Abstract
Background
Severe atopic conditions associated with elevated serum IgE are heterogeneous with few known causes. Nearly every patient with autosomal-dominant hyper-IgE syndrome (AD-HIES) due to signal transducer and activator of transcription 3 (STAT3) mutations has a history of eczematous dermatitis and elevated IgE; however, clinical atopy has never been systematically studied.
Objective
Understanding of genetic determinants of allergic disease may lead to novel therapies in controlling allergic disease.
Methods
We conducted clinical evaluation of the rates of food allergies and anaphylaxis in patients with AD-HIES, a cohort of patients with no STAT3 mutation but with similar histories of elevated IgE and atopic dermatitis, and healthy volunteers with no history of atopy. Morphine skin prick testing, ImmunoCAP assays for allergen-specific IgE, and basophil activation were measured. A model of systemic anaphylaxis was studied in transgenic mice carrying an AD-HIES mutation. STAT3 was silenced in LAD2 and primary human mast cells to study the role of STAT3 in signaling and degranulation after IgE cross-linking.
Results
Food allergies and anaphylaxis were markedly diminished in patients with AD-HIES compared with a cohort of patients with no STAT3 mutation but with similar histories of elevated IgE and atopic dermatitis. Morphine skin prick testing and basophil activation were diminished in patients with AD-HIES, whereas mice carrying an AD-HIES mutation were hyporesponsive to systemic anaphylaxis models. Rapid mast cell STAT3 serine727 phosphorylation was noted after IgE cross-linking, and inhibition of STAT3 signaling in mast cells lead to impaired FcεRI-mediated proximal and distal signaling, as well as reduced degranulation.
Conclusion
This study serves as an example for how mutations in specific atopic pathways can lead to discrete allergic phenotypes, encompassing increased risk of some phenotypes but a relative protection from others.
Keywords: Autosomal dominant hyper-IgE syndrome, Job syndrome, signal transducer and activator of transcription 3, mast cell, degranulation
Studies of allergy in the context of monogenic diseases can help shed mechanistic light on the contribution of select defined genes and pathways to the pathogenesis of atopy. Understanding the effect of disruptions of these pathways can provide insight into the diagnosis and management of more common allergic disease.
Patients with autosomal-dominant hyper-IgE syndrome (AD-HIES) carry dominant-negative signal transducer and activator of transcription 3 (STAT3) mutations, develop frequent skin and lung infections, and also have a variety of nonimmunologic manifestations that affect bones and connective tissue.1,2 In addition, almost all have an eczematous rash3 present early in life, as well as the markedly elevated serum IgE levels that give the disease its name.4 Of note, one-third of patients in the broader population with atopic dermatitis develop food allergies.5 Despite these observations, the susceptibility of patients with AD-HIES to specific food allergies has not been carefully examined. We have found that fewer patients with AD-HIES develop food allergies and anaphylaxis than patients with marked IgE elevations and eczema without STAT3 mutations. This is at least in part because of the effects of defective STAT3 signaling on mast cell degranulation.
METHODS
Human subjects and clinical studies
All studies that involved human subjects were performed in accordance with protocols 10-I-0148 and 00-I-0159, approved by the National Institute of Allergy and Infectious Diseases Institutional Review Board. The study population included healthy volunteers (N = 41), patients with severe atopic disease referred to the National Institutes of Health (NIH; N = 65), and patients with AD-HIES (N = 71). All patients referred to the National Institute of Allergy and Infectious Diseases with severe atopy were screened for enrollment in the study, and those patients consenting to inclusion were examined by an allergist/immunologist. Patients with AD-HIES were healthy and visited the NIH for annual wellness examinations. Most patients were taking antimicrobials at the time of the study, and none were on systemic immunosuppressants or mast cell stabilizers. Further, a subset of patients and healthy volunteers agreed to skin prick testing and blood draw that required withdrawal of long-acting antihistamine therapy for 5 to 7 days. ImmunoCAP assays were performed for patients whose serum was available. The STAT3 locus of atopic control and patients with AD-HIES was sequenced to confirm phenotypic diagnoses. Within the surveyed group, 42% of patients with AD-HIES had mutations in the DNA binding domain, 55% in the SH2 region, and 2.5% in the transactivation region of STAT3. Serum, plasma, and peripheral blood mononuclear cells were prepared from venous blood and preserved.
Subjects were either directly examined by, or their history and physical were supervised by, clinicians trained in allergy and immunology to make an assessment of the presence or absence of atopic dermatitis and IgE-mediated food allergy. Because of the subjects’ severe atopy and the distinct clinical features of AD-HIES, examining clinicians were not blinded to the patients’ cohorts. A childhood history of outgrown atopic disease was recorded as a positive response. Symptoms judged not to be IgE mediated, for example, oral allergy syndrome, were recorded as a negative response. Diagnoses were based on published consensus guidelines for the diagnosis of atopic dermatitis, food allergy, and anaphylaxis.6,7 Significance was determined by a 2-tailed χ2 test with the use of GraphPad Prism 5.0 software (GraphPad Inc, San Diego, Calif).
Morphine sulfate at concentrations of 0.1, 0.3, 1, 3, and 10 mg/mL was used as a means of direct mast cell degranulation. Positive histamine and negative diluent controls were used. Reactions were recorded at 15 minutes as the length equating to the widest point on the wheal and the width measuring perpendicular to the widest point. A wheal of 3 mm greater than the negative control was considered positive.
ImmunoCAP assay
Sera or plasma were analyzed for allergen-specific IgE antibodies by using the Phadia ImmunoCAP System (Phadia AB, Uppsala, Sweden) as detailed by the manufacturer. Briefly, venous blood samples were obtained at the time of initial evaluation, and sera/plasma were stored at −80°C until analyzed. Standards and samples were added to the solid matrix (ImmunoCAP) and incubated. After appropriate washing, enzyme-labeled anti-IgE was added to the ImmunoCAP and incubated. After a washing step, bound anti-IgE was detected with appropriate reagents, and fluorescence was determined and compared with the standard curve to determine allergen-specific antibody concentration. Sera/plasma were analyzed for the following food allergens: egg white, milk, and peanut.
Basophil activation test
Peripheral blood was collected in heparinized tubes and incubated on ice before the assay. Whole blood (100 µL) was divided into aliquots in Eppendorf tubes, and 20 µL of basophil stimulation buffer (BSB; 20 mmol/ L HEPES, 125 mmol/L NaCl, 5 mmol/L KCl, 2.4 mmol/L CaCl2•2H20, 1 mmol/L MgCl2•2H20, 0.5 mmol/L glucose, 0.1% BSA, pH 7.4 filter sterilized) with 10 ng/mL recombinant human IL-3 was added to each tube. Cells were incubated with PBS, BSB with increasing concentrations of anti-IgE, or 0.002 mmol/L f-Met-Leu-Phe (Sigma-Aldrich, St Louis, Mo) in BSB for 15 minutes at 37°C. Degranulation was halted by transferring samples to an ice bath. Cells were immediately stained with Lin1-fluorescein isothiocyanate (BD Biosciences, San Jose, Calif), CD63-phycoerythrin (PE; BD Biosciences), CD123-PE/cyanine 5 (BD Biosciences), and CD203c-allophycocyanin (BioLegend, San Diego, Calif) for 20 minutes on ice. Red blood cells were lysed and PBMCs were fixed with fluorescence-activated cell sorting lysing solution (BD Biosciences) at room temperature for 15 minutes. After removal of fluorescence-activated cell sorting lysing solution, samples were frozen in 10% dimethyl sulfoxide/PBS and acquired together on a Fortessa flow cytometer with the use of Diva software (BD Biosciences). All flow cytometric data were analyzed with FlowJo software (TreeStar Inc, Ashland, Ore).
mut-Stat3 mice
mut-Stat3 mice were a generous gift from John O’Shea (S. Steward-Tharp, A. Laurence, and J. O’Shea, manuscript in preparation) and were housed and bred in accordance with the guidelines of the NIH protocol LAD10E. The mut-Stat3 transgene encodes a deletion of V463 in the DNA binding domain of STAT3, a mutation found in a number of patients with AD-HIES. The mice carry 2 copies of a bacterial artificial chromosome that contains the transgene and were crossed onto a C57Bl/6 background. All comparisons were to wild-type littermate controls. After euthanasia, peritoneal exudate cells were isolated by peritoneal lavage. Cells were counted and blocked with PBS/1% BSA that contained 100 µg/mL rat-IgG1 (Jackson ImmunoResearch Laboratories Inc, West Grove, Pa), 100 µg/mL hamster-IgG (Jackson ImmunoResearch Laboratories Inc), and 100 µg/mL anti-CD16/32 (FcBlock; BD Biosciences). Mast cells were detected by staining with anti–IgE-PE (eBioscience, San Diego, Calif), anti–FcεRI(MAR-1)-PE (eBioscience), CD117-allophycocyanin (BD Biosciences), and anti–Lin-V450 (BD Biosciences). Transgene negative littermates were used as controls in all mouse experiments.
Passive systemic anaphylaxis assay
Mice were sensitized intravenously with 3 µg of 2,4-dinitrophenol (DNP)-specific IgE (H1-DNP-e−26) in 200 mL of PBS and were challenged intravenously after 24 hours with 0.6 µg of rat anti-mouse IgE in 200 µL of PBS (BD Pharmingen, San Diego, Calif). Before injection into mice, the anti-mouse IgE antibodies were dialyzed against PBS to eliminate the sodium azide in the preparation and then were ultracentrifuged to remove potential aggregates. Protein concentration was determined after dialysis and centrifugation. Alternatively, anaphylaxis was induced by intravenous injection of compound 48/80 (Sigma-Aldrich) at a sublethal concentration (50 µg in 200 µL of PBS). Concentrations ≥100 µg of compound 48/80 were lethal. Implantable electronic transponders (Bio Medic Data Systems Inc, Seaford, Del) were inserted under the dorsal skin of isoflurane-anesthetized mice at least 24 hours before the start of the anaphylaxis experiments as previously described.8 Basal body temperatures before induction of anaphylaxis and temperature changes during anaphylaxis were monitored with an electronic scanner (Bio Medic Data Systems Inc). Graphs depict means ± SEMs. Significance was determined by 2-way ANOVA with the use of GraphPad Prism 5.0 software (GraphPad Inc). Transgene negative littermates were used as wild-type controls.
Mast cell transduction and degranulation assay
The following STAT3-targeted short hairpin RNAs (shRNAs) were purchased from Sigma-Aldrich: CCGGGCTGACCAACAATCCCAAGAA CTCGAGTTCTTGGGATTGTTGGTCAGCTTTTT (TRCN0000020840), human mast cells, LAD2; CCGGGCACAATCTACGAAGAATCAACTC GAGTTGATTCTTCGTAGATTGTGCTTTTT (TRCN0000020842), human mast cells, LAD2; CCGGGCAAAGAATCACATGCCACTTCTCGA GAAGTGGCATGTGATTCTTTGCTTTTT (TRCN0000020843), LAD2; CCGGCATCTGAAACTACTAACTTTGCTCGAGCAAAGTTAGTAGTTT CAGATGTTTTTG (TRCN0000329811), LAD2; CCGGGCAAAGAA TCACATGCCACTTCTCGAGAAGTGGCATGTGATTCTTTGCTTTTTG (TRCN0000329886), LAD2; and CCGGCAACAAGATGAAGAGCAC CAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT (SHC002, control nontarget control vector).
The Mission Lentiviral packaging mix (Sigma-Aldrich) and the pLKO1 transfer vectors with shSTAT3 or control shRNA (3.4 µg) were cotransfected into 293T cells (4 × 106 cells) with FuGENE6 transfection reagent (Roche, Indianapolis, Ind) in Opti-MEM. The transfected 293T cells were grown in Dulbecco modified Eagle medium that contained FBS (10%), l-glutamine (4 mmol/L). Sixteen to 19 hours after transfection, the media were removed and replaced with fresh Dulbecco modified Eagle medium that included penicillin (100 U/mL) and streptomycin (100 µg/mL). After 62 to 65 hours, the lentivirus was collected by centrifugation (25,000 rpm for 1 hour 40 minutes at 4°C), and the resulting pellet was resuspended in 3 mL of prewarmed complete StemPro medium (Invitrogen, Carlsbad, Calif). Transduction of human mast cells was conducted by transferring the 3 mL of resuspended virus to a T75 culture flask that contained 1 to 3 × 106 human mast cells in 15 mL of complete StemPro medium. Two days after transduction, the medium was changed to virus-free complete StemPro medium, and antibiotic selection was initiated (0.2 µg/mL puromycin; Sigma-Aldrich). For experiments in LAD2 human mast cells, less lentivirus (10%) was added, and cells were not selected for puromycin. With this method, LAD2 cells were used at 5 days after infection. On day 5 or 7 after transduction, the human mast cells were incubated overnight with biotinylated human-IgE (100 ng/mL),9 rinsed with HEPES buffer that contained 0.04% BSA (Sigma-Aldrich), and then triggered in the same buffer with the indicated concentrations of stimuli for 30 minutes. Degranulation was determined by release of β-hexosaminidase from the mast cell granules as calculated as a percentage of the total amount present in the cells and supernatant fluid.
Flow cytometry for STAT3 phosphorylation
LAD2 mast cells were maintained in complete StemPro medium that was supplemented with stem cell factor (100 ng/mL). Primary human mast cells were derived from CD34+ cells from healthy volunteers. Primary human mast cells were maintained in complete StemPro medium with IL-6 (100 ng/mL) and stem cell factor (100 ng/mL). Mast cells were starved of cytokines overnight before experiments and were incubated with biotinylated human-IgE (100 ng/mL). Cells were washed with HEPES buffer that contained 0.04% BSA and were equilibrated at 37°C for 15 minutes before activation. IgE was cross-linked with 100 ng/mL streptavidin for 2, 5, and 15 minutes. Reactions were terminated by fixation with 1.6% paraformaldehyde for 10 minutes at room temperature. Cells were permeabilized with ice-cold 100% methanol at 4°C for at least 20 minutes, washed with PBS/ 0.5% BSA, and stained for flow cytometry with antibodies specific for phosphorylated (p)STAT3-Serine727-PE or pSTAT3-Tyrosine705-PE, and phosphorylated extracellular signal-regulated kinase (pERK)-Alexa Fluor 647 (all from BD Biosciences). Signaling was determined relative to an unstimulated control.
Immunoblot analysis
For cell signaling and immunoblot analysis, LAD2 human mast cells were transduced with shRNA constructs as described in the Mast cell transduction and degranulation section. At 5 days after infection, cells were sensitized with IgE as described for degranulation assays. Cells were stimulated at a concentration of 5 × 106 cells/mL in HEPES buffer, and immunoblot analysis was performed as described.10
RESULTS
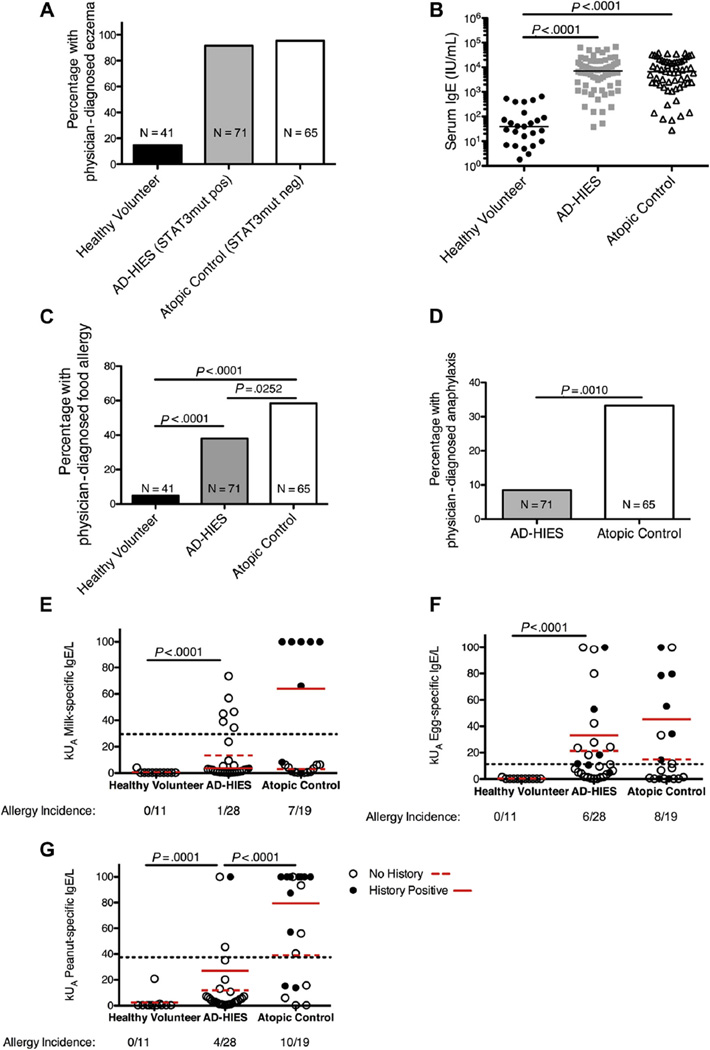
We determined the lifetime frequency and severity of food allergies among our cohort of patients with AD-HIES and a cohort of patients with severe atopy, comparably elevated IgE, but no STAT3 mutation. In addition, we examined an unselected cohort of healthy volunteers (Table I; Fig 1, A and B). The median concentrations of serum IgE in healthy volunteers, patients with AD-HIES, and atopic controls were 39.6, 7079, and 6628 IU/ mL, respectively. The healthy controls and patients with AD-HIES had a 10-year difference in age (34.51 vs 24.69 years; P =.0011), whereas the atopic control patients were significantly younger (14.98 years; P >.0001 vs healthy volunteers or patients with AD-HIES; Table I). Thus, the lifetime prevalence of allergy incidence in the atopic control group is likely to be underestimated in relation to the other patient groups. More than one-third (38%) of patients with STAT3 mutation had immediate hypersensitivity to food, significantly less than the 58.3% observed in patients with atopy without a STAT3 mutation (Fig 1, C). Far fewer patients with AD-HIES had anaphylaxis to a food allergen than atopic controls (8.5% vs 33.3%; Fig 1, D). Given that some food allergies do not present until later in life, as the control patients with atopy age, their allergy incidence may increase, thus amplifying the difference between the 2 atopic groups.
TABLE I.
Frequencies of allergies within study cohorts
| Physician-diagnosed allergy, % (frequency) |
||||||
|---|---|---|---|---|---|---|
| Cohort | No. of subjects | Female, % | Mean age, y (range) | Egg | Milk | Peanut |
| Healthy volunteer | 41 | 61 | 34.51 (18–66) | 0 | 2.6 (1/39) | 0 |
| Patients with AD-HIES | 71 | 54 | 24.69 (2–58) | 14.1 (10/71) | 4.2 (3/71) | 12.2 (9/71) |
| Atopic control | 65 | 40 | 14.98 (1–58) | 21.5 (14/65) | 24.6 (16/65) | 30.8 (20/65) |
FIG. 1.
STAT3-mutant (AD-HIES) patients have reduced incidence of food allergies compared with patients with similar degrees of atopic dermatitis, high IgE, and no STAT3 mutation. A, Physician-diagnosed eczema in healthy volunteer control, patients with AD-HIES (STAT3-mutant), and atopic (no STAT3 mutation) control cohorts. B, Serum IgE levels in healthy volunteer controls (N = 25), patients with AD-HIES (N = 72), and atopic controls (N = 60). Each symbol represents a single patient. Line represents the median concentration of serum IgE. C, Physician-diagnosed food allergies in healthy volunteers, patients with AD-HIES, and atopic control patients were determined by interview. D, Incidence of physician-diagnosed anaphylaxis in patents with AD-HIES and control patients with atopy. Significance for panels C and D as determined by a 2-tailed χ2 test. E–G, Allergen-specific IgE in serum or plasma was measured by ImmunoCAP assay for 3 common food allergens: egg, milk, and peanut. Dotted line represents the 95% positive predictive value of each allergen as previously reported.5 P values between healthy volunteers and atopic controls are P = .0013 for milk, P = .0001 for egg, and P = .0001 for peanut, respectively. Significance was determined by 2-tailed Mann-Whitney test.
One explanation for the lower incidence of allergies in patients with AD-HIES could be impairments in the production of allergen-specific IgE. Other antigen-specific antibody responses are impaired in many patients with AD-HIES11 as are the frequency of memory B cells and IL-21–dependent plasma cell differentiation.12,13 The generation of T follicular helper cells, which contribute to germinal center formation14–16 and are important for the development of high-affinity antibody, including IgE,17,18 is STAT3 dependent19 and impaired in AD-HIES,20 as is class switching to IgE in certain in vitro models.21 However, IgE class switching can occur outside of the germinal center reaction, and this pathway may be unaffected in patients with AD-HIES.18 We therefore measured allergen-specific IgE in serum from healthy volunteers, patients with AD-HIES, and atopic controls with the use of the ImmunoCAP system, which can predict the likelihood of clinical food allergy on the basis of the level of allergen-reactive IgE.22 We found that many patients with AD-HIES were capable of producing allergen-specific IgE in amounts above the level that has been validated to correlate with food allergy 95% of the time (Fig 1, E–G). Despite a higher prevalence of egg, milk, and peanut allergies in atopic controls than in patients with AD-HIES (Table I), no significant difference was found in the levels of egg- and milk-specific IgE between the 2 groups (Fig 1). Levels of peanut-specific IgE were higher in the atopic control population, and it appears that IgE levels were more well correlated to clinical allergy in the atopic controls than in the patients with AD-HIES (Fig 1, E–G; Table I). Peanut allergies accounted for a large portion of the reported food allergies (18% in patients with AD-HIES and 19% in atopic controls); however, whereas 55% of the cases of anaphylaxis in atopic controls was to peanut, there was no peanut-specific anaphylaxis in the patients with AD-HIES. Theoretically, peanut-specific anaphylactic responses could be disproportionately affected by STAT3 mutation; however, more cases of allergy and anaphylaxis to other foods in both groups would be required to reach statistical significance. Although it is still possible that STAT3-dependent differences in allergen-specific IgE may play some role in protection from food allergies and anaphylaxis in AD-HIES, patients were capable of producing high levels of multiple food allergen-specific serum IgE.
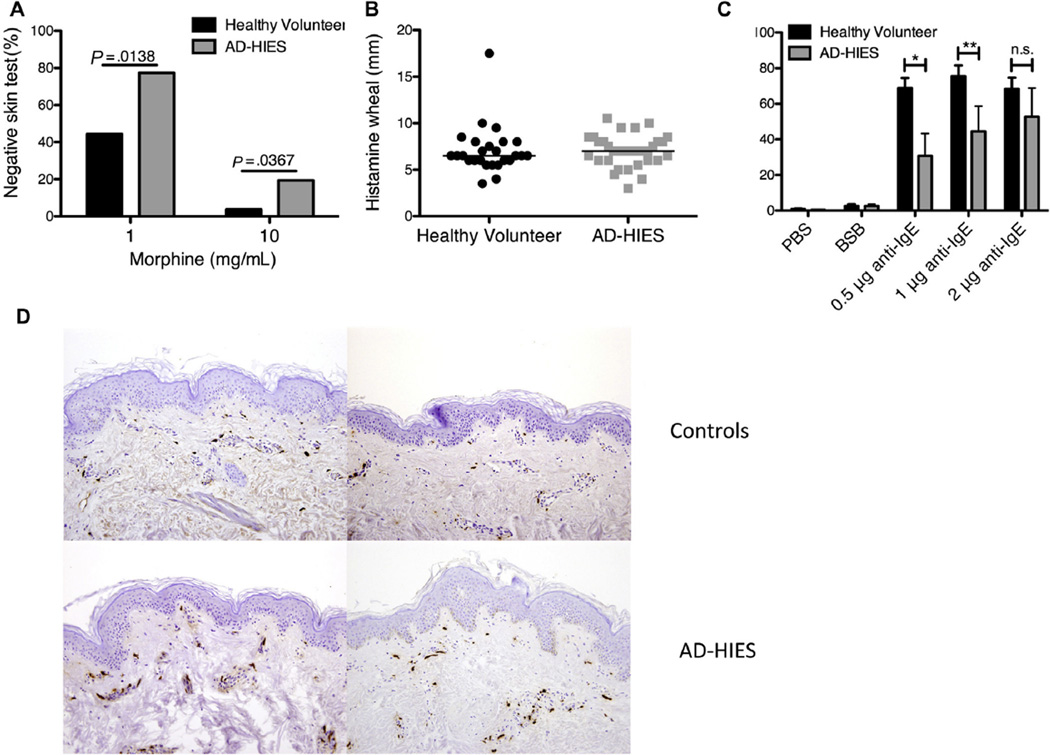
Because anaphylaxis appeared to be the phenotype that most strikingly differed between patients with AD-HIES and atopic control subjects, we investigated the pathway of mast cell degranulation. Mast cells are key mediators of immediate allergic responses and anaphylaxis, releasing histamine and other inflammatory mediators of inflammation when FcεRI-bound IgE is cross-linked by its cognate antigen. Subcutaneous morphine can elicit mast cell degranulation via G protein-coupled receptors independent of surface IgE,23 which permits interrogation of mast cell degranulation without the need for a specific allergen. We therefore performed subcutaneous morphine prick testing in healthy volunteers and patients with AD-HIES. We found that patients with AD-HIES failed to develop a skin wheal to 2 different doses of morphine at a substantially higher rate than healthy volunteers (Fig 2, A). Median wheal size in response to histamine was similar among both groups, ruling out a differential tissue response to histamine release in patients with AD-HIES (Fig 2, B). Opiate skin testing could not be performed on a sufficiently large group of atopic controls; however, previous studies have noted that atopic patients’ responses to opiates are not impaired, and if anything increased.24
FIG. 2.
Reduced mast cell degranulation in STAT3-mutant humans and mice. A, Healthy volunteers (N = 26) and patients with AD-HIES (N = 31) were skin prick tested for morphine reactivity with increasing doses of morphine. Scores of 0 or 1 were considered nonreactive. Error was determined by 1-tailed Mann-Whitney test. B, Histamine reactivity in healthy volunteers (N = 26) and patients with AD-HIES (N = 31). C, A basophil activation test was performed on healthy volunteers (N = 5) and patients with AD-HIES (N = 5) with increasing concentrations of anti-IgE. Activated basophils were gated on Lin1– CD123+ CD203c+ CD63high. Error was determined by 1- tailed Mann-Whitney test (*P = .0278, **P = .0476). D, Immunohistochemical staining of tryptase (magnification, ×20) from skin biopsies of human flank in 2 controls (top) and 2 patients with AD-HIES (bottom).
We next examined ex vivo basophil activation after IgE stimulation as a surrogate for mast cell activation because basophils are also activated via IgE cross-linking and are readily detectable in peripheral blood by flow cytometry, although they are not reported to play a prominent role in IgE-mediated anaphylaxis in mice.25 Basophils from patients with AD-HIES were found to be less sensitive to IgE cross-linking than basophils from healthy controls despite higher levels of FcεRIα on basophils from patients with AD-HIES than from nonallergic subjects (Fig 2, C).26 In contrast, we observed no significant difference in basophil activation between atopic control patients and healthy volunteers (see Fig E1, A, in this article’s Online Repository at www.jacionline.org). No difference was found in basophil activation in response to f-Met-Leu-Phe between either patients with AD-HIES or atopic control patients and healthy volunteers (Fig E1, B and C). Tryptase staining of skin biopsies from the flank of 2 STAT3-mutant patients and healthy volunteers showed a similar distribution of mast cells (Fig 2, D) between the 2 groups, arguing against a defect in mast cell trafficking or survival to explain the different responses.
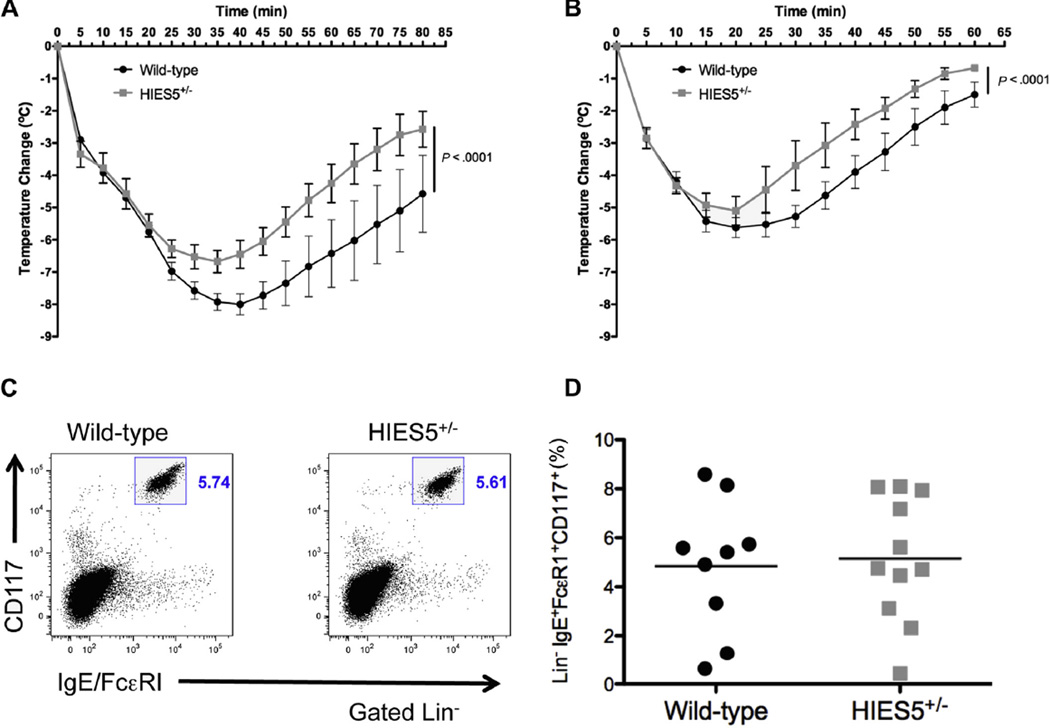
To further study in vivo mast cell function and anaphylaxis, we used the mut-Stat3 transgenic mouse that carries 2 copies of a bacterial artificial chromosome that encodes a deletion of V463 in the DNA binding domain of STAT3, a mutation found in a number of patients with AD-HIES (S. Steward-Tharp, A. Laurence, and J. O’Shea, manuscript in preparation). An IgE-mediated passive systemic anaphylaxis model was used on mut-Stat3 and wild-type littermate control mice 24 hours after injection with DNP-specific IgE. When challenged with mouse anti-IgE, the body temperature of the mut-Stat3 mice did not fall as low as the wild-type controls and remained higher during the recovery phase (P <.05 beginning at 25 minutes after challenge), indicating a lesser degree of response to mast cell-mediated anaphylaxis (Fig 3, A). Systemic anaphylaxis triggered by the mast cell secre-tagogue compound 48/80, which does not use surface IgE for degranulation, led to a similar response (P < .05 beginning at 30 minutes; Fig 3, B). Similar to our examination of human skin biopsies, the frequency and distribution of mast cells in mut-Stat3 mice in the skin and peritoneum were not disturbed nor was surface IgE expression (Fig 3, C and D). Together these data suggest that STAT3 mutations lead to impaired in vivo mast cell degranulation, which may contribute to the decreased incidence of food allergy and anaphylaxis.
FIG. 3.
Reduced mast cell degranulation in STAT3-mutant mice. A, IgE-mediated systemic anaphylaxis in mut-Stat3 mice compared with wild-type littermate controls. Data are representative of 2 independent experiments with 4 to 5 mice per group. B, Compound 48/80 was used to induce IgE-independent anaphylaxis in mut-Stat3 and wild-type mice. Data are representative of 4 mice per group. C, Dot plot of surface FcεRI expression versus CD117 expression. D, Percentage of Lin−IgE+FcεRI+CD117+ peritoneal mast cells. Data are cumulative of 2 independent experiments of age- and sex-matched animals with 4 to 6 mice per group.
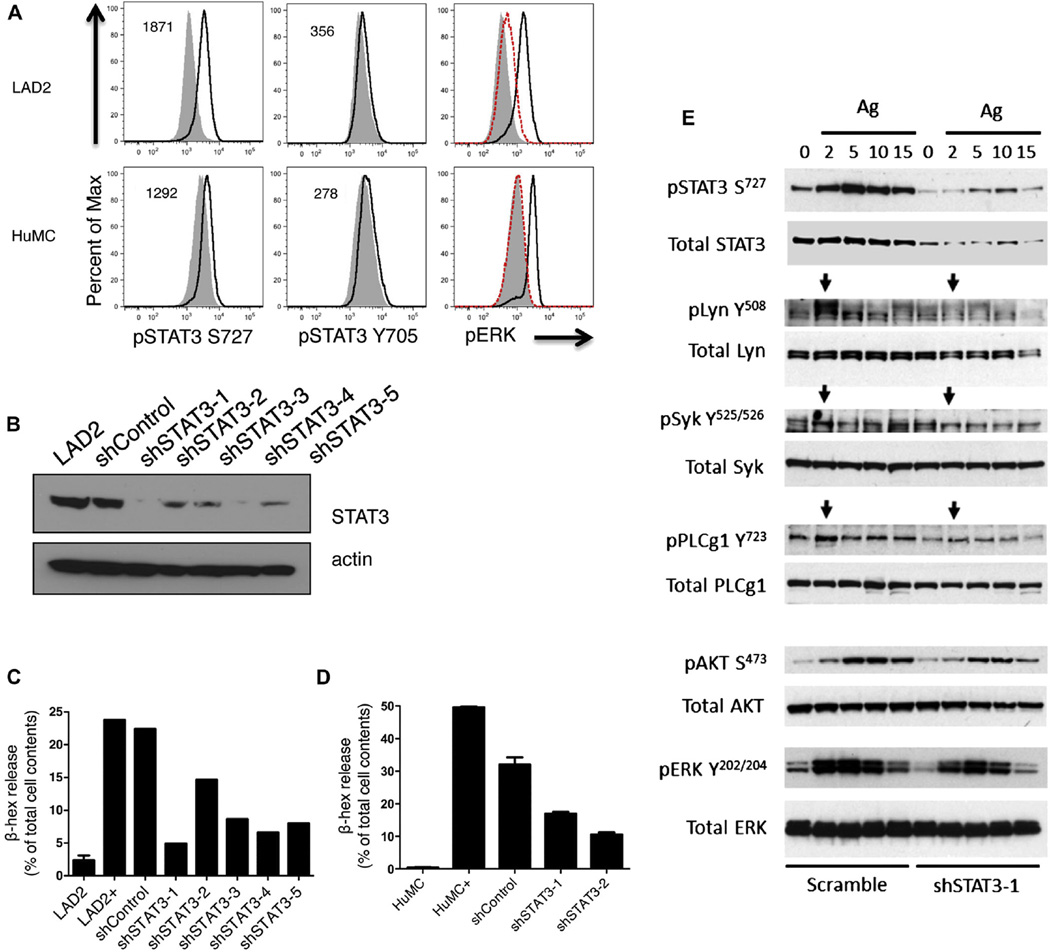
To establish a direct role for STAT3 in mast cell degranulation, we cross-linked IgE receptors on LAD2 and normal human mast cell lines and noted rapid STAT3 phosphorylation at serine727 after 2 minutes of IgE cross-linking (Fig 4, A; see also Fig E2, A, in this article’s Online Repository at www.jacionline.org), which preceded ERK phosphorylation. Minimal phosphorylation was observed at tyrosine705 on STAT3 in both LAD2 and primary human mast cell lines (Fig 4, A). To further define the role of STAT3 in mast cell biology, LAD2 mast cells were transduced with lentiviruses that encoded STAT3 shRNA constructs (Fig 4, B; Fig E2, B). Silencing of STAT3 expression inhibited mast cell degranulation after IgE cross-linking in direct proportion to the degree of silencing of STAT3 in LAD2 cells (Fig E2, C). Similarly, silencing of STAT3 in primary human mast cells lead to decreased IgE-mediated mast cell degranulation (Fig 4, D). Although knockdown of STAT3 in LAD2 mast cells did not lead to a reduction in surface FcεRI expression (see Fig E3, E, in this article’s Online Repository at www.jacionline.org), FcεRI cross-linking of the STAT3-silenced mast cells lead to reduced phosphorylation of proximal signaling molecules Lyn and Syk and more profound decreases further downstream in phosphorylated phospholipase C γ 1, Akt, and ERK (Fig 4, E; Fig E3). C3a- and compound 48/80-mediated degranulation were subtly inhibited by silencing of STAT3, whereas thapsigargin stimulation of granulation was not affected by the loss of STAT3 (Fig E3, B–D, and F). Therefore, STAT3 plays an important role in the proximal signaling involved in IgE-dependent mast cell degranulation, whereas the role of STAT3 in IgE-independent degranulation is less profound.
FIG. 4.
STAT3 contributes to human mast cell proximal and distal FcεRI signaling events. A, STAT3 phosphorylation on Serine727 and Tyrosine705 after 2 minutes of IgE cross-linking in LAD2 and primary human mast cells (HuMCs). The change in mean fluorescence intensity between stimulated (black line) and unstimulated (gray solid) cells is given in the upper left corner. As a positive control, ERK phosphorylation at 0 (solid gray), 2 (black line), and 15 (dashed red line) minutes after cross-linking was measured by flow cytometry. Data are representative of 2 independent experiments with LAD2 cells and 1 experiment in primary human mast cells. B, Western blot of lysates from the human mast cell line LAD2 transduced with 5 different shRNAs directed against STAT3. C, Mast cell degranulation was measured by FcεRI cross-linking and subsequent β-hexosaminidase (β-hex) release in LAD2 cells transduced with 5 different shRNAs against STAT3. Data are representative of 2 independent experiments. LAD2, unstimulated control; LAD+, FcεRI cross-linking. D, Mast cell degranulation was measured by FcεRI cross-linking and subsequent β-hexosaminidase release in primary human mast cells transduced with 2 different shRNAs against STAT3. HuMC, unstimulated control; HuMC+, FcεRI cross-linking. E, Silencing of STAT3 reduced proximal and distal FcεRI-induced mast cell signaling. Phosphorylation of STAT3 (S727) was evident with antigen stimulation that was much less marked with shSTAT3. Early and proximal FcεRI signaling occurs through Lyn and Syk kinases and is evident at 2 minutes (arrows) in the scramble control cells. However, silencing of STAT3 reduces phosphorylation at 2 minutes. FcεRI signaling defects were also evident with phospholipase C, γ 1 (PLCγ1) phosphorylation at 2 minutes. Defects in distal FcεRI signaling (AKT and ERK), which occur later, were also evident with STAT3 silencing. Ag, Antigen.
DISCUSSION
Although STAT3 and cytokines that use STAT3-mediated signaling have been shown to be important in mast cell survival, cytokine production, and IgE-mediated transcription,27–31 the clinical, in vivo, and in vitro studies show that mast cell degranulation itself depends on STAT3 function. Roles for STAT3 in proximal receptor-mediated signaling have been established in a variety of settings, including as scaffold protein in platelets.32 Future study is required to identify the proximal target of STAT3 in FcεRI signaling and perhaps other cell types, whereas in vivo studies of STAT3-mutated mice will hopefully determine the extent to which other mechanisms may contribute to the observed reduction in anaphylaxis in patients and mice.
This study serves as model for using monogenic diseases to better understand the pathogenesis of atopy and suggests that suppression of the STAT3 pathway could aid in control and treatment of mast cell–mediated atopic conditions.
Supplementary Material
Key messages.
Patients with AD-HIES have fewer lifetime allergies than patients with a similar degree of atopy and high levels of IgE and no STAT3 mutation.
Patients with AD-HIES have compromised mast cell degranulation and basophil activation.
Knockdown of STAT3 in mast cells leads to changes in FcεRI-mediated signaling and impaired mast cell degranulation.
Acknowledgments
We thank the patients and their families at the NIH Clinical Center for their commitment to clinical research and for generously agreeing to participate in this study. mut-Stat3 mice were a kind gift from Scott Steward-Tharp, Arian Laurence, and John O’Shea. Erica Brittain and Jeff Skinner at NIH provided statistical expertise. We thank the members of the Milner laboratory for their helpful suggestions about this work.
Supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases and National Institute of Arthritis and Musculoskeletal and Skin Diseases within the National Institutes of Health.
Abbreviations used
- AD-HIES
Autosomal dominant hyper-IgE syndrome
- BSB
Basophile stimulation buffer
- DNP
2,4-Dinitrophenol
- ERK
Extracellular signal-regulated kinase
- NIH
National Institutes of Health
- PE
Phycoerythrin
- shRNA
Short hairpin RNA
- STAT3
Signal transducer and activator of transcription 3
Footnotes
Disclosure of potential conflict of interest: N. Jones has been supported by one or more grants from SAIC. M. O’Brien owns stock/stock options in PFE. J. Rivera is a member of the University of Singapore Scientific Advisory Board, is chair of the INFLAMEX LABEX Center of Excellence-INSERM SAB, and has one or more patents (planned, pending, or issued) with the National Institutes of Health. A. M. Siegel has been supported by one or more grants from the National Institute of Allergy and Infectious Diseases (Intramural Funding) and is employed by and owns stock/stock options in Amplimmune, Inc. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 2.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 3.Sowerwine KJ, Holland SM, Freeman AF. Hyper-IgE syndrome update. Ann N Y Acad Sci. 2012;1250:25–32. doi: 10.1111/j.1749-6632.2011.06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley RH, Wray BB, Belmaker EZ. Extreme hyperimmunoglobulinemia E and undue susceptibility to infection. Pediatrics. 1972;49:59–70. [PubMed] [Google Scholar]
- 5.Eigenmann PA, Sicherer SH, Borkowski TA, Cohen BA, Sampson HA. Prevalence of IgE-mediated food allergy among children with atopic dermatitis. Pediatrics. 1998;101:E8. doi: 10.1542/peds.101.3.e8. [DOI] [PubMed] [Google Scholar]
- 6.Boyce JA, Assa’ad A, Burks AW, Jones SM, Sampson HA, Wood RA, et al. Guidelines for the diagnosis and management of food allergy in the United States: Summary of the NIAID-sponsored Expert Panel Report. J Allergy Clin Immunol. 2010;126:1105–1118. doi: 10.1016/j.jaci.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieberman P, Nicklas RA, Oppenheimer J, Kemp SF, Lang DM, Bernstein DI, et al. The diagnosis and management of anaphylaxis practice parameter: 2010 update. J Allergy Clin Immunol. 2010;126:477–480. doi: 10.1016/j.jaci.2010.06.022. e1–42. [DOI] [PubMed] [Google Scholar]
- 8.Olivera A, Eisner C, Kitamura Y, Dillahunt S, Allende L, Tuymetova G, et al. Sphingosine kinase 1 and sphingosine-1-phosphate receptor 2 are vital to recovery from anaphylactic shock in mice. J Clin Invest. 2010;120:1429–1440. doi: 10.1172/JCI40659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuehn HS, Radinger M, Gilfillan AM. Measuring mast cell mediator release. Curr Protoc Immunol. 2010:38. doi: 10.1002/0471142735.im0738s91. Chapter 7: Unit7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruse G, Beaven MA, Ashmole I, Bradding P, Gilfillan AM, Metcalfe DD. A truncated splice-variant of the FcepsilonRIbeta receptor subunit is critical for microtu-bule formation and degranulation in mast cells. Immunity. 2013;38:906–917. doi: 10.1016/j.immuni.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckley RH. The hyper-IgE syndrome. Clin Rev Allergy Immunol. 2001;20:139–154. doi: 10.1385/CRIAI:20:1:139. [DOI] [PubMed] [Google Scholar]
- 12.Avery DT, Deenick EK, Ma CS, Suryani S, Simpson N, Chew GY, et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med. 2010;207:155–171. doi: 10.1084/jem.20091706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speckmann C, Enders A, Woellner C, Thiel D, Rensing-Ehl A, Schlesier M, et al. Reduced memory B cells in patients with hyper IgE syndrome. Clin Immunol. 2008;129:448–454. doi: 10.1016/j.clim.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong H, Dolpady J, Wabl M, Curotto de Lafaille MA, Lafaille JJ. Sequential class switching is required for the generation of high affinity IgE antibodies. J Exp Med. 2012;209:353–364. doi: 10.1084/jem.20111941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erazo A, Kutchukhidze N, Leung M, Christ AP, Urban JF, Jr, Curotto de Lafaille MA, et al. Unique maturation program of the IgE response in vivo. Immunity. 2007;26:191–203. doi: 10.1016/j.immuni.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma CS, Avery DT, Chan A, Batten M, Bustamante J, Boisson-Dupuis S, et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012;119:3997–4008. doi: 10.1182/blood-2011-11-392985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avery DT, Ma CS, Bryant VL, Santner-Nanan B, Nanan R, Wong M, et al. STAT3 is required for IL-21-induced secretion of IgE from human naive B cells. Blood. 2008;112:1784–1793. doi: 10.1182/blood-2008-02-142745. [DOI] [PubMed] [Google Scholar]
- 22.Sampson HA, Ho DG. Relationship between food-specific IgE concentrations and the risk of positive food challenges in children and adolescents. J Allergy Clin Immunol. 1997;100:444–451. doi: 10.1016/s0091-6749(97)70133-7. [DOI] [PubMed] [Google Scholar]
- 23.Klinker JF, Seifert R. Morphine and muscle relaxants are receptor-independent G-protein activators and cromolyn is an inhibitor of stimulated G-protein activity. Inflamm Res. 1997;46:46–50. doi: 10.1007/s000110050058. [DOI] [PubMed] [Google Scholar]
- 24.Cohen RW, Rosenstreich DL. Discrimination between urticaria-prone and other allergic patients by intradermal skin testing with codeine. J Allergy Clin Immunol. 1986;77:802–807. doi: 10.1016/0091-6749(86)90377-5. [DOI] [PubMed] [Google Scholar]
- 25.Tsujimura Y, Obata K, Mukai K, Shindou H, Yoshida M, Nishikado H, et al. Basophils play a pivotal role in immunoglobulin-G-mediated but not immunoglobulin-E-mediated systemic anaphylaxis. Immunity. 2008;28:581–589. doi: 10.1016/j.immuni.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Saini SS, Klion AD, Holland SM, Hamilton RG, Bochner BS, Macglashan DW., Jr The relationship between serum IgE and surface levels of FcepsilonR on human leukocytes in various diseases: correlation of expression with FcepsilonRI on basophils but not on monocytes or eosinophils. J Allergy Clin Immunol. 2000;106:514–520. doi: 10.1067/mai.2000.108431. [DOI] [PubMed] [Google Scholar]
- 27.Traum D, Timothee P, Silver J, Rose-John S, Ernst M, LaRosa DF. IL-10-induced gp130 expression in mouse mast cells permits IL-6 trans-signaling. J Leukoc Biol. 2011;91:427–435. doi: 10.1189/jlb.0411209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnenblick A, Levy C, Razin E. Immunological trigger of mast cells by mono-meric IgE: effect on microphthalmia transcription factor, STAT3 network of interactions. J Immunol. 2005;175:1450–1455. doi: 10.4049/jimmunol.175.3.1450. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki Y, Yoshimaru T, Inoue T, Ra C. Mitochondrial Ca2+ flux is a critical determinant of the Ca2+ dependence of mast cell degranulation. J Leukoc Biol. 2006;79:508–518. doi: 10.1189/jlb.0705412. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita T, Sawai N, Hidaka E, Yamashita T, Koike K. Interleukin-6 directly modulates stem cell factor-dependent development of human mast cells derived from CD34(+) cord blood cells. Blood. 1999;94:496–508. [PubMed] [Google Scholar]
- 31.Drube S, Heink S, Walter S, Lohn T, Grusser M, Gerbaulet A, et al. The receptor tyrosine kinase c-Kit controls IL-33 receptor signaling in mast cells. Blood. 2010;115:3899–3906. doi: 10.1182/blood-2009-10-247411. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Gushiken FC, Bolgiano D, Salsbery BJ, Aghakasiri N, Jing N, et al. Signal transducer and activator of transcription 3 (STAT3) regulates collagen-induced platelet aggregation independently of its transcription factor activity. Circulation. 2013;127:476–485. doi: 10.1161/CIRCULATIONAHA.112.132126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.