Abstract
The early detection of microbes is the responsibility of the innate immune system which has evolved to sense pathogen derived molecules such as lipopolysaccharides and non-self nucleic acid, to trigger host defense countermeasures. These sensors include the RIG-I-like helicase family (RLH) that specifically recognizes viral RNA, as well as the cytoplasmic, nucleotide binding oligermerization domain (NOD)-like receptor and Toll-like receptor (TLR) pathways that sense a variety of microbial derived molecules. Comprehending how the cell senses foreign DNA, generated by certain viruses, bacteria and possibly parasites has proven elusive but is of significant importance since such information could shed insight into the causes of microbial related disease, including viral associated cancers and autoimmune disorders. Plasmacytoid dendritic cells are known to utilize TLR9 to detect pathogen-associated DNA and to trigger the production of type I interferon (IFN), as well as other cytokines, although alternate key DNA detecting sensors remain to be identified. Recently however, a molecule referred to as AIM2 (absent in melanoma 2) was found to be essential for mediating inflammatory reactions triggered by cytoplasmic DNA. In addition, an endoplasmic reticulum associated protein referred to as STING (for Stimulator of Interferon Genes) was demonstrated as being pivotal for facilitating IFN production in response to intracelleular DNA and a variety of DNA pathogens. Here, we review recent discoveries relating to the detection of foreign DNA, including the importance of the STING and AIM2 and the activation of innate signaling pathways.
Introduction
The innate immune response comprises a series of cellular sensors and signaling pathways that activates host defense mechanisms in response to microbial invasion. These immediate cellular reactions lead to the production of type I interferon (IFN), which exerts anti-microbial effects by switching on the transcription of numerous cellular anti-pathogen genes, as well as other cytokines that induce inflammatory responses [1-3]. Innate immune stimulation also galvanizes the adaptive immune response involving antibody production and cytotoxic T cell activity, critical for ensuring complete eradication of infectious agents from the host [1-3]. Unraveling the molecular mechanisms of innate immune activation has proven to be complex, although significant progress has been made in the last decade, especially relating to our understanding of the Toll-like receptor (TLR) pathway. For example, it is now known that nucleic acid constituting microbial RNA and DNA (referred to as PAMPs- pathogen associated molecular patterns) are potent activators of innate immune signaling pathways [1-3]. It is also apparent that a number of sensors have evolved in a variety of cell-types to detect pathogen-generated nucleic acid. Most cells are able to recognize foreign nucleic acid species and can directly trigger the production of type I IFN. In addition, hematopeotic cells, including macrophages and dendritic cells (DC's) produce IFN and other cytokines which can result in a pro-inflammatory response by engulfing infected apoptotic cell debris that contains viral antigen and nucleic acid [4]. Here, we will describe innate immune pathways that focus on the detection of microbial DNA, generated from viruses, bacteria and possibly fungi and parasites.
The Toll-like Pathway
Cellular pattern recognition receptors (PRR's) such as the TLR's are responsible for sensing a selection of microbial derived molecules such as viral RNA's and bacterial surface molecules. There are approximately 10 human TLRs, and the different members recognize a variety of microbial PAMPs such as lipopolysaccharides (TLR4) common on bacterial cell walls, viral dsRNA (TLR3), viral ssRNA (TLR7/8), as well as viral or bacterial unmethylated DNA (TLR9; see below) all of which results in the production of IFN as well as other cytokines [1-3]. TLR3, 7/8 and 9 reside within the endoplasmic reticulum and traffic to endosomal compartments in response to the presence of cytoplasmic nucleic acid enclosed in such structures[1-3]. TLR3 recruits an adaptor referred to as TRIF which associates with tumor necrosis factor receptor (TNFR)- associated factor 3 (TRAF3) and 6 as well as receptor interacting protein 1 (RIP1) perhaps through a RIP homotypic interaction motif (RHIM) [3]. This signaling complex also involves association with Fas associated death domain containing proteins (FADD) and the TNFR associated death domain containing protein (TRADD) [1-3]. These events lead to the ubiquitination of RIP1 and to the activation of nuclear factor of kappa light polypeptide gene enhancer in B-cells (NF-κB). A further series of ubiquitination events involving TRAF3 facilitates the activation of mitogen activated protein kinases (MAPK) and two IKK-related kinases referred to as Tank-binding kinase 1 (TBK1) and IKK-i which phosphorylate interferon regulatory factor 3 (IRF3) and IRF7. Dimerized IRF3 and IRF7 translocate to the nucleus and associate with NF-kB on the IFN β promoter, to activate transcription. Secreted type I IFN binds to interferon receptors (IFNARs) present on nearly all cell types and triggers the production of numerous anti-viral genes through the JAK/STAT pathway [3]. Cells exposed to IFN exhibit pronounced resistance to virus replication and animals defective in IFN production exhibit exquisite sensitivity to virus infection[5].
In plasmacytoid dendritic cells (pDC's), which are major sources of IFN, another mechanism has evolved to trigger the production of type I IFN in response to infection by RNA and DNA viruses[6]. For example, following RNA virus infection, or engulfment of apoptotic cell debris containing viral ssRNA, pathogen nucleic is detected by TLR7 which causes association with MyD88 (myeloid differentiation primary response gene 88) and members of the IRAK (interleukin-1 receptor associated kinase), TRAF3 and 6[3]. These events rapidly and potently lead to phosphorylation of IRF7 which as described above, induces dimerization and translocation to the nucleus to activate the production of type I IFN [5].
It became clear however, that TLR3 or 7 deficient animals, while exhibiting susceptibility to certain types of virus infection remained able to produce type I IFN in response to RNA species[1-3]. These observations lead to the hunt for TLR-independent pathways that could plausibly explain alternate mechanisms of viral RNA-mediated IFN induction. These innate pathways turned out to be dependent on DExD/H box RNA helicase sensors called retinoic acid inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and LGP2 as well as the death domain containing proteins FADD and RIP1[7]. RIGI and MDA5 helicases are largely responsible for the production of IFN in all cell types, in response to RNA virus infection, except for pDC's which are TLR7 dependent[3]. Helicase interaction with viral RNA induces the recruitment of a mitochondrial and peroxisome associated molecule referred to as IPS-1 (also known as VISA, Cardif or MAVS) and through TRAF3, FADD and RIP1 leads to the activation of MAP kinases, TBK1 and IKK-i which enable the transcription factors AP1, IRF3 and 7 and NF-κB to translocate into the nucleus and bind to and activate the IFNβ promoter, as described [1-3].
Thus, significant progress has been made in unraveling mechanisms responsible for recognizing PAMPs including RNA species derived from virus infection. However, less is known relating to how foreign DNA, derived from cancer causing viruses, bacteria, fungus and parasites, is sensed by the cell to trigger innate immune responses including the production of type I IFN. This is of significance since microbial derived or endogenous self-DNA may be responsible for stimulating our own innate immune pathways, which includes the overproduction of type I IFN and which can provoke autoimmune disease[4]. DNA sensing pathways have also been implicated in facilitating innate and adaptive immune pathways mediated by plasmid-based vaccines [8]. Such vaccines can be manufactured quickly and economically although they suffer from not being highly immunogenic. Understanding the mechanisms of DNA-dependent innate signaling may therefore lead to improvements in the effectiveness of DNA vaccine procedures.
However, recently progress has been made in elucidating those pathways controlling the triggering of immune signaling in response to microbial DNA. TLR9 for example, is known as being able to stimulate type I IFN production mainly in pDC'S in response to non -methylated CpG DNA[8]. In addition, it has recently been shown that PYD and HIN200 (hematopoietic interferon-inducible nuclear antigens with 200 amino acid repeats) domain containing protein referred to as AIM2 is responsible for triggering inflammatory responses in response to cytoplasmic DNA [9-12]. An essential pathway that governs the production of type I IFN by foreign DNA has also been recently discovered and shown to be dependent on a molecule referred to as STING (Stimulator of IFN Genes) [13,14]. Here we review these recent discoveries that may help to explain mechanisms of DNA-mediated innate immune signaling.
TLR-dependent DNA sensing mechanisms
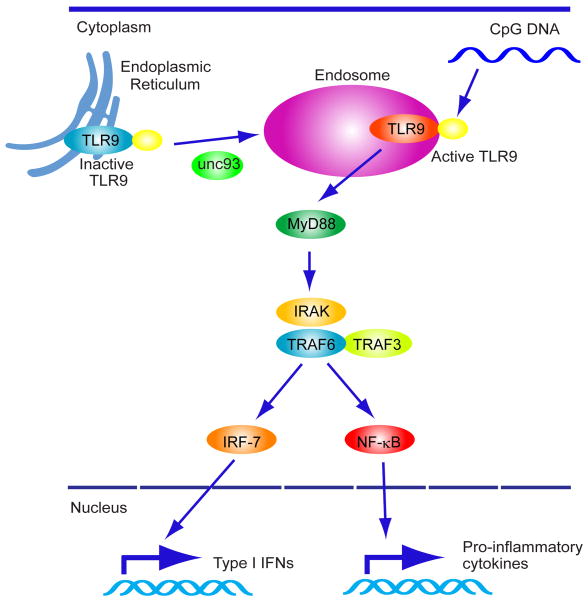
TLR9 was one of the first sensors found to play a role in the triggering innate immune signaling in response to foreign DNA [8]. This receptor is mainly expressed in pDCs and B cells and recognizes CpG (cytidine-phosphate-guanosine) DNA motifs that are commonly found in bacteria and viruses, but which is not usually found in vertebrates. TLR9 harbors a leucine rich repeat (LRR) motif, a Toll/IL-1R homology domain and is considered a type I integral membrane glycoprotein [3,8]. Inactive TLR9 is observed to associate with the endoplasmic reticulum (ER) in unstimulated pDCs. However, CpG DNA, internalized via a clatherin-dependent endocytic pathway, traffics to lysosomal compartments to associate with pre-processed, active TLR9 that has also localized from the ER to these regions. The trafficking of TLR9 is controlled by UNC93B, a 12-membrane-spanning ER protein which directly interacts with TLR9 [15]. As mentioned, proteolytic cleavage of TLR9 in the endolysosomal compartment is likely required for TLR9 activation in response to CpG DNA [16]. However, the full mechanism of how TLR9 is triggered to move to CpG containing lysosomes is still unclear. Upon recognition of CpG DNA in endosomes, TLR9 interacts with myeloid differentiation primary response gene [88] (MyD88) which contains a TIR domain and a death domain [1-3]. MyD88 interacts with IRAK-1 (IL-1R associated kinase 1), IRAK-4 and IRF-7. This event leads to recruitment of TRAF3 and 6, which activates the TAK1 (transforming growth factor β-activated kinase 1), MAPK as well as NF-κB. IRAK1 directly interacts with and phosphorylates IRF7 which as mentioned is required for IFNβ transcriptional activity (Figure 1) [1-3].
Fig 1. TLR9-dependent innate immune signaling.
Latent TLR9 resides in the ER and traffics to endosomes where it is processed to become active. Trafficking is UNC-93B-dependent. Cytoplasmic CpG DNA engulfed in endolysosomes associates with TLR9 which triggers recruitment of a signaling complex consisting of MyD88, IRAK4 and TRAF6. TRAF6 in turn activates TAK1, and subsequently MAPK and the IKK complex (IKKα, IKKβ, and IKKγ) to activate NF-κB. NF-κB and MAPK regulates inflammatory cytokine expression. IRAK4 also activates TRAF3 and IRAK1, which catalyzes IRF7 phosphorylation to induce type I IFNs expression.
Several studies using TLR9 deficient mice have emphasized a role for this receptor in host innate immune responses against DNA viruses such as herpes simplex virus and CMV [3,8,17-19]. Reports have also indicated that TLR9 may facilitate plasmid based immunization protocols [8]. In this light, the use of TLR ligands has been used to improve the immunogenicity of a variety of antigens used in vaccines strategies. For example, synthetic oligodeoxynucleotide (ODN) ligands, such as CpG ODN for TLR9 have been reported to be a potent adjuvant which enhances immunization procedures [20]. Engagement of the TLR's can stimulate the production of a variety of co-stimulatory molecules, as well as IFN-induced genes that can facilitate T and B cell activation. Aside from exhibiting strong adjuvant properties, CpG monotherapy has also been demonstrated to have prophylactic efficacy against a wide range of viral, bacterial and parasitic pathogens. Finally, TLR9 agonists have been utilized as treatments for hematologic malignancies, skin cancers and glioblastoma[21].
TLR9 has been also implicated in the development of autoimmune diseases such as systemic lupus erythematosus (SLE) [4,22,23]. Considerable evidence indicates that self nucleic acids can act as endogenous ligands for DNA receptors which raise serum levels of IFN and which correlate with both SLE disease activity and severity [4]. For example, anti-DNA antibodies, found in serum of patients with SLE (Systemic lupus erythematosus), bind self-DNA have been reported to induce type I IFN via a cooperative interaction between TLR9 and CD32 in pDC's [24]. Chimeric TLR9, which localizes to the cell surface, can respond to synthetic TLR9 ligands but not to viral DNA, suggesting intracellular localization of TLR9 is required to prevent the recognition of self DNA [25]. Thus, the unraveling the molecular mechanisms of DNA triggered innate immune signaling pathways may have important implications in our understanding of autoimmune disease and could lead to the development of new therapeutics to treat such disorders.
It became clear however, that despite such progress, alternate DNA sensors likely existed to facilitate innate immune signaling. For example, the induction of immune responses following plasmid-DNA immunization could still occur in TLR9-deficient mice [26]. In addition, the production of type I IFNs remained unaffected in a number of TLR9 deficient cell-types infected with DNA pathogens such as herpes simplex virus 1 (HSV1), or intracellular bacteria such as Listeria monocytogenes, which are known to introduce DNA into cytosol during infection[18,27]. The transfection of TLR9 lacking cells with synthetic double-stranded DNA (dsDNA) did also not affect the induction of type I IFN [27]. Finally, DNA derived from engulfed apoptotic cells was also found to induce type I IFN expression in a TLR9-independent manner [28]. Thus, while TLR9 signaling constitutes an important component of our innate immune system, considerable evidence also indicated the existence of alternate DNA sensing pathways, the discovery of which we describe next.
The STING Pathway and Recognition of Intracellular DNA
In an attempt to identify components of innate signaling, we developed a screen to isolate molecules able to activate the IFN-β promoter [13]. These experiments lead to the identification of a molecule referred to as STING (Stimulator of Interferon genes,) a transmembrane containing protein that was found to localize in the endoplasmic reticulum (ER) of numerous cell types such as macrophage, dendritic cells as well as endothelial and epithelial cells [13,14]. Overexpression of STING, also known as MITA/MPYS/ERIS, induced the activation of both NF-κB and IRF3 to stimulate type I IFN production [13,29-31]. STING deficient mice were found to be viable and early investigations using STING deficient MEFs indicated that negative-stranded RNA viruses such as vesicular stomatitis virus (VSV) or Sendai Virus (SV) required STING for effective type I IFN production [13]. Indeed STING-deficient mice were exquisitely sensitive to VSV infection. However, synthetic dsRNA (polyIC) was not affected in its ability to produce IFN, in the absence of STING. This suggested that STING played an important role in RIG-I mediated signaling, which senses RNA viruses such as VSV and SV, but not MDA5, responsible for facilitating poly IC signaling[13,14]. RIG-I signaling is also known to be dependent on IPS-1, a mitochondrial and peroxisome-associated protein [7]. Our data indicated that ER-associated STING likely associates in close proximity with mitochondria associated with the ER (mitochondria associated membrane- MAM) [13,14]. Mitochondria form close contacts with the ER to obtain calcium for oxidative phosphorlyation purposes [32]. Studies further indicated that STING associated with SSR2/TRAPβ, a member of the translocon associated protein (TRAP) complex comprising four subunits (α–Δ) that facilitate translocation of proteins from ER associated ribosomes, into the ER lumen following translation [13,33]. The translocon associated protein (TRAP) complex is known to associate with the translocon, comprising three subunits, SEC61α, SEC61β and SEC61γ [33]. The translocon complex is critical for protein folding and secretion. Given this, one plausible model could include that viral RNA associated with ER-attached ribosomes may activate RIG-I and subsequently IPS-1 associated with MAM's. STING, associated with the translocon clearly facilitates this process since cells lacking STING are defective in RNA-virus mediated IFN induction. Although it is not clear how signaling from STING/translocon complexes to IRF3/NF-κB occurs, it has been reported that the translocon may physically associate with exocyst complexes which are conserved octameric complexes that tether secretory vesicles to membranes, and which facilitate protein synthesis and secretion[34]. Recently, Sec5, one of the components of exocyst complex, was found to recruit and activate TBK1 which, as mentioned, is essential for IRF3/7 activation and type I IFN induction in response to RNA and DNA pathogens [35]. Preliminary analysis indicated that STING could associate with TBK1. Thus, STING may link cytosolic DNA-mediated signaling to TBK1 activation through MAM/ER/translocon and exocyst interactions.
As part of these studies, we also noticed that MEFs derived from STING deficient mice failed to induce type I IFNs in response to infection with HSV-1 or Listeria monocytogenes, or transfection of interferon stimulatory DNA, ISD (which comprises double-stranded 45 base pair oligonucleotides lacking CpG sequences) [13,14]. However, STING did not exhibit any predicted DNA-binding motifs and did not appear to directly associate with DNA, indicating the likely existence of upstream DNA binding sensors responsible for STING activation. STING was also essential for intracellular DNA mediated activation of type I IFN in macrophages and in conventional DC's [14]. Indeed, both HSV-1 and the gram-positive intracellular bacterium Listeria failed to induce type I IFN in macrophages as well as DC's lacking STING[13,14]. STING has also been shown to be important for gram-negative bacterium (F. tularensis) triggered IFN production in macrophages and for recognizing human immunodeficiency virus (HIV-1) infection [36,37]. However, while the production of type I IFN by transfected DNA was ablated in the absence of STING, IL1β production was not affected (which has been shown to be AIM2 dependent, see below) [14]. Thus, STING functions independent of AIM2 and is not critical for inflammasome activation. Finally, our studies indicated that STING was important for intracellular DNA-mediated and HSV-1 activated type I IFN production in pDC's, although the presence of TLR9 in these cells was found to partially compensate for loss of STING in response to Listeria infection[14]. Perhaps unsurprisingly, we observed that STING deficient animals succumbed to lethal HSV-1 infection due to a lack of type I IFN production, which facilitated robust virus replication [14]. Recently, a mouse model, referred to as Goldenticket, was developed using N-ethyl-N-nitrosourea. This animal exhibited pronounced defects in the recognition of cyclic di-nucleotides generated by Listeria, ligands responsible for triggering type I IFN induction. Further analysis indicated that Goldenticket had acquired a point mutation in the STING gene, further confirming the importance of STING in controlling nucleotide and intracellular bacteria- based triggering of innate immune responses[38].
Following infection with HSV-1, but not RNA viruses, STING rapidly trafficked from the MAM/ER region through to the Golgi to reside in a distinct perinuclear region, via mechanisms that presently remain unclear [14]. This process is perhaps associated by STING ubiquitination [39]. The endosomal compartments that comprise STING and TBK1 following trafficking in response to intracellular DNA remain to be fully characterized although were reported to involve autophagy-related gene 9a (Atg9a) [40]. Thus, STING trafficking to perinuclear endosomal compartments may be important for the stimulation of innate immune pathways involving the production of type I IFN, at least in response to intracellular DNA (Figure 2,3).
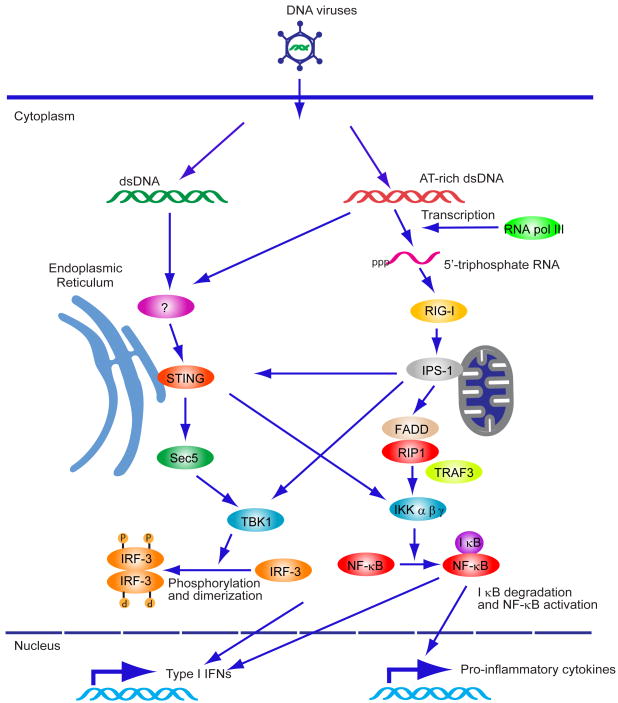
Fig 2. Cytoplasmic DNA-mediated IFN production.
Cytosolic AT-rich dsDNA is recognized by RNA PolIII and can be transcribed into 5′triphosphate RNA which activates RIG-I/IPS-1 signaling. IPS1 interacts with FADD/RIP1 which activate NF-κB. IPS-1 also activates TBK1 which phosphorylates IRF-3 and induces type I IFN expression. AT-rich dsDNA or non-AT-rich dsDNA is also recognized by unknown receptor(s) and activates STING-dependent signaling. STING localizes in the ER with the translocon complex and activates TBK1 to induce type I IFN expression. STING also activates NF-κB to induce pro-inflammatory cytokine responses.
Fig 3. STING is associated with the translocon.
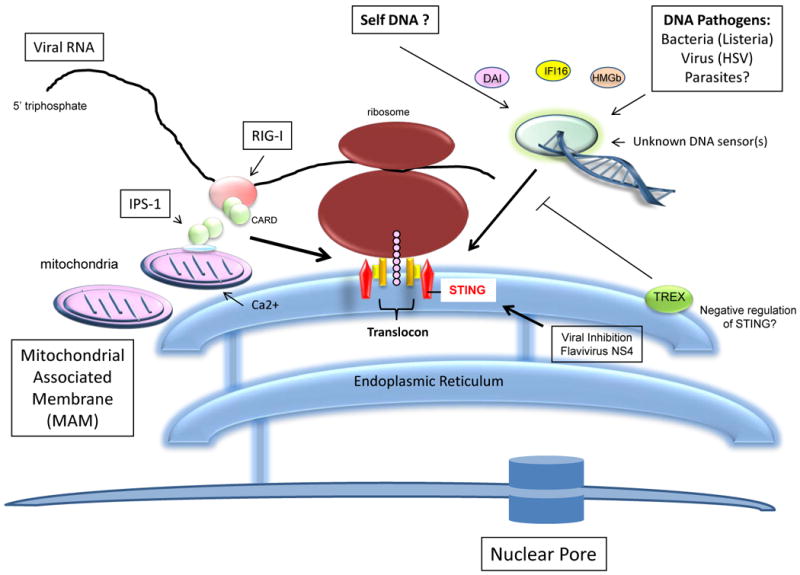
STING associates with the translocon associated protein (TRAP) complex comprising four subunits (TRAP α–Δ) responsible for facilitating translocation of proteins from ER associated ribosomes, into the ER lumen following translation. The TRAP complex is known to associate with three subunits, SEC61α, SEC61β and SEC61γ. TRAP associated STING is essential for DNA-mediated cytoplasmic IFN production triggered by DNA sensor(s). STING also facilitates RIG-I-mediated signaling perhaps through IPS-1 associated with mitochondria bound to the ER (MAM). STING function was found to be inhibited by certain flavivirus proteins.
Finally, it is worth noting that previous work has shown the importance of TBK-1 in stimulating plasmid DNA-based immune responses[41]. Plausibly, TBK1 dependent pathways are triggered by intracellular plasmid B-form DNA which augments immune responses by stimulating type I IFN and NF-κB-dependent pathways, similar to agonists of the TLR9 pathway [41]. Since we observed that STING functions upstream of TBK-1 and is required for TBK-1 activity in response to intracellular DNA we investigated whether STING was important for the adjuvant effects of plasmid DNA. We observed that loss of STING indeed affected adaptive immune responses, such as those stimulated by plasmid DNA vaccination as well DNA virus infection such as with vaccinia virus[14]. Thus, the stimulation of STING regulated pathways may provide a mechanism to augment DNA-based plasmid immunization strategies in the future.
The importance of STING in facilitating innate immune responses following infection with either RNA or DNA viruses may also make this pathway a key target for viral subversion. Indeed, STING was found to exhibit homology with flavivirus proteins (Dengue Virus, Yellow Fever Virus and Hepatitis C Virus; NS4) which are known to reside in the ER [14]. Thus, flaviviruses, and possibly other viruses, may target the STING pathway to inactivate host defense capabilities. Further identification of STING-associated molecules will no doubt further help elucidate mechanisms of DNA-mediated innate immune signaling processes in the cell.
TREX1: A negative regulator of intracellular DNA-mediated innate signaling?
Type I IFN is an essential factor for host defense against evading pathogens, but inappropriate production of type I IFN leads to autoimmune diseases such as SLE, as earlier mentioned [4,42]. In eukaryotes, localization of self DNA is restricted to the nucleus and mitochondria, thereby sequestering self DNA from cytoplasmic DNA sensing mechanisms which may activate pro-inflammatory cytokine pathways. Cellular DNases eliminate aberrant self DNA found in apoptotic bodies, extracellular space, cytosol, and endosomes [4]. Several studies have shown that defective clearance of self-DNA leads to inappropriate activation of type I IFNs production through a TLR-independent innate immune signaling pathway, which is tightly linked to autoimmune diseases. For example, DNase I deficiency or mutations are associated with lupus-like syndrome in mice and humans [4,43,44]. In addition, DNase II deficient mice accumulate incompletely digested DNA, which causes TLR-independent type I IFN production, inflammatory responses and early death, [42,45]. Crossing susceptible mice with mice deficient in the type I IFN receptor abrogated lethality indicating the importance of excessive IFN production in pathogenesis [42]. While it has been shown that TLR9 may play some role in functioning as a DNA sensor to facilitate SLE, it is likely that other major sensor(s) remain to be identified. It also remains to be seen whether STING is involved in mediating cytoplasmic DNA-triggered, IFN-dependent SLE.
Recently, several studies have reported that Trex1, 3′-repair exonuclease 1, regulates DNA homeostasis and is involved in autoimmune diseases. For example, mutations in the human Trex1 gene are known to cause SLE and AGS (Aicardi-Goutieres syndrome) [46-48]. In mice, Trex1-deficiency manifests defective G1/S transition and chronic ATM-dependent checkpoint activation. This correlates with the accumulation of ssDNA molecules in the ER, produced during S phase [48]. Such animals develop lethal autoimmunity via elevated production of type I IFNs and auto-antibodies [49]. Genetic ablation of IRF3 or the type I IFN receptor rescued Trex1-deficient mice from mortality, suggesting that IRF3-dependent IFN production was indeed responsible for the observed Trex1-dependent autoimmune disorder. Cytosolic ssDNA derived from endogenous retroelements or aberrant replication intermediates was found to accumulate in the hearts of Trex1-deficient mice leading to cardiomyopathy [48,49]. Similar to STING, Trex1 has also been shown to localize in the ER. Collectively, these observations suggest that Trex1 is required for the prevention of autoimmune disorders that could otherwise be generated by cell-intrinsic DNA substrates [48,49].
The identity of the ssDNA sensor(s) responsible for inducing IFN and causing Trex1-dependent autoimmunity remain to be identified. It is further unclear whether these disorders are STING-dependent. However, it is plausible that Trex1 may play a role in negatively regulating the STING pathway. For example, a recent report indicated that reverse transcribed human immunodeficiency virus type I (HIV1) DNA can stimulate the STING pathway, an affect that could suppress viral replication[37]. HIV was found to use Trex1 to rapidly digest excess viral DNA to avoid STING activation. It will be of interest to evaluate whether other retroviruses/retroelements trigger the STING pathway, or whether viruses other than HIV and possibly flaviviruses suppress this pathway to avoid host defense countermeasures. Unraveling these pathways may also shed light into causes of autoimmune disease.
The Nod-like receptor (NLR Pathway): AIM2 and cytosolic DNA-mediated inflammasome-dependent innate signaling
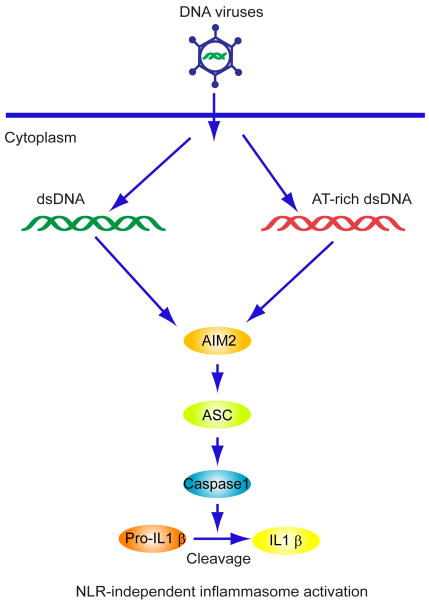
The NLR pathway has also been shown to be important in sensing cytosolic DNA and triggering inflammasome-dependent innate immune signaling[50]. This involves caspase-1 dependent processing of cytokines such as IL-1β, a key pro-inflammatory mediator that can stimulate recruitment of macrophages and DC's to sites of infection or injury. These events generally do not involve the induction of IFN and are independent of TBK1. The NLR's comprise 22 genes, in humans, and are formed into 4 inflammasome sub-families (NLRP1, NLRP3, IPAF and AIM2) [50]. NLRP1 (NACHT-leucine-rich repeat-PYD containing protein 1) was first described and has been reported to play a role in the susceptibility to select forms of bacteria such as Bacillus anthracis, although its mechanisms of activation are unclear. NLRP3, in contrast has been reported to trigger the production of IL-1β in response to a variety of stimuli (PAMPs, Danger associated molecular patterns-DAMPS, and environmental stresses). For example, adenovirus infection has been reported to activate ASC (Apoptosis-associated speck-like protein containing a caspase recruitment domain)/caspase1-mediated secretion of IL-1β. Thus, inflammatory responses induced by DNA pathogens could, in part, involve the NLRP3 pathway. IPAF (Ice-protease activating factor) is activated by gram-negative bacteria and can directly associate with caspase-1 independent of ASC [50]. However, the fourth family (AIM2- absent in melanoma 2) has recently been identified as a major regulator of DNA-mediated inflammatory responses [9-12]. Indeed, four different groups identified AIM2, a PYD, HIN-200 containing family member as the cytoplasmic dsDNA sensor that activates ASC/caspase 1-mediated secretion of IL-1β. Intracellular DNA species, for example from select bacteria binds to AIM2's HIN-200 region. This causes the N-terminal pyrin domains (PYD's) to recruit ASC and activate caspase-1. The PYD/HIN-200 (PYHIN) family consists of four members: IFIX, IFI16, MNDA as well as AIM2. Inflammasome formations involving AIM2 are NLR-independent and the term AIM2-like receptors (which also includes IFI16- see below) has been coined to distinguish this mechanism of action. Importantly, while the AIM2 inflammasome is essential for caspase-1 activation it is completely dispensable for type I IFN production in response to cytosolic dsDNA (Figure 4) [9-12]. This underscores that cytosolic DNA-mediated AIM2 -dependent inflammatory signaling is distinct from type I IFN-dependent innate signaling, which is largely dependent on STING. Thus, different DNA sensors exist to trigger pro-inflammatory or type I IFN production in response to infection. It is unclear whether AIM2 may play a role in autoimmune disease, although no doubt experimentation is ongoing designed to address this issue.
Figure 4.
AIM2-dependent signaling and the inflammatory responses: Cytosolic DNA binds to the HIN-200 domains of AIM2 and activates the ASC/Caspase1 inflammasome pathway to produce IL-1β.
2-2. Cytosolic DNA-mediated RNA polymerase III/RIG-I/IPS-1-dependent innate signaling
Recent studies have demonstrated the existence of a novel cytosolic DNA sensing mechanism that is able to induce type I IFN in a DNA-dependent RNA polymerase III manner[51,52]. Transfected Poly(dA:dT) (synthetic AT-rich dsDNA), but not other types of DNA including poly(dG:dC) (synthetic GC-rich dsDNA), calf thymus DNA, PCR fragments, or plasmid DNA, have been found capable of producing dsRNA containing 5′triphosphate ends and to activate RIG-I/IPS-1 dependent innate signaling. RNAi abrogation of POL3RF, a component of RNA polymerase III, or pharmacological inhibition of RNA polymerase III was found to inhibit production of dsRNA from poly(dA:dT) [51]. As we have mentioned, STING also appears to be involved in RIG-I/IPS-1-mediated signaling, although STING deficient cells remain partially capable of producing IFNβ in response to infection with negative stranded RNA viruses such as Sendai virus (SV) and VSV [13,14]. It was also noted that while IFNβ production mediated by ISD, HSV-1, Listeria, or transfected plasmid DNA were completely abrogated in STING deficient MEFs, such cells produced detectable amounts of IFNβ after transfection with poly(dA:dT) [13,14]. These observations indicate that AT-rich DNA may induce both STING-dependent and STING-independent, RNA polymerase III/RIG-I/IPS-1-regulated innate immune signaling pathways. The physiological importance of these processes awaits clarification though emphasizes that the cell has evolved a number of mechanisms to detect aberrant DNA species.
Other sensors of intracellular DNA
While AIM2 appears to be the main sensor for intracellular DNA-mediated, IL1-β dependent inflammasome related responses, identification of molecules responsible for associating with DNA and triggering IFN production await full discovery. While STING seems to be pivotal for cytoplasmic DNA-mediated IFN production, STING does not directly associate with DNA. Thus, other sensor(s) must exist to trigger this process and regulate the STING pathway. Molecules that have been reported to play a role in the sensing of intracellular DNA to produce IFN include DAI (DNA-dependent activator of IRFs, also referred to as Z DNA binding-binding protein-1 [ZBP-1]) [53]. DAI overexpression was shown to produce IFN in L929 cells and it remains to be determined if DAI functions through the STING pathway [14]. However, cells derived from DAI deficient mice induced type I IFN normally in response to cytosolic DNA[41]. DAI deficient mice were also found to elicit a normal immune response to DNA based vaccines [41]. This data would suggests that DAI is not solely responsible for the recognition of foreign DNA.
Other molecules reported to play a role in sensing intracellular DNA species includes the high mobility group box (HMGB) proteins [54]. Cells lacking HMGB1 exhibited defects in both intracellular DNA and poly IC mediated IFN production, but not LPS. In contrast, cells lacking HMGB2 exhibited defects in IFN production only in response to intracellular DNA. Combined HMGB1-3 loss manifested defects in AIM2-dependent IL-β production. It was thus suggested that HMBG's are required for full-blown activation of innate immune responses by cytosolic nucleic acids (i.e regulate the RLR, TLR, AIM2 and STING pathways [54]. The physiological importance of the HMGB's in initiating innate signaling awaits clarification as does how these proteins may interact with various downstream signaling molecules.
Other proteins have also been implicated in being able to sense cytoplasmic DNA and to regulate innate signaling responses. For example, IFI16, which is an IFN-inducible protein and member of the PYHIN protein family that contains a pryin domain and two DNA binding HIN domains has recently been shown to play a role in detecting intracellular DNA and triggering IFN production [55]. RNAi studies indicated that IFI16 depletion reduces the induction of IFN by synthetic DNA and HSV-1 and is dependent on STING. Further analysis of animals lacking IFI16 will underscore the importance of this molecule in innate signaling pathways. Finally, although is highly likely that other candidate DNA sensors will no doubt be reported, DEAH/RNA helicase A helicases have also been implicated in sensing microbial DNA in pDC's. Collectively, the host appears to have evolved a variety of such sensors to detect microbial DNA species, many of which could be cell-specific and which have evolved to cater to different microbes [56].
Concluding remark
Significant progress has now been made into unraveling mechanisms of intracellular DNA-mediated innate responses, such as TLR9-mediated induction of type I IFN as well as AIM2 production of IL-1β. In addition, the discovery of STING as a key modulator of both DNA and RNA triggered innate immune signaling should additionally facilitate our understanding of these innate signaling processes. Given these observations, it will be of interest to evaluate the importance of STING in stimulating host defenses against other types of DNA pathogens including parasites such as malaria. Key issues that remain to be elucidated also include the clarification of key cytosolic DNA receptor(s) which activate the STING pathway to induce IFN expression. It will also be important to determine the role of STING-dependent and AIM2-dependent signaling in autoimmune diseases and to see if such pathways can be targeted to treat such disorders. In contrast, stimulating these pathways may facilitate vaccination or anti-pathogen prophylactic strategies. Finally, it is still unclear as where these sensors recognize DNA and trigger subsequent signaling events. The finding that STING is present in the ER, as well as key TLR's such as 3, 7 and 9 make this part of the cell an important component of innate immune signaling. The role of the translocon in innate signaling also remains to be further studied, the findings of which may facilitate the treatment of nucleic acid based diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 7.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 8.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]; **This manuscript describes the identification of TLR9 as the sensor for CpG DNA.
- 9.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]; **Manuscripts 9-12 demonstrate AIM2, a HIN-200/PYD domain containing protein, as the sensor for cytoplasmic DNA-mediated formation of an inflammasome and production of IL1β.
- 10.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 12.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Manuscripts 13-14 identify a new molecule, STING as a regulating pathway that activates type I interferon in response to cytosolic DNA and a variety of DNA pathogens.
- 14.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 16.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This manuscript shows that latent TLR9 resides in the ER and following trafficking is processed in endolysosomes to become active and receptive as a sensor to CpG DNA triggered innate immune responses.
- 17.Hochrein H, Schlatter B, O'Keeffe M, Wagner C, Schmitz F, Schiemann M, Bauer S, Suter M, Wagner H. Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent and -independent pathways. Proc Natl Acad Sci U S A. 2004;101:11416–11421. doi: 10.1073/pnas.0403555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, Mudd S, Shamel L, Sovath S, Goode J, et al. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci U S A. 2004;101:3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson HL, Dar A, Napper SK, Marianela Lopez A, Babiuk LA, Mutwiri GK. Immune mechanisms and therapeutic potential of CpG oligodeoxynucleotides. Int Rev Immunol. 2006;25:183–213. doi: 10.1080/08830180600785868. [DOI] [PubMed] [Google Scholar]
- 21.Krieg AM. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008;27:161–167. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- 22.Santiago-Raber ML, Baudino L, Izui S. Emerging roles of TLR7 and TLR9 in murine SLE. J Autoimmun. 2009;33:231–238. doi: 10.1016/j.jaut.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13:543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- 24.Ehlers M, Fukuyama H, McGaha TL, Aderem A, Ravetch JV. TLR9/MyD88 signaling is required for class switching to pathogenic IgG2a and 2b autoantibodies in SLE. J Exp Med. 2006;203:553–561. doi: 10.1084/jem.20052438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 26.Spies B, Hochrein H, Vabulas M, Huster K, Busch DH, Schmitz F, Heit A, Wagner H. Vaccination with plasmid DNA activates dendritic cells via Toll-like receptor 9 (TLR9) but functions in TLR9-deficient mice. J Immunol. 2003;171:5908–5912. doi: 10.4049/jimmunol.171.11.5908. [DOI] [PubMed] [Google Scholar]
- 27.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]; **The manuscript presents evidence that cytosolic DNA activates type I interferon responses independent of the TLR pathway.
- 28.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]; **Manuscripts 29-31 describe the indentification of MITA/ERIS and MPYS (STING homologues) and show a role in innate signaling and apoptosis.
- 30.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartmann E, Gorlich D, Kostka S, Otto A, Kraft R, Knespel S, Burger E, Rapoport TA, Prehn S. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem. 1993;214:375–381. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- 34.Guo W, Novick P. The exocyst meets the translocon: a regulatory circuit for secretion and protein synthesis? Trends Cell Biol. 2004;14:61–63. doi: 10.1016/j.tcb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M, Jr, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]; **The group implicates TBK1 in exocyst function and the exocyst in innate immune signaling.
- 36.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O'Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010 doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, et al. The ENU-induced Goldenticket (Gt) mouse mutant reveals an essential function of Sting (Tmem173, Mita, Mpys, Eris) in the in vivo interferon response to Listeria monocytogenes and cyclic-di-nucleotides. Infect Immun. 2010 doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Saitoh T, Fujita N, Yoshimori T, Akira S. Regulation of dsDNA-induced innate immune responses by membrane trafficking. Autophagy. 2010;6:430–432. doi: 10.4161/auto.6.3.11611. [DOI] [PubMed] [Google Scholar]
- 41.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]; **This manuscript shows that TBK1 is important for plasmid DNA-mediated innate immune responses.
- 42.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]; **This manuscript emphasizes that DNA-mediated autoimmune disease involves the overstimulation of type I interferon production.
- 43.Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 44.Yasutomo K, Horiuchi T, Kagami S, Tsukamoto H, Hashimura C, Urushihara M, Kuroda Y. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 45.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, Nagata S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 46.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]; **Manuscripts 46-48 demonstrate a role for the 3′-5′ exonuclease TREX1 in preventing intracellular DNA from over stimulating innate immune signaling pathways and contributing to autoimmune disease.
- 47.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 48.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 49.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrilli V, Dostert C, Muruve DA, Tschopp J. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The group indicates that DNA-dependent RNA polymerase III can synthesize 5′-ppp RNA from certain cytosolic DNA species such as poly (dA-dT) and activate RIG-I signaling.
- 52.Choi MK, Wang Z, Ban T, Yanai H, Lu Y, Koshiba R, Nakaima Y, Hangai S, Savitsky D, Nakasato M, et al. A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proc Natl Acad Sci U S A. 2009;106:17870–17875. doi: 10.1073/pnas.0909545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]; **The ZBP-binding protein DAI may be a cytosolic DNA sensor in certain cells.
- 54.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]; **Mobility group box (HMGB) proteins 1,2 and 3 were shown to function as sentinels for nucleic acid-mediated innate immune signaling.
- 55.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The DNA binding HIN-200/PYD containing protein IFI16, a member of the same family as AIM2, was shown to be a sensor for intracellular DNA-mediated type I IFN production.
- 56.Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]