SUMMARY
Activation of the STING (Stimulator of Interferon Genes) pathway by microbial or self-DNA, as well as cyclic di nucleotides (CDN), results in the induction of numerous genes that suppress pathogen replication and facilitate adaptive immunity. However, sustained gene transcription is rigidly prevented to avoid lethal STING-dependent pro-inflammatory disease by mechanisms that remain unknown. We demonstrate here that after autophagy-dependent STING delivery of TBK1 (TANK-binding kinase 1) to endosomal/lysosomal compartments and activation of transcription factors IRF3 (interferon regulatory factors 3) and NF-κB (nuclear factor kappa beta), that STING is subsequently phosphorylated by serine/threonine UNC-51-like kinase (ULK1/ATG1) and IRF3 function is suppressed. ULK1 activation occurred following disassociation from its repressor adenine monophosphate activated protein kinase (AMPK), and was elicited by CDN’s generated by the cGAMP synthase, cGAS. Thus, while CDN’s may initially facilitate STING function, they subsequently trigger negative-feedback control of STING activity, thus preventing the persistent transcription of innate immune genes.
INTRODUCTION
Host cells have evolved a variety of mechanisms to recognize and eliminate invading microbes, including developing the ability to recognize pathogen associated proteins and nucleic acid and subsequently invoke powerful cellular signaling events that stimulate the production of innate immune genes (Blasius and Beutler, 2010; Kawai and Akira, 2011; Tamura et al., 2008). Such defenses include the Toll-like receptors (TLR), RIG-I (RLR) family of receptors and nucleotide-binding domain and leucine-rich repeat-containing (NLR) receptors that sense microbial molecules such as CpG DNA, viral RNA’s and lipopolysaccharides (Blasius and Beutler, 2010; Kawai and Akira, 2011; Tamura et al., 2008). In addition, an endoplasmic reticulum (ER) associated molecule referred to as STING (for stimulator of interferon genes) has recently been shown to control a new sensing pathway which is essential for detecting aberrant cytosolic DNA species and for triggering the production of host defense genes such as type I interferon (IFN) (Ishikawa and Barber, 2008; Ishikawa et al., 2009).
The activation of STING (also referred to as TMEM 173/ MPYS/MITA/ERIS) may involve direct association with cytosolic DNA species as well as with cyclic di nucleotides (cyclic di guanosine monophosphate or adenosine monophosphate; cyclic di GMP or AMP) generated directly from certain intracellular bacteria or via a DNA binding protein cGAS (cGAMP synthase, also known as male abnormal 21 domain containing 1 [Mab-21 Domain Containing1/MBD21D] or C6orf150) (Burdette et al., 2011; Diner et al., 2013; Jin et al., 2008; Sun et al., 2013; Sun et al., 2009; Woodward et al., 2010; Zhong et al., 2008). However, following the detection of cytosolic DNA, cGAS utilizes GTP and ATP to generate non-canonical 2’–3’- cyclic GMP-AMP (cGAMP), rather than 3’–5’- canonical cyclic di nucleotide species generally generated by bacteria (Ablasser et al., 2013; Civril et al., 2013; Gao et al., 2013; Kranzusch et al., 2013; Zhang et al., 2013). Activated STING, accompanied by TANK-binding kinase 1 (TBK1) then undergoes dramatic autophagy-related trafficking involving ATG9 and associates with endosomes containing the transcription factors IRF3 (interferon regulatory factors 3) and NF-κB (nuclear factor-kappa B) (Ishikawa et al., 2009; Saitoh et al., 2009). Phosphorylated IRF3 and activated NF-κB translocate to the nucleus to initiate the transcription of numerous innate immune genes, including IFN and members of the IFIT family (Abe et al., 2013).
However, while STING has been shown to be essential for the protection of the host against DNA pathogens, sustained STING stimulation, such as by self DNA has also been shown to be responsible for lethal inflammatory disease in at least two murine models (DNaseII−/− and DNaseIII/TREX1−/−) and plausibly may therefore play a key role in inflammatory/autoimmune disease in humans (Ahn et al., 2012; Gall et al., 2012). Thus, while STING is essential for initiating host defense counter measures, chronic STING activity needs to be controlled to avoid the deleterious consequences that sustained innate immune gene induction would have upon the host. Here, we demonstrate that after activation and trafficking, STING is phosphorylated by UNC-51-like kinase (ULK1). This occurs following ULK1 dissociation from its repressor adenine monophosphate activated protein kinase (AMPK) and was found to be triggered by cGAS generated CDN’s. Therefore, while CDN’s may initially facilitate STING activity, they also initiate a negative-feedback control mechanism, to thwart prolonged innate immune gene transcription, and prevent inflammatory disorders.
RESULTS
Phosphorylation of S366 Inhibits STING Function
Previously, we observed that STING activation by DNA invoked trafficking that resembled autophagy and resulted in the delivery of STING/TBK1 to endosomal/lysosomal regions to activate the transcription factors NF-κB and IRF3/7 (Ishikawa et al., 2009; Saitoh et al., 2009). As an extension of these findings, we have now determined that these events lead to an increase in the molecular weight of STING and subsequently to the degradation of STING within 12 hours (Figure 1A and 1B). The observed shift in molecular weight was likely due to phosphorylation since phosphatase treatment eliminated this modification (Figure S1A and S1B). Phosphorylation likely occurred after trafficking from the ER through the Golgi since Brefeldin A (BFA), which inhibited STING movement, also prevented the phosphorylation event (Figure 1C and 1D, Figure S1C). Degradation of STING occurred in the lysosomal compartment since chloroquine, which inhibits lysosomal degradation, prevented STING degradation (Figure S1D). To further evaluate the autophagy process, we suppressed VPS34 (Class III phosphatylinositol 3-kinase, PI3K), Beclin-1, or the serine/threonine protein kinase ULK1 (ATG1) expression which are considered important components required for the autophagy process in response to nutrient depravation (Tooze et al., 2010). We noted that RNAi suppression of VPS34 but not ULK1 or Beclin-1 inhibited STING trafficking indicating ULK1-independent processes reminiscent of non-canonical autophagy reported for TLR9 activation in response to phagocytosed DNA associated with autoantibodies (Figure 1E and 1F, Figure S1E) (Henault et al., 2012). Accordingly, suppression of VPS34, but not ULK1 inhibited the ability of dsDNA to activate STING-dependent type I IFN induction (Figure 1F and 1G). Indeed, we observed that suppression of ULK1 appeared to augment the production of type I IFN. These observations indicate that STING recognition of dsDNA triggers a transient non-conventional autophagy-related process required for IRF3/7 and NF-κB activation.
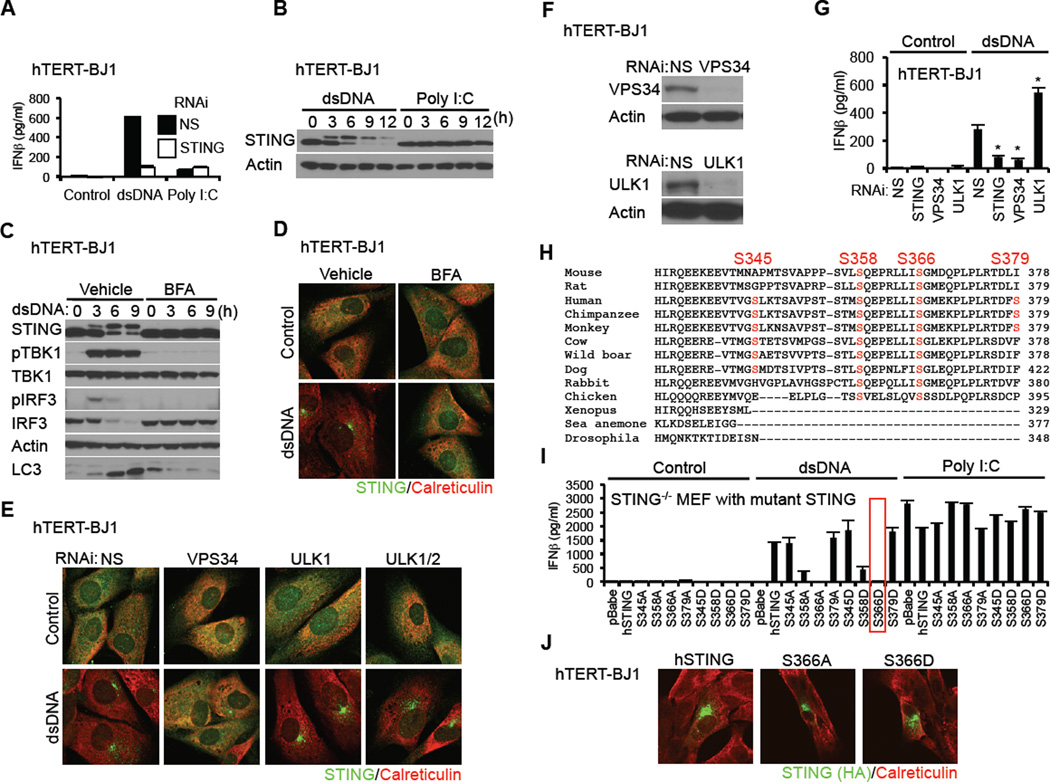
Figure 1. Phosphorylation of STING S366 Inhibits Type I IFN Production in dsDNA Signaling.
(A) hTERT-BJ1 cells were transfected with dsDNA (4 µg/ml) or poly I:C (4 µg/ml) using lipofectamine 2000 for 16 hr. IFNβ level was measured by ELISA.
(B) hTERT-BJ1 cells were transfected with dsDNA or poly I:C as described in Figure 1A for the indicated times and STING was detected by immunoblot.
(C and D) hTERT-BJ1 cells were incubated with ethanol (vehicle) or BFA (0.05 µg/ml) for 1 hr and then transfected with dsDNA (4 µg/ml) for the indicated times (C) or 6 hr (D). Immunoblot (C) or immunostaining (D) was performed with the indicated antibodies; lack of LC3 processing (bottom band) indicated loss of autophagy.
(E) hTERT-BJ1 cells were treated with siRNA (NS: Non-specific siRNA) for 3 days and then transfected with dsDNA (4 µg/ml) for 6 hr. Immunostaining was performed with the indicated antibodies.
(F) Immunoblot was performed to confirm knockdown efficiency of ULK1 and VPS34.
(G) siRNA-treated hTERT-BJ1 cells were transfected with dsDNA (4 µg/ml) for 16 hr and IFNβ was measured by ELISA.
(H) Alignment of STING amino acid sequences. Highlighted amino acids indicate dsDNA-dependent phosphorylated serines, as detected by mass spectrometry.
(I) Primary STING−/− MEF cells were reconstituted with STING variants using retroviruses. The cells were transfected with dsDNA (4 µg/ml) or poly I:C (4 µg/ml) for 16 hr and IFNβ measured by ELISA.
(J) hTERT-BJ1 cells were transfected with HA-tagged mutant hSTING for 36 hr and immuno-stained with the indicated antibodies.
Asterisks indicate significant difference (P < 0.05) compared to NS determined by Student’s t-test. Error bars indicate sd. See also Figure S1.
However, after autophagy, little is known relating to the identity of kinases involved in STING phosphorylation and regulation. To start to evaluate the role of phosphorylation in STING function, we purified STING from dsDNA-treated and untreated hTERT-BJ1 cells using affinity purification processes. Non-active STING or phosphorylated STING was examined by mass spectrometry. These experiments indicated that STING purified only from dsDNA-treated cells was phosphorylated in vivo, on 4 sites (S345, S358, S366, S379) (Figure 1H, Figure S1F and S1G). STING phosphorylation was not observed in resting, untreated cells (data not shown). Of the phosphorylation sites, S358 and S366 appeared to be highly conserved in mammalian cells (Figure 1H). To evaluate the role of these residues in STING function, each serine was individually or collectively substituted for alanine or aspartic acid, and transduced by retrovirus into STING−/− MEFs prior to treatment with dsDNA. This analysis confirmed that S366A had greatly reduced ability to induce type I IFN (Figure 1I, Figure S2A and S2B) (Tanaka and Chen, 2012). However, we additionally observed that substitution of S366 to S366D, to mimic constitutive phosphorylation also rendered STING unable to stimulate the production of type I IFN (Figure 1I). Substitution of the other serines to aspartic acid did not appreciably affect STING function accept partially in the case of S358D. We further noted that S366A or S366D was able to traffic normally in response to cytosolic dsDNA, suggesting that phosphorylation of S366 does not effect STING trafficking, and likely occurs post-Golgi trafficking (Figure 1J). We conclude that phosphorylation of S366 (S365 in murine STING) occurs after autophagy and may negatively regulate STING activity to prevent sustained type I IFN production following activation by cytosolic dsDNA.
The IRF3 but not NF-κB Pathway is Preferentially Inhibited by STING S366D
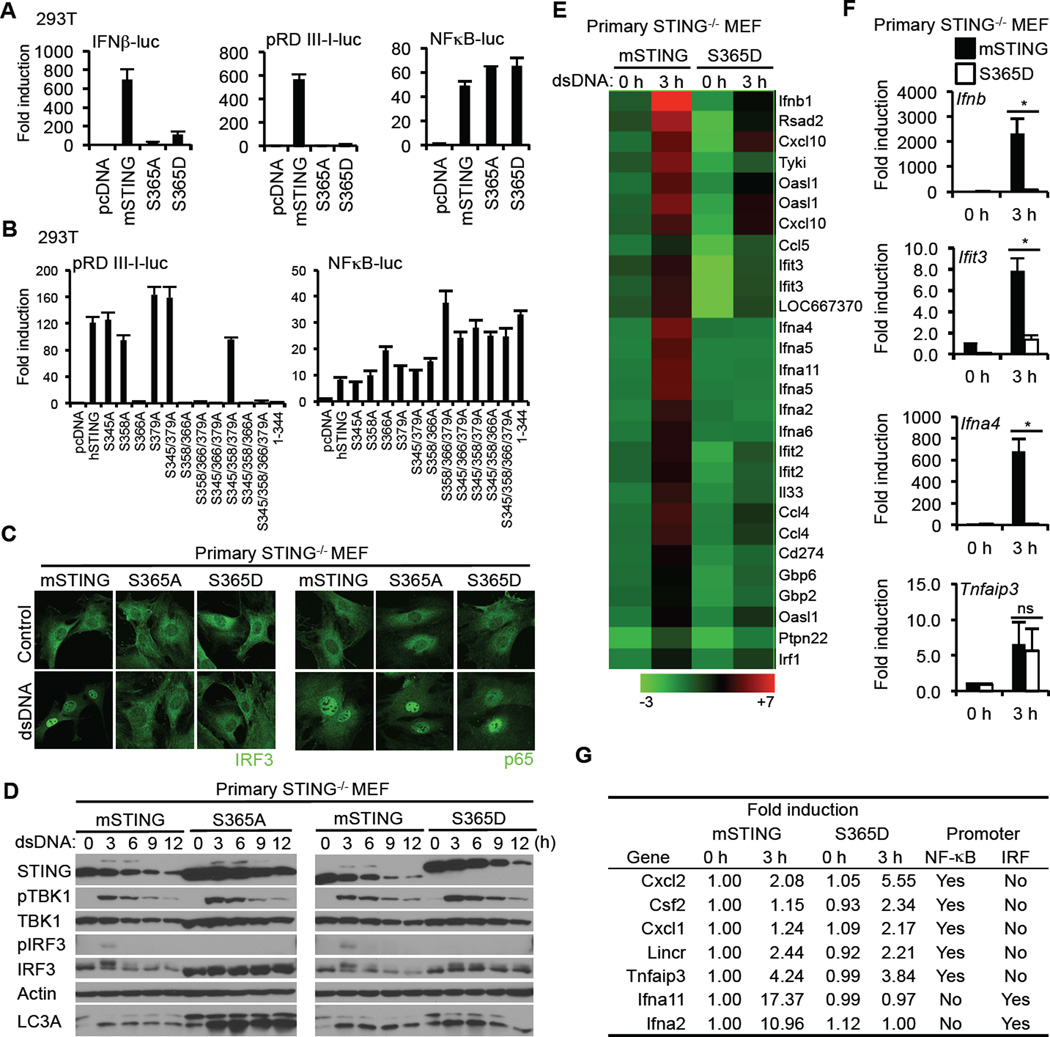
Type I IFN production requires the coordinated activation of a number of transcription factors, such as NF-κB as well as the IRF3 (Tamura et al., 2008). To evaluate whether both the IRF3 as well as the NF-κB pathways were affected by phosphorylation of S366, wild type murine STING, STING-S365A or STING-S365D were transfected into 293T cells with luciferase reporter constructs under control of IFNβ, NF-κB or IRF3 (pRD III-I) responsive promoter elements. This study indicated that phosphomimetic STING-S365D failed to activate the IFNβ promoter and IRF3 promoter but not the NF-κB promoter (Figure 2A). Similarly, only STING variants with an S366 substitution failed to activate the pRD III-I promoter, but not the NF-κB promoter, indicating the importance of S366 in IRF3 stimulation (Figure 2B). The mutational change appeared to be S366 specific since residues substituted either side of S366 (I365A and G367A) did not influence STING function (Figure S2C). Accordingly, immunohistochemical analysis indicated the translocation of the p65 subunit of NF-κB, but not IRF3, into the nucleus of STING−/− MEF cells when reconstituted with S365D and treated with dsDNA (Figure 2C). STING is required to transport TBK1 to endosomal regions for association with IRF3 (Ishikawa et al., 2009). Immunoblot analysis of reconstituted STING−/− MEF cells treated with dsDNA demonstrated TBK1 phosphorylation but not phosphorylation of IRF3 (Figure 2D). Immunoblot analysis also indicated that murine S365D electrophoresed at a similar size to that of naturally phosphorylated STING, suggesting that the change in molecular weight occurring as a result of dsDNA activation was partly related to S365 phosphorylation (Figure 2D). Thus, S365 phosphorylation does not appear to affect TBK1 activity, but rather IRF3 phosphorylation. To extend this analysis further, we stably reconstituted STING−/− MEFs with wild type STING or STING S365D and treated the cells with dsDNA before carrying out a microarray analysis. This analysis confirmed significant suppression of type I IFN production as well as other cytosolic DNA mediated, STING-dependent genes including members of the IFIT family (Figure 2E and 2F). However, the expression of a number genes such as TNFAIP3 remained unaffected by phosphorylation of STING on S366 in the presence of cytosolic dsDNA, presumably since they only require NF-κB for transcriptional activation and not both IRF3/7 and NF-κB as does the transcription of type I IFN (Figure 2G). Compared to the S365D variant, STING S365A was considerably inactive, suggesting that the former does not simply function as a phosphomutant (Figure S2D). Our data confirms that phosphorylation of STING on S366 strongly prevents the transcriptional activity of IRF3.
Figure 2. Phosphorylation of S366 of STING Inhibits IRF3 but not NF-κB Activity.
(A and B) HEK293T cells were transfected with plasmids encoding the luciferase gene under control of the indicated promoter with STING variants. After 36 hr, luciferase activity was measured.
(C) Primary STING−/− MEF cells were reconstituted with mSTING variants using retroviruses. The cells were transfected with dsDNA (4 µg/ml) for 3 hr and then stained with the indicated antibodies.
(D) Reconstituted STING−/− MEF cells were transfected with dsDNA (4 µg/ml) for the indicated times and immunoblots performed.
(E) Reconstituted STING−/− MEF cells were transfected with dsDNA (4 µg/ml) for 3 hr. RNA was purified and examined for gene expression with Illumina Sentrix BeadChip Array (Mouse WG6 version2). Pseudo colors indicate transcript levels below, equal to, or above the mean (green, black, and red, respectively). The scale represents the intensity of gene expression (log10 scale). The results shown here are representative of three independent experiments.
(F) Realtime PCR was carried out with the indicated probes to confirm gene array analysis shown in Figure 2E. Total RNA was extracted from reconstituted STING−/− MEF cells after dsDNA treatment for 3 hr and then cDNA was synthesized.
(G) The expression of NF-κB target genes in STING−/− MEF with S365D was comparable with that in STING−/− MEF with mSTING. Promoter sequence of listed genes (−1000 to +200) was obtained through DBTSS (http://dbtss.hgc.jp) and analyzed by TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) at threshold score 85. The results shown here are the average of three independent experiments.
Asterisks indicate significant difference (P < 0.05) determined by Student’s t-test. ns means not significant. Error bars indicate sd. See also Figure S2.
ULK1 and 2 are Responsible for Phosphorylating STING on S366
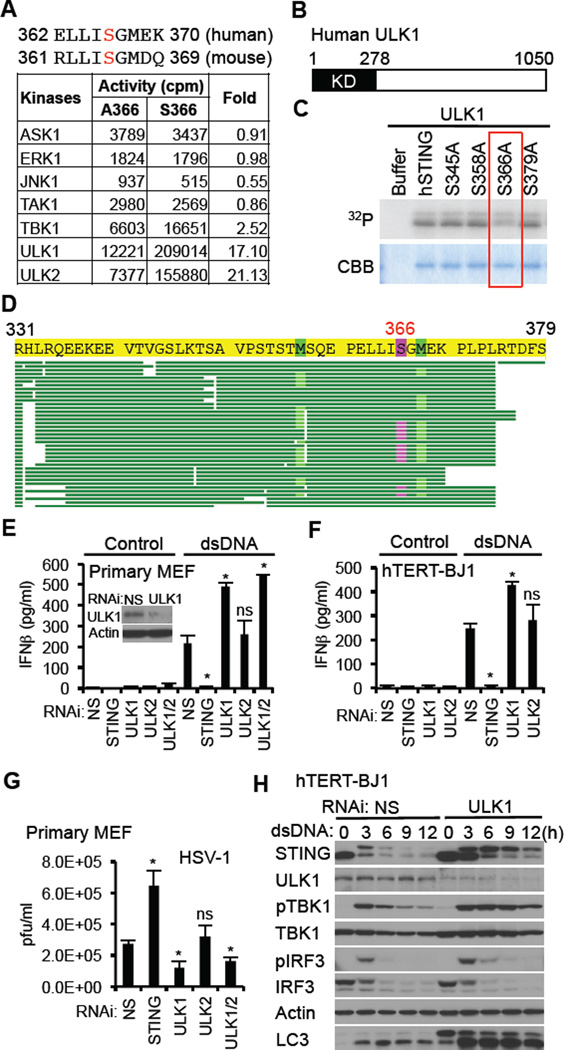
To attempt to identify the kinase responsible for phosphorylation of S366, we utilized a high throughput approach and analyzed the ability of 272 purified serine/threonine kinases to in vitro phosphorylate a 9 amino acid peptide containing this residue (ELLISGMEK) (Figure 3A). We found that ULK1 and ULK2, key serine/threonine kinase involved in autophagy regulation phosphorylated S366 to high specificity compared to all other kinases examined (17.1 and 21.13-fold respectively) (Mizushima, 2010). To confirm this finding, we carried out an in vitro kinase assay using purified STING and ULK1 and examined STING-related phosphorylation events by autoradiography following SDS-PAGE electrophoresis. Phosphorylation of STING was reduced using a STING S336A variant (Figure 3B and 3C). To additionally verify this, we isolated ULK1 phosphorylated STING from SDS-PAGE gels and determined the phosphorylation site(s) by mass spectrometry. This approach confirmed phosphorylation of STING by ULK1 and additionally demonstrated that this event only occurred on S366 and on no other serine or threonine residues (Figure 3D). As a control, we similarly examined the ability of TBK1 to posttranslationally modify STING and did not observe any phosphorylation events, indicating that STING is likely not a substrate for TBK1, at least in vitro (Figure S3A–C). Collectively, these data indicate that ULK1/2 can phosphorylate STING on S366.
Figure 3. ULK1 Negatively Regulates IFNβ Production by Phosphorylating STING S366.
(A) ELLISGMEK which includes Ser366 was used as substrate to identify ULK1 and ULK2.
(B) Schematic of human ULK1. KD is kinase domain.
(C) In vitro kinase assay was performed with recombinant ULK1 protein using recombinant hSTING protein as substrate. The indicated serine sites that were identified by mass spectrometry were substituted with alanine.
(D) Recombinant hSTING protein was incubated with recombinant ULK1 in presence of ATP for 15 min. Phosphorylated STING was analyzed by mass spectrometry. Highlighted amino acid (S366) was identified as the only phosphorylation site.
(E and F) Primary MEF cells (E) or hTERT-BJ1 cells (F) were treated with siRNA as indicated (NS: nonspecific siRNA) and then transfected with dsDNA (4 µg/ml) for 16 hr. IFNβ was measured by ELISA. Knockdown efficiency of ULK1 in primary MEF cells was confirmed by immunoblot.
(G) siRNA-treated primary MEF cells were infected with HSV-1 (MOI = 0.1) for 24 h and then plaque assay was performed.
(H) siRNA-treated hTERT-BJ1 cells were transfected with dsDNA (4 µg/ml) for the indicated times and then immunoblot was performed.
Asterisks indicate significant difference (P < 0.05) compared to NS determined by Student’s t-test. ns means not significant. Error bars indicate sd. See also Figure S3.
To evaluate these findings further, we RNAi suppressed ULK1, ULK2 or both in MEF cells treated with dsDNA and measured type I IFN production. We found using MEF cells that suppression of STING expression significantly abrogated IFN production in response to dsDNA signaling, and confirmed that ULK1 suppression lead to elevated levels of type I IFN production presumably since IRF3 function is maintained (Figure 3E). Similar observations were seen when suppressing ULK1 expression in hTERT-BJ1 cells (Figure 3F). Suppression of ULK2 had less effect, probably since we did not observe significant ULK2 expression, at least in hTERT-BJ1 or MEF cells (Figure S3D and S3E). ULK1 and 2 were also observed to be robustly expressed in primary human dendritic cells (Figure S3F). Suppression of ULK1 expression in MEF cells was also seen to inhibit herpes simplex virus type I (HSV1) replication, likely due to elevated type I IFN exerting anti-viral effects (Figure 3G, Figure S3G). Immunoblot analysis of ULK1 specific RNAi treated hTERT-BJ1 cells indicated that STING was not degraded as efficiently as in control cells, suggesting that phosphorylation on this site may also play a role in facilitating STING degradation (Figure 3H). STING also shifted in size due to alternate phosphorylation events, plausibly involving the other three serine sites that we discovered by proteomic approaches (Figure 1H). Slightly sustained TBK1 and IRF3 phosphorylation was also observed in the absence of ULK1 perhaps indicating that the presence of STING facilitates TBK1 stability. As shown earlier, loss of ULK1 did not affect STING-dependentautophagy, as additionally emphasized by evident LC3-II conversion (Figure 1C and 3H) Our data indicates that ULK1 and perhaps ULK2, depending on whether a particular cell expresses ULK2, is able to phosphorylate STING on S366, to perhaps facilitate STING degradation as well as prevent IRF3 translocation.
AMPK and not mTOR is the Master Regulator of ULK1/2 Activity
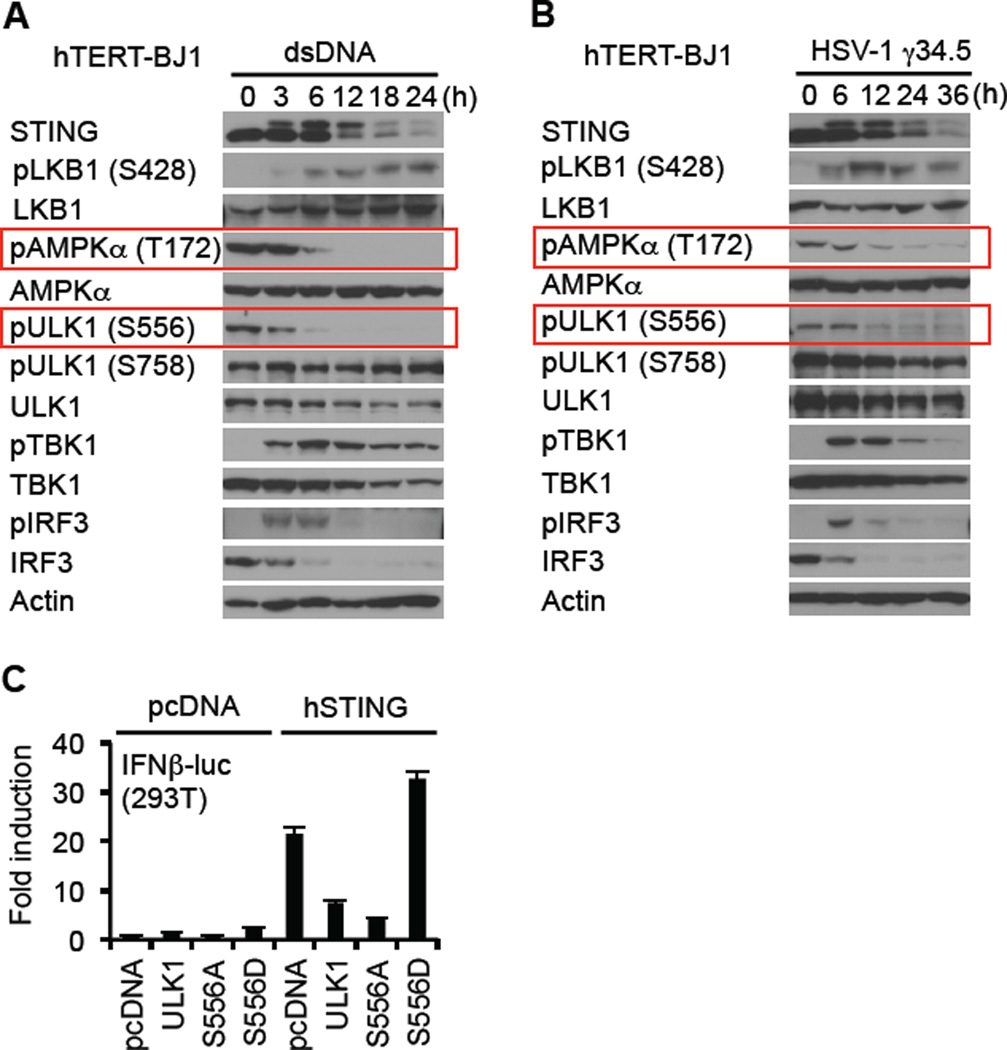
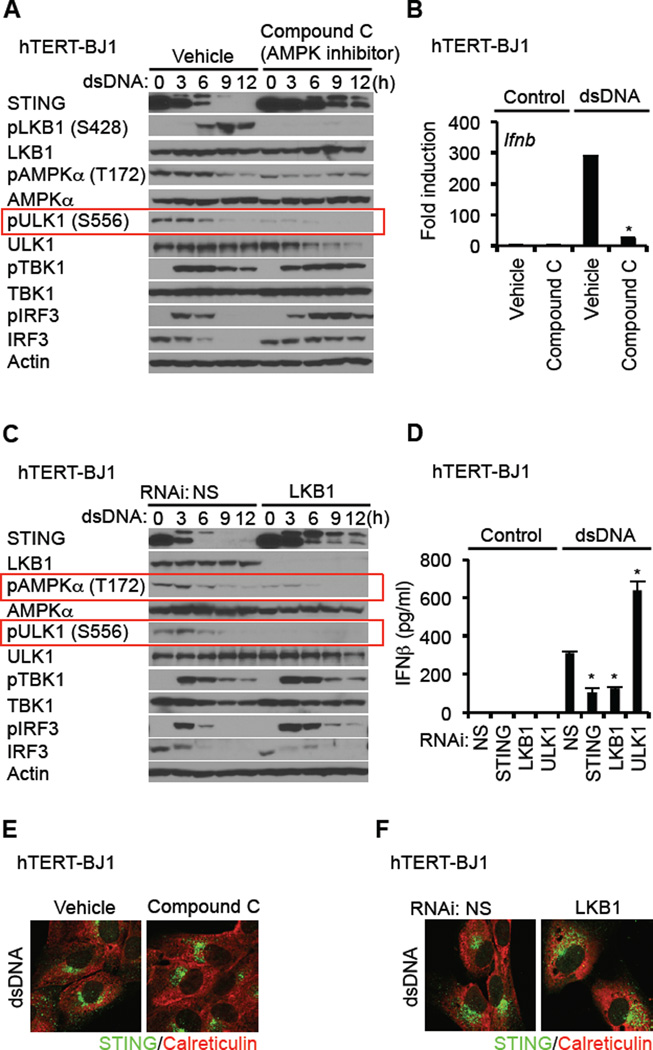
The mechanisms whereby ULK1 is regulated are complex, though involve control by the 5’ adenosine monophosphate [AMP]-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) (Alers et al., 2012). AMPK is a heterotrimeric protein and key energy sensing kinase that is activated by environmental stress such as Ca2+ efflux or ATP consumption. Under non stressful conditions, AMPK T172 is maintained in a phosphorylated state by upstream AMPK kinases such as LKB1 and Ca2+ - calmodulin dependent protein kinase-kinase-beta (CaMKK beta) (Alexander and Walker, 2011). Constitutively phosphorylated AMPK in turn phosphorylates ULK1 on S556, which maintains ULK1 in an inactive state. However, additional post-translational modifications of AMPK following cell stress, releases it from association with ULK1, with concomitant loss of ULK1 S556 phosphorylation (Alers et al., 2012). This enables ULK1 to phosphorylate target proteins. Similarly, mTOR, which can also be activated by amino acid depravation likewise regulates ULK1 activity through phosphorylation, but targets S758 (Mizushima, 2010). Given this, we examined the control of ULK1 phosphorylation by these two pathways. When treated with dsDNA, we noted that phosphorylation of ULK1 S758 was relatively unaffected following dsDNA treatment (Figure 4A). However, we observed that phosphorylation of AMPKα T172 was dramatically reduced in the presence of cytosolic DNA which corresponded to a reduction of S556 on ULK1 (Figure 4A). The effect induced by dsDNA was also seen following infection with HSV1 (Figure 4B). In contrast, vesicular stomatitis virus (VSV) did not induce dephosphorylation of AMPK T172 or ULK1 S556 (Figure S4A). Constitutively active ULK1 (S556D) was observed to enhance the ability of STING to stimulate transcription of the IFN promoter, unlike ULK1 S556A, presumably since ULK1 S556D is maintained in an inactive, phophorylated state, which fails to impede IRF3 function (Figure 4C). The kinase activity of ULK1 was required for the regulation of STING. (Figure S4B). Inhibition of AMPK by Compound C confirmed that ULK1 S556 phosphorylation was an AMPK-dependent event since this process was completely negated by this drug (Figure 5A). Accordingly, compound C thus suppressed dsDNA-dependent but not significantly dsRNA-dependent type I IFN induction (Figure 5A and B, Figure S5A). We observed that AMPK inhibition facilitated STING stability, similar to loss of ULK1, again indicating that phosphorylation of STING effects its degradation (Figure 5A). Moreover, levels of IRF3 were maintained and did not undergo rapid degradation presumably since phosphorylated IRF3 did not translocate and was not efficiently degraded (Figure 5A, Figure S5B). Conversely, activation of AMPK was also seen to modestly enhanced type I IFN production (Figure S6D).
Figure 4. dsDNA Induces the Dephosphorylation of AMPK T172 and ULK1 S556.
(A and B) hTERT-BJ1 cells were transfected with dsDNA (4 µg/ml) (A) or infected with HSV-1 γ34.5 (MOI = 10) (B) for the indicated times and then immunoblot carried out with the indicated antibodies.
(C) HEK293T cells were co-transfected with plasmids encoding luciferase driven by the IFNβ promoter as well as hSTING, and ULK1 variants. After 36 hr, luciferase activity was measured. See also Figure S4.
Figure 5. LKB1 Phosphorylation of AMPK is Disrupted by Cytosolic dsDNA.
(A) hTERT-BJ1 cells were treated with DMSO (vehicle) or compound C (10 µM) for 1 hr prior to dsDNA transfection (4 µg/ml) for the indicated times. Immunoblot was performed with the indicated antibodies.
(B) hTERT-BJ1 cells were treated with DMSO or compound C as described in Figure 5A and then transfected with dsDNA (4 µg/ml) for 6 hr. cDNA was synthesized from total RNA and then realtime PCR was performed with a probe for Ifnb.
(C) hTERT-BJ1 cells were treated with siRNA for NS (Non-specific siRNA) or LKB1 and then transfected with dsDNA (4 µg/ml) for the indicated times. Immunoblot was performed with the indicated antibodies.
(D) siRNA-treated hTERT-BJ1 cells were transfected with dsDNA (4 µg/ml) for 16 hr. IFNβ was measured by ELISA.
(E and F) hTERT-BJ1 cells were treated with compound C as described in Figure 5A (E) or siRNA for LKB1 as described in Figure 5C (F). Confocal analysis of STING trafficking in response to dsDNA (4 µg/ml) was carried out using anti-STING antibody.
Asterisks indicate significant difference (P < 0.05) compared to vehicle (B) or NS (D) determined by Student’s t-test. Error bars indicate sd. See also Figure S5.
Given these findings, we next evaluated the roles of the AMPK kinases, CaMKK beta and LKB1 in cytosolic dsDNA signaling. We observed that suppression of CaMKK beta somewhat reduced the phosphorylation status of AMPK T172, but ULK1 S556 phosphorylation was maintained under resting conditions and underwent de-phosphorylation in the presence of cytosolic DNA suggesting that a kinase other than CaMKK beta may be predominantly involved in the regulation of AMPK T172 (Figure S5D and S5E). In addition, we noted that cytosolic DNA also induced the phosphorylation of LKB1 on S428, and that this was also dependent on AMPK (Figure 5A, Figure S5C). To further investigate the role of LKB1 in AMPK/ULK1 activity, we used RNAi to suppress LKB1 expression. Loss of LKB1 completely eliminated ULK1 S556 phosphorylation under resting conditions indicating that LKB1 was largely responsible for maintaining AMPK T172 phosphorylation and for limiting ULK1 activity (Figure 5C). Loss of LKB1 was also observed to inhibit dsDNA-dependent type I IFN induction, similar to compound C (Figure 5D). These events were predominantly dsDNA specific, and did not occur using lipofectamine alone (Figure S5F). It was also observed that loss of LKB1 or AMPK activity did not affect STING autophagy, suggesting that AMPK control of ULK1 occurs in autophagosome/endosomal regions after STING trafficking (Figure 5E and 5F). Our data indicates that LKB1 predominantly maintains phosphorylation of AMPK on T172, an event that is disrupted by the presence of cytosolic DNA leading to ULK1 activation and STING phosphorylation.
Cyclic Di Nucleotides (cGAMP) Regulate AMPK
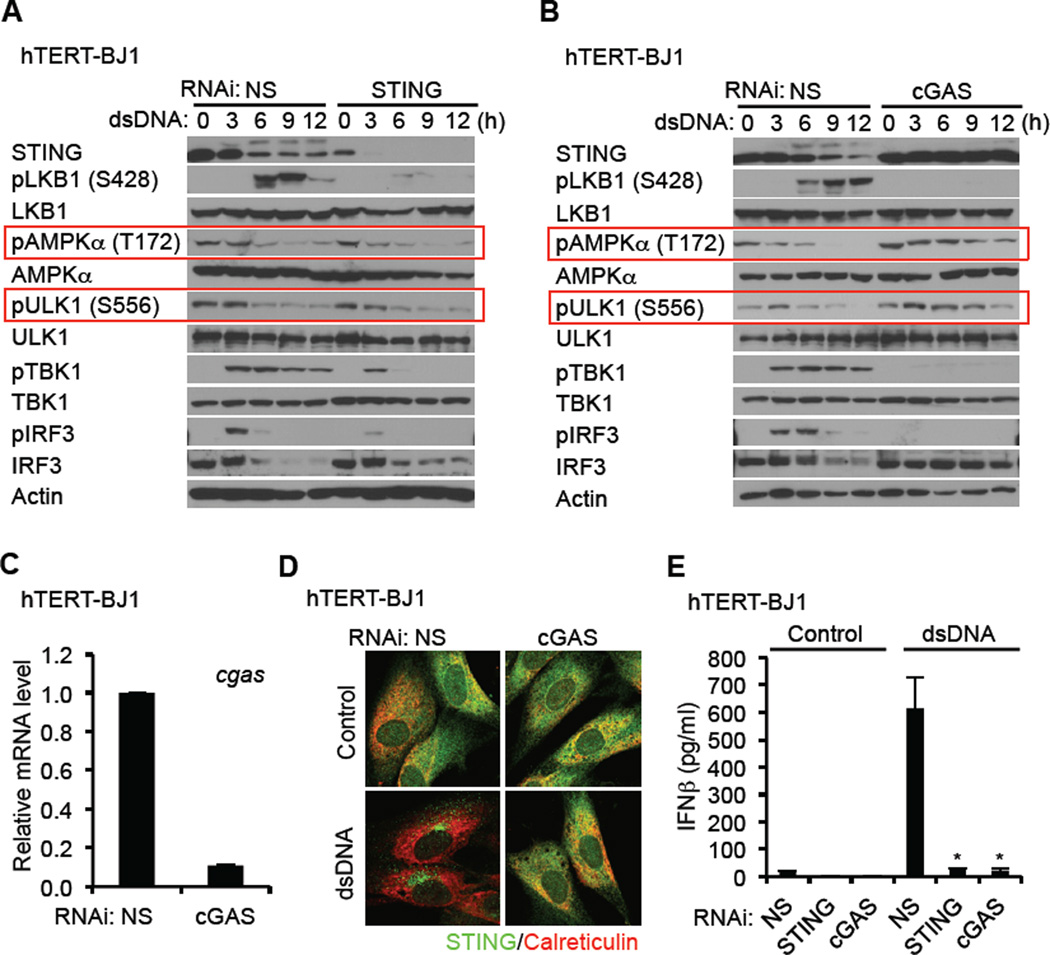
The activation of AMPK has been reported to be dependent on nutrient depravation, fluctuations in calcium levels and/or increases in AMP/ATP ratios. However, we did not notice significant fluctuations in the levels of these regulators (data not shown). We also observed that loss of STING in hTERT-BJ1 cells did not affect the phosphorylation of AMPK on T172 or ULK1 on S556, indicating that the control of ULK1 phosphorylation by cytosolic DNA was a STING independent event (Figure 6A). However, loss of STING impeded cytosolic DNA-mediated autophagy and prevented LKB1 phosphorylation by AMPK on S428. BFA also prevented LKB1 phosphorylation by AMPK by similarly impeding autophagy related trafficking (Figure S6A). We confirmed that while STING was required for cytosolic DNA-mediated autophagy, it was not necessary for rapamycin-dependent autophagy (Figure S6B).
Figure 6. cGAS is Required for the Negative Regulation of STING through the AMPK-ULK1 Axis.
(A and B) hTERT-BJ1 cells were treated with siRNA for NS (Non-specific), STING (A), and cGAS (B) for three days and then transfected with dsDNA (4 µg/ml) for the indicated times. Immunoblot was performed with the indicated antibodies.
(C) Knock down efficiency of cGAS in hTERT-BJ1 cells were confirmed by realtime PCR.
(D) siRNA-treated hTERT-BJ1 cells were transfected with dsDNA for 6 hr as described in Figure 6A. STING trafficking was observed by confocal microscopy.
(E) siRNA-treated hTERT-BJ1 cells were transfected with dsDNA (4 µg/ml) for 16 hr. IFNβ was measured by ELISA.
Asterisks indicate significant difference (P < 0.05) compared to NS determined by Student’s t-test. Error bars indicate sd. See also Figure S6.
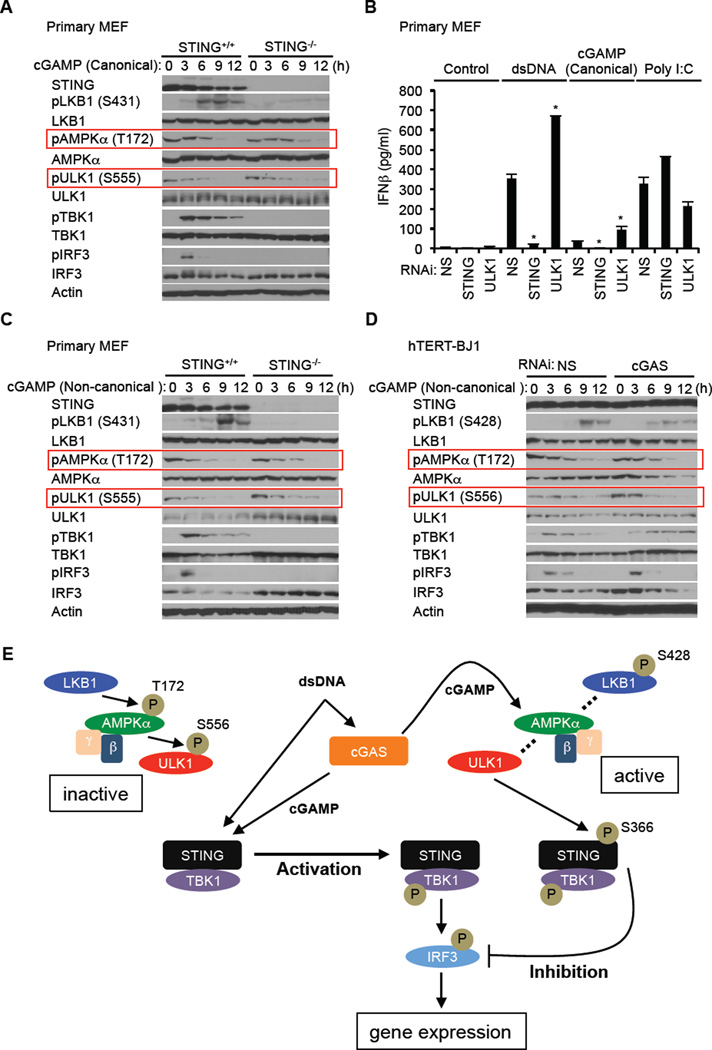
To further understand the triggering of AMPK/ULK1 activity, we used RNAi to suppress the synthase cGAS, which has been reported to associate with cytoplasmic DNA and to generate cyclic di nucleotides which are capable of binding to and facilitating STING function. Surprisingly, we observed that loss of cGAS prevented AMPK T172 dephosphorylation and subsequently ULK1 S566 dephosphorylation (Figure 6B and 6C). STING-dependent autophagy was also prevented by loss of cGAS in hTERT-BJ1 cells, in response to dsDNA (Figure 6D and 6E). These results suggested a role for cGAS and the generation of cyclic di nucleotides in the regulation of AMPK activity. To extend our studies, we next examined whether cyclic di nucleotides (cGAMP) could activate the AMPK/ULK1 pathway in MEF cells. This analysis confirmed that canonical and non-canonical cGAMP alone could trigger the dephosphorylation of AMPK and ULK1 on T172 and S555 respectively (Figure 7A–C). This event did not require STING as previously shown (Figure 6A and 7A). Similar to our observations with dsDNA, loss of ULK1 enhanced cGAMPs ability to induce type I IFN. However, we notice that cyclic dinucleotides are not as robust activators of STING dependent innate immune signaling when compared to dsDNA (Figure 7B). We next treated hTERT-BJ1 cells with non-canonical cGAMP to evaluate whether these di nucleotides could similarly activate the AMPK/ULK1 regulatory pathway. In hTERT-BJ1 cells, the amino acid sequence of STING is H232 which cannot bind readily to canonical cyclic di nucleotides such as c-di-AMP, c-di-GMP or cGAMP (Ablasser et al., 2013; Civril et al., 2013; Gao et al., 2013; Kranzusch et al., 2013; Zhang et al., 2013). In most other types of human cells, such as THP-1, the amino acid sequence of STING is R232, which can bind readily to canonical cyclic di nucleotides. This analysis demonstrated that comparable to MEF cells, non-canonical cGAMP triggered the dephosphorylation of AMPK T172 and ULK1 S556 in hTERT-BJ1 cells (Figure 7D). In addition, non-canonical cGAMP also stimulated the phosphorylation of TBK1 and IRF3 in hTERT-BJ1 cells albeit weaker than in comparison with dsDNA (Figure 7D). Non-canonical cGAMP was observed to induce STING trafficking in both primary MEF cells and hTERT-BJ1 cells while canonical cGAMP activated STING only in MEF cells (Figure S6C). Collectively, our data indicates that cGAS is essential for the activation of AMPK/ULK1 suppression of STING (Figure 6B) and that cGAMPs are responsible for triggering the dephosphorylation of AMPK T172 and activation of ULK1 which phosphorylates STING on S366 to impede its activity (Figure 7E). Thus, cyclic di nucleotides while facilitating STING function, provide a means to control STING via a negative-feedback loop.
Figure 7. cGAMPs Activate AMPK-ULK1 Control of STING.
(A) Primary STING+/+ and STING−/− MEF cells were transfected with canonical cGAMP (3’–5’ cGAMP) (8 µg/ml) using lipofectamine 2000 for the indicated times. Immunoblot was performed with the indicated antibodies.
(B) Primary MEF cells were treated with siRNA for NS (Non-specific), STING, or ULK1 for three days and then transfected with dsDNA (4 µg/ml), canonical cGAMP (8 µg/ml) or poly I:C (4 µg/ml) as described in Figure 7A for 16 hr. IFNβ was measured by ELISA.
(C) Primary STING+/+ and STING−/− MEF cells were transfected with non-canonical cGAMP (2’–3’ cGAMP) (8 µg/ml) as described in Figure 7A for the indicated times. Western blot was performed with the indicated antibodies.
(D) hTERT-BJ1 cells were treated with siRNA for NS or cGAS for three days and then transfected with non-canonical cGAMP (8 µg/ml) as described in Figure 7A for the indicated times. Immunoblot was performed with the indicated antibodies.
(E) Model of STING activation by cytosolic DNA and subsequent inhibition by ULK1.
Asterisks indicate significant difference (P < 0.05) compared to NS determined by Student’s t-test. Error bars indicate sd. See also Figure S6
DISCUSSION
Our data demonstrates that in the presence of cytosolic DNA, STING rapidly traffics with TBK1 via VPS34-related autophagosomes to associate with endosomal compartments containing NF-κB and IRF3. This triggers the production of numerous cytokines and chemokines that can regulate the adaptive immune response (Abe et al., 2013; Ishikawa and Barber, 2008; Ishikawa et al., 2009). This event appears to be independent of ULK1, and is reminiscent of a non-canonical autophagy related process involving plasmacytoid dendritic cell (pDC’s)-mediated phagocytosis of anti-nuclear antibody bound to CpG DNA (DNA-immune complexes; DNA-IC) (Henault et al., 2012). Following DNA-IC engulfment, autophagy facilitates DNA-dependent TLR9 trafficking to cellular compartments harboring IRF3 and NF-κB. Our data here indicates a similar mechanism of type I IFN induction that is dependent on STING and which is triggered by non-CpG DNA species. Aside from being ULK1-independent, STING-dependent autophagosomes do not have double-membrane structures and are dependent on ATG9a (Ishikawa et al., 2009; Saitoh et al., 2009). After trafficking, STING is phosphorylated and degraded, presumably to avoid the deleterious consequences that sustained innate immune gene induction can have upon the host (Ahn et al., 2012). Our data here indicates that the phosphorylation of STING occurs after the formation of autophagosomes and trafficking through the Golgi, and predominantly occurs on four serine residues as determined by mass spectrophotometry. Subsequent studies indicated that phosphorylation of S366 was found to inhibit STING-dependent IRF3 activity, but not robustly NF-κB. Indeed, the mechanisms whereby STING controls NF-κB activity remain to be determined. We note that phosphorylation of S366 may also facilitate STING degradation, to additionally prevent sustained function. It is not clear why the three remaining serine sites are targeted for phosphorylation. However, our data suggest that they do not strongly affect STING function, but more likely influence STING turnover likely to ensure that the presence of STING is not lasting. It was also noted that STING longevity (for example, achieved by expression of STING S366A that could not be phosphorylated, or suppression of ULK1) corresponded with a decrease in the degradation of phosphorylated TBK1. Thus, STING appears to be phosphorylated in autophagosome/endosomal compartments and degraded, a consequence that enables the proteolytic degradation of phosphorylated TBK1 after it has phosphorylated IRF3. A STING variant S366A, unable to be phosphorylated, in contrast, appears to shield TBK1 from degradation and to largely prevent the phosphorylation and translocation of IRF3, by mechanisms that remain to be clarified. However, it is possible that phosphorylated-TBK1 may require release from STING to be able to target IRF3. The mechanisms of how STING/VPS34 pre-autophagosomal structures (PAS) are initiated are also unknown and will be focus of further study.
Using a serine/threonine kinase library, we determined that after trafficking, STING S366 was predominantly phosphorylated by the mammalian C. elegans uncoordinated-51 (UNC-51) like kinase ULK1 (ATG1) and ULK2, first identified as important components of the autophagy process in yeast (Alers et al., 2012; Mizushima, 2010; Tooze et al., 2010). Mammalian ULK1 shares approximately 50% homology with ULK2 and the loss of both completely block amino acid starvation-induced autophagy in murine embryonic fibroblasts (Alers et al., 2012). ULK1 appears to be more ubiquitously expressed than ULK2 and exists in a complex with ATG13 and RB1CC1. The major energy sensor MTOR, as a complex with RAPTOR and MLST8 (mammalian target of rapamycin complex 1; MTORC1) directly phosphorylates ULK1 on S758 under nutrient rich conditions, an event that likely sequesters ULK1 in an inactive state through association with RAPTOR. Inhibition of the MTOR complex, for example by rapamycin is sufficient to induce autophagy (Alers et al., 2012; Mizushima, 2010). However, we did not see any dephosphorylation of ULK1 S758 in cytosolic DNA treated cells. Moreover, rapamycin-dependent autophagy was observed to occur independent of STING, unlike cytosolic DNA-mediated autophagy, which did require STING. In contrast, we did observe dephosphorylation of ULK1 S556 in hTERT-BJ1 and MEF cells which is controlled by another key energy sensor, AMPK (Hardie et al., 2012). In this situation, AMPK, which exists as a heterotrimeric protein (α, β, γ) can be targeted by a number of kinases such as CaMKK beta, which responds to fluctuations in calcium levels, or by liver kinase B1 (LKB1; also referred to as serine/threonine kinase 11; STK11), which responds to fluctuations in ADP and AMP. LKB1 similarly exists in a trimeric complex with the pseudokinase STRAD and the adaptor protein MO25 (Alexander and Walker, 2011). Phosphorylation of AMPK by CaMMK beta or LKB1 cause the dephosphorylation of T172 and prevents AMPK from phosphorylating ULK1 on S556. However, our data indicated that in response to cytosolic DNA, relatively low fluctuations in Ca2+ or AMP levels were measurable, suggesting that other modes of AMPK T172 dephosphorylation were responsible. Nevertheless, we did note that LKB1 was essential for AMPK phosphorylation on T172. This leads us to consider alternate mechanisms of LKB1/AMPK/ULK1 control and to investigate the role of cyclic di nucleotides in this process. First, we found a key role for the synthase cGAS in AMPK dephosphorylation in response to cytosolic DNA. Concomitantly, we observed that canonical or non-canonical cyclic di nucleotides can themselves directly cause the dephosphorylation of AMPK T172 and ULK1 S556 in a STING or cGAS-independent manner. Thus, cGAS is likely responsible for generating cyclic di nucleotides from cytosolic DNA which facilitates STING activity. In addition, cGAS generated cGAMPs provide a mechanism for the subsequent negative control of STING, that occurs after trafficking and delivery of TBK1 to cellular compartments containing IRF3 and NF-κB. It is not yet clear how cyclic di nucleotides activate the LKB1/AMPK/ULK1 axis. However, the cyclic nucleotide cAMP has been reported to regulate the Epac (exchange protein directly activated by cAMP) family, which are guanine nucleotide exchange factors (GEFs) able to influence LKB1-mediated control of AMPK T172 phosphorylation (Fu et al., 2011). Relatively few cyclic di nucleotide binding proteins have thus far been reported, and so the identification of the cGAMP regulators of AMPK activity remain to elucidated. In summary, cyclic di nucleotide control of AMPK/ULK1 and STING helps ensure that sustained production of STING-dependent pro-inflammatory genes is prevented, events that may otherwise lead to inflammation and autoimmune disorders (Ahn et al., 2012; Gall et al., 2012). Our data provides information on the control of DNA-mediated innate immune signaling pathways which could conceivably be therapeutically targeted to help prevent a variety of self-DNA triggered disorders.
EXPERIMENTAL PROCEDURE
Cells, Reagents, and Viruses
Primary STING+/+ and STING−/− MEF cells were prepared as described (Ishikawa and Barber, 2008). TBK1−/− MEF cells were kindly provided by W. C. Yeh. HEK293T cells, Platinum-E retroviral packaging cell line, hTERT-BJ1 cells, and primary human dendritic cells were purchased from the American Type Cell culture (ATCC), Cell biolabs, Clontech, and Stemcell, respectively. MEF cells, HEK293T cells, and Platinum-E cells were cultured in DMEM supplemented with 10% FBS and antibiotics. hTERT-BJ1 cells were cultured in a 4:1 ratio of DMEM : Medium 199 with 10% FBS, 1 mM sodium pyruvate, and 4 mM L-glutamine. Poly I:C was obtained from American Biosciences. ISD (90-mer), used as dsDNA in this study, was prepared as described previously (Ishikawa and Barber, 2008). Canonical cGAMP (3’–5’ cGAMP) and Non-canonical cGAMP (2’–3’ cGAMP) were purchased from Invivogen and BioLog, respectively. BFA, compound C, rapamycin, and STO-609 were purchased from SIGMA. Non-specific siRNA (NS), siRNA for STING, ULK1, ULK2, LKB1, cGAS, VPS34, and Beclin-1 were purchased from Dharmacon. Lipofectamine 2000 and lipofectamine RNAiMAX were purchased from Invitrogen. Anti-STING rabbit polyclonal antibody was prepared as described previously (Ishikawa and Barber, 2008). Other antibodies used in this paper were as follows: Anti-β-actin (SIGMA, A5441); Anti-Calreticulin (Abcam, ab14234); Anti-IRF3 (Invitrogen, 39–2700, used for murine IRF3); Anti-IRF3 (Santa Cruz Biotechnology, sc-9082); Anti-phospho-IRF3 (Cell signaling, 4947); Anti-p65 (Cell signaling, 3987); Anti-TBK1 (Abcam, ab40676); Anti-phospho-TBK1 (Cell signaling, 5483); Anti-LC3A (Cell signaling, 4599); Anti-ULK1 (Cell signaling, 4773); Anti-phospho-ULK1 (Ser556) (Cell signaling, 5869); Anti-phospho-ULK1 (Ser758) (Cell signaling, 6888); Anti-ULK2 (Abcam, ab97695); Anti-AMPKα (Cell signaling, 5832); Anti-phospho-AMPKα (Thr172) (Cell signaling, 2535); Anti-VPS34 (Cell signaling, 4263); Anti-Beclin-1 (Cell signaling, 3495); Anti-LKB1 (Cell signaling, 3047); Anti-phospho LKB1 (Ser428) (Abcam, ab63473); Anti-HA (SIGMA, H9658). HSV-1 (KOS strain) was purchased from ATCC. HSV-1 γ34.5 was kindly provided by Bernard Roizman. HSV-1 luc was kindly provided by David Leib. Purified ULK1 and TBK1 were purchased from SIGMA and Millipore, respectively. Probe for Ulk1, Ulk2, Infb, Ifit3, Ifna4, Tnfaip3, and cgas was purchased from Applied Biosystems.
Proteomics
hTERT-BJ1 cells in ten 15 cm dishes were used for untreated samples, dsDNA-treated samples, and negative controls (untreated and precipitated with mouse IgG), respectively. The cells were transfected with 5 µ g/ml of dsDNA for 6 hr. After that the cells were lysed in TNE buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40) with protease and phosphatase inhibitors and then subjected to immunoprecipitation with monoclonal anti-STING antibody covalently conjugated beads for 18 hours at 4°C. After washing with TNE buffer, STING protein was eluted using Elution buffer (pH 2.8, Thermo) and then neutralized by Tris-HCl (pH 9.5). The samples were boiled in SDS-sample buffer and then separated in a 7.5% acrylamide gel. The gel was stained with CBB staining kit (Thermo) and the visualized bands including STING were analyzed by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (µ LC/MS/MS) at the Harvard Mass Spectrometry and Proteomics Resource Laboratory.
Reconstitution of STING−/− MEF Cells
Platinum-E retroviral packaging cells were transfected with pBabe-STING-puro using Lipofectamine 2000. After 2 days, the supernatants containing retrovirus were collected. STING−/− MEF cells were incubated with the supernatants in the presence of 10 µ g/ml of polybrene (SIGMA) for 6 hr. After another 2 days, 2 µ g/ml of puromycin (SIGMA) was added to culture media to remove uninfected cells.
In vitro Kinase Assay
181–379 of hSTING was cloned into pET-26b and expressed in BL21 (DE3). Recombinant hSTING protein was isolated using HisPur Ni-NTA Resin (Thermo) and eluted with imidazole. 60 ng of hSTING protein was incubated with 20 ng of GST-ULK1 in 20 mM Tris-HCl (pH 7.5), 25 mM KCl, 0.1 mM EDTA, 1 mM DTT, 5% glycerol, 2 mM MgCl2, 2 mM MnCl2, 1.25 µM ATP, and 0.3 mg/ml BSA in presence of [γ-32P] ATP (Perkin Elmer) for 15 min at 30°C. The samples were boiled in SDS-sample buffer and separated in a 7.5% acrylamide gel. After running the gel, gel was fixed in 40% methanol solution with 10% acetic acid and stained with CBB staining kit (Thermo). After destaining, the dried gel was exposed to BioMax Light Film (Kodak).
Kinase Screening
To identify the kinase that phosphorylates residue Ser366 of STING, a screen of 272 recombinant protein kinases was performed at KINEXUS (Vancouver, Canada). Briefly, peptide (S366, ELLISGMEK) and (A366, ELLIAGMEK) were mixed with individual protein kinases in presence of [γ-33P] ATP. After removing unreacted [γ-33P] ATP from the reaction, radioactivity was quantified in a scintillation counter. The radioactivity of S366 was compared with that of A366.
Supplementary Material
HIGHLIGHTS.
Cytosolic DNA generated cyclic di nucleotides facilitate innate signaling via STING.
Cyclic di nucleotides subsequently activate the AMPK/ULK1 (ATG1) pathway.
Activated ULK1 targets STING on serine 366 to impede IRF3 activity.
Phosphorylation of STING averts chronic inflammatory cytokine production.
ACKNOWLEDGMENTS
We thank Delia Gutman for technical assistance, Biju Issac for gene array analysis, and George McNamara for confocal microscopy analysis. This work was funded in part by grants R01AI079336 and UO1AI083015. H.K. carried out most of the experiments, K.K. purified STING. G.N.B. wrote the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
The Gene Expression Omnibus accession number for microarray data is [pending].
SUPPLEMENTAL INFORMATION
Supplementary information includes Extended Experimental Procedures and Six Figures can be found with this article online at.
REFERENCES
- Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2'–5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander A, Walker CL. The role of LKB1 and AMPK in cellular responses to stress and damage. FEBS Lett. 2011;585:952–957. doi: 10.1016/j.febslet.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Wakabayashi Y, Lippincott-Schwartz J, Arias IM. Bile acid stimulates hepatocyte polarization through a cAMP-Epac-MEK-LKB1-AMPK pathway. Proc Natl Acad Sci U S A. 2011;108:1403–1408. doi: 10.1073/pnas.1018376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMP-activated protein kinase: a target for drugs both ancient and modern. Chem Biol. 2012;19:1222–1236. doi: 10.1016/j.chembiol.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–997. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Jefferies HB, Kalie E, Longatti A, McAlpine FE, McKnight NC, Orsi A, Polson HE, Razi M, Robinson DJ, et al. Trafficking and signaling in mammalian autophagy. IUBMB Life. 2010;62:503–508. doi: 10.1002/iub.334. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.