Summary
How the cell recognizes cytosolic DNA including DNA based microbes to trigger host defense related gene activation remains to be fully resolved. Here, we demonstrate that STING (for Stimulator of Interferon Genes), an endoplasmic reticulum (ER) translocon associated transmembrane protein, acts to detect cytoplasmic DNA species. STING homodimers were able to complex with self (apoptotic, necrotic) or pathogen related ssDNA and dsDNA and were indispensible for HSV-1-mediated transcriptional activation of a wide array of innate immune and pro-inflammatory genes in addition to type I IFN. Our data indicates that STING instigates cytoplasmic DNA-mediated cellular defense gene transcription and facilitates adoptive responses that are required for protection of the host. In contrast, chronic STING activation may manifest inflammatory responses and possibly autoimmune disease triggered by self-DNA.
Introduction
Potent activators of cellular innate responses are known to include microbial nucleic acid, derived from the genomes of viruses as well as bacteria (Kumar et al.; Schenten and Medzhitov). For example, RNA viruses can trigger the production of innate immune genes, such as type I interferon (IFN), through their nucleic acid being recognized by the cytoplasmic RNA sensors retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation antigen 5 (MDA5) (Yoneyama and Fujita). In addition, members of the Toll-like receptor (TLR) family, such as TLR3 and 7 have similarly evolved to be able to recognize viral RNA and to initiate the production of type I IFN. While the cellular molecular mechanisms responsible for sensing viral RNA have become clarified, less is known relating to how the cell senses microbial DNA species to trigger host defense associated gene regulation. It is established that TLR9 recognizes pathogen derived CpG DNA to trigger innate immune signaling predominantly in plasmacytoid dendritic cells (pDCs) (Hemmi et al., 2000). Moreover, Absent in melanoma 2 (AIM2), a HIN-200 domain containing protein is known to be able to recognize cytoplasmic DNA species and trigger inflammasome-dependent IL-β synthesis (Alnemri, 2010; Schroder and Tschopp, 2010). However, we recently reported the isolation of a transmembrane component of the endoplasmic reticulum (ER), referred to as STING (Stimulator of Interferon Genes), which we demonstrated was essential for the production of type I IFN in fibroblasts, macrophages and dendritic cells (DCs) in response to cytoplasmic dsDNA as well as select DNA viruses and intracellular bacteria, although the mechanisms remained to be fully elucidated (Ishikawa and Barber, 2008; Ishikawa et al., 2009). Here, we report that STING accomplishes these events by associating with aberrant cytoplasmic DNA species, including self ssDNA and dsDNA, to trigger host defense related gene transcription. Our data indicates that STING is essential for innate responses triggered by intracellular DNA pathogens, while chronic activation may contribute towards DNA activated inflammatory disease.
Results
STING triggers the expression of multiple primary innate immune and pro-inflammatory genes in response to intracellular ssDNA as well as dsDNA
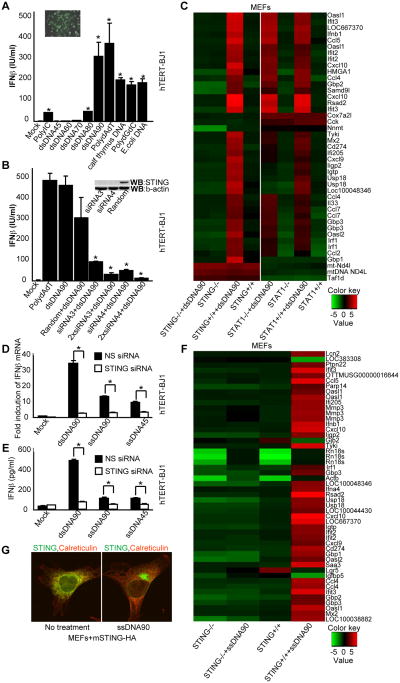
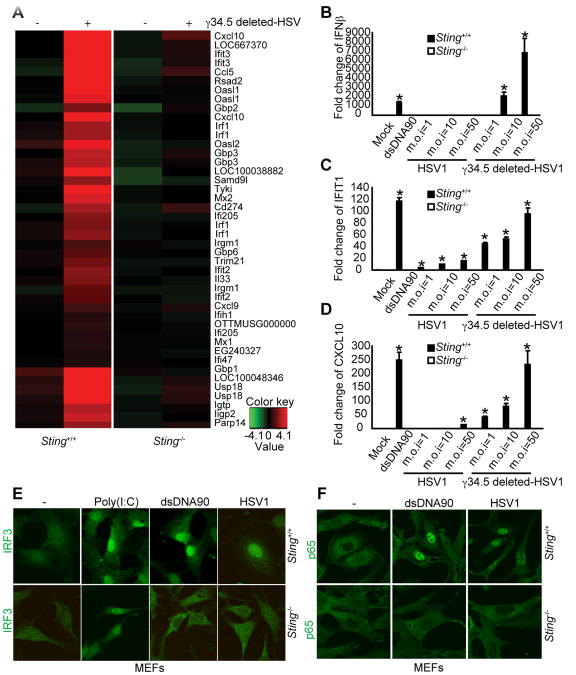
The minimum size of dsDNA optimally required to activate STING-dependent type I IFN signaling in murine cells was noted to be approximately 45 base pairs (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Stetson and Medzhitov, 2006). In normal human cells (hTERT-BJ1), however, we observed that dsDNA of approximately 90 base pairs (referred to herein as dsDNA90) was more efficient at activating type I IFN following in vitro transfection, although smaller sizes also remained capable of facilitating these events to a lesser degree (Figure 1A). Using RNAi knockdown procedures, we additionally confirmed that STING (also referred to as MPYS/MITA) (Jin et al., 2008; Zhong et al., 2008) is indeed essential for the production of type I IFN in hTERT-BJ1 cells (Figure 1B). Further analysis using microarray procedures to measure mRNA expression confirmed that cytoplasmic dsDNA can induce a wide array of innate immune genes, in addition to type I IFN, in hTERT-BJ1s (Figure S1A). The induction of these innate genes which included members of the IFIT family appeared to be STING-dependent since RNAi knockdown of STING in hTERT-BJ1s greatly eliminated their stimulation by cytoplasmic dsDNA (Figure S1B-F). That cytoplasmic dsDNA induced a variety of innate immune genes in a STING-dependent manner was confirmed using Sting+/+ or Sting-/- murine embryonic fibroblasts (MEFs) (Figure 1C). To confirm that the induction of these mRNAs were STING-dependent genes (SDG), and not stimulated through type I IFN dependent autocrine or paracrine signaling, we similarly treated type I IFN-signaling defective Stat1-/- MEFs with dsDNA and verified that the production of the SDGs remained largely unaffected (Figure 1C). Real time PCR analysis confirmed our array results (Figure S1G and data not shown). We noted that ssDNA of 45 nucleotides (ssDNA45) weakly induced innate immune gene production in hTERT-BJ1s and less so in MEFs (Ishikawa and Barber, 2008; Ishikawa et al., 2009). However, transfected ssDNA comprising 90 nucleotides (ssDNA90) was observed to more robustly activate an array of genes, including type I IFN in hTERT-BJ1s and MEFs (Figure 1D-F and Figure S1B, G). We observed that STING likely resides as a homodimer in the ER of both human and murine cells, and migrates from the ER to perinuclear regions in the presence of cytoplasmic ssDNA or dsDNA ligands or HSV1 infection to activate type I IFN-dependent transcription factors (Ishikawa et al., 2009) (Figure 1G and Figure S1H-K). A defective HSV1 mutant lacking the γ34.5 gene, which is responsible for preventing translational inhibition mediated by the double-stranded dependent protein kinase (PKR), was similarly observed to activate innate immune gene production in a STING dependent manner (Figure 2A-2D and data not shown). The translocation of IRF3 and 7 as well as NF-κB into the nucleus was observed following HSV1 infection and shown to be dependent on STING (Figure 2E, F). Accordingly, it was confirmed that many of the SDGs contained IRF3/7 and NF-κB binding sites in their promoter region (data not shown). Interestingly, cyclic di-GMP, reported to bind STING, did not robustly activate STING in primary human or mouse cells similar to DNA (Figure S6). Thus, cytoplasmic ssDNA or dsDNA that includes transfected plasmid DNA can potently induce the transcription of a wide array of innate immune related genes that is dependent on STING.
Figure 1. STING controls cytosolic ssDNA and dsDNA innate signaling. See also Figure S1.
(A) Human Telomerase Fibroblasts (hTERT-BJ1) were transfected with various nucleotides (3 μg/ml) for 16h. Endogenous IFNβ levels were measured. hTERT-BJ1 cells were transfected with FITC conjugated dsDNA90 and was examined by fluorescent microscopy to ensure efficient transfection.
(B) hTERT-BJ1 cells were transfected with mock, random or two independent human STING siRNAs (siRNA 3 or 4) for 3 days followed by dsDNA90 transfection (3 μg/ml) for 16 hours. Endogenous IFNβ levels were measured. Silencing of hSTING protein was demonstrated by immunoblotting, with β-actin serving as a loading control.
(C) Primary Sting+/+, Sting-/-, Stat1+/+ or Stat1-/- MEFs were transfected with or without dsDNA90 (3 μg/ml) for 3 hours. Total RNA was purified and examined for gene expression by Illumina Sentrix BeadChip Array (Mouse WG6 version 2). Most variable genes were selected. Rows represent individual genes; columns represent individual samples. Pseudo-colors indicate transcript levels below, equal to, or above the mean (green, black, and red, respectively). The scale represents the intensity of gene expression (log10 scale ranges between -5 and 5).
(D) hTERT-BJ1 cells were treated with NS or STING siRNA. After 3 days, cells were treated with dsDNA90, ssDNA90 or ssDNA45 (3 μg/ml). IFNβ mRNA levels were measured by real time RT-PCR after 16 hours.
(E) hTERT-BJ1 cells were treated with NS or STING siRNA. At 3 days, cells were treated with dsDNA90, ssDNA90 or ssDNA45 (3 μg/ml). IFNβ mRNA levels were measured after 16 hours.
(F) Primary Sting+/+ or Sting-/- MEFs were transfected with or without ssDNA90 (3 μg/ml). After 3h, the same as c.
(G) Primary MEFs with mSTING-HA were treated with or without ssDNA90 (3 μg/ml) for 3 hours and stained with anti-HA antibody (green) and calreticulin (red) as an ER marker. *P<0.05, Student's t-test. Error bars indicated s.d. Data are representative of at least two independent experiments.
Figure 2. STING is essential for HSV1-mediated innate immune signaling. See also Figure S6.
(A) MEFs were infected with γ34.5 deleted-HSV1 (m.o.i=1) for 3h. Total RNA was purified and examined for gene expression using Illumina Sentrix BeadChip Array (Mouse WG6 version2). Most variable genes were selected. Rows represent individual genes; columns represent individual samples. Pseudo-colors indicate transcript levels below, equal to, or above the mean (green, black, and red, respectively). The scale represents the intensity of gene expression (log10 scale ranges between -4.1 and 4.1).
(B) Sting+/+ or Sting-/- MEFs were treated with or without dsDNA, HSV1 or γ34.5 deleted-HSV1 for 3h. Total RNAs were purified and examined by real time PCR for IFNβ(B), IFIT1(C) or CXCL10(D). Error bars indicate s.d.
(E) Sting+/+ or Sting-/- MEFs were treated with poly(I:C), dsDNA90 or HSV1 and cells were stained by anti-IRF3 antibody. PolyIC is RIG-I/MDA5 dependent and STING independent.
(F) Sting+/+ or Sting-/- MEFs were treated with dsDNA90 or HSV1 and cells were stained by anit-p65 antibody at 3 hrs PI.
STING Complexes with Intracellular DNA
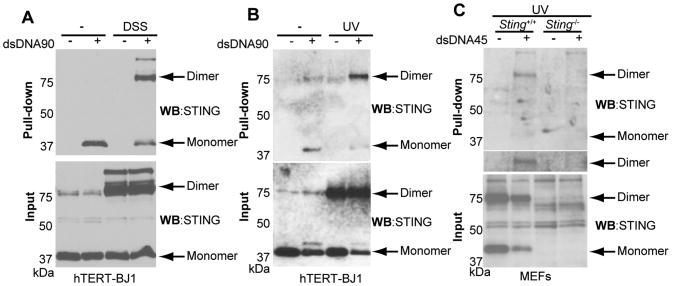
Given the importance of STING in regulating cytoplasmic DNA signaling events, it remained plausible that STING itself could associate with DNA species. To evaluate whether STING bound cytoplasmic DNA, we first transfected hTERT-BJ1s with biotinylated dsDNA90 and subsequently treated them with or without the irreversible protein cross-linker disuccinimidyl suberate (DSS). After cells lysis, extracts were mixed with streptavidin beads. This experiment indicated that STING could be pulled down in both DSS untreated as well as treated cells, likely as a dimer (Figure 3A). Similar results were obtained using hTERT-BJ1s transfected with biotinylated dsDNA90 and subsequently UV treated to cross link DNA associating proteins (Figure 3B). RNAi knockdown of STING in hTERT-BJ1s eliminated the observed binding and STING-DNA complexes were also only observed in Sting+/+ MEFs but not Sting-/- MEFs (Figure 3C and Figure S2A). Of interest is that we similarly observed that transfected STING could also bind to ssDNA (Figure S2B-D). It has previously been reported that IFI16 a member of the HIN-200 family of proteins or helicase DDX41 may be involved with sensing cytoplasmic DNA to initiate type I IFN production through STING. However, knock down of IFI16 or DDX41 in hTERT cells did not affect the binding of STING to DNA. Neither did loss of IFI16 or DDX41 appear to affect STING trafficking or dsDNA-mediated translocation of IRF3/NF-κB by dsDNA or HSV1, or baculovirus which cannot encode any viral products in mammalian cells but can stimulate cytosolic DNA-mediated innate immune pathways (Figure S3). However, loss of IFI16 did appear to influence the production of HSV1 confirming a role for this IFN-induced protein in antiviral activity (Figure S3B, C) (Gariano et al., 2012; Kerur et al., 2011; Unterholzner et al., 2010; Zhang et al., 2011).
Figure 3. STING binds to DNA in vivo. See also Figure S2 and S3.
(A) hTERT-BJ1 cells were transfected with biotin conjugated dsDNA90 (3 μg/ml) for 6h and treated with DSS. Lysates were precipitated using streptavidin agarose beads and analyzed by immunoblotting using anti-HA antibody.
(B) hTERT-BJ1 cells were transfected with biotin conjugated dsDNA90 (3 μg/ml) for 6h and treated with UV. Same as A.
(C) Sting+/+ or Sting-/- MEFs were transfected with biotin conjugated dsDNA45 and crosslinked by UV. Lysates were precipitated by streptavidin agarose beads and analyzed by immunoblotting.
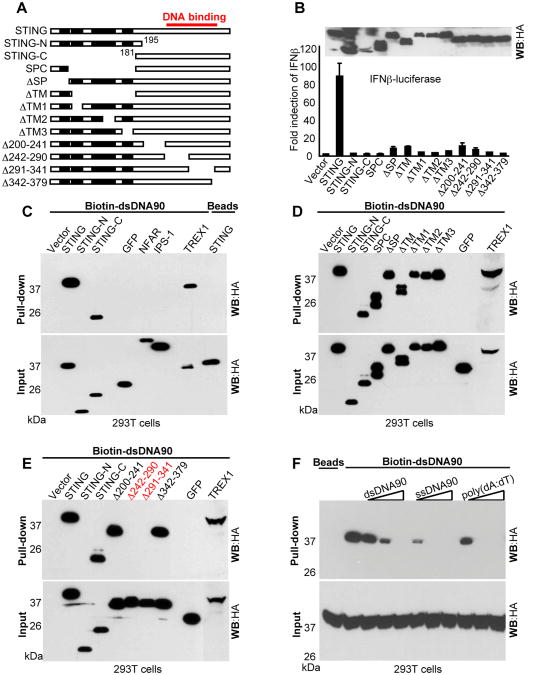
Our analysis was complemented by transfecting 293T cells, which do not normally express STING with a variety of STING deletion constructs (Figure 4A). After 24 hours, cell lysates were mixed with biotinylated dsDNA90 agarose beads and STING-DNA binding analyzed. This study complemented our above analysis and similarly indicated that STING was capable of binding dsDNA (Figure 4C). Further study indicated that the C-terminal regions of STING (aa 181-379) was sufficient for this association. In contrast, the N-terminal region of STING (aa 1-195) and three similarly HA-tagged controls (GFP, NFAR1 and IPS1) were not found to exhibit any binding to dsDNA90 (Figure 4C and 4D). The DNA binding exonuclease TREX1 served as a positive control (Figure 4C). A further series of extensive studies indicated that amino acid region 242-341 of STING was likely involved in binding dsDNA (Figure 4E). Competition experiments indicated that ssDNA as well as poly(dA:dT) could effectively compete with dsDNA90 for STING binding (Figure 4F). PolyIC did not bind to STING or prevent STING from associating with DNA (Figure S2E-G). All STING variants analyzed, but not the wild type, lacked the ability to activate the type I IFN promoter when expressed in 293T cells indicating the importance of STING's authentic conformation for effective function (Figure 4B). We were also able to rescue dsDNA-dependent signaling in a variety of cells following reconstitution of STING, including Sting-/- MEFs and 293T cells (Figure S4A-E).
Figure 4. STING binds to DNA in vitro. See also Figure S4.
(A) Schematic of STING variants.
(B) 293T cells were transfected with IFNβ-luciferase and STING variants and luciferase activity were measured.
(C-E) 293T cells were transfected with the indicated plasmids. Cell lysates were precipitated with biotin conjugated dsDNA90 agarose beads and analyzed by immunoblotting using anti-HA antibody.
(F) Full length STING-HA was expressed in 293T cells and lysates were incubated with biotin conjugated dsDNA90 agarose beads in the presence of dsDNA90, ssDNA90 or Poly(dA:dT) and analyzed by immunoblotting using anti-HA antibody.
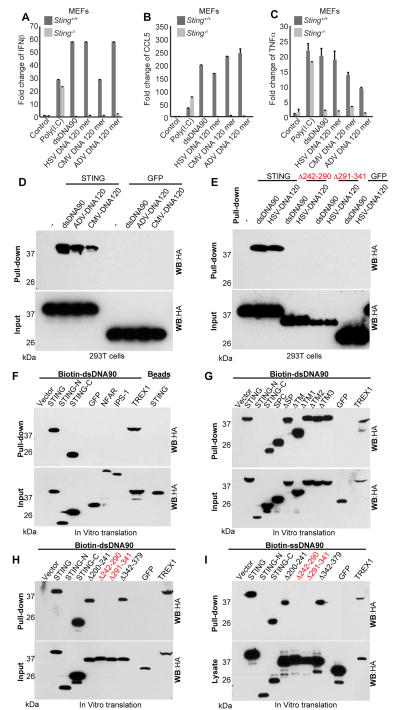
To extend these studies further, we transfected dsDNA90 into hTERT-BJ1s or MEFs and treated them with formaldehyde to crosslink the cellular proteins to the nucleic acid. Subsequent ChIP analysis following STING pull down further confirmed that transfected DNA can directly associate with STING as determined using dsDNA90 specific primers (Figure S5A, B). STING was also observed to bind to dsDNA in ELISA based analysis (Figure S5C-E). Finally, we complemented our above analysis by designing biotinylated dsDNA (120-mers) representing the genomes of HSV1, cytomegalovirus (CMV) as well as adenovirus (ADV). Transfection of the 120-mers into MEF cells confirmed that they were potently able to induce the production of a variety of cytokines, such as type I IFN, CCL5 and TNFα in a STING-dependent manner (Figures 5A-5C and Figure S5F). Subsequently, these viral DNA's were incubated with 293T cell lysates previously transfected with STING and mixed with streptavidin beads. This approach indicated that these viral nucleic acids could also robustly bind to full length STING, but not STING Δ242-290 or STING Δ291-341, indicating the importance of region 242-341 in nucleic acid association (Figure 5D, E). Collectively, our data demonstrates by multiple methods that STING is capable of associating with DNA.
Figure 5. STING binds to DNA. See also Figure S5.
(A-C) Sting+/+ or Sting-/- MEFs were treated with poly(I:C), dsDNA90, HSV DNA 120mer, CMV DNA 120mer or ADV DNA 120mer for 3 hours. Total RNA was purified and examined by real time PCR for gene expression of IFNβ (A), CCL5(B) or TNFα(C). Error bars indicate s.d.
(D-E) 293T cells were transfected with indicated plasmids. Cell lysates were precipitated with biotin-dsDNA90, biotin-ADV DNA 120mer, biotin-CMV DNA 120mer (D) or biotin-HSV DNA 120mer (E) agarose beads and analyzed by immunoblotting using anti-HA antibody.
(F-H) In vitro translation products were incubated with biotin-dsDNA90 agarose beads and analyzed by immunoblotting using anti-HA antibody.
(I) In vitro translation products were incubated with biotin-ssDNA90 agarose beads and analyzed by immunoblotting using anti-HA antibody.
STING binds ssDNA and dsDNA without a requirement for accessory molecules
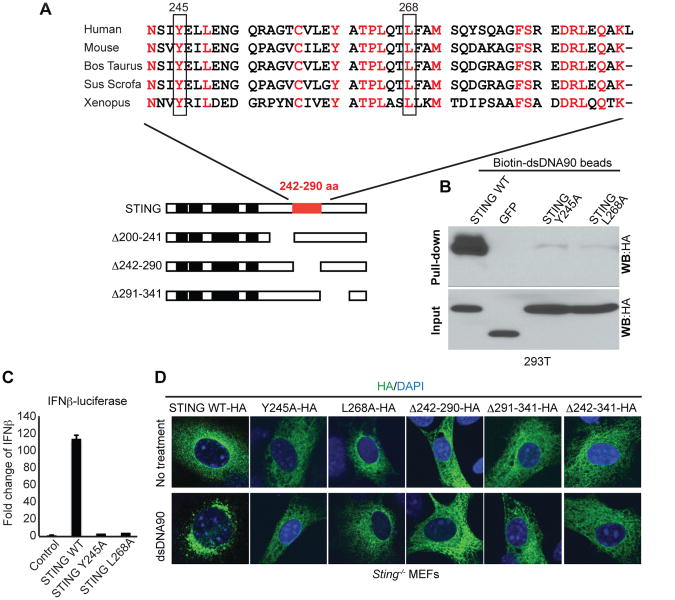
To evaluate whether STING could associate with dsDNA alone, or require accessory molecules, we extended our studies using in vitro transcribed STING and biotinylated dsDNA90 agarose beads. These experiments confirmed that the C-terminal STING (aa 181-379) could associate with dsDNA90, and suggested that this association did not likely require co-factors (Figure 5F-H). As in our 293T experiments, region 242-341 was observed to be required for STING-dsDNA interaction. Given that we had observed that ssDNA could also trigger STING function and compete with dsDNA90, we next elaborated on whether ssDNA could comparably bind to STING. Our results indicated that ssDNA90 was indeed able to complex to the same regions of STING as dsDNA90 (Figure 5I). To further refine our studies, we generated point mutations focusing on conserved residues noted to be within the region of STING (aa 242-290) considered important for DNA binding (Figure 6A). STING variants with amino acids 245 or 268 changed to alanine (Y245A and L268A) were expressed in 293T cells and were not observed to be efficiently precipitated using biotinylated DNA. Such STING variants did not exhibit any ability to activate innate immune gene expression (Figure 6B, C). Similarly, STING K150R variant, reportedly deficient in lysine 63-linked ubiquitination via Trim56, was also capable of binding DNA (Figure S7). Further, reconstitution of these STING variants unto STING-/- MEFs indicated that these variants did not traffic following transfection with dsDNA90. Larger STING variants (STINGΔ242-341, Δ242-290, Δ291-341) similarly did not appear to exhibit function, indicating the importance of the cytoplasmic tail in the interaction with DNA (Figure 6D). Thus, STING is likely able to interact with ssDNA species as well as dsDNA and STING variants unable to bind DNA exhibit little activity.
Figure 6. Y245 and L268 are important for STING binding DNA. See also Figure S7.
(A) Alignment of amino acid sequence of STING in 242-290aa.
(B) 293T cells were transfected with STING WT-HA, GFP-HA, STING Y245A-HA or STING L268A-HA for 24 hours. Cell lysates were precipitated with biotin-dsDNA90 beads and analyzed by immunoblotting with anti HA antibody.
(C) 293T cells were transfected with IFNβ-luciferase plasmid and plasmid encoding STING or mutants. After 24 hours, luciferase activity was measured. Error bars indicate s.d.
(D) STING-/- MEFs were transfected with plasmid encoding STING or mutants using an Amaxa nucleofector apparatus (program A-023) with Amaxa MEF nucleofector kit 1 according to the manufacturer' s instructions. After 24 hours, the cells were treated with dsDNA90 for 6 hours and then stained using anti-HA (green) antibody and DAPI (blue).
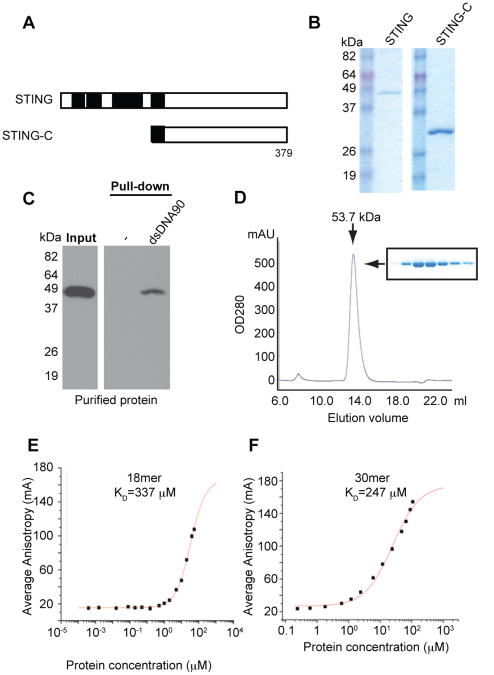
To confirm these observations, we purified full length STING or GFP control protein from 293T cells and the C-terminal region of STING (181-379) from E.coli to greater than 95% homogeneity (Figure 7A,B and S5G-I). Full length STING purified from 293T cells using affinity chromatography was noted to bind to DNA under relatively high salt and detergent conditions (Figure 7C). However, we noted that full length STING was insoluble following purification from E.coli unlike the carboxyl region of STING (aa 181-379), which remained soluble. We confirmed that the C-terminal region of STING, but not purified control GFP, could associate with biotinylated dsDNA90 or ssDNA90 (Figure S5G-I). To evaluate this interaction further we used Surface Plasmon Resonance (SPR) analysis to demonstrate that soluble purified STING (aa 181-379) bound to dsDNA90 with a calculated dissociation constant (KD) of 13.7 μM as determined by this method (Figure S5I). To complement our analysis we purified an extended version of the C-terminal region of STING (aa 152-379) from E.coli, which we found remained soluble throughout the purification process (Figure 7 B and D). Coomassie gel analysis and size exclusion chromatography again confirmed the high level of purity of STING and confirmed that STING exists as a dimer with a calculated molecular weight of approximately 53,000 Daltons, as previously reported and also indicated by our earlier studies (Figure 7D) (Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012; Yin et al., 2012). To complement our SPR studies, we used fluorescence anisotropy assays to start to determine the minimum size of DNA that could associate with STING. This approach confirmed our previous observations and demonstrated the binding of the C-terminal region of STING (aa 152-379) to dsDNA 30-mers or 18-mers with an affinity of KD of 247μM and 347μM respectively (Figure 7E, F). Collectively, these findings support the notion that STING can associate directly with DNA species.
Figure 7. STING-DNA binding analysis. See also Figure S5.
(A) Schematic of STING variants.
(B) Coomassie brilliant blue staining of purified hSTING-HA protein from 293T cells or purified hSTING-C-6xHis (aa152-379) protein from E.coli.
(C) Purified hSTING-HA proteins were precipitated with biotin-conjugated dsDNA90 and analyzed by immunoblotting using STING antibody.
(D) hSTING-C was highly purified by N-affinity chromatography and subjected to Superdex 200 GL 10/30 size-exclusion column. Our analysis indicated that purified hSTING-C has a molecular weight of 53.7 kDa, which is consistent with that predicted size of a dimer.
(E-F) Direct binding of purified hSTING-C with 18mer (E) or 30mer (F) FAM labeled dsDNA oligonucleotide was determined using Fluorescence Polarization (FP) assays. The raw data of average fluorescence anisotropy is shown in a sigmoidal binding curve, with a calculated dissociation constant (KD) of 337 ± 26 μM and 247 ± 38 μM respectively.
Discussion
Our data indicates that STING can control the activation of the transcription factors NF-κB and IRF3/7 by cytoplasmic DNA species to initiate the production of a wide variety of primary innate immune genes that includes Type I IFN as well as members of the IFIT family and select chemokines. Studies here and elsewhere demonstrate that STING resides in the ER of the cell, almost certainly as a dimer (Ishikawa and Barber, 2008; Ishikawa et al., 2009). We have also previously provided evidence that STING forms part of the translocon complex. It is possible that endocytosed or phagocytosed viruses or even infected cell debris containing inappropriately digested viral or cellular DNA fuse with the ER where STING is located. Our data demonstrate by multiple methods that STING can complex with both ssDNA as well as dsDNA, which would include plasmid-based DNA and gene therapy vectors. Indeed, the transfection of plasmid DNA or introduction of gene therapy vectors in the cell triggers the production of a wide variety of primary innate immune genes that is dependent on STING. Our studies indicate that the association of STING with aberrant cytoplasmic DNA, induces STING trafficking through the Golgi, in perhaps pre-autophagosome complexes to associate with transcription factors responsible for activation of innate immune responses (Figure S4H) (Ishikawa et al., 2009). While the mechanisms controlling these events remain unclear, STING has been shown to traffic with autophagosome markers ATG9 after the recognition of DNA species (Saitoh et al., 2009; Watson et al., 2012). Moreover, STING appears essential for escorting TBK1 to endosomal compartments for activation of IRF3/7 (Ishikawa et al., 2009). Following delivery of TBK1, STING is degraded to presumably avoid the sustained production of innate immune related pro-inflammatory genes that could have deleterious effects upon the host (Ahn et al., 2012).
STING has been shown to play a key role in activating innate immune signaling in response to a variety of DNA pathogens including parasites and bacteria (Lippmann et al., 2011; Nazmi et al., 2012; Sharma et al., 2011). In addition, STING appears important for host defense against RNA viruses by mechanisms that remain to be clarified (Gall et al., 2012; Ishikawa and Barber, 2008; Ishikawa et al., 2009; Yan et al., 2010). For example, STING-deficient cells and mice are sensitive to RNA virus infection, but such viruses do not robustly stimulate STING trafficking or STING-dependent innate immune gene activation. Possibly, the mechanisms of STING activity against RNA viruses may involve the RIG-I pathway or other pathways. A growing number of reports indicate that STING may be commonly targeted for inactivation by a variety of pathogens, likely to thwart host defense countermeasures (Ishikawa et al., 2009; Sun et al., 2012).
STING may also be important for triggering inflammatory disease such as systemic lupus erythematosus (SLE), Aicardi-Goutieres syndrome (AGS) and/or polyarthritis (Ahn et al., 2012; Gall et al., 2012). Such disorders are characterized by the overproduction of cytokines such as type I IFN brought about by stimulation of host innate immune responses, speculatively by chronic infection or self nucleic acids from inappropriately apoptosed cells or even necrotic cells (Ahn et al., 2012; Nagata and Kawane, 2011). Indeed, our data confirms that STING is able to associate with self apoptotic or necrotic DNA suggesting a putative role for this transmembrane protein in inflammatory disease (Figure S4F, G). Loss of STING rescues lethality observed in DNase II-/- or TREX1-/- mice, which are regarded as models for self DNA-initiated inflammatory disease. Collectively, our data indicates that STING is critically important for early host defense and for initiating adaptive immune responses in response to DNA and perhaps RNA pathogens. However, chronic STING activation may lead to inflammatory disorders (Ahn et al., 2012).
Aside from our findings that STING can complex DNA, it also been reported that cyclic di-GMP produced from bacteria can bind to STING (Burdette et al., 2011; Woodward et al., 2010). Thus, STING may be a sensor not only for pathogen derived ssDNA and dsDNA, but also cyclic di-GMP. However, we did not notice as robust STING trafficking or cyclic di-GMP-induced gene induction in mammalian cells compared to that invoked by cytoplasmic DNA, for reasons that remain unclear (Figure S6). Crystal structures indicate that cyclic di-GMP binds to STING within the region aa 242-290 (Ouyang et al., 2012; Shu et al., 2012; Yin et al., 2012) (Huang et al., 2012; Shang et al., 2012). Our own data indicates that DNA similarly binds to these regions, although further crystallographic analysis will be required for a comparative analysis. We also noticed that region 291-341 was also important for STING binding and it is plausible that effective DNA association may require both of these regions for stabile interactions. We noted that while dsDNA greater than 45 bp were required for full activation of STING in the cell, smaller pieces of DNA were able to associate with purified STING in vitro. Thus, it is possible that a certain number of STING molecules may be required to associate with DNA in the cell to initiate autophagosome-like innate signaling events. Plausibly, only longer pieces of dsDNA or ssDNA may be eligible to trigger innate immunity. Post-translational modifications such as phosphorylation and ubiquitination likely regulate STING function, although we observed that TRIM56 was not required for STING to bind DNA (Figure S7) (Tsuchida et al., 2010). A number of alternate, putative DNA sensors have also been reported to regulate innate immune signaling such as IFI16 and DDX41 (Unterholzner et al., 2010; Zhang et al., 2011). However, our data indicate that STING alone is sufficient and necessary to bind DNA and facilitate cytoplasmic DNA-mediated innate immune signaling (Figure S3, 7). Our data is in agreement with Brunette et al., who also showed that STING activity is independent of IFI16 (Brunette et al., 2012). Perhaps such molecules exhibit importance in cell-types other than the ones portrayed here and the establishment of murine models that lack such genes may shed further light into their function. In summary, our data indicates that STING is essential for detecting aberrant cytoplasmic DNA species and for stimulating innate immune signaling events. However, chronic STING activation may be responsible for DNA-triggered inflammatory disease (Ahn et al., 2012). Understanding STING function may conceivably lead to the development of potent adjuvants for vaccine development or conversely therapeutics that could control inflammation aggravated disease.
Experimental Procedures
Cells and viruses
293T cells were obtained from ATCC and were maintained in DMEM (Invitrogen) supplemented with 10% FBS (GEMINI Bio-Products). hTERT-BJ1 cells were purchased from Clontech and cultured in 4:1 ratio of DMEM: Medium 199 (Invitrogen: Sigma Aldrich) supplemented with 10% FBS, 1 mM sodium pyruvate and 4 mM L-glutamine (Invitrogen). STAT1+/+ and STAT1-/- MEFs were provided by David Levy.
Antibodies
Rabbit polyclonal antibody against STING was described previously (Ishikawa and Barber, 2008). Other antibodies were obtained from the following sources: HA (Sigma Aldrich), His (Sigma Aldrich), Flag (Sigma Aldrich), β-actin (Sigma Aldrich), IRF3 (Santa Cruz Biotechnology) and p65 (Cell signaling), IFI16 (Santa Cruz Biotechnology), TRIM56 (Abcam), DDX41 (Santa Cruz Biotechnology).
RNA interference
hTERT-BJ1 cells were transfected with corresponding siRNAs (Dharmacon) using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's instruction. At 72h after siRNA transfection, cells were used for further experiments. Experiments were done in duplicate or triplicate using more than one RNAi to each gene.
Gene array analysis
Total RNA was isolated from cells using the RNeasy RNA extraction kit (Qiagen). Total RNA was analyzed by Bioanalyzer RNA 6000 Nano (Agilent Technologies). Gene array analysis was examined by Illumina Sentrix BeadChip Array (Mouse WG6 version 2 for RNA extracted from MEFs, and Human HT-12_V4_Bead Chip for RNA extracted from human cell lines) (Affymetrix) at the Oncogenomics Core Facility, University of Miami. Microarray analysis was performed at the Center of Computational Science, University of Miami.
Real time PCR
RNA was converted into cDNA using QuantiTect Reverse Transcription Kit (Qiagen), and real time PCR was performed using TaqMan® gene Expression Assay (Applied Biosystems).
Confocal Microscopy
Cell were fixed with 4% paraformaldehyde in DMEM for 15 min at 37°C and were permeabilized with 0.2% Triton X-100. Fixed and permeabilized cells were pre-incubated with 0.1%BSA in PBS and were then incubated with primary antibodies in 0.1%BSA in PBS. Cells were then incubated with fluorophore-conjugated secondary antibodies.
Virus infection
MEFs cells were seeded in 24-well plates and grown to 70% confluence. After washing with PBS, the cells were infected with HSV1 (KOS strain) or γ34.5 deleted-HSV1 at the indicated multiplicity of infection (MOI). The cells were incubated with virus for 1h at 37°C in serum-free DMEM (Invitrogen). The cells were washed with PBS twice, and complete medium was added to the cells
Biotin-dsDNA or ssDNA agarose beads
10μl of Streptavidin agarose beads (Thermo Scientific) were incubated with 0.1 nmole of biotin-conjugated dsDNA or ssDNA (Sigma Aldrich) in PBS at 4°C for 1h. After incubation, the agarose beads were washed three times and resuspended in lysis buffer.
DNA90 sequences
ssDNA-sense strand containing biotin label at the 5′ end (TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACATACAGATCTACTAGTGA TCTATGACTGATCTGTACATGATCTACA) was annealed to ssDNA90-antisense, to create dsDNA90.
Reporter analysis
293T cells were transiently transfected with 250 ng of firefly luciferase reporter plasmid together with a total of 250 ng of various expression plasmids, empty plasmids, or 125 ng of pRT-TK (internal control) using Lipofectamine2000 (Invitrogen) following the manufacturer's instruction. 24h later, the luciferase activity in the total cell lysate was measured.
In vivo DNA binding analysis
For endogenous STING pull-down assay, hTERT-BJ1 cells were transfected biotin conjugated dsDNA90 or ssDNA90 using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. After 6h, cells were crosslinked with disuccinimidyl suberate (DSS) (2 mM) (Thermo Scientific) or UV (UVC, 254nm) using a Hoefer UV Crosslinker (Hoefer) for 3 min (12 × 104 μJ/cm2). Cell lysis was performed with CHAPS buffer (10 mM CHAPS in PBS, 1 mM NaF, 1 mM Na3VO4 and protease inhibitors) for 1h at 4°C. Cell lysates were incubated with 10μl of streptavidin agarose beads (Thermo Scientific) at 4°C overnight. The complexes were washed five times with CHAPS lysis buffer. The agarose beads were boiled in SDS sample buffer. Images were obtained using Supersignal West Femto Maximum Sensitivity Substrate (Thermo Scientific). 293T cells were transfected with corresponding plasmids by calcium phosphate method. After 24h, cells were lysed in CHAPS lysis buffer (10 mM CHAPS in PBS, 1 mM NaF, 1 mM Na3VO4 and protease inhibitors) and incubated for 1h at 4°C. After centrifugation, cell lysates were precipitated with 10μl of biotin conjugated dsDNA90 agarose beads at 4°C for 3h. The agarose beads were washed five times by wash buffer (10 mM CHAPS in PBS, 300 mM NaCl, 1 mM NaF and 1 mM Na3VO4) and proteins were eluted with SDS sample buffer by boiling for 10 min. Western blot was developed by Supersignal West Pico Chemiluminescent Substrate (Thermo Scientific).
In vitro DNA binding analysis
Proteins were produced by TNT Coupled Wheat Germ Extract System (Promega). Proteins were diluted with TNE buffer (50 mM Tris pH7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and protease inhibitors). Lysates were precipitated with 10μl of biotin conjugated dsDNA90 or ssDNA agarose beads at 4°C overnight. The agarose beads were washed five times with wash buffer (50 mM Tris pH7.5, 300 mM NaCl, 1 mM EDTA, 1% NP-40) and proteins were eluted with SDS sample buffer by boiling for 10 min. Western blot analysis of pull down materials were detected using Supersignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
ChIP assay
ChIP assay was performed using Chromatin Immunoprecipitation (ChIP) Assay kit (Millipore) following the manufacturer's instructions. DNA was purified by phenol/chloroform extraction and PCR was performed using Vent DNA polymerase (NEB). The following primers were used for dsDNA90 PCR: forward, 5′-CTAAGGGTGTGGCCCTTCCGCATAGAACTGTACAGATCTACTAGTGATCT-3′; reverse, 5′-CCCTGGAAGATGGAAGCGTTTTGCAACCGCATGTAGATCATGTACAGATC-3′.
Competition assay
In vitro translation products were incubated with dsDNA90 (Sigma Aldrich), ssDNA90 (Sigma Aldrich) or Poly(dA:dT) (Sigma Aldrich) (0.01 mg/ml, 0.1 mg/ml or 1.0 mg/ml, respectively) in TNE buffer (50 mM Tris pH7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and protease inhibitors) and biotin conjugated dsDNA90 agarose beads at 4°C. After 2h, the agarose beads were washed five times with wash buffer (50 mM Tris pH7.5, 300 mM NaCl, 1 mM EDTA and 1% NP-40) and precipitated with SDS sample buffer by boiling for 10 min.
Protein purification and DNA binding assay
293T cell were transiently transfected with STING-HA or GFP-HA by calcium phosphate method. After 24h, cells were washed with PBS and lysed in TNE lysis buffer (50 mM Tris pH7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and protease inhibitors). Supernatants were precipitated by anti-HA resin (Sigma Aldrich) for overnight at 4°C and washed with TNE buffer. Resin was suspended by elution buffer (TNE lysis buffer and 100 μg/ml HA peptide (Sigma Aldrich) and protease inhibitors). For STING c-term (6×His) (aa 152-379) purification from E.coli, a single colony was inoculated in LB medium with kanamycin and grown at 37°C until OD of 0.5. IPTG (a final concentration 0.2 mM) was added and culture was incubated at 16°C overnight. Culture were pelleted and resuspended in lysis buffer (20mM Tris, pH7.5, 300mM NaCl, 10% glycerol, 5mM DTT and protease inhibitor). The suspension was lysed using French Press (AVESTIN, Inc.). After centrifugation, supernatants were added with 50mM imidazole and transferred to HisTrap HP (GE Healthcare), washed with washing buffer (lysis buffer with 50mM imidazole), and eluted in elution buffer (lysis buffer with 300 mM imidazole). For STING c-term (6×His) (aa 181-379) purification from E.coli, a single colony was inoculated in TB medium with kanamycin and grown at 37°C until OD of 0.7 and then IPTG (a final concentration 1 mM) was added. After 3h, culture were pelleted and resuspended in phosphate buffer (20 mM sodium phosphate buffer, pH8.0, 50 mM imidazole and protease inhibitor). The suspension was sonicated. After centrifugation, supernatants were transferred to HisTrap HP (17-5247-01, GE Healthcare, Piscataway, NJ), washed with phosphate buffer, and eluted in elution buffer (20 mM sodium phosphate buffer, pH8.0 and 250 mM imidazole). For purified protein pull-down, 50ng of purified proteins were diluted with 10μl of TNE buffer, and incubated with 0.1nmole of biotin conjugated dsDNA90 at 4°C for 2 hours. Protein and DNA complex were then added to 10μl of streptavidin beads prewashed in TNE buffer, and incubated at 4°C for 2 hours. The beads were washed five times with TNE buffer and proteins were eluted with SDS sample buffer by boiling for 10 min. Images were obtained using Supersignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Surface Plasmon Resonance (SPR) assay
SPR assay was performed by Affina Biotechnologies, Inc, Stamford, CT. using the BIAcore 3000 system (GE Healthcare,). The certification grade streptavidin chip (SA5) (GE Healthcare) was equilibrated with HBS-EP buffer (10 mM Hepes, 150 mM NaCl, 3 mM EDTA, 1mg/ml BSA and 0.05% surfactant P-20). Biotinylated DNA90 was immobilized at the flow rate of 30 μl/min to the level of 530 RU dsDNA90. Solutions of C-term region of STING in the HBS-EP were injected across the surface at the flow rate of 30 μl/min at 25°C. The sample injection was followed by 5-60 min dissociation phase at the same flow rate. The sensor chips were regenerated under 6M Urea and 1M NaCl in the HBS-EP buffer. BIAnalysis evaluation software (BIAEval 3.2) was used for calculation of the rate and equilibrium constants. The equilibrium binding constant, KD, was determined as the ratio of the dissociation rate constant, kdiss, and kass, the association rate constant.
Fluorescence anisotropy assay
C-term region of STING (aa 152-379) protein stock was diluted in 20 mM sodium phosphate buffer pH 8.0 to a consistent volume of 90 ul. To each protein concentration 10 ul of the 3 nM FAM-DNA (30mer: FAM-TACAGATCTACTAGTGATCTATGACTGATC; 18 mer: FAM-TACAGATCTACTAGTGAT) was added yielding a final FAM-DNA concentration of 300 pM. The fluorescence polarization was measured using a Beacon 2000 equipped with a 490 nm bandpass filter for excitation and a 535 nm filter for emission collection. The mean and standard deviation of the anistropy were calculated in Excel for each protein concentration and exported to Origin for plotting. The Kd is estimated from a nonlinear least squares fit to the function y = Amin + (Amax - Amin) × [Protein]/(Kd + [Protein]).
Statistics
Student's t-test was used to analyze data. P values of 0.05 or less were considered to donate significance.
Supplementary Material
Highlights.
Activation of cytosolic DNA signaling by STING.
Acknowledgments
We thank David Levy for Stat1-/- MEFs, David Leib for HSV-Luc and Vladlen Slepak for BIAcore analysis, Arun Malhotra for assistance with purification, Arabela Grigorescu for FP analysis, Biju Issac and Daria Salyakina for gene array analysis, and George McNamara for technical assistance.
This work was funded in part by R01AI079336 and UO1AI083015.
A.H, TA and TX carried out most of the experiments, D.G assisted in DNA binding analysis, T.X and A.M performed DNA array and STING-dependent gene analysis, H.K carried out STING dimerization and trafficking analysis, K.K purified STING. JA did HSV studies. G.N.B wrote the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnemri ES. Sensing cytoplasmic danger signals by the inflammasome. J Clin Immunol. 2010;30:512–519. doi: 10.1007/s10875-010-9419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity Initiates in Nonhematopoietic Cells and Progresses via Lymphocytes in an Interferon-Dependent Autoimmune Disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012;8:e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Lippmann J, Muller HC, Naujoks J, Tabeling C, Shin S, Witzenrath M, Hellwig K, Kirschning CJ, Taylor GA, Barchet W, et al. Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell Microbiol. 2011;13:1668–1682. doi: 10.1111/j.1462-5822.2011.01646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Kawane K. Autoinflammation by endogenous DNA. Advances in immunology. 2011;110:139–161. doi: 10.1016/B978-0-12-387663-8.00004-1. [DOI] [PubMed] [Google Scholar]
- Nazmi A, Mukhopadhyay R, Dutta K, Basu A. STING Mediates Neuronal Innate Immune Response Following Japanese Encephalitis Virus Infection. Sci Rep. 2012;2:347. doi: 10.1038/srep00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Medzhitov R. The control of adaptive immune responses by the innate immune system. Advances in immunology. 2011;109:87–124. doi: 10.1016/B978-0-12-387664-5.00003-0. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, Gu L. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, Chan J, Bartholomeu DC, Lauw F, Hall JP, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, Clementz MA, Banach BS, Li K, Baker SC, Chen Z. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA Targets Bacteria for Autophagy by Activating the Host DNA-Sensing Pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nature immunology. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Molecular cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.