene, the ortholog of human RAS, and that the 3' UTRs of human RAS genes contain multiple let-7 complimentary sites. They also found that reducing the activity of let-7 in human cell lines led to increased expression of RAS. Paired analysis of RAS protein and let-7 expression from human lung cancer and normal adjacent tissue samples showed low levels of let-7 and high levels of RAS in lung cancer tissues 39.
In addition, Chin and colleagues found that a single nucleotide polymorphism (SNP) in a let-7 complementary site of KRAS mRNA (LCS6) is associated with increased risk of NSCLC in moderate smokers 40. The LCS6 SNP was found in 18.1 to 20.3% of lung cancer cases and only 5.8% of healthy controls tested. Based on in vitro experiments and analyses of patient samples the authors concluded that this SNP alters the ability of let-7 to regulate translation of KRAS, leading to overexpression of KRAS and increased lung cancer risk.
Let-7 family miRNAs seem to play a significant role in human lung cancer, at least in part through the regulation of RAS.
Other miRNAs may also interact with RAS. For instance, Wang and colleagues 41 found that miR-451 is downregulated in NSCLC, and that low expression correlated with poor survival. The authors were able to show that miR-451 inhibits the expression of ras-related protein 14 (RAB14), suggesting that lower expression of miR-451 may allow this oncogene to escape regulation.
P53
The p53 gene on chromosome 17p13 has long been recognised as a relevant tumour suppressor, with loss of function in many types of cancer including lung cancer. p53 is mutated in a large number of lung cancer cell lines and tumour specimens from patients with lung cancer 42; 43. There is growing evidence that the tumour suppressive activity of p53 is at least in part moderated by miRNAs. In the lung cancer cell line H1299 expression of p53 resulted in upregulation of miR-34a. The overexpression of miR-34a suppressed proliferation of lung cancer cells in vitro and promoted apoptosis 44. Several groups have shown that p53 directly regulates the expression of miR-34 family members, and that these miRNAs result in the downregulation of genes associated with cell cycle control in cultured tumour cells 45; 46; 47 including the lung cancer cell line A549 45. Further miRNAs, including miR-125a, have more recently also been linked to p53-regulated apoptosis in lung cancer cells 48.
miRNAs as prognostic biomarkers in Lung Cancer
The first evidence that alterations in miRNA expression correlate with prognosis in lung cancer was provided by work from Takamizawa and colleagues, published in 2004 49. Expression of the miRNA let-7 was measured in 143 tumour samples from patients who had undergone curative resection for stage I-III NSCLC. The investigators also measured let-7 expression in 20 lung cancer cell lines. Compared to expression levels in normal lung tissue, expression of let-7 was significantly reduced in many of the lung cancer cell lines and patient samples. Patients whose tumours showed low let-7 expression had poorer overall survival than those with higher expression of let-7. Although the low let-7 group also had more advanced stage disease at the time of surgery, multivariate analysis showed let-7 expression to be an independent prognostic factor.
Further evidence for the role of let-7 in lung cancer was provided by Kumar and colleagues 50who, after expressing let-7g miRNA in murine KRAS positive lung adenocarcinoma cells, found a significant decrease in cell proliferation and increase in apoptosis. The same authors found that increased let-7g expression reduced tumour growth in an in vivo mouse model of lung cancer.
In addition to let-7, a number of other miRNAs have been suggested to be of prognostic significance in lung cancer. Yanaihara and colleagues 51 analyzed miRNA expression in 104 pairs of lung cancers and corresponding noncancerous lung tissues and identified miRNAs which showed statistically significant differences in expression. There were significant differences in the miRNA expression pattern in adenocarcinoma compared with squamous cell carcinoma. In patients with adenocarcinoma high hsa-mir-155 or reduced hsa-let-7a-2 expression correlated with poor prognosis; however in multivariate analysis only high hsa-mir-155 expression remained of prognostic significance.
Yu and colleagues analysed 122 NSCLC samples and identified a panel of five miRNAs which correlated with prognosis 52. While hsa-miR-221 and hsa-let-7a expression correlated with improved survival, hsa-miR-137, hsa-miR-372 and hsa-miR-182 correlated with poor survival. In both the training and test collectives, NSCLC patients with a high-risk miRNA constellation had shorter median overall survival and shorter time to relapse than patients with a low-risk miRNA signature.
Patnaik and colleagues 53 described miRNA expression profiles which were associated with lung cancer recurrence after curative resection of stage I NSCLC. The investigators examined archived tumour tissue from 77 patients and compared miRNA expression patterns in patients who went on to recur with those who did not. They found that 47% of the 279 miRNAs identified were differently expressed in the two groups. Duncavage and colleagues 29 also retrospectively examined tumour samples from patients with resected stage I NSCLC. These investigators focused on a panel of six miRNAs (let-7a, miR-7, miR-21, miR-155, miR-210 and miR-221) in 46 patients and found significantly lower miR-221 expression in samples from tumours which recurred compared with those without recurrence.
Alterations in miRNA processing also appear to be of prognostic relevance in lung cancer. A Japanese group found that levels of Dicer, an enzyme which cleaves hairpin-shaped pre-miRNAs in the cytoplasm, are reduced in a subset of tumour samples from patients with lung cancer. The authors showed that patients whose tumours expressed lower levels of Dicer experience significantly shorter survival after surgery for stage I-III NSCLC than those with high Dicer expression. They also found a correlation between low levels of Drosha, an enzyme which performs a cleavage step in the nucleus to form pre-miRNAs, and poor survival; however, this association did not reach statistical significance 54.
Not all studies have shown a correlation between miRNA expression and lung cancer prognosis, and studies in similar patient populations have produced some conflicting results. The results of a large retrospective analysis of tumour samples from the randomised International Adjuvant Lung Cancer Trial (IALT) did not confirm the prognostic significance of miRNAs previously identified as markers. Tumour samples from 639 patients with stage I-III NSCLC who had been randomised to receive adjuvant cisplatin-based chemotherapy or follow-up without adjuvant were analysed for the expression of a panel of miRNAs selected because of evidence for their prognostic or predictive importance: miR-21, miR-29b, miR-34a/b/c, miR-155 and let-7a 55. The expression of individual miRNAs was found to correlate significantly with histology and with lymphoid infiltration. Expression of miR-21, miR-29b, miR-34a and miR-155 was associated with lymphoid infiltration, or, for miR-34a, lymphatic invasion. There was a non-significant trend towards poorer prognosis in patients with negative miR-21 expression. However, none of the miRNAs analysed were found to be predictive of benefit from cisplatin-based adjuvant chemotherapy.
While many studies have identified correlations between individual miRNAs or miRNA signatures and lung cancer survival, the inconsistencies in the existing data suggest that our understanding of the pathophysiology and regulatory mechanisms at play is far from complete.
One complicating factor seems to be that the diversity in lung cancer is mirrored by diversity in miRNA-signatures. The various histological forms and aetiological subgroups of lung cancer also differ in their changes in miRNA expression. For instance, the miRNA profiles of various histological subgroups of lung cancer appear to be distinct. Landi and colleagues examined tumour samples from 165 patients with adenocarcinoma and 125 patients with squamous cell carcinoma who had been enrolled in the Italian EAGLE (Environmental And Genetics in Lung cancer Etiology) and found that miRNA expression profiles differ significantly between these groups 56. Rather than including all subgroups of NSCLC, Raponi and colleagues compared miRNA expression in 61 squamous cell tumours to normal lung tissue and identified prognostic miRNAs 57. In addition, the miRNA expression in tumours from never-smokers appears to differ from that in tumours from smokers 58. Studies which fail to recognise and characterise the diversity of lung cancer are likely to produce conflicting results.
In addition to hypothesis-building retrospective studies, well designed prospective studies in well-defined patient populations are needed to investigate the utility of miRNAs as prognostic and perhaps also predictive markers in lung cancer.
MiRNA Profiles in Early Detection and Lung Cancer Screening
Early detection of lung cancer is a clinically important goal. The recently published results of large trials of CT-screening are encouraging, showing that yearly CT-screening in smokers identifies lung cancer at an earlier and thus more curable stage, and lowers the risk of lung cancer death. However, CT-screening also finds a multitude of non-cancerous lung nodules, and the expense and risk associated with following and correctly diagnosing these nodules is enormous. Screening all smokers for lung cancer using CT alone is not feasible at this time. The widespread implementation of lung cancer screening will likely depend on the development of multimodal models of lung cancer risk (including lung function, biomarkers and imaging results) to select individuals for screening, coupled with a risk-adapted approach to patient follow-up. A biomarker found in easily accessible materials such as sputum, exhaled air or peripheral blood could help focus screening on high risk patients. MiRNAs taken from the peripheral blood seem to remain intact and stable and are detectable with simple assays like quantitative real-time PCR (qRT-PCR). In addition, changes in miRNA expression have been shown to occur at early stages of carcinogenesis, making these markers potentially suitable for the detection of early stage tumours 59, 60. MiRNA profiles may therefore have a role in the early detection and screening of lung cancer. An Italian group 61 examined the utility of miRNA signatures in lung cancer screening using blood samples from two CT screening trials, one published by Pastorino and colleagues 62 and the Multicentric Italian Lung Detection (MILD) trial, The authors performed extensive miRNA profiling of primary lung tumours, paired normal lung tissues, and multiple plasma samples collected before and at the time of disease. They were able to define a plasma miRNA signature which was associated with increased risk of lung cancer. Several other groups have also integrated miRNA profiles into CT-screening programs. Table 1 in the review from Boeri and Colleagues 63 summarizes ten recent studies which used a range of techniques including q-RT-PCR and microfluidic cards to compare miRNA expression in blood samples from lung cancer patients with controls in this context.
In vitro miRNA Imaging in Lung Cancer
The use of imaging techniques to assess for the presence and distribution of specific molecules or molecular changes in lung cancer patients promises to greatly improve our ability to detect and monitor clinically relevant aspects of tumour biology before and during treatment. One exciting current example of this type of technology is the use of positron emission tomography (PET) tracers which bind to the EGF-receptor. Bahce and colleagues 64 recently reported on the development of [11 C] erlotinib PET, in which tracer is preferentially taken up by tumours harbouring EGFR mutations. Further development of this technique may allow for non-invasive, predictive assessment of patients' EGFR-mutation status.
Similarly, as the clinical relevance of alterations in miRNA expression in lung cancer increases, techniques to quantify clinically relevant miRNAs in vivo may prove to be of prognostic and predictive value in patient care. Fluorescence-based imaging 65 and bioluminescence-based imaging using luciferase 66 have been established to image miRNAs in cell culture and small animal experiments. However, the translation of such techniques into the setting of clinical medicine is limited by several factors including the limited penetration of optical signals though tissue layers and the difficulty in delivering the reporter molecule safely and predictably to the cells of interest in human patients 67.
MiRNAs in lung cancer treatment
The role of miRNAs in regulating oncogenic pathways makes them attractive targets for lung cancer therapy. Artificially increasing or decreasing the abundance of specific miRNAs may be a means of modifying tumour biology and thus changing clinical outcomes. Several strategies to manipulate miRNAs in vivo are under investigation. Antagomirs are antisense sequences which inhibit target miRNAs 68. Through the down-regulation of specific miRNAs, antagomirs have been shown to up-regulate mRNAs in in vivo models 69.
The overexpression of specific miRNAs in order to reduce the expression of target proteins has also been investigated. For instance, Hossain and colleagues showed that Mir-17-5p expression decreases translation of AIB1 mRNA to AIB1 protein in vitro 70.
In addition to the direct upregulation or downregulation of specific oncogenic miRNAs, the indirect modification of miRNA expression patterns through the use of novel substances administered systemically is under investigation. A thorough review of strategies to manipulate miRNA biology for the treatment of cancer is provided by Rossbach 71.
MiRNAs to Prevent Metastasis
Most patients with lung cancer present at an advanced stage of disease and cannot be offered curative treatment. However, even those with early stage or locally advanced disease who are treated with surgery or multimodal approaches often go on to develop metastases. The rate of disease relapse with distant metastases has been described to be 15-40% in patients operated for early stage disease, and 60% in patients operated in stage IIIa 72. Adjuvant chemotherapy is associated with significant morbidity and mortality, and only slightly improves survival 73. A treatment to effectively prevent metastasis would therefore be of great clinical value. There is early evidence suggesting that the manipulation of miRNAs may be one means of preventing metastasis. Ma and colleagues 74 demonstrated that miR-9 promotes the development of metastases by downregulating E-cadherin. Using a mouse model of breast cancer, the investigators showed that knocking down miR-9 expression reduced the formation of lung metastases. The size of the primary tumour remained unchanged.
A complete list of miRNAs thought to be involved in metastasis is provided in a recent review by Schoof et al. 75.
MiRNAs to overcome chemotherapy resistance
Despite excitement over new drugs for specific molecularly defined patient subgroups, such as gefitinib and erlotinib for EGFR mutated tumours and crizotinib for tumours with EML4-ALK fusion, the majority of lung cancer patients are treated with platin-based chemotherapy. Cisplatin and carboplatin form the backbone of most adjuvant, multimodal and palliative systemic treatment regimes. Resistance to platin is an important clinical problem, resulting in treatment failure and disease progression in many patients. The molecular mechanisms of platin resistance have yet to be fully understood; however, it seems that miRNAs may regulate some genes which contribute to resistance to these drugs. Wang and colleagues 76 identified altered expression of 14 miRNAs in cisplatin resistant lung cancer cells in A549 lung cancer cells compared to in cisplatin sensitive cells. Cells were more sensitive to cisplatin when miR-138 was upregulated. Downregulation of the excision repair cross-complementation group 1 (ERCC1) by miR-138 seemed to be a possible mechanism.
Zhang and colleagues also investigated the role of miRNAs in cisplatin resistant A549 cells, and found that transfection of cisplatin resistant cells with miR-513a-3p resulted in increased sensitivity to cisplatin. MiR-513a-3p downregulated the glutathione S-transferase P1 (GSTP1), which promotes cisplatin resistance 77.
MiRNAs and radiation therapy
Radiation is a mainstay of lung cancer treatment. In locally advanced disease it can be combined with chemotherapy and in some situations with surgical resection to cure disease. In patients with advanced lung cancer it is of palliative value in treating symptomatic metastases or local complications related to the primary tumour such as superior vena cava syndrome. However, the response to radiation is heterogeneous, with some patients benefiting from treatment and others going on to progress with only side effects. The mechanisms underlying this heterogeneity have yet to be fully understood, however some evidence points to differences in gene expression between lung cancers which respond to radiation and those which do not 78; 79. There is also growing evidence that miRNAs may be differently expressed in tumours which respond to radiation compared with those which do not. Lynam-Lennon and colleagues found that miR-31 expression is reduced in oesophagus tumours which do not respond well to radiation, and linked expression of miR-31 to the regulation of DNA repair genes 80. Weidhaas and colleagues were able to demonstrate that the overexpression of let-7 in lung cancer cell lines improves radiosensitivity 81.
Summary
The biology of lung cancer is increasingly complex. Our ability to generate information risks outpacing our ability to process and sort our collective knowledge. A major challenge for the years to come will be to develop strategies to extract clinically meaningful conclusions from the data we generate. MiRNAs show promise as an adjunct to lung cancer diagnosis and treatment. The stability of miRNAs, and their availability in minimally invasive sampling of blood and lavage, makes them clinically appealing markers. The integration of changes in miRNA signatures into lung cancer screening algorithms may be able to increase the specificity of screening and lower the morbidity and cost associated with high false positive rates. And the ability to predict, or even to optimise, response to established therapies such as cisplatin-based chemotherapy and radiation could lower rates of treatment failure and patient exposure to the side effects of ineffective therapies. MiRNAs are exciting molecular players with the potential to change our approach to lung cancer.
References
- 1.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2013. Ann Oncol. 2013 Mar;24(3):792–800. doi: 10.1093/annonc/mdt010. PubMed PMID: 23402763. Epub 2013/02/14. [DOI] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V. et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012 Dec 15;380(9859):2095–128. doi: 10.1016/S0140-6736(12)61728-0. PubMed PMID: 23245604. Epub 2012/12/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberg A, Ford J, Samet J. Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:29S–55S. doi: 10.1378/chest.07-1347. PubMed PMID: doi:10.1378/chest.07-1347. [DOI] [PubMed] [Google Scholar]
- 4.Piperi C, Vlastos F, Farmaki E, Martinet N, Papavassiliou AG. Epigenetic effects of lung cancer predisposing factors impact on clinical diagnosis and prognosis. Journal of cellular and molecular medicine. 2008;12(5a):1495–501. doi: 10.1111/j.1582-4934.2008.00309.x. PubMed PMID: 18363846. Epub 2008/03/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C. et al. Phase III Study Comparing Cisplatin Plus Gemcitabine With Cisplatin Plus Pemetrexed in Chemotherapy-Naive Patients With Advanced-Stage Non-Small-Cell Lung Cancer. Journal of Clinical Oncology. 2008;26(21):3543–51. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 6.Hanna N, Shepherd FA, Fossella FV, Pereira JR, De Marinis F, von Pawel J. et al. Randomized Phase III Trial of Pemetrexed Versus Docetaxel in Patients With Non-Small-Cell Lung Cancer Previously Treated With Chemotherapy. Journal of Clinical Oncology. 2004;22(9):1589–97. doi: 10.1200/JCO.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 7.Sandler AB, Schiller JH, Gray R, Dimery I, Brahmer J, Samant M. et al. Retrospective Evaluation of the Clinical and Radiographic Risk Factors Associated With Severe Pulmonary Hemorrhage in First-Line Advanced, Unresectable Non-Small-Cell Lung Cancer Treated With Carboplatin and Paclitaxel Plus Bevacizumab. Journal of Clinical Oncology. 2009;27(9):1405–12. doi: 10.1200/JCO.2008.16.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramanian J, Govindan R. Lung Cancer in Never Smokers: A Review. Journal of Clinical Oncology. 2007;25(5):561–70. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 9.Becker-Santos DD, Vucic EA, Lam S, Lam WL, Martinez VD. Abstract B15: Genomic and epigenomic events in arsenic-related lung squamous cell carcinomas from smokers and never smokers. Clinical Cancer Research. 2012;18(3 Supplement):B15. [Google Scholar]
- 10.Ciuleanu T, Brodowicz T, Zielinski C, Kim JH, Krzakowski M, Laack E. et al. Maintenance pemetrexed plus best supportive care versus placebo plus best supportive care for non-small-cell lung cancer: a randomised, double-blind, phase 3 study. The Lancet. 2009;374(9699):1432–40. doi: 10.1016/S0140-6736(09)61497-5. [DOI] [PubMed] [Google Scholar]
- 11.Ardizzoni A, Boni L, Tiseo M, Fossella FV, Schiller JH, Paesmans M. et al. Cisplatin- Versus Carboplatin-Based Chemotherapy in First-Line Treatment of Advanced Non-Small-Cell Lung Cancer: An Individual Patient Data Meta-analysis. Journal of the National Cancer Institute. 2007;99(11):847–57. doi: 10.1093/jnci/djk196. [DOI] [PubMed] [Google Scholar]
- 12.Lynch T, Bell D, Sordella R, Gurubhagavatula S, Okimoto R, Brannigan B. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004 May 20. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Kris MG, Johnson BE, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Aronson SL. et al. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: The NCI's Lung Cancer Mutation Consortium (LCMC) ASCO Meeting Abstracts. 2011;29(18 suppl):CRA7506. [Google Scholar]
- 14.Hammerman P, Hayes D, Wilkerson M, Schultz N, Bose R, Chu A. et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 September 27. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berghmans T, Paesmans M, Sculier JP. Prognostic factors in stage III non-small cell lung cancer: a review of conventional, metabolic and new biological variables. Ther Adv Med Oncol. 2011 May;3(3):127–38. doi: 10.1177/1758834011401951. PubMed PMID: 21904576. Pubmed Central PMCID: Pmc3150064. Epub 2011/09/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y. et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011 Feb;6(2):244–85. doi: 10.1097/JTO.0b013e318206a221. PubMed PMID: 21252716. Epub 2011/01/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scoccianti C, Vesin A, Martel G, Olivier M, Brambilla E, Timsit JF. et al. Prognostic value of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: the EUELC cohort. Eur Respir J. 2012 Jul;40(1):177–84. doi: 10.1183/09031936.00097311. PubMed PMID: 22267755. Epub 2012/01/24. [DOI] [PubMed] [Google Scholar]
- 18.Scagliotti G, Hanna N, Fossella F, Sugarman K, Blatter J, Peterson P. et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two Phase III studies. Oncologist. 2009 Mar;14(3):253–63. doi: 10.1634/theoncologist.2008-0232. PubMed PMID: 19221167. Epub 2009/02/18. [DOI] [PubMed] [Google Scholar]
- 19.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009 Sep 3;361(10):947–57. doi: 10.1056/NEJMoa0810699. PubMed PMID: 19692680. Epub 2009/08/21. [DOI] [PubMed] [Google Scholar]
- 20.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E. et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012 Mar;13(3):239–46. doi: 10.1016/S1470-2045(11)70393-X. PubMed PMID: 22285168. Epub 2012/01/31. [DOI] [PubMed] [Google Scholar]
- 21.Kim D-W, Ahn M-J, Shi Y, De Pas TM, Yang P-C, Riely GJ. et al. Results of a global phase II study with crizotinib in advanced ALK-positive non-small cell lung cancer (NSCLC) ASCO Meeting Abstracts. 2012;30(15_suppl):7533. - [Google Scholar]
- 22.Eismann U, Oberschmidt O, Ehnert M, Fleeth J, Ludtke F, Struck S. et al. Thymidylate synthase gene expression in solid tumors predicts for response to pemetrexed in vitro. ASCO Meeting Abstracts. 2006;24(18_suppl):13058. [Google Scholar]
- 23.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V. et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006 Sep 7;355(10):983–91. doi: 10.1056/NEJMoa060570. PubMed PMID: 16957145. Epub 2006/09/08. [DOI] [PubMed] [Google Scholar]
- 24.Friboulet L, Olaussen KA, Pignon J-P, Shepherd FA, Tsao M-S, Graziano S. et al. ERCC1 Isoform Expression and DNA Repair in Non-Small-Cell Lung Cancer. New England Journal of Medicine. 2013;368(12):1101–10. doi: 10.1056/NEJMoa1214271. PubMed PMID: 23514287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM. et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011 Aug 4;365(5):395–409. doi: 10.1056/NEJMoa1102873. PubMed PMID: 21714641. Epub 2011/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.AMBROS V, BARTEL B, BARTEL DP, BURGE CB, CARRINGTON JC, CHEN X. et al. A uniform system for microRNA annotation. RNA. 2003;9(3):277–9. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimson A. A targeted approach to miRNA target identification. Nat Methods. 2010;7(10):795–7. doi: 10.1038/nmeth1010-795. [DOI] [PubMed] [Google Scholar]
- 28.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 29.Duncavage E, Goodgame B, Sezhiyan A, Govindan R, Pfeifer J. Use of MicroRNA Expression Levels to Predict Outcomes in Resected Stage I Non-small Cell Lung Cancer. Journal of Thoracic Oncology. 2010;5(11):1755–63. doi: 10.1097/JTO.0b013e3181f3909d. 10.097/JTO.0b013e3181f909d. [DOI] [PubMed] [Google Scholar]
- 30.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ. et al. The microRNA spectrum in 12 body fluids. Clinical chemistry. 2010 Nov;56(11):1733–41. doi: 10.1373/clinchem.2010.147405. PubMed PMID: 20847327. Epub 2010/09/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina-Pinelo S, Suarez R, Pastor MD, Nogal A, Marquez-Martin E, Martin-Juan J. et al. Association between the miRNA signatures in plasma and bronchoalveolar fluid in respiratory pathologies. Disease markers. 2012;32(4):221–30. doi: 10.3233/DMA-2011-0882. PubMed PMID: 22430188. Epub 2012/03/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esquela-Kerscher A, Slack FJ. Oncomirs [mdash] microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 34.Iorio M, Croce C. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012 March 1. 2012;4(3):143–59. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010. [DOI] [PMC free article] [PubMed]
- 37.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS. et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005 Sep 1;23(25):5900–9. doi: 10.1200/JCO.2005.02.857. PubMed PMID: 16043828. Epub 2005/07/27. [DOI] [PubMed] [Google Scholar]
- 38.Huncharek M, Muscat J, Geschwind J-F. K-ras oncogene mutation as a prognostic marker in non-small cell lung cancer: a combined analysis of 881 cases. Carcinogenesis. 1999;20(8):1507–10. doi: 10.1093/carcin/20.8.1507. [DOI] [PubMed] [Google Scholar]
- 39.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A. et al. RAS Is Regulated by the let-7 MicroRNA Family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I. et al. A SNP in a let-7 microRNA Complementary Site in the KRAS 3′ Untranslated Region Increases Non-Small Cell Lung Cancer Risk. Cancer Research. 2008;68(20):8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14) Oncogene. 2011;30(23):2644–58. doi: 10.1038/onc.2010.642. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi T, Nau M, Chiba I, Birrer M, Rosenberg R, Vinocour M. et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246(4929):491–4. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 43.Ding L, Getz G, Wheeler D, Mardis E, McLellan M, Cibulskis K. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N. et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 45.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y. et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee Kwang H. et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Molecular cell. 2007;26(5):745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c Are Targets of p53 and Cooperate in Control of Cell Proliferation and Adhesion-Independent Growth. Cancer Research. 2007;67(18):8433–8. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 48.Jiang L, Huang Q, Chang J, Wang E, Qiu X. MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in lung cancer cells. Experimental Lung Research. 2011;37(7):387–98. doi: 10.3109/01902148.2010.492068. PubMed PMID: 21777146. [DOI] [PubMed] [Google Scholar]
- 49.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H. et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Research. 2004;64(11):3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 50.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA. et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proceedings of the National Academy of Sciences. 2008;105(10):3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 52.Yu S, Chen H, Chang G, Chen C, Chen H, Singh S. et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of MicroRNA Expression Profiles That May Predict Recurrence of Localized Stage I Non-Small Cell Lung Cancer after Surgical Resection. Cancer Research. 2010;70(1):36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 54.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K. et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Science. 2005;96(2):111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, Saito M. et al. MicroRNA Expression and Clinical Outcomes in Patients Treated with Adjuvant Chemotherapy after Complete Resection of Non-Small Cell Lung Carcinoma. Cancer Research. 2010;70(21):8288–98. doi: 10.1158/0008-5472.CAN-10-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW. et al. MicroRNA Expression Differentiates Histology and Predicts Survival of Lung Cancer. Clinical Cancer Research. 2010;16(2):430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H. et al. MicroRNA Classifiers for Predicting Prognosis of Squamous Cell Lung Cancer. Cancer Research. 2009;69(14):5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 58.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I. et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proceedings of the National Academy of Sciences. 2009;106(29):12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mascaux C, Laes JF, Anthoine G, Haller A, Ninane V, Burny A. et al. Evolution of microRNA expression during human bronchial squamous carcinogenesis. European Respiratory Journal. 2009 February 1. 2009;33(2):352–9. doi: 10.1183/09031936.00084108. [DOI] [PubMed] [Google Scholar]
- 60.Heegaard N, Schetter A, Welsh J, Yoneda M, Bowman E, Harris C. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer. 2012 March 15. 2012;130(6):1378–86. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F. et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proceedings of the National Academy of Sciences. 2011 March 1. 2011;108(9):3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pastorino U, Bellomi M, Landoni C, De Fiori E, Arnaldi P, Picchio M. et al. Early lung-cancer detection with spiral CT and positron emission tomography in heavy smokers: 2-year results. Lancet. 2003 August 23. 2003;362(9384):593–7. doi: 10.1016/S0140-6736(03)14188-8. [DOI] [PubMed] [Google Scholar]
- 63.Boeri M, Pastorino U, Sozzi G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J. 2012;18(3):268–74. doi: 10.1097/PPO.0b013e318258b743. [DOI] [PubMed] [Google Scholar]
- 64.Bahce I, Smit EF, Lubberink M, van der Veldt AAM, Yaqub M, Windhorst AD. et al. Development of [11C]erlotinib Positron Emission Tomography for In Vivo Evaluation of EGF Receptor Mutational Status. Clinical Cancer Research. 2013;19(1):183–93. doi: 10.1158/1078-0432.CCR-12-0289. [DOI] [PubMed] [Google Scholar]
- 65.Kato Y, Miyaki S, Yokoyama S, Omori S, Inoue A, Horiuchi M. et al. Real-time functional imaging for monitoring miR-133 during myogenic differentiation. Int J Biochem Cell Biol. 2009;41(11):2225–31. doi: 10.1016/j.biocel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee JY, Kim S, Hwang DW, Jeong JM, Chung J-K, Lee MC. et al. Development of a Dual-Luciferase Reporter System for In Vivo Visualization of MicroRNA Biogenesis and Posttranscriptional Regulation. Journal of Nuclear Medicine. 2008;49(2):285–94. doi: 10.2967/jnumed.107.042507. [DOI] [PubMed] [Google Scholar]
- 67.Wang F, Niu G, Chen X, Cao F. Molecular imaging of microRNAs. Eur J Nucl Med Mol Imaging. 2011;38(8):1572–9. doi: 10.1007/s00259-011-1786-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krutzfeldt J, Rajewsky N, Braich R, Rajeev K, Tuschl T, Manoharan M. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438(7068):685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 69.Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A. et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Research. 2008;36(4):1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hossain A, Kuo MT, Saunders GF. Mir-17-5p Regulates Breast Cancer Cell Proliferation by Inhibiting Translation of AIB1 mRNA. Molecular and Cellular Biology. 2006;26(21):8191–201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rossbach M. Therapeutic implications of microRNAs in human cancer. Journal of Nucleic Acids Investigation. 2011;2:e3. (1):17-22. [Google Scholar]
- 72.Pisters KM, Le Chevalier T. Adjuvant chemotherapy in completely resected non-small-cell lung cancer. J Clin Oncol. 2005 May 10;23(14):3270–8. doi: 10.1200/JCO.2005.11.478. PubMed PMID: 15886314. Epub 2005/05/12. [DOI] [PubMed] [Google Scholar]
- 73.Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ. et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008 Jul 20;26(21):3552–9. doi: 10.1200/JCO.2007.13.9030. PubMed PMID: 18506026. Epub 2008/05/29. [DOI] [PubMed] [Google Scholar]
- 74.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D. et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schoof C, Botelho E, Izzotti A, Vasques LR. MicroRNAs in cancer treatment and prognosis. Am J Cancer Res. 2012;2(4):414–33. [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q, Zhong M, Liu W, Li J, Huang J, Zheng L. Alterations of microRNAs in cisplatin-resistant human non-small cell lung cancer cells (A549/DDP) Exp Lung Res. 2011 Sep;37(7):427–34. doi: 10.3109/01902148.2011.584263. PubMed PMID: 21787234. Epub 2011/07/27. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X, Zhu J, Xing R, Tie Y, Fu H, Zheng X. et al. miR-513a-3p sensitizes human lung adenocarcinoma cells to chemotherapy by targeting GSTP1. Lung Cancer. 2012 September 1. 2012;77(3):488–94. doi: 10.1016/j.lungcan.2012.05.107. [DOI] [PubMed] [Google Scholar]
- 78.Lee Y, Oh J, Yoon S, Kwon M, Song C, Kim K. et al. Differential gene expression profiles of radioresistant non-small-cell lung cancer cell lines established by fractionated irradiation: tumor protein p53-inducible protein 3 confers sensitivity to ionizing radiation. Int J Radiat Oncol Biol Phys. 2010;77(3):858–66. doi: 10.1016/j.ijrobp.2009.12.076. [DOI] [PubMed] [Google Scholar]
- 79.Kraus A, Ferber I, Bachmann S, Specht H, Wimmel A, Gross M. et al. In vitro chemo- and radio-resistance in small cell lung cancer correlates with cell adhesion and constitutive activation of AKT and MAP kinase pathways. Oncogene. 2002;21(57):8683–95. doi: 10.1038/sj.onc.1205939. [DOI] [PubMed] [Google Scholar]
- 80.Lynam-Lennon N, Reynolds J, Marignol L, Sheils O, Pidgeon G, Maher S. MicroRNA-31 modulates tumour sensitivity to radiation in oesophageal adenocarcinoma. J Mol Med (Berl) 2012;90(12):1449–58. doi: 10.1007/s00109-012-0924-x. [DOI] [PubMed] [Google Scholar]
- 81.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M. et al. MicroRNAs as Potential Agents to Alter Resistance to Cytotoxic Anticancer Therapy. Cancer Research. 2007;67(23):11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]