Abstract
As Acute Myeloid Leukemia (AML) patient response to cytarabine-based standard-of-care treatment is variable, stratification into subgroups by biomarker-predicted response may lead to improved clinical outcomes. Here we assess cell mitochondrial depolarization to pro-apoptotic signaling BH3-only peptides as a surrogate for the function of Bcl-2 family proteins to address clinical response to cytarabine-based therapy in AML patients (n=62). Peripheral blood mononuclear cell (PBMC) or bone marrow aspirate (BM) specimens were obtained from newly diagnosed AML patients, viably preserved, and assayed by flow cytometry following BH3 profile assay with individual BH3 peptides. Mann-Whitney analysis indicates biomarker correlation with response to induction therapy: notably BIM priming was highly significant (p=2×10−6) with a compelling sensitivity/specificity profile (AUC=0.83; CI[0.73,0.94]; p=2×10−10). Multivariate analysis indicates improved profiles for BIM readout + patient age (AUC=0.89; CI[0.81,0.97])and BIM + patient age +cytogenetic status (AUC=0.91; CI[0.83,0.98]). When patients were stratified by cytogenetic status, BIM readout was significant for both, intermediate (p=0.0017; AUC=0.88; CI[0.71,1.04]) and for unfavorable (p=0.023; AUC=0.79; CI[0.58,1.00]) risk groups, demonstrating predictive power independent of cytogenetics. Additional analyses of secondary clinical endpoints displayed correlation between overall survival (OS; p=0.037) and event-free survival (EFS; p=0.044) when patients were stratified into tertiles by BIM peptide response. Taken together, these results highlight the potential utility of BH3 profiling in personalized diagnostics of AML by offering actionable information for patient management decisions.
Introduction
Acute myeloid leukemia (AML) is the second most common leukemia, with approximately 14,600 newly diagnosed cases and 10,400 deaths annually in the US (1, 2). Response rates generally are inverse to patient age; the outcomes for the majority of patients treated with standard-of-care regimens (cytarabine+anthracycline) remains poor with approximately 25% of patients surviving 3 or more years (2, 3). Although aggressive treatments have improved outcome in young patients, patients over 60 comprising the majority of AML cases, remain a therapeutic enigma. The development of personalized diagnostic tests that could identify patients that will benefit from conventional cytarabine+anthracycline regimens, and conversely direct those unlikely to benefit to alternative therapies, could potentially improve response rates and minimize toxicity.
Prognostic markers for AML have been identified including age and performance status but by themselves these are not therapeutically leverageable. A number of prognostic molecular events have been identified in AML including translocations and mutations in MLL, AML/ETO, Flt3-ITD, NPM1, CEBPalpha, IDH1, IDH2, RUNX1 and WT1 and in epigenetic modifying genes such as TET2 and ASXL1 (4–6), and changes in cell signaling protein profiles (7, 8). Though these events carry prognostic significance, the heterogeneity of patient response with a given molecular event demonstrates that other factors must be involved in regulating the biology of the leukemic blast and consequently the relative sensitivity to a given therapy. Impairment of apoptosis is a hallmark of AML and Bcl-2 family proteins comprise key modulators of such at the mitochondrial level. It has been proposed that steady state expression levels of these proteins would confer prognostic information in AML. To date however, these measurements have not provided a predictive biomarker for incorporation into routine clinical use due to conflicting outcomes relevance data (9–11). Differential expression in AML subtypes has been cited as a confounding factor limiting clinical utility of this approach (11).
The study of pathways in the context of constituent component expression and measured changes in response to perturbation has demonstrated to yield important prognostic information (12, 13). The underlying principle of BH3 profiling is that mitochondrial depolarization following BH3 peptide exposure serves as a functional biomarker for cellular response to pro-apoptotic cues (14–17). Early conceptual investigations into mitochondrial profiling have drawn correlations between therapeutic efficacy and BH3 peptide-derived metrics (18–21). The current study offers translational and statistical evidence for clinical utility of BH3 profiling in discriminating response to standard-of-care-based therapeutic management of AML.
Materials and Methods
AML Patient Cohort
Newly diagnosed AML patient samples were obtained from peripheral blood draw or bone marrow aspirate (BM) collection prior to induction chemotherapy administration at The University of Texas M. D. Anderson Cancer Center (MDACC) between September 1999 and March 2007 (22). Specimens were acquired during routine diagnostic assessments in accordance with the regulations and protocols (Lab 01–473) approved by the investigational review board of MDACC. Informed consent was obtained in accordance with Declaration of Helsinki. Patients were selected for inclusion in this study on the basis of availability of cryopreserved cells from the larger pool of 511 cases from a population treated with cytarabine-based regimens. Following Ficoll purification, CD3/CD19 cell depletion removed contaminating T and B cells and cells were cryopreserved in liquid N2.
Patient Treatment
All 62 patients were treated with high-dose ara-C (HDAC)-based chemotherapy (cytarabine + anthracycline [n=48], cytarabine + non-anthracycline [n=7], and cytarabine + fludarabine [n=8]; one patient received cytarabine + non-anthacycline and cytarabine + fludarabine on subsequent cycles [no response on either cycle]). Complete Response (CR) = Normal bone marrow morphology, absolute neutrophil count greater than 1,000, platelet count >100K and rising hemoglobin. Relapse is >5% blasts in the marrow or blasts in the peripheral blood in a patient formerly in CR. Primary refractory (No Response; NR) = residual leukemia after 2 cycles of induction chemotherapy. For statistical analyses, CR= patients who exhibited response, with or without subsequent relapse; NR=primary refractory.
Cytogenetic Risk Status Determination
Cytogenetic risk determination was performed by a CLIA-certified cytogenetics laboratory at MDACC. Patient risk-group assignment was carried out according to Cancer and Leukemia Group B (CALGB) criteria: Favorable = inv16, t(8:21), T(15;17) intermediate = diploid, -y, insufficient metaphases, Unfavorable = all others, −5, −7, +8, t(6;9), 11q, PH1+, misc.
BH3 Profiling
AML specimens were stained with antibodies CD45-V450 (BD Biosciences, San Jose CA), CD3-Biotin (BD Bioscience, San Jose CA), and CD20-Biotin (eBiosciences, San Diego CA). Secondary antibody was Streptavidin-APC (BD Biosciences, San Jose CA). Specimens were permeabilized with digitonin (Sigma-Aldrich, St Louis MO) and incubated with JC-1 mitochondrial dye (Enzo Life Sciences, Farmingdale NY) and peptides (BIM 100μM, BIM 0.1μM, PUMA 100μM, PUMA 10μM, NOXA 100μM, BAD 100μM, BMF 100μM, HRK 100μM, or PUMA2A 100μM) or dimethyl sulfoxide (DMSO [(1%]) or Carbonyl cyanide m-chlorophenyl hydrazone (CCCP [10μM]) at room temperature. Samples were run in duplicate except in cases where insufficient viable cells were available.
Samples were analyzed on a FACS CantoII (BD Biosciences, San Jose CA) using BD FACS Diva software. The blast population was identified as CD45 dim, CD3 and CD20 negative. Intensely stained CD45 cells (mature lymphocytes) were excluded from analyses as described previously (20, 21). The quantifiable propensity of a pro-apoptotic peptide to induce mitochondrial depolarization relative to an uncoupling reagent control is referred to as percent priming. For the blast population this was calculated using the median signal intensity of the PE channel normalized for DMSO as background (negative control) and CCCP served as 100% priming (positive control). For calculation of % priming, the following formula is utilized:
Statistical Analysis
Biomarkers were analyzed by testing the association between the biomarker status (% priming) and responder or non-responder classification. Univariate comparisons were made using Mann-Whitney test; all reported p-values are two-sided. The threshold for significance was p < .05/7 = .007 for the primary analysis to account for multiple comparisons (7 biomarkers). Secondary analyses used a threshold of p <.05 and are only reported when the primary analysis for a marker was significant. The predictive ability of markers was assessed using the area under the curve (AUC) statistic. Multivariate analyses were performed using logistic regression and significant adjustment variables from Table 1 (age, cytogenetic risk status.) OS and EFS were tested for significant correlation with % priming by logrank test for trend. Patients having received stem cell transplant were censured from OS analyses. Analyses were performed using SAS software, version 9.2 (SAS Institute Inc., Cary, NC), R version 2.14.2 (R Core Team; Vienna, Austria), and/or Graphpad Prism version 5.04 (La Jolla, CA).
Table 1.
Clinicopathologic variables for patient cohort.
| Responders (n=40) | Non-Responders (n=22) | p-value | ||
|---|---|---|---|---|
| Age (range 17.4–85.5 yr) | Mean (SD) | 53.1 (15.0) | 63.3 (12.7) | 0.008 |
| Treatment regimes | Anthra-HDAC | 33 (53.2%) | 15 (24.2%) | 0.0001 |
| Flu-HDAC | 7 (11.3%) | 0 (0.0%) | ||
| HDAC-plus non Anthra | 0 (0.0%) | 7 (11.3%) | ||
| Gender | Male | 16 (25.8%) | 10 (16.1%) | 0.68 |
| Female | 24 (38.7%) | 12 (19.4%) | ||
| FAB (French-American- British classification) | M0 | 0 (0.0%) | 2 (3.3%) | 0.36 |
| M1 | 7 (11.5%) | 2 (3.3%) | ||
| M2 | 9 (14.8%) | 9 (14.8%) | ||
| M4 | 10 (16.4%) | 6 (9.8%) | ||
| M5 | 11 (18.0%) | 2 (3.3%) | ||
| M7 | 1 (1.6%) | 0 (0.0%) | ||
| RAEBT | 1 (1.6%) | 1 (1.6%) | ||
| Cyto-genetics | Favorable | 4 (6.5%) | 0 (0.0%) | 0.003 |
| Intermediate | 26 (41.9%) | 8 (12.9%) | ||
| Unfavorable | 9 (14.5%) | 14 (22.6%) | ||
| NA | 1 (1.6%) | 0 (0.0%) | ||
| Performance Status | 0 | 4 (6.4%) | 7 (11.3%) | 0.18 |
| 1 | 22 (35.5%) | 12 (19.4%) | ||
| 2+ | 14 (22.6%) | 3 (4.8%) | ||
| NPM1 | Mutation | 6 (9.7%) | 4 (6.5%) | 0.47 |
| Wild type | 11 (17.7%) | 14 (22.6%) | ||
| NA | 23 (37.1%) | 4 (6.5%) | ||
| FLT3 ITD | Positive | 11 (17.7%) | 6 (9.7%) | 0.92 |
| Negative | 24 (38.7%) | 14 (22.6%) | ||
| NA | 5 (8.1%) | 2 (3.2%) | ||
| FLT3 D835 | Positive | 3 (4.8%) | 1 (1.6%) | 0.89 |
| Negative | 33 (53.2%) | 19 (30.7%) | ||
| NA | 4 (6.5%) | 2 (3.2%) | ||
| Ras mutation | Positive | 4 (6.5%) | 4 (6.5%) | 0.47 |
| Negative | 19 (30.6%) | 12 (19.3%) | ||
| NA | 17 (27.4%) | 6 (9.7%) |
Comparisons between responders and non-responders on patient characteristics are made by Chi-square tests (Fisher’s exact tests if sample size for any cell of table is less than 5) for categorical variables, and Mann-Whitney tests for continuous variables. Of these variables only age, cyto-genetic status, and treatment were statistically significant for responders (CR) versus non-responders (NR). p value<0.01.
RPPA methodology
Proteomic profiling was performed on AML patient samples using validated methods described previously (7, 23). Patient samples were printed in 5 serial dilutions onto slides along with normalization and expression controls. Slides were probed with a validated primary Ab against total BCL2L11 (Epitomics, Burlingame, CA) at a 1:500 dilution and a secondary Ab to amplify the signal at a dilution of 1:15000 (24). The stained slides were analyzed using Vigene Tech Microvigene Version 2.9 software (Carlisle, MA) to produce quantified data.
Results
Patient Cohort Characteristics
AML patients were stratified by cytarabine-based regimen response status relative to clinical pathologic variables (Table 1). Mann-Whitney analyses were performed to test for association between clinical variables and chemotherapeutic response. Patient age, cyto-genetic risk, and treatment displayed statistically significant association relative to response (p=0.008, p=0.003, and p=0.0001, respectively). Age and cyto-genetic status were subsequently utilized in multivariate analyses for BH3 profiling biomarkers.
BH3 Profiling of Patient Specimens
Of 62 viably preserved AML patient specimens that were BH3 profiled, 61 provided analyzable data (overall technical success rate of 98.4%). The one sample that was eliminated from consideration prior to statistical analysis contained insufficient viable cells by Trypan Blue exclusion to continue with analysis. Representative data is shown in Supplementary Figure 1 of two NR low priming of biomarker panel) and two CR patients (high priming of biomarker panel). Note that the overall Coefficient of Variation (CV) for repeat samples from individual patients is generally 3–5%, indicative of a technically robust assay with limited run-to-run variability.
Among the biomarker peptides assayed, BIM(0.1) elicited priming scores correlated with response with pronounced significance (p=1.8×10−6). It is worth noting that the site from which the specimen was drawn (PB or BM) did not influence analysis as priming scores were significantly associated with response for PB and BM specimens analyzed independently as subsets (data not shown). Analyses of other BH3 Profiling biomarkers assayed are indicated in Table 2. In addition to BIM(0.1), PUMA(10) displayed (borderline) significant association with response (p=0.0064), (Supplementary Figure 2.) NOXA, BAD, HRK, BMF and PUMA2A did not display significant correlation (P>0.007) with response.
Table 2.
BH3 Profiling assayed biomarkers and respective significances in response discrimination.
| BH3 Peptide | Mean %Priming +/− SD | p-value* | AUC [95%CI] | |
|---|---|---|---|---|
|
| ||||
| NR | CR | |||
| BIM | 70.1 ± 32.6 | 88.2 ± 17.6 | 0.023 | 0.68 [0.53,0.82] |
| BIM(0.1) | 13.8 ± 13.4 | 36.8 ± 21.2 | 0.0000018 | 0.83 [0.72,0.93] |
| PUMA | 44.7 ± 29.5 | 64.0 ± 22.7 | 0.017 | 0.69 [0.54,0.84] |
| PUMA(10) | 33.3 ± 24.3 | 50.4 ± 23.7 | 0.0064 | 0.71 [0.58,0.85] |
| NOXA | 26.3 ± 15.5 | 35.1 ± 24.9 | 0.20 | 0.60 [0.46,0.75] |
| BAD | 33.2 ± 29.6 | 52.7 ± 24.3 | 0.014 | 0.70 [0.55,0.85] |
| BMF | 45.6 ± 31.6 | 64.4 ± 24.9 | 0.016 | 0.69 [0.53,0.84] |
| HRK | 20.8 ± 22.4 | 36.6 ± 22.4 | 0.010 | 0.72 [0.57,0.88] |
| PUMA2A | 10.8 ± 19.5 | 16.1 ± 18.9 | 0.16 | 0.61 [0.45,0.78] |
Summary of the mean % priming (±S.D), p-values and areas under the curve (AUC) for all profiling biomarkers analyzed for non-response (NR) and clinical response (CR) AML patient specimens. BIM(0.1) was identified as a highly significant biomarker (p<0.001 and AUC>0.80) PUMA(10) was also (borderline) statistically significant (p<0.007). Statistical significance was by Mann-Whitney Analysis.
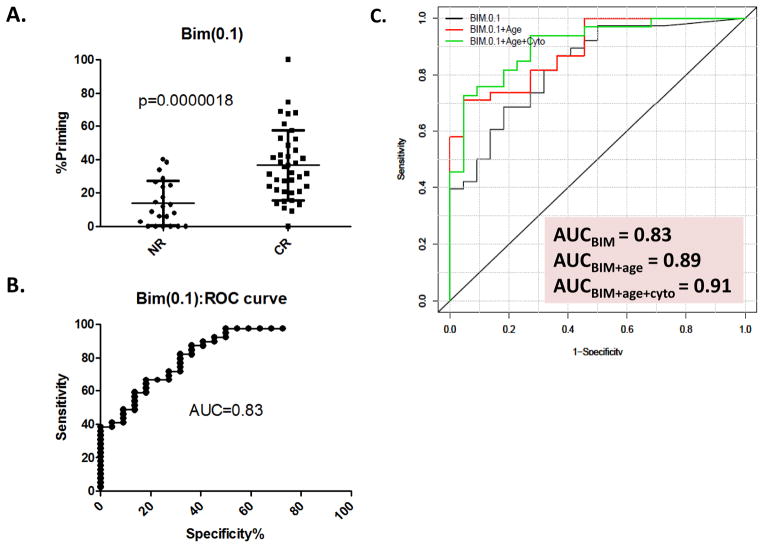
When the BIM(0.1) priming scores of individual patients are segregated into responder and non-responder groups (Figure 1A), a clear trend emerges. AML patients likely to exhibit response to cytarabine-based therapy display higher BIM(0.1) readout (% priming = 36.8 ± 21.2[SD]) than patients not likely to respond (%priming = 13.2 ± 13.4[SD]). In establishing sensitivity and specificity of this biomarker, receiver operator characteristic (ROC) plot depiction indicates an AUC of 0.83 (95%CI[0.73,0.94]) (Figure 1B), an indication of the ability of the biomarker to correctly discriminate individual specimens. Interestingly, a single biomarker may identify 89.7% of responders while at the same time 59.1% of those patients unlikely to respond. With a more stringent sensitivity cut-off of 92.3%, specificity still identifies 54.6% of unlikely responders.
Figure 1. Dot-plot and ROC-plot depictions of BIM patient response discrimination.
A. Dot-plot for the mean %priming (± S.D.) of BIM(0.1) comparing the 22 non-response (NR) and 39 clinical response (CR) patients. Higher BIM(0.1)%primed patients have statistically better clinical response. Statistical significance was determined using the Mann-Whitney Analysis B. ROC-plot of the sensitivity and specificity of BIM(0.1) as a predictor of clinical outcome as determined by the ROC curve (AUC=0.83) using the 61 patients for which there was available data for this marker. C. Multivariate analysis ROC curve for BIM(0.1) alone (Black), or BIM(0.1) combined with significant adjustment clinicopathological variables (+patient age [Red] or +patient age+cytogenetic status [Green]).
Age and cytogenetics have been recognized as prognostic factors in AML and that was true in this dataset as well (Table 1). To determine if the addition of BIM(0.1) % priming biomarker added prognostic information beyond that of age and cytogenetics, each were serially added to BIM(0.1) % priming in multivariate analyses. The addition of patient age to BIM(0.1) yields an increase in AUC to 0.89 from previous BIM(0.1) AUC=0.83 alone (Figure 1C). Further, when BIM(0.1) is adjusted for patient age and cytogenetic risk, then AUC further increases to 0.91. Within this latter adjustment, >90% sensitivity is achieved with identification concurrent with segregation of >70% of the likely non-responders (Figure 1C).
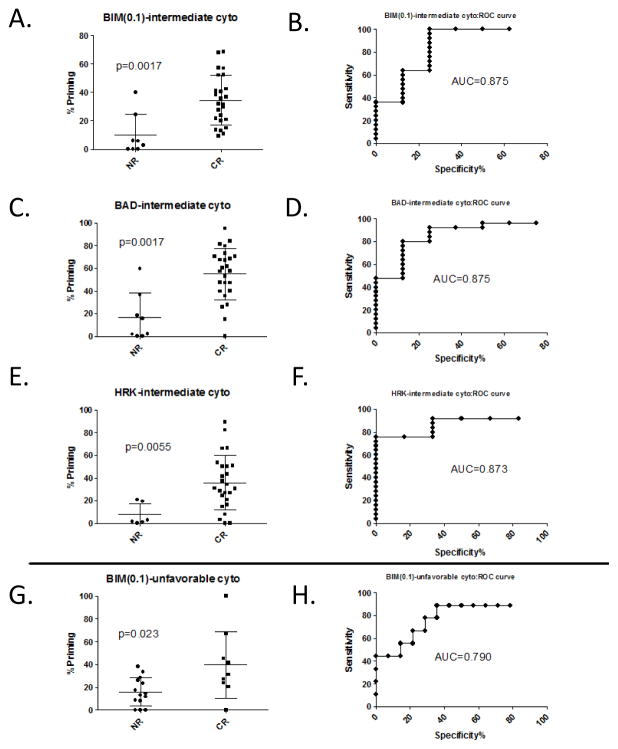
Patients were stratified by cytogenetic risk status and analyzed by Mann-Whitney for response discrimination. In the intermediate risk sub-group, BIM(0.1) was significantly associated (n=33[8 NR, 25 CR), p=0.0017; AUC=0.88, CI[0.71, 1.04]) with further discriminating response and in the unfavorable group BIM(0.1) was also significant (n=23[14 NR, 9 CR], p=0.023; AUC=0.79, CI[0.58, 1.00]) (Figure 2, A, B and G, H). The p-values are somewhat diminished relative to BIM(0.1) analysis of the combined cohort due to reduced statistical power from the sub-grouped number of patients. Interestingly, both BAD and HRK analysis yielded significant p-values in response discrimination (n=33 [8NR, 25 CR], p=0.0017; AUC=88, CI[0.74, 1.00] and p=0.0055; AUC=0.87; CI[0.75, 1.00], respectively); however, this was only observed for the intermediate risk group (Figure 2, C and E). While sensitivity and specificity assessment by ROC analyses of these biomarkers in response discrimination gives AUCs of 0.88 for BIM(0.1), 0.88 for BAD, and 0.87 for HRK in the intermediate group and 0.79 for BIM(0.1) for the unfavorable group (Figure 2, B, D, F, and H, respectively), these AUCs may benefit from somewhat imbalanced subgroupings for responders versus non-responders in the independent sub-groups. Statistical analysis was not possible for favorable patients due to low patient numbers (n=5).
Figure 2. BH3 peptides response prediction stratified by cytogenetic status.
Response prediction stratified by intermediate cytogenetic status (A–F) depicted in dot-plots (A, C, and E) and ROC-plots (B, D, and F) for BIM(0.1) (A and B), BAD (C and D), or HRK (E and F) or unfavorable cytogenetic status for BIM(0.1) as a dot-plot (G) and ROC-plot(H). BIM(0.1), BAD, and HRK were all statistically significant for intermediate cytogenetic status (p value <0.01 and AUC >0.85). Statistical significance was by Mann-Whitney Analysis. (Mean ± S.D. for each set is indicated on dot-plots).
Comparison of BIM(0.1) BH3 Profiling Percent Priming and BIM (BCL2L11) Protein Levels
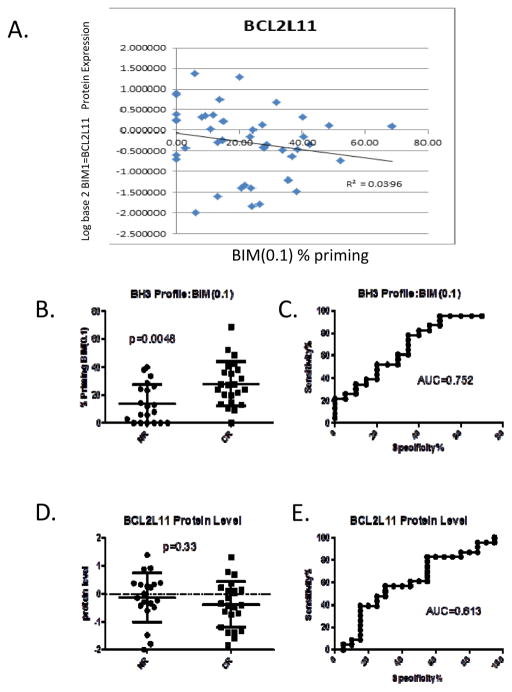
We sought to assess whether BIM BH3 profiling response discrimination is merely redundant with BIM protein levels in AML patient specimens within this study. We find that no correlation exists between BIM protein level and % priming (Figure 3, panel A) yielding an R2 =0.04. Samples analyzed were limited to those having data for both BCL2L22 protein and BH3 profiling BIM(0.1) (n=43; 20 NR, 23 CR).
Figure 3. Correlation of BIM (0.1) Priming and BIM (BCL2L11) Protein Levels and Response Prediction.
BIM protein levels (BCL2L11) determined by RPPA assay were compared to BIM(0.1) BH3 profiling % priming yielding an R2=0.0396 indicating that the two metrics are distinct from one another (A). When statistical analysis is restricted to only patients for which there is BH3 profiling and corresponding BIM RPPA data, BIM(0.1) BH3 profiling remains significant predictor of response (B and C) while BIM protein levels are not (D and E). (Mean ± S.D. for each set is indicated on dot-plots).
BH3 profiling of BIM(0.1) maintains a significant p-value (p=0.0048) for response discrimination with a notable AUC=0.75 (CI[0.60, 0.90]) (Figure 3, panels B and C) in this subset for which both BH3 profiling and RPPA data exist from the total patients cohort. The power of the analysis is reduced relative to our earlier analyses as sample size is diminished from n=62 to n=43 and many of the samples that did not have RPPA data were among the highest scoring BH3 Profiling specimens. The p-value and AUC for response discrimination for this same subset of specimens for BCL2L11 protein level is p=0.33; AUC=0.61 (CI[0.44, 0.79]) (Figure 3, panels D and E). These data provide strong evidence that BH3 profiling is not correlated with overall protein levels and that BH3 profiling may offer a new paradigm by which to predict cytarabine response in AML patients.
Secondary Clinical Endpoints: Overall Survival and Event-free Survival
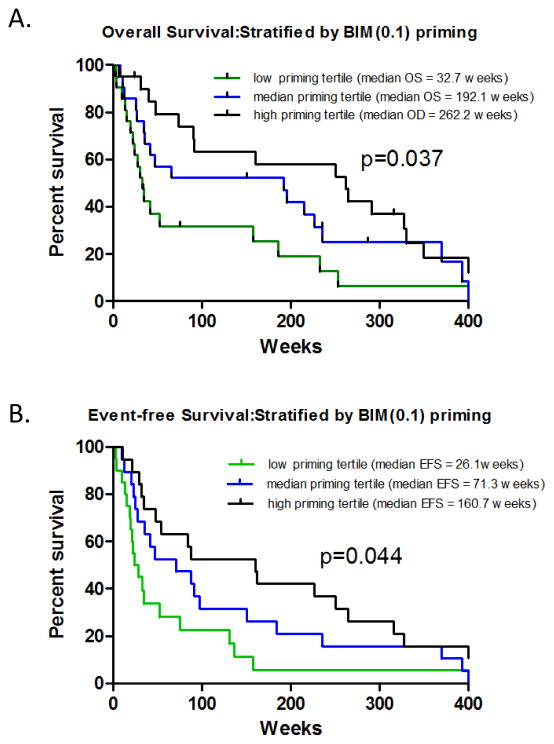
BH3 profiling biomarkers were also analyzed for correlation to the secondary clinical endpoints overall survival (OS) and Event-Free Survival (EFS). Interestingly, when the patient cohort is divided into tertiles by BIM % priming (High, Intermediate, Low), corresponding OS yielded a median of 262.2, 192.1 and 32.7 weeks, respectively (p=0.037,) (Figure 4A). When analysis of tertiles was conducted for EFS, median EFS was 26.1, 71.3, and 160.7 weeks for low priming, intermediate priming, and high priming tertiles, respectively (p=0.044) (Figure 4B). All other peptides displayed non-significant association on tertile segregated patients (all p>0.1, data not shown). These data are consistent with our earlier assessment of the importance of BIM(0.1) is discriminating clinical outcomes for cytarabine-treated AML patients.
Figure 4. OS and EFS vs. AML Patients Subgrouped by BIM % Priming Tertiles.
AML patients were stratified by low, medium, and high BIM % priming scores and then analyzed for OS (panel A) and EFS (panel B). Logrank analyses indicates borderline significant associations between OS and BIM(0.1) priming (p=0.037), and EFS and BIM(0.1) priming (p=0.044). For OS, low, medium, and high priming tertiles comprised 19, 18, and 18 patients respectively (total n=55 for which OS data was available; patients who received stem cell transplant were censured). For EFS, low, medium, and high priming tertiles comprised 20, 20, and 21 patients respectively (total n=61 for which EFS data was available).
Discussion
The current study highlights the potential clinical utility of BH3 profiling biomarkers in discriminating patient response to standard-of care (cytarabine-based) chemotherapeutic regimens for AML patients. Namely, a single BH3 profiling biomarker (BIM[0.1]) utilized in conjunction with patient age and cytogenetic status delivers a sensitivity and specificity profile with an AUC>0.9. Interestingly, the two most interesting biomarkers in the current study (BIM [p=10-6] and potentially PUMA [p=0.0064]) display functional roles in the BH3 proteins “activator” class, directly binding to and modulating Bax/Bak, The other biomarkers assessed here (NOXA, BAD, HRK) are classified as “sensitizers” by engaging specific anti-apoptotoic proteins (MCL-1, BCL-2, BCL-xL) are not significantly associated with outcomes.
While the statistics achieved with a limited number of patients are impressive, the current study is limited in several aspects that must be addressed in future studies. The current study utilized patient specimens that were collected and viably cryopreserved. However, analytic scrutiny of prospective collection versus viably frozen technically has indicated that the test will be utilized to direct treatments of patients as they present in real time. Further, a skewed dataset with more long-term CR and more primary refractory cases was selected for this study. The range of priming and correlation with clinical endpoints will need to be assessed in randomized cohorts more representative of an unselected population including intermediate response endpoints (i.e. CRi, CRp). While the current cohort comprises a first test set that provided notable predictive capability, additional studies are required to confirm the present findings. Still, even with a 62 patients cohort, the BIM (0.1) p-value is notable (p=1.8×10−6). When placed in the context of patient age and cytogenetic status (p=0.008 and p-0.003, respectively), BIM(0.1) significance is striking juxtaposed to clinicopathlogic variables already accepted to be associated with clinical outcomes.
A key question addressed here was whether a significant BH3 profiling metric is merely redundant with BH3-only protein expression. Our results indicate BH3 profiling and protein expression are decidedly not correlated and response is only predicted by the more functional of the two metrics (BH3 profiling). Protein levels alone do not assess the context in which these expressions occur, including phosphorylation state, subcellular location, or the broader context of similar measurements for directly or indirectly co-operating proteins. Still while the analyses here were directed to response, it is plausible that BH3 protein levels may trend coincident with other clinical endpoints (SM Kornblau, unpublished observations).
While static protein levels have not been conclusively shown to correlate with clinical outcomes, other investigational studies highlight the importance of BIM as a key regulatory node in apoptosis following chemotherapy in AML. Zhang et al. (25) showed that sorafenib treatment resulted in BIM upregulation and activation of the intrinsic apoptotic pathway. Conversely, decreasing BIM expression significantly abrogated sorafenib-induced apoptosis. In separate studies, intrinsic apoptosis pathway response to MEK inhibitors and FLT3 inhibitors was regulated through levels of both BIM and PUMA (26, 27) although at least in the case of FLT3 inhibitors, BIM apparently plays the more key role. Finally, Grocek et al. have reported that cytarabine-mediated apoptosis in AML following vitamin D3 treatment occurs through modulation of miR-23 and subsequently BIM levels (28). Taken together, these studies underscore the functional importance of BIM (not directly assessed by static expression levels) as addressed by BH3 profiling for assessing pro-apoptotic response to AML treatment. Recent studies have addressed the potential utility of BH3 profiling for predicting chemotherapeutic response in AML patients. Ni Chonghaile et al. (20) utilized a small cohort of 15 patients (6 NR, 9 CR). While this cohort was of statistically low power and specific therapeutic treatment of patients was not addressed, BIM BH3 profiling was significant for general chemotherapeutic response. Vo et al. (21) found correlation between BH3 profiling of CD34+ stem cells and response to induction chemotherapy comprising daunorubicin, etoposide or mitoxantrone in AML patients and BH3 profiling of BIM was used to identify patients that could likely benefit from allogenic stem cell transplant. In this study, no significant correlation was observed between BH3 profiling and cytarabine efficacy in cell lines or patient-derived specimens. As cytarabine remains a key component of standard-of-care treatment of AML patients, our current study sought to focus on defining the medical utility of mitochondrial response as actionable information for patient management using a statistically powered cohort of patients uniformly treated with cytarabine-based therapy. BH3 profiling cut-points from this cohort come into focus and alongside additional patient information such as age and cytogenetic risk status, improved sensitivity and specificity profiles herein serve as a harbinger for the potential clinical application for such a diagnostic.
Application of personalized medicine approaches in patient management decisions carries certain considerations that cannot be discounted. When the default is a patient will receive therapy, one may want to avoid targeting a specific biomarker status whereby a patient likely to benefit from treatment is mis-classifed even if there is improvement in the overall clinical endpoints of the patient group overall. Resolved that a 10% false negative rate is acceptable, the question to be addressed is how many of the patients unlikely to respond can be moved to a different treatment? Based on the current data, theoretically applying a preliminary cut-point for patients with BIM(0.1) priming at ≥15% would identify ≥90% of likely responders who should receive cytarabine while 55%–60% of likely non-responders would be spared the treatment (negative predictive value). Further, when age and cytogenetic status are considered in the context of BIM(0.1) priming, then ≥90% sensitivity may be achieved concurrently with classifying ≥70% of non-responders. Additional BH3 profiling biomarkers and algorithms may have application in discriminating response to alternative therapies for patients not deemed suitable for cytarabine-based regimens.
Supplementary Material
Acknowledgments
The authors would like to thank David Richard for helpful discussions during manuscript preparation. This work was funded in part by NCI-SBIR #HHSN261201200039C and #HHSN261201299985C contracts to Eutropics Pharmaceuticals and by grants from NIH (CA016672 and CA55164) to MA and SK.
Footnotes
Conflict-of-interest disclosure.
William Pierceall, Nicole Carlson, Ryan Lena, Noel Blake, Michael Elashoff, and Michael Cardone are employees of Eutropics, Inc. The remaining authors declare no conflicts-of-interest.
References
- 1.NCI cancer statistics. ( www.cancer.gov/cancertopics/types/leukemia)
- 2.American Cancer Society. Cancer Facts and Figures 2013. Atlanta, Ga: American Cancer Society; 2013. [Last accessed May 2, 2013]. Available online. [Google Scholar]
- 3.Bishop JF. The treatment of adult acute myeloid leukemia. Semin Oncol. 1997;24:57–69. [PubMed] [Google Scholar]
- 4.Grimwade D, Walker H, Harrison G, Oliver F, Chatters S, Harrison CJ, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–20. doi: 10.1182/blood.v98.5.1312. [DOI] [PubMed] [Google Scholar]
- 5.Baldus CD, Mrózek K, Marcucci G, Bloomfield CD. Clinical outcome of de novo acute myeloid leukaemia patients with normal cytogenetics is affected by molecular genetic alterations: a concise review. Br J Haematol. 2007;137:387–400. doi: 10.1111/j.1365-2141.2007.06566.x. [DOI] [PubMed] [Google Scholar]
- 6.Licht JD. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene. 2001;20:5660–79. doi: 10.1038/sj.onc.1204593. [DOI] [PubMed] [Google Scholar]
- 7.Kornblau SM, Tibes R, Qiu Y, Chen W, Kantarjian HM, Andreeff M, et al. Functional proteomic profiling of AML predicts response and survival. Blood. 2009;113:154–64. doi: 10.1182/blood-2007-10-119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornblau SM, Minden MD, Rosen DB, Putta S, Cohen A, Covey T, et al. Dynamic single-cell network profiles in acute myelogenous leukemia are associated with patient response to standard induction therapy. Clin Cancer Res. 2010;16:3721–33. doi: 10.1158/1078-0432.CCR-10-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wuchter C, Karawajew L, Ruppert V, Schrappe M, Harbott J, Ratei R, et al. Clinical significance of CD95, Bcl-2 and Bax expression and CD95 function in adult de novo acute myeloid leukemia in context of P-glycoprotein function, maturation stage, and cytogeneticis. Leukemia. 1999;13:1943–53. doi: 10.1038/sj.leu.2401605. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann SH, Karp JE, Svingen PA, Krajewski S, Burke PJ, Gore SD, Reed JC. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood. 1998;91:991–1000. [PubMed] [Google Scholar]
- 11.Kornblau SM, Thall PF, Estrov Z, Walterscheid M, Patel S, Theriault A, et al. The prognostic impact of BCL-2 protein expression in acute myelogemous leukemia varies with cytogenetics. Clin Cancer Res. 1999;5:1758–66. [PubMed] [Google Scholar]
- 12.Danna EA, Nolan GP. Transcending the biomarker mindset: deciphering disease mechanisms at the single-cell level. Curr Opin Chem Biol. 2006;10:20–7. doi: 10.1016/j.cbpa.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 13.Rosen DB, Minden MD, Kornblau SM, Cohen A, Gayko U, Putta S, et al. Functional characterization of FLT3 receptor signaling deregulation in acute myeloid leukemia by single cell network profiling (SCNP) PLoS One. 2010;5:e13543. doi: 10.1371/journal.pone.0013543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12:171–85. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Letai A. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–32. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- 16.Ryan JA, Brunelle JK, Letai A. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proc Natl Acad Sci USA. 2010;107:12895–900. doi: 10.1073/pnas.0914878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinstein B, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–80. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 18.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–65. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 19.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest. 2007;117:112–21. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore Vdel G, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334:1129–33. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vo TT, Ryan J, Carrasco R, Neuberg D, Rossi DJ, Stone RM, et al. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151:344–55. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornblau SM, Qui YH, Zhang N, Singh N, Faderi S, Ferrajoli A, et al. Abnormal expression of FLI1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011;118:5604–12. doi: 10.1182/blood-2011-04-348052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibes R, Qiu YH, Lu Y, Hennessey B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array (RPPA): validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoetic stem cells (HSC) Mol Cancer Ther. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 24.Hunyady B, Krempels K, Harta G, Mezey E. Immunohistochemical signal amplification by catalyzed reporter deposition and its application in double immunostaining. J Histochem Cytochem. 1996;44:1353–62. doi: 10.1177/44.12.8985127. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Konopleva M, Ruvolo VR, McQueen T, Evan RL, Bomman WG, et al. Sorafenib induces apoptosis of AML cells via Bim-mediated activation of the intrinsic apoptosis pathway. Leukemia. 2008;22:808–18. doi: 10.1038/sj.leu.2405098. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, Konopleva M, Burks JK, Dywer KC, Schober WD, Yang JY, et al. Blockade of mitogen-activated protein kinase/extracellular signal-related kinase kinase and murine double minute synergistically induces apoptosis in acute myeloid leukemia via BH3-only proteins Puma and Bim. Cancer Res. 2010;70:2424–34. doi: 10.1158/0008-5472.CAN-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordigarden A, Kraft M, Elliasson P, Labi V, Lam EW, Villunger A, Jonsson AI. BH3-only protein Bim more critical than Puma in tyrosine kinase inhibitor-induced apoptosis of human leukemic cells and transduced hematopoetic progenitors carrying oncogeneic FLT3. Blood. 2009;113:2302–11. doi: 10.1182/blood-2008-07-167023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grocek E, Wang X, Liu X, Liu CG, Studzinski GP. MicroRNA-32 upregulation by 1,25-dihydroxyvitamin D3 in human myeloid leukemia cells leads to Bim targeting and inhibition of AraC-induced apoptosis. Cancer Res. 2011;71:6230–9. doi: 10.1158/0008-5472.CAN-11-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.