Abstract
The family Anostomidae is an interesting model for studies of repetitive elements, mainly because of the presence of high numbers of heterochromatic segments related to a peculiar system of female heterogamety, which is restricted to a few species of Leporinus genus. Thus, cytogenetic mapping of the retrotransposable elements Rex1, Rex3, and Rex6 was performed in six Leporinus species, to elucidate the genomic organization of this genus. The sequencing of the Rex1 and Rex3 elements detected different base pair compositions in these elements among species, whereas the Rex6 element was not identified in the genomes of these species. FISH analysis using Rex1 detected different distribution patterns, L. elongatus, L. macrocephalus, and L. obtusidens had clusters in the terminal regions, whereas the signals were dispersed throughout all of the chromosomes with some signals in the terminal position in other species. The Rex3 signals were found mainly in the terminal positions in all the chromosomes of all species. The W chromosomes of L. elongatus, L. macrocephalus, and L. obtusidens contained the Rex1 and Rex3 signal in an interstitial position. These results suggest the emergence of different activity levels for these elements during the evolution of the species analyzed. Despite the conserved karyotype macrostructure species Leporinus often discussed, our results show some variation in hybridization patterns, particularly between the species with specific patterns in their sex chromosomes and species without this differentiated system.
Keywords: transposable elements, mobile DNA, repetitive sequences, sex chromosome, FISH
Introduction
Large portions of eukaryote genomes comprise repetitive DNA sequences. These sequences may be related to specific functions and structures, such as rDNA synthesis, segregation of centromeric regions, and the protection of telomeric regions. These sequences may have been produced by changes in the genome, which affected evolutionary trajectories and led to the production of regulatory and coding segments.1,2 Repetitive sequences can be organized into two classes: tandem repeats, such as satellites, minisatellites, and microsatellites; and repeats dispersed throughout the genome as transposable elements.3 Transposable elements can move around the genome in two ways: sequence excision mediated by a transposase enzyme (transposons) or via intermediate RNA produced by reverse transcriptase (retrotransposons).4
Transposable elements have important roles in genomic diversity and evolution,5 and these elements may affect the evolutionary trajectory of the host organism in different ways, such as causing changes in gene functions via insertions and inducing chromosome rearrangements.6 From a cytogenetic perspective, these elements may accumulate in specific sites in chromosomes, such as terminal, pericentromeric, and other heterochromatic regions where the recombination rate is reduced,7 or in small euchromatic segments, such as those found in Drosophila melanogaster chromosomes.8 Thus, these elements may be associated with mechanisms that lead to speciation events,4 which confirms that these elements have very important roles in the genomic and biological diversity of vertebrates.
The retrotransposable elements in the Rex family have been studied widely in some fish groups, including Cichlidae,9-14 Tetraodontidae,15-17 Nototheniidae, Artedidraconidae, Bathydraconidae, Bovichtidae,18 and Loricariidae.19 The Rex family elements have a high diversity of chromosome locations in previously studied fish species, and it has been hypothesized that they may have roles in genome differentiation and evolution.9-17,20 However, the characteristics and dynamics of these fish retroelements, which occur mainly in the order Characiformes (a large fish order), still need to be elucidated.
The family Anostomidae is considered to be an interesting model for studies of repetitive elements. Cytogenetic studies of several species in this family have detected a conserved karyotypic structure in the 41 species that have already been karyotyped. The diploid number of 54 comprises metacentric and submetacentric chromosomes, and the karyotypes of most species carry a single NOR bearing pair.21-24 The genus Leporinus has been well studied and 25 of the 87 species in this genus have been karyotyped. The presence of a peculiar sex chromosome system with female heterogamety is restricted to a few species in this genus, i.e., L. elongatus, L. macrocephalus, L. obtusidens, L. reinhardtii, L. silvetrii, L. conirostris, and L. trifasciatus.25 Recent studies of Leporinus have isolated, characterized, and correlated repetitive elements in these species to study the evolution of sex chromosomes, as well as to discriminate hybrid species.26-32 Other studies have shown that repetitive sequences such as 5S rDNA may have different distributions among Leporinus species with or without heteromorphic sex chromosomes.33
The high heterochromatic content in the chromosomes of Leporinus species, especially the sex chromosomes, demonstrates the presence of high numbers of repetitive sequences in the genomes of these species.28-32,34 In the present study, cytogenetic mapping of the retrotransposable elements Rex1, Rex3, and Rex6 was performed in the chromosomes of Leporinus species to elucidate the activity and dispersion levels of these elements in genome of this genus. This analysis determined whether these elements are related to the heterochromatic regions of autosomes and sex chromosomes and whether their distributions are compatible with the hypothetical maintenance of a stable macrostructure in the karyotype of this genus.
Results
Isolation of Rex1, Rex3, and Rex6 elements, and sequence analysis
The amplification and sequencing of Rex1 and Rex3 elements detected sequences with different base pair numbers among Leporinus species. The sequence alignment detected a high similarity among the sequences that corresponded to the Rex1 element, where the scores ranged from 97.93% to 100.0%. However, the Rex3 sequences were more variable among species, with scores ranging from 48.12% to 100.0%. The similarity levels of Rex1 with others sequences in Gen Bank were 79–83% and Rex3 were 83–88%. The amplified segments corresponded to the coding domains of the reverse transcriptase gene. The base pair numbers and similarity levels for each species are shown in Table 1.
Table 1. Base pare number and similarity levels of Rex1 and Rex3 in Leporinus species.
| Specie | Retrotransposable element | Base pair | Similarity level |
|---|---|---|---|
|
L. elongatus |
Rex1 |
498 |
79%, Pterophyllum scalare (JX576338.1) |
|
Rex3 |
411 |
88%, Dissostichus mawsoni (AY331110.1) |
|
|
L. friderici |
Rex1 |
482 |
82%, Pterophyllum scalare (JX576338.1) |
|
Rex3 |
429 |
83%, Cichla monoculus (KF131684.1) |
|
|
L. lacustris |
Rex1 |
483 |
82%, Pterophyllum scalare (JX576338.1) |
|
Rex3 |
426 |
88%, Dissostichus mawsoni (AY331110.1) |
|
|
L. obtusidens |
Rex1 |
477 |
82%, Pterophyllum scalare (JX576338.1) |
|
Rex3 |
426 |
88%, Notothenia coriiceps (AY331103.1) |
|
|
L. macrocephalus |
Rex1 |
500 |
80%, Pterophyllum scalare (JX576338.1) |
|
Rex3 |
429 |
88%, Dissostichus mawsoni (AY331110.1) |
|
| L. striatus |
Rex1 |
456 |
83%, Pterophyllum scalare (JX576338.1) |
| Rex3 | 426 | 88%, Dissostichus mawsoni (AY331110.1) |
Sequencing of the amplified fragments generated using the Rex6 primer set in different PCR conditions, showed that these fragments did not correspond to the Rex6 element. The sequences contained 1019 base pairs and their maximum shared identity was with fragments of a chromosome 13 clone from Mus musculus (accession number: AC034285.6).
Chromosome mapping of the Rex1 element
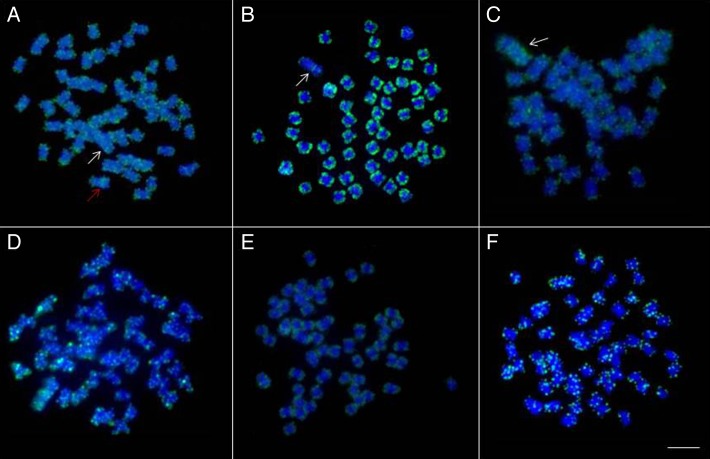
Fluorescent in situ hybridization analysis (FISH) using the PCR fragments of the Rex1 element as a probe detected different mapping patterns in the study species. L. elongatus (Fig. 1A), L. macrocephalus (Fig. 1B), and L. obtusidens (Fig. 1C) had isolated clusters in the terminal regions of most chromosomes, although the W chromosome contained signals in the interstitial region of the long arm. L. elongatus had an interstitial cluster in the W1 chromosome (Fig. 1A). In L. friderici (Fig. 1D), L. lacustris (Fig. 1E), and L. striatus (Fig. 1F), the majority of the clusters were dispersed throughout all of the chromosomes and some of the signals were in terminal positions in male and female specimens.
Figure 1. Chromosome mapping of Rex1 element in metaphases of female specimens of (A) Leporinus elongatus, (B) L. macrocephalus, (C) L. obtusidens, (D) L. friderici, (E) L. lacustris, and (F) L. striatus. The arrows indicate the W chromosome, red arrow indicate W1 chromosome. Scale bar: 10 µm.
Chromosome mapping of the Rex3 element
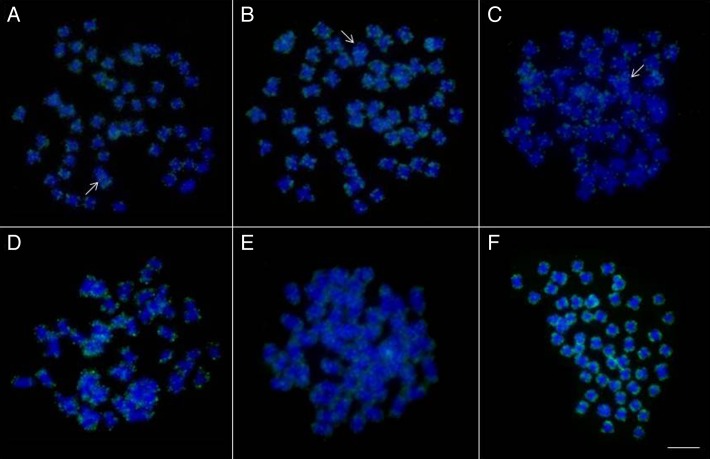
The results obtained using the PCR fragments of Rex3 as a probe detected the same mapping pattern in all species (Fig. 2). The clusters were isolated in the terminal regions of all chromosomes and some clusters were dispersed in the short and long arms of male and female specimens. In L. elongatus, L. macrocephalus, and L. obtusidens, the W chromosome had signals in terminal positions and the interstitial regions of the long arms.
Figure 2. Chromosome mapping of Rex3 element in metaphases of female specimens of (A) Leporinus elongatus, (B) L. macrocephalus, (C) L. obtusidens, (D) L. friderici, (E) L. lacustris, and (F) L. striatus. The arrows indicate the W chromosome. Scale bar: 10 µm.
Discussion
The analysis of Rex1 and Rex3 generated sequences of 456–500 base pairs and 411–429 base pairs, respectively, which were similar to these sequences in other fish groups from the Antarctic,18 and cichlid fishes.10 This was confirmed by the BLASTn analysis where the Rex1 and Rex3 sequences shared high similarity with those of other fish groups, which demonstrates that these elements are fairly well conserved in different families. Previous researchers have suggested that the Rex6 element may have been lost or diverged greatly, depending on the rate of host evolution. The loss of Rex6 elements has been reported in Antarctic fishes,18 and may have occurred in Leporinus species because none were identified in the species analyzed in the present study.
The chromosomal mapping of retrotransposable elements in fish has demonstrated their remarkable diversity.20 In the present study, the Leporinus species had different hybridization signals with dispersed clusters and signals in the terminal regions. These patterns were similar to those observed in the Bryconinae,35 Erythrinidae,36 Sternopygidae,37 Artedidraconidae,18 Bathydraconidae,18 Bovichtidae,18 Channichthyidae,18 Nototheniidae,18 Loricariidae,19 Pimelodidae,38 and Tetraodontidae.17 Species in the family Cichlidae are exceptional because the Rex elements are frequently distributed in the pericentromeric regions.9-14
In particular, Rex1 had different chromosome locations in Leporinus species. Leporinus elongatus, L. macrocephalus, and L. obtusidens had isolated signals in terminal positions in all chromosomes. In other species, there was hybridization with clusters in the short and long arms of all chromosomes. It is known that the distribution patterns of transposable elements are nonrandom and are related to the specific characteristics of different sites in the genome.5 Specific distribution patterns have also been observed in Drosophila, where some euchromatic regions have been replaced by retrotransposable elements.39 Furthermore, a correlation between chromosome rearrangement and retrotransposon activity was reported by Ozouf-Costaz et al.18 in notothenioid species. Thus, the differences in the Rex1 hybridization sites in Leporinus species may be related to small rearrangements in specific regions of their genomes.
The distributions of Rex3 elements were similar to those of Rex1 in L. elongatus, L. macrocephalus, and L. obtusidens. These similar patterns suggest the compartmentalization of sequences in the terminal positions of the chromosomes in these species. Thus, is it likely that similar evolutionary mechanisms account for the distribution of these elements in Leporinus species and those in other fish families, such as Cichlidae,10,11 and Nototheniidae.18 In L. friderici, L. lacustris, L. obtusidens, and L. striatus, the Rex3 elements were present as isolated signals in terminal positions in the short and long arms of chromosomes.
Many studies of Anostomidae species have highlighted the correlations between heterochromatic regions in chromosomes and repetitive sequences.28,30-32 Many repetitive sequences have been isolated from heterochromatic segments, mainly in Leporinus species, such as those present in the W chromosome, pericentromeric positions, and NOR regions.26-32 In the present study, retrotransposable elements in the Rex family were also abundant in other heterochromatic regions in Leporinus chromosomes, such as terminal positions and some interstitial segments. This distribution reflects the pattern expected for transposable elements distribution described by Kidwell,7 related to a low rate of recombination. L. friderici and L. striatus possessed a few signals in the euchromatic regions of chromosomes. Other studies have suggested that transposable elements can accumulate in euchromatic segments, such as those found in Drosophila melanogaster where 3.8% of the euchromatic genome comprises transposable elements.40
The presence of specific signals for retrotransposable elements in sex chromosomes has been poorly reported in fish. Ozouf-Costaz et al.18 identified Rex3 element hybridization signals in an interstitial position in the short arm of the Y-chromosome of Chionodraco hamatus (Channichthyidae). The interstitial markers in this chromosome may indicate the activity of fusion rearrangement mechanisms during the differentiation of this family.41 The interesting localizations of Rex1 and Rex3 elements in the W chromosomes of Leporinus elongatus, L. macrocephalus, and L. obtusidens may be related to the suppression of recombination during the sex chromosome differentiation process, due mainly to an increase in heterochromatic segments. Suppression is a critical component of this process and there is a natural bias toward the accumulation of repetitive elements.3
An increase in heterochromatic segments because of the differentiation of W chromosomes in Leporinus was proposed by Galetti and Foresti.42 These researchers suggested that the heterochromatin concentration had a specific role in the differentiation of sex chromosomes, thereby indicating a common origin of the chromosomal sex system found in different species of Leporinus. Recent studies have supported a hypothesis based on repetitive sequences related to the W chromosome,28,30-32 where these sequences are related strictly to species with W chromosomes. Another study demonstrated synteny among the W chromosomes of Leporinus species, which supports the hypothesis of a common origin for this chromosome.43 In the present study, the similar distribution patterns of the Rex1 and Rex3 elements in the sex chromosomes of three Leporinus species, i.e., L. elongatus, L. macrocephalus, and L. obtusidens, also supports the hypothesis of a common origin of the W chromosome in Leporinus species.
The karyotypic macrostructures of Leporinus species have been studied many times and a fairly high level of conservation has been reported,25,27,34,44 but our results show variations in the hybridization signal patterns, particularly between species with sex chromosomes and species without this differentiated system. The retroelements analyzed in the present study reflected the same patterns observed in other repetitive elements,28,30 reinforced that heterochromatic regions of the sex chromosomes had distinct differences among autosomes of the Leporinus species.
Material and Methods
Material
Fish material, chromosome preparation, and DNA extraction
Six Leporinus species were analyzed in this study: L. elongatus (Valenciennes, 1849) from Centro Nacional de Pesquisa e Conservação de Peixes Continentais (CEPTA), Pirassununga, São Paulo state; L. obtusidens (Valenciennes, 1847) from Leme fish farm, São Paulo state; and L. macrocephalus (Garavello and Britiski, 1988), L. striatus (Kner, 1858), L. lacustris (Amaral Campos, 1945), and L. friderici (Bloch, 1794) were collected from the Paraguay River basin, Mato Grosso State-Brazil. In total, 12 specimens were analyzed (one male and one female of each species). The chromosomal preparations were obtained using kidney cells, as described by Foresti et al.45 Genomic DNA was extracted from the liver using the phenol-chloroform-isoamyl alcohol technique.46 The specimens were collected in accordance with collection license issued by Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (19833-1), and the material was processed according to Colégio Brasileiro de Experimentação Animal (016/04—CEEA).
Methods
Isolation of Rex1, Rex3, and Rex6 elements
PCR of the Rex1, Rex3, and Rex6 elements used a reaction mixture with a volume of 13.5 µL, which contained 6.25 µL of PCR Mix (Qiagen), 5.25 µL of Milli-Q water, 0.5 μL primer F (10 μM), 0.5 µL of primer R (10 μM), and 1.0 µL of template DNA (200 ng). PCR was performed in a thermocycler (Eppendorf Mastercycler) with the following cycling conditions: initial denaturation at 95 °C for 5 min, followed by 34 cycles at 95 °C for 40 s, annealing at 55 °C for 40 s, and chain elongation at 72 °C for 5 min, with a final extension at 72 °C for 5 min.
Specific primers were designed based on the partial sequences of Rex1 in L. elongatus, which were obtained using the primer set Rex1F—5′-TTCTCCAGTG CCTTCAACACC and Rex1R—5′-TCCCTCAGCA GAAAGAGTCT GCTC.47,48 This set was used in the PCR reaction and was subsequently sequenced by MacroGen, Korea. The sequence was used to design a specific primer for Leporinus with the tool OligoPerfect™ Designer (http://tools.invitrogen.com), which produced the following sequences: Rex1EL—F 5′-AGCAAGCTAG AGAGTGCTGG and Rex1EL—R 5′-ACAGAGCGTG TGTGTTGTCC. Rex3 and Rex6 amplification used the following primer sets: Rex3F—5′-CGGTGAYAAA GGGCAGCCCTG and Rex3R—5′-TGGCAGACNG GGGTGGTGGT, and Rex6F—5′-TAAAGCATAC ATGGAGCGCC AC and Rex6R—5′-GGTCCTCTAC CAGAGGCCTG GG.47,48
Cloning, sequencing, and sequence analysis
The amplified DNA was cloned with a Pgem-T kit (Promega) and used to transform competent Escherichia coli (Promega, Jm109) cells. Clones containing the digested DNA fragments were stored before nucleotide sequencing and used as probes in the chromosomal FISH experiments. The DNA fragments were purified using ExoSAP-IT enzyme and sequenced (MacroGen Inc). The sequences were edited using the Pregep4 program, aligned with CLUSTALW,49 in DAMBE5,50 and analyzed with the BLASTn tool,51 via the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/blast) to determine the similarities among sequences.
FISH and chromosome analysis
The FISH analyses were based on the method of Pinkel et al.52 with several modifications proposed by Silva et al.31 where the retrotransposable elements Rex1 and Rex3 were used as the probes. The probes were labeled via nick translation with biotin-14-dATP (Invitrogen) to facilitate chromosomal mapping.
The hybridization signals were detected using appropriate antibody sets based on anti-avidin, which was followed by the application of avidin-FITC to enhance the signals of the biotin-labeled probes. The chromosomes were counterstained with DAPI, mounted with antifade solution, and observed using an Olympus BX51 microscope coupled to an Olympus digital camera (model D71). The chromosome images were captured using the DP Controller program.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP-2012/01437-0) for financial support, and to Bioedit® for helping with the English language. They also thank to Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA) for providing the collection license (19833-1).
Citation: de Borba RS, da Silva EL, Parise-Maltempi PP. Chromosome mapping of retrotransposable elements Rex1 and Rex3 in Leporinus Spix, 1829 species (Characiformes: Anostomidae) and its relationships among heterochromatic segments and W sex chromosome. Mobile Genetic Elements 2013; 3:e27460; 10.4161/mge.27460
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/27460
References
- 1.Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–22. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 2.Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–62. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–20. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 4.Böhne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008;16:203–15. doi: 10.1007/s10577-007-1202-6. [DOI] [PubMed] [Google Scholar]
- 5.Kidwell MG, Lisch DR. Transposable elements and host genome evolution. Trends Ecol Evol. 2000;15:95–9. doi: 10.1016/S0169-5347(99)01817-0. [DOI] [PubMed] [Google Scholar]
- 6.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–68. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kidwell MG. Transposable elements, in Gregory, T. (ed). The Evolution of the genome. Elsevier Academic Press, Burlington, MA, 2005; p.165-223 [Google Scholar]
- 8.Bartolomé C, Maside X, Charlesworth B. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol. 2002;19:926–37. doi: 10.1093/oxfordjournals.molbev.a004150. [DOI] [PubMed] [Google Scholar]
- 9.Mazzuchelli J, Martins C. Genomic organization of repetitive DNAs in the cichlid fish Astronotus ocellatus. Genetica. 2009;136:461–9. doi: 10.1007/s10709-008-9346-7. [DOI] [PubMed] [Google Scholar]
- 10.Teixeira WG, Ferreira IA, Cabral-de-Mello DC, Mazzuchelli J, Valente GT, Pinhal D, Poletto AB, Venere PC, Martins C. Organization of repeated DNA elements in the genome of the cichlid fish Cichla kelberi and its contributions to the knowledge of fish genomes. Cytogenet Genome Res. 2009;125:224–34. doi: 10.1159/000230006. [DOI] [PubMed] [Google Scholar]
- 11.Valente GT, Mazzuchelli J, Ferreira IA, Poletto AB, Fantinatti BEA, Martins C. Cytogenetic mapping of the retroelements Rex1, Rex3 and Rex6 among cichlid fish: new insights on the chromosomal distribution of transposable elements. Cytogenet Genome Res. 2011;133:34–42. doi: 10.1159/000322888. [DOI] [PubMed] [Google Scholar]
- 12.Gross MC, Schneider CH, Valente GT, Porto JIR, Martins C, Feldberg E. Comparative cytogenetic analysis of the genus symphysodon (discus fishes, cichlidae): chromosomal characteristics of retrotransposons and minor ribosomal DNA. Cytogenet Genome Res. 2009;127:43–53. doi: 10.1159/000279443. [DOI] [PubMed] [Google Scholar]
- 13.Fantinatti BEA, Mazzuchelli J, Valente GT, Cabral-de-Mello DC, Martins C. Genomic content and new insights on the origin of the B chromosome of the cichlid fish Astatotilapia latifasciata. Genetica. 2011;139:1273–82. doi: 10.1007/s10709-012-9629-x. [DOI] [PubMed] [Google Scholar]
- 14.Schneider CH, Gross MC, Terencio ML, do Carmo EJ, Martins C, Feldberg E. Evolutionary dynamics of retrotransposable elements Rex1, Rex3 and Rex6 in neotropical cichlid genomes. BMC Evol Biol. 2013;13:152. doi: 10.1186/1471-2148-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasilva C, Hadji H, Ozouf-Costaz C, Nicaud S, Jaillon O, Weissenbach J, Roest Crollius H. Remarkable compartmentalization of transposable elements and pseudogenes in the heterochromatin of the Tetraodon nigroviridis genome. Proc Natl Acad Sci U S A. 2002;99:13636–41. doi: 10.1073/pnas.202284199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouneau L, Fischer C, Ozouf-Costaz C, Froschauer A, Jaillon O, Coutanceau JP, Körting C, Weissenbach J, Bernot A, Volff JN. An active non-LTR retrotransposon with tandem structure in the compact genome of the pufferfish Tetraodon nigroviridis. Genome Res. 2003;13:1686–95. doi: 10.1101/gr.726003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer C, Bouneau L, Coutanceau JP, Weissenbach J, Volff JN, Ozouf-Costaz C. Global heterochromatic colocalization of transposable elements with minisatellites in the compact genome of the pufferfish Tetraodon nigroviridis. Gene. 2004;336:175–83. doi: 10.1016/j.gene.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Ozouf-Costaz C, Brandt J, Korting C, Pisano E, Bonillo C, Coutanceau JP, Volff JN. Genome dynamics and chromosomal localization of the non-LTR retrotransposons Rex1 and Rex3 in Antartic fish. Antarct Sci. 2004;16:51–7. doi: 10.1017/S0954102004001816. [DOI] [Google Scholar]
- 19.Ferreira DC, Oliveira C, Foresti F. Chromosome mapping of retrotransposable elements Rex1 and Rex3 in three fish species in the subfamily Hypoptopomatinae (Teleostei, Siluriformes, Loricariidae) Cytogenet Genome Res. 2011;132:64–70. doi: 10.1159/000319620. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira DC, Porto-Foresti F, Oliveira C, Foresti F. Transposable elements as a potential source for understanding the fish genome. Mob Genet Elements. 2011;1:112–7. doi: 10.4161/mge.1.2.16731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galetti PM, Jr., Foresti F, Bertollo LAC, Moreira Filho O. Heteromorphic sex chromosomes in three species of the genus Leporinus (Pisces, Anostomidae) Cytogenet Cell Genet. 1981;29:138–42. doi: 10.1159/000131562. [DOI] [PubMed] [Google Scholar]
- 22.Galetti PM, Foresti F, Bertollo LAC, Moreira-Filho O. Characterization of eight species of Anostomidae (Cypriniformes) fish on the basis of the nucleolar organizing region. Caryologia. 1984;37:401–6. [Google Scholar]
- 23.Molina WF, Galetti PM., Jr. Early replication banding in Leporinus species (Osteichthyes, Characiformes) bearing differentiated sex chromosomes (ZW) Genetica. 2007;130:153–60. doi: 10.1007/s10709-006-9002-z. [DOI] [PubMed] [Google Scholar]
- 24.Silva EL. Estudo da organização estrutural de elementos repetitivos isolados do genoma de Leporinus elongatus em diferentes espécies da família Anostomidae (Teleostei, Characiformes) Ph D. Dissertation, Universidade Estadual Paulista, Botucatu, SP, 2012 [Google Scholar]
- 25.Galetti PM, Lima NRW, Venere PC. A monophyletic ZW sex chromosome system in Leporinus (Anostomidae, Characiformes) Cytologia (Tokyo) 1995;60:375–82. doi: 10.1508/cytologia.60.375. [DOI] [Google Scholar]
- 26.Nakayama I, Foresti F, Tewari R, Schartl M, Chourrout D. Sex chromosome polymorphism and heterogametic males revealed by two cloned DNA probes in the ZW/ZZ fish Leporinus elongatus. Chromosoma. 1994;103:31–9. doi: 10.1007/BF00364723. [DOI] [PubMed] [Google Scholar]
- 27.Koehler MR, Haaf T, Guttenbach M, Schartl M, Schmid M. Cytogenetics of the genus Leporinus (Pisces, Anostomidae). II. Molecular cytogenetics, organization and evolutionary conservation of a chromosome-specific satellite DNA from Leporinus obtusidens. Chromosome Res. 1997;5:325–31. doi: 10.1023/B:CHRO.0000038763.52875.48. [DOI] [PubMed] [Google Scholar]
- 28.Parise-Maltempi PP, Martins C, Oliveira C, Foresti F. Identification of a new repetitive element in the sex chromosomes of Leporinus elongatus (Teleostei: Characiformes: Anostomidae): new insights into the sex chromosomes of Leporinus. Cytogenet Genome Res. 2007;116:218–23. doi: 10.1159/000098190. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto DT, Parise-Maltempi PP, Laudicina A, Bortolozzi J, Senhorini JA, Foresti F, Porto-Foresti F. Repetitive DNA probe linked to sex chromosomes in hybrids between Neotropical fish Leporinus macrocephalus and Leporinus elongatus (Characiformes, Anostomidae) Cytogenet Genome Res. 2009;124:151–7. doi: 10.1159/000207523. [DOI] [PubMed] [Google Scholar]
- 30.Marreta ME, Faldoni FLC, Parise-Maltempi PP. Cytogenetic mapping of the W chromosome in the genus Leporinus (Teleostei, Anostomidae) using a highly repetitive DNA sequence. J Fish Biol. 2012;80:630–7. doi: 10.1111/j.1095-8649.2011.03199.x. [DOI] [PubMed] [Google Scholar]
- 31.da Silva EL, de Borba RS, Parise-Maltempi PP. Chromosome mapping of repetitive sequences in Anostomidae species: implications for genomic and sex chromosome evolution. Mol Cytogenet. 2012;5:45. doi: 10.1186/1755-8166-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.da Silva EL, Busso AF, Parise-Maltempi PP. Characterization and genome organization of a repetitive element associated with the nucleolus organizer region in Leporinus elongatus (Anostomidae: Characiformes) Cytogenet Genome Res. 2013;139:22–8. doi: 10.1159/000342957. [DOI] [PubMed] [Google Scholar]
- 33.Ferreira IA, Oliveira C, Venere PC, Galetti PM, Jr., Martins C. 5S rDNA variation and its phylogenetic inference in the genus Leporinus (Characiformes: Anostomidae) Genetica. 2007;129:253–7. doi: 10.1007/s10709-006-0005-6. [DOI] [PubMed] [Google Scholar]
- 34.Galetti PM, César ACG, Venere PC. Heterochromatin and NORs variability in Leporinus fish (Anostomidae, Characiformes) Caryologia. 1991;44:287–92. [Google Scholar]
- 35.Silva JR. Isolamento e caracterização de elementos transponíveis em espécies do gênero Brycon (Characidae, Bryconinae). Ph D. Dissertation, Universidade Estadual Paulista, Botucatu, SP, 2012 [Google Scholar]
- 36.Cioffi MB, Martins C, Vicari MR, Rebordinos L, Bertollo LAC. Differentiation of the XY sex chromosomes in the fish Hoplias malabaricus (Characiformes, Erythrinidae): unusual accumulation of repetitive sequences on the X chromosome. Sex Dev. 2010;4:176–85. doi: 10.1159/000309726. [DOI] [PubMed] [Google Scholar]
- 37.Sene VF. Citogenetica molecular e caracterização cromossômica no gênero Eigenmannia (Teleostei, Gymnotiformes, Sternopygidae). Ph D. Dissertation, Universidade Estadual Paulista, Botucatu, SP, 2011 [Google Scholar]
- 38.Matoso DA, Val VMFA, Fonseca VM, Silva M, Moraes-Neto A, Almeida MC, Vicari MR, Moreira-Filho O, Artoni RF. Chromosomal polymorphism in Steindachneridion melanodermatum Garavello, 2005 (Siluriformes, Pimelodidae): a reappraisal the existence of sex chromosome system in the species. Rev Fish Biol Fish. 2005;21:497–508. doi: 10.1007/s11160-011-9201-2. [DOI] [Google Scholar]
- 39.Busseau I, Chaboissier MC, Pélisson A, Bucheton A. I factors in Drosophila melanogaster: transposition under control. Genetica. 1994;93:101–16. doi: 10.1007/BF01435243. [DOI] [PubMed] [Google Scholar]
- 40.Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3:H0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pisano E, Ozouf-Costaz C. Chromosome change and the evolution in the Antarctic fish suborder Notothenioidei. Antarct Sci. 2000;12:334–42. doi: 10.1017/S0954102000000390. [DOI] [Google Scholar]
- 42.Galetti PM, Foresti F. Evolution of the ZZ/ZW system in Leporinus (Pisces, Anostomidae): the role of constitutive heterochromatin. Cytogenet Cell Genet. 1986;43:43–6. doi: 10.1159/000132296. [DOI] [Google Scholar]
- 43.Parise-Maltempi PP, da Silva EL, Rens W, Dearden F, O’Brien PCM, Trifonov V, Ferguson-Smith MA. Comparative analysis of sex chromosomes in Leporinus species (Teleostei, Characiformes) using chromosome painting. BMC Genet. 2013;14:60. doi: 10.1186/1471-2156-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galetti Júnior PM, Mestriner CA, Venere PC, Foresti F. Heterochromatin and karyotype reorganization in fish of the family Anostomidae (Characiformes) Cytogenet Cell Genet. 1991;56:116–21. doi: 10.1159/000133063. [DOI] [PubMed] [Google Scholar]
- 45.Foresti F, Oliveira C, Almeida-Toledo LF. A method for chromosome preparations from large specimens of fishes using in vitro shortterm treatment with colchicine. Experientia. 1993;49:810–3. doi: 10.1007/BF01923555. [DOI] [Google Scholar]
- 46.Sambrook J, Russel DW. Molecular cloning. A laboratory manual. (Cold New York: Spring Harbor Laboratory Press. Third Edition, 2001 [Google Scholar]
- 47.Volff JN, Körting C, Sweeney K, Schartl M. The non-LTR retrotransposon Rex3 from the fish Xiphophorus is widespread among teleosts. Mol Biol Evol. 1999;16:1427–38. doi: 10.1093/oxfordjournals.molbev.a026055. [DOI] [PubMed] [Google Scholar]
- 48.Volff JN, Körting C, Schartl M. Multiple lineages of the non-LTR retrotransposon Rex1 with varying success in invading fish genomes. Mol Biol Evol. 2000;17:1673–84. doi: 10.1093/oxfordjournals.molbev.a026266. [DOI] [PubMed] [Google Scholar]
- 49.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia X. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 2013;30:1720–8. doi: 10.1093/molbev/mst064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A. 1986;83:2934–8. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]