Abstract
The proteasome is a multimeric and multicatalytic intracellular protease responsible for the degradation of proteins involved in cell cycle control, various signaling processes, antigen presentation, and control of protein synthesis. The central catalytic complex of the proteasome is called the 20S core particle. The majority of these are flanked on one or both sides by regulatory units. Most common among these units is the 19S regulatory unit. When coupled to the 19S unit, the complex is termed the asymmetric or symmetric 26S proteasome depending on whether one or both sides are coupled to the 19S unit, respectively. The 26S proteasome recognizes poly-ubiquitinylated substrates targeted for proteolysis. Targeted proteins interact with the 19S unit where they are deubiquitinylated, unfolded, and translocated to the 20S catalytic chamber for degradation. The 26S proteasome is responsible for the degradation of major proteins involved in the regulation of the cellular cycle, antigen presentation and control of protein synthesis. Alternatively, the proteasome is also active when dissociated from regulatory units. This free pool of 20S proteasome is described in yeast to mammalian cells. The free 20S proteasome degrades proteins by a process independent of poly-ubiquitinylation and ATP consumption. Oxidatively modified proteins and other substrates are degraded in this manner. The 20S proteasome comprises two central heptamers (β-rings) where the catalytic sites are located and two external heptamers (α-rings) that are responsible for proteasomal gating. Because the 20S proteasome lacks regulatory units, it is unclear what mechanisms regulate the gating of α-rings between open and closed forms. In the present review, we discuss 20S proteasomal gating modulation through a redox mechanism, namely, S-glutathionylation of cysteine residues located in the α-rings, and the consequence of this post-translational modification on 20S proteasomal function.
Abbreviations: GSH, reduced glutathione; GSSG, oxidized glutathione; ROS, reactive oxygen species; SAXS, small angle X-rays scattering; 20SPT, 20S proteasome core particle; 26SPT, 26S proteasome; TEM, transmission electron microscopy
Keywords: Proteasome, S-glutathionylation, Oxidized proteins, Proteasomal gating, Redox regulation
Graphical abstract
Highlights
-
•
Four main molecular mechanisms are implicated in glucose-mediated vascular damage.
-
•
Impaired antioxidant defense contributes to T2DM and related complications.
-
•
SNPs in antioxidant enzymes are associated with pathogenesis of type 2 diabetes.
-
•
Genotyping of gene variants in populations will help identify individuals at risk.
The proteasome
The proteasome is a ubiquitous, intracellular, multimeric, and multicatalytic protease responsible for the degradation of intracellular proteins [1]. It is composed of a central unit called 20SPT. This unit is most often flanked on one or both sides by regulatory units such as 19S, 11S, and/or PA200. The most abundant regulatory complex is the 19S unit, which recognizes poly-ubiquitinylated proteins. When coupled to the 19S unit, the proteasome is termed 26SPT. The 26SPT is responsible for quality control of protein synthesis and the degradation of proteins involved in major metabolic pathways related to cell cycle regulation, antigen presentation, and most signaling processes. The poly-ubiquitinylated proteins bind to specific subunits of the 19S complex, are deubiquitinylated by enzymes present in the 19S unit, are unfolded, and are translocated to the catalytic chamber through the ATPase activity of a hexameric ring located in the base of the 19S unit. The 20SPT comprises two α and two β heptameric rings. The internal β rings are responsible for catalysis, while the peripheral α rings are responsible for the gating of the 20SPT.
Because 20S proteasome free pool lacks regulatory units, the regulation of the gating mechanism between open and closed forms is unclear. We have presented evidence that S-glutathionylation of cysteine residues located in the α-rings may represent a redox post-translational regulation of the 20S proteasome.
Protein S-glutathionylation
The term S-glutathionylation was adopted in this review by taking into account the conclusive criticism on the alternative nomenclature (S-glutathiolation), as discussed by Mieyal & Chock [2].
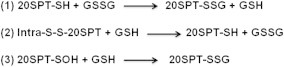
S-glutathionylation is the formation of mixed disulfides between glutathione and cysteine residues of proteins. Originally described as a result of oxidative stress [3], S-glutathionylation was later recognized as a post-translational modification that can play major regulatory functions [3–6]. In the last 10 years, the investigation on protein S-glutathionylation was intensified because of the increased understanding on ROS and NO derived species role in signal transduction and most likely by the reversibility of protein S-glutathionylation. Either protein thiols or glutathione (GSH) can be activated by oxidants to further react with other sulfhydryls. Therefore, protein S-glutathionylation might transduce redox signals generated by ROS and NO derived species [7,8]. Because the intracellular environment is highly reducing, except in some organelles, such as the endoplasmic reticulum, very few protein thiols are prone to be S-glutathionylated. Therefore, it is arguable that S-glutathionylation occurs in a site- and protein-specific manner. The global susceptibility and site-specific specificity of proteins for S-glutathionylation is dependent on the reactivity of their thiol groups. Two relevant factors in protein susceptibility for S-glutathionylation may be thiol steric accessibility and thiol pKa; these factors depend upon protein folding and vicinity to side chains of basic amino acids, respectively. The specificity of protein S-glutathionylation remains an active field of research [5,6,9]. The chemical mechanisms of protein S-glutathionylation proposed to date [5,10] are based on either thiol protein or GSH pool redox modifications (Fig. 1). These reactions may be triggered through ROS or NO derived species. The most cited and explored mechanisms of protein S-glutathionylation are based on either thiol–disulfide exchange through protein thiolate and glutathione disulfide (GSSG) or the reaction between an oxidized thiol to sulfenic acid with the reduced form of GSH.
Fig. 1.

Major mechanisms of proteinS-glutathionylation. Route 1 describes the classical mechanism of protein S-glutathionylation. This mechanism causes modification through thiol–disulfide exchange. Formation of adduct may be triggered by an increased GSSG pool mediated by intensified ROS formation or the oxidized glutathione species GSOH and GS(O)SG. Route 2 describes mechanisms based on protein sulfenic acid formation through reaction with peroxides or peroxynitrite followed by reaction with GSH. Other physiological oxidants, e.g., hypochlorous acid or chloramine derivatives can also oxidize thiol groups to the sulfenic form [15]. Route 3 describes mechanisms based on nitrosylated protein or gluathionyl derivatives that are formed through reaction with intermediates of NO radical metabolism.
Data obtained by new methodologies based on encoded fluorescent protein-glutaredoxin conjugates indicated that intracellular GSH/GSSG ratios could be two orders magnitude higher compared to those previously described [11,12]. Cells most likely can maintain such highly reducing intracellular conditions through elimination of excess GSSG by either extrusion [13] or vacuole uptake [12]. A GSH/GSSG in the range of 10,000 would suggest that S-glutathionylation through thiol–disulfide exchange is highly specific to selected proteins or even improbable. However, investigation on the unintended consequences on cell biology of adding a genetically encoded fluorescent probe is required. Indeed, expression of some of these probes in fibroblasts resulted in the appearance of a dimer form that exhibited an intermediate excitation fluorescence spectrum between those of oxidized and reduced monomers [14].
Other mechanisms of protein S-glutathionylation are based on protein thiol activation by oxidants such as hydrogen peroxide, peroxynitrite, and others through formation of sulfenic acid (P-SOH; [15]), followed by reaction with GSH (Scheme 1, reaction 2; [16]). However, proteins such as peroxiredoxins and glutathione peroxidases are one million times more reactive towards hydroperoxides than other thiol-proteins, such as protein tyrosine phosphatases [17,18]. Therefore, peroxiredoxins and glutathione peroxidases are the most probable biological sensors of hydroperoxides. These proteins would then have to transmit peroxide binding signals to other redox players, such as thioredoxin [19,20]. However, the mechanisms that connect thiol oxidation by hydroperoxides to protein S-glutathionylation are still elusive.
Scheme 1.
Proposed mechanisms for the S-glutathionylation of mammalian and yeast 20SPT .
Many examples of substrates undergoing S-glutathionylation by other specific mechanisms have already been reported [21]; however, this is not the focus of present review. Because non-enzymatic thiol–disulfide reactions are slow [17,22], a challenge is the identification of proteins that add and remove the glutathione moiety. Indeed, some candidates for protein S-glutathionylation, such as glutathione-S-transferase [23] or alternative enzyme-assisted thiol–disulfide exchange [8], have been proposed. Regarding the reversal of protein S-glutathionylation, glutaredoxins (Grxs) are frequently considered as oxidoreductases that catalyze the reduction of protein mixed disulfides. These proposed pathways strengthen the notion that protein S-glutathionylation is either a regulatory post-translational modification and/or a protective mechanism of thiol homeostasis [4,24]. We have accumulated evidence that protein S-glutathionylation is a post-translational modification that regulates 20S proteasome activity, as will be discussed below.
Redox control of 20SPT gating
The first observation of 20SPT redox modification through S-glutathionylation was in preparations of the human proteasome incubated with increasing concentrations of both redox forms of glutathione [25]. In those studies, the peptidolytic chymotrypsin-like (ChT-L) proteasomal activity was shown to be modulated by both glutathione redox forms: micromolar concentrations activated the ChT-L activity and millimolar concentrations inhibited it. Interestingly, it was also observed that inhibition of the 20SPT by irreversible inhibitors increased the incorporation of G[35S]H into purified human 20SPT preparations. Initially, the thiol–disulfide exchange mechanism for S-glutathionylation (reactions 1 and 2, Scheme 1) was considered because the human 20SPT possesses two S–S bonds. However, it is also possible that GSH reacted with oxidized proteasomal thiol groups (reaction 3), not predicted at that time.
Further studies with preparations of 20SPT purified from Saccharomyces cerevisiae demonstrated inhibition of the ChT-L activity only after proteasomal incubation with millimolar concentrations of GSH. A pathway involving the oxidation of proteasomal thiol groups to sulfenic acid by hydrogen peroxide followed by reaction with GSH to form S-glutathionylated proteasomes was supported by data from these studies (Scheme 1, reaction 3; [26]). An increased pool of S-glutathionylated 20SPT was observed in yeast cells incubated with H2O2 [26], unraveling the notion of proteasomal S-glutathionylation as a post-translational regulatory mechanism.
20SPT S-glutathionylation is a widespread post-translational modification that has been corroborated through proteomic approaches in various species including plants and mammals [27–29].
S-glutathionylated 20SPT from S. cerevisiae was recently investigated by two-dimensional electrophoretic separation of the 14 proteosomal subunits followed by LC-Q-ToF-MS analysis [30]. Two Cys residues of the α5-subunit were found to be S-glutathionylated in the 20SPT preparations purified from yeast cells grown to stationary phase in fermentative medium (glucose as the carbon source). When the same proteasome preparations were incubated in vitro in the presence of GSH, four additional S-glutathionylated Cys residues were observed: one each in subunits α5 and α6 and two in subunit α7. Remarkably, five out of the six Cys residues prone to S-glutathionylation were also found through LC-Q-ToF-MS analyses either in the reduced or hyperoxidized sulfinic acid form, evidencing their susceptibility towards oxidation. Additionally, these findings suggested that S-glutathionylation occurs after oxidation of Cys residues to the sulfenic form followed by reaction with reduced glutathione (Scheme 1, reaction 3). Two of these Cys residues (α5-C76 and α7-C42) are highly conserved, from S. cerevisiae to humans.
Large-scale structural movements were verified through small angle X-ray scattering (SAXS) experiments that compared yeast proteasomal preparations where both Cys residues in the α5-subunit were S-glutathionylated or unmodified (Fig. 2 and Ref. [30]. A profound structural re-modeling was observed during transition of the S-glutathionylated to the DTT-reduced 20SPT form. When S-glutathionylated, the 20SPT is in the open gate conformation whereas after treatment with sulfhydryl reductants, such as DTT, its gate is closed. These results were confirmed by TEM evaluation of the same 20SPT preparations (Fig. 3 and Ref. [30].
Fig. 2.

Modeling of the 20SPT redox forms according to SAXS analyzes. (A) Top (left) and front (right) views of the 20SPT purified from yeast cells grown under conditions that promote fermentation; (B) same preparations after treatment with DTT. Internal and external diameters of the catalytic chamber (upper models) and the length of the 20SPT (botton left models) in both redox conditions were obtained through SAXS measurements [30]. Modeling of S-glutathionylated 20SPT (A) indicated a decreased length and a concave surface. The opposite conformation was deduced from DTT-treated 20SPT (B). Colored models are alternative modelings highlighting the gate conformation.
Fig. 3.

TEM images of the yeast 20SPT top view. (A)–(C) are panels representative of the closed conformation of 20SPT purified from (A) cells grown in respiratory medium where the closed conformation prevails (Demasi et al., unpublished); (B) strains carrying the α5−C76S mutation and, (C) cells grown in fermentative conditions after treatment with DTT that reduces S-glutathionylated or oxidized Cys residues to the sulfenic acid. (D) and (E) are images representative of the 20SPT open conformation: (D) preparations obtained from strains carrying the mutated α5-C221S 20SPT; (E) image of the 20SPT obtained from the α5-C221S strain (left) and wild type 20SPT from cells grown in fermentative glucose-rich (middle) and -synthetic media (right).
Unpublished findings from our group involving yeast strains harboring either the α5-C76S or the α5-C221S mutations have revealed that the conserved α5-C76 residue is directly involved in gating regulation, as the closed conformation is prevalent in 20SPT preparations from the α5-C76S strain. However, over 90% of 20SPT are in the open conformation in the α5-C221A strain. It seems that α5-C221, the only Cys residue located on the surface of the yeast 20SPT [30], is responsible for negative regulation of the gating mechanism. This residue most likely functions as a firsthand sensor of loss of the intracellular reducing ability regarding proteasomal functionality.
Growing yeast in either fermentative or respiratory media using glucose or glycerol/ethanol, respectively, promotes different intracellular redox conditions [31]. Glucose represses genes related to mitochondrial biogenesis and antioxidant defenses, while growth under respiratory conditions increases the expression of genes related to antioxidant defenses despite an increased production of ROS. Purified 20SPT from yeast cells grown in either fermentative or respiratory media was analyzed by SDS-PAGE followed by immunoblotting with anti-GSH antibody. The results of this experiment indicated that 20SPT in cells grown in glucose-rich medium were preferentially S-glutathionylated [32]. The labeling of the α5-subunit in proteasome preparations from cells grown in glucose, but not of those grown in glycerol/ethanol, was confirmed by the separation of the 20SPT subunits by 2D electrophoresis followed by anti-GSH immunoblotting (Demasi et al., unpublished). By comparing both preparations through TEM, 85% of the 20SPT purified from cells grown in glycerol/ethanol were in the closed conformation compared with 30% of the 20SPT from cells grown in glucose. Taken together, these results indicate S-glutathionylation of the α5-Cys residues depends on intracellular redox conditions.
Variable diameters of the open 20SPT gate were observed by TEM analysis of wild-type 20SPT extracted from yeast cells grown in glucose-rich, synthetic, or glycerol/ethanol media or α5-C221S and α5-C76S mutants extracted from yeast cells grown in glucose-rich medium (Fig. 3E; unpublished results). Most likely, gate opening is a stepwise process starting with oxidation of specific sulfhydryl groups followed by S-glutathionylation of a few Cys residues. It seems that the maximum diameter of the open conformation is achieved when the α5-Cys76 residue is S-glutathionylated. Analysis of 20SPT dimensions by SAXS is underway by our group to confirm these results.
S-glutathionylation of α5-C221 and –C76 residues as observed in extracts from yeast cells grown to stationary phase in glucose-rich medium, degraded oxidized proteins at an increased rate [30]. Decreased protein degradation was observed in 20SPT preparations obtained by treatment with sulfhydryl reductants (DTT or TCEP). Moreover, yeast cells presented a decreased pool of oxidized proteins, a decreased ratio of 20S to 19S complexes and increased pool of the 20SPT in open configuration when grown in medium that triggers fermentative metabolism. Opposite results were observed in cells presenting increased reductive ability, such as those grown in respiratory conditions (Demasi et al., unpublished).
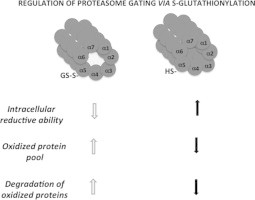
The data obtained thus far supports the hypothesis that 20SPT S-glutathionylation is a post-translational modification that modulates proteasome activity toward oxidized proteins. The gate opening of the free 20SPT pool would increase the degradation of oxidized proteins when cells transiently lose their reductive ability (Fig. 4).
Fig. 4.

The regulation of the 20SPT S-glutathionylation inside cells and the degradation of oxidized proteins. According to our hypothesis, when cells go through an oxidative imbalance, which results in the loss of their reducing ability, the pool of oxidized proteins increases and free 20SPT is S-glutathionylated. This modification of the 20SPT allows gate opening which increases the degradation of oxidized proteins (left). Opposite conditions are likely when cells possess increased reducing ability (right).
The deglutathionylation of the yeast 20SPT was investigated in vitro with the yeast recombinant oxidoreductases Grx2, Trx1 and Trx2 as potential deglutathionylating enzymes [32]. These three enzymes were equally competent for 20SPT deglutathionylation, as identical molar ratios of 20SPT:oxidoreductases produced similar results. However, these enzymes are easily degraded in vitro by the 20SPT. Because the substrate moieties of the oxidoreductases are located on the external proteasomal surface (α5-C221), in the α-pocket (α5-C76), and out of the catalytic chamber (internal β-rings), their activity as proteasomal deglutathionylating enzymes is reliable. However, the physiological meaning of their role in the regulation of 20SPT redox modulation needs further investigation.
The 19S regulatory unit was also shown to go through redox modification via S-glutathionylation. The S-glutathionylation of the Rpn1 and Rpn2 subunits of the 19S regulatory complex was shown to induce the loss of ChT-L proteasomal catalytic activity in mammalian cells [33]. Interestingly, oxidoreductases associate with the 19S complex in both fission yeast Schizosaccharomyces pombe (Txl1) and mammalian (Txnl1) cells; however, this process is not yet well understood [34,35]. It is unclear whether these proteins function as disulfide reductants of proteasomal substrates, of the proteasome itself, or of proteasomal co-factors.
Proteasome channel gating
The relationship between 20SPT channel gating and the coupling of regulatory units 19S, 11S and the PA200 has been extensively studied [36–38]. Of the regulatory units, the 19S is most abundant. Binding of this unit to the 20S core particle results in gate opening and substrate degradation. Proteolysis is a coordinate process involving binding of the poly-ubiquitinylated substrate, deubiquitinylation by 19S activity and binding of ATP to the 19S ATPase hexameric ring. Gating is dependent on the coupling of both units (20S and 19S) and on binding of ATP to the hexameric ring located in the base of the 19S unit. However, stabilization of the 20SPT open conformation and substrate unfolding and translocation are dependent on the poly-ubiquitinylated substrate binding to the 19S unit and ATP hydrolysis, respectively. The 19S regulatory particle and 20SPT interact through the 19S ATPase hexameric ring and the 20SPT α-heptameric ring. A perfect fitting between these rings is impossible because of their diverse symmetries. The 19S unit undergoes conformational changes depending on the nucleotide state of its ATPase subunits allowing interaction with the 20SPT α-ring [38]. Pockets between α-subunits on the 20SPT participate in interactions between 19S and 20SPT.
In the closed 20SPT conformation, the N-termini of the α-subunits form an intricate lattice of interactions that block access to the catalytic chamber through the so called α-annulus, located just below the surface of the α-heptameric ring. This conformation supposedly maintains a fixed opening of 13–20 Å allowing the entrance of only small peptides [39]. The N-terminal of the α3 subunit of yeast proteasome was shown to be essential for the closed conformation as it causes the stabilization of the neighboring tails of the α-ring. Deletion of the α3 N-terminal causes gate opening as deduced from its activation whereas deletion of the N-termini of both α3 and α7 was shown to promote permanent open conformation of the 20SPT [40]. It has been shown that α3-subunit knockout yeast strains incubated in high levels of the pro-oxidant cadmium induced the formation of an alternative α-ring structure composed of a double α4-subunit [41]. Remarkably, these mutated strains grew more robustly than the wild type in the presence of cadmium, suggesting that the alternative conformation provides a selective advantage under oxidative stress. A possible explanation for this phenotype might be that the absence of the α3-subunit facilitates formation of the 20SPT open conformation.
Some treatments such as exposure to minute amounts of detergents, hydrophobic peptides, and medium at low ionic strength promote disordering of the blocking α-N-termini allowing substrate entry [42].
Interestingly, 20SPT represents 20–30% of the total proteasome pool in mammal and yeast cells [43–46]. However, there is no systematic study addressing the gate conformation of the free physiological 20SPT pool. As will be discussed below, the free 20SPT pool is likely crucial for the degradation of oxidized proteins.
20SPT as the main player in the removal of oxidized proteins
Proteins can oxidize by ROS and NO derived species through a variety of pathways [47]. Oxidative protein damage results in chemical modification of the side chain of amino acid residues [48]. Proteins are poorly repaired in comparison to other macromolecules. Only the sulfur-containing amino acids Met and Cys can be reduced from sulfoxide and some oxidized forms, respectively. Since the middle 1980s, protein degradation has been proposed as the main mechanism of cells to cope with oxidative modification [49,50]. This mechanism was first observed in the ATP-independent degradation of oxidatively damaged proteins in extracts of red blood cells, whereas the non-oxidized forms of the model proteins were not degraded under same conditions [49]. Moreover, the degradation of mildly oxidized proteins by 20SPT was extensively investigated in vitro [51]. Since that time, the 20SPT has been proposed to be the preferential system to degrade oxidized proteins. The degradation of oxidized proteins was demonstrated in mammal and yeast cellular models to be independent of poly-ubiquitinylation. These results precluded degradation by the 26SPT pool [52–54].
Indeed, the most important data to date linking degradation of oxidized proteins to a free pool of 20SPT inside cells is indirect: uncoupling between the 19S and 20S particles during oxidative stress [54–56], transient inactivation of enzymes involved in the ubiquitylation and deubiquitylation of proteins [57–59], higher susceptibility of the 19S to oxidation as compared to the 20S [55,60], and no preference for the ubiquitinylation of oxidized proteins [61]. In addition, 26SPT decoupling was shown to be dependent on the proteasome-interacting protein Ecm29 in budding yeast [54]. Strains lacking this protein are more sensitive to oxidative stress. Their recovery from it is also delayed. In mammalian cells, a similar mechanism of decoupling under oxidative challenge was described as being mediated by HSP70 [56]. These data suggest that when cells experience oxidative imbalances, there is an adaptive response to increase the pool of free 20SPT.
An important criticism against degradation of oxidized proteins by free 20SPT is that the closed gate does not support the entrance of folded proteins [42]. However, it is known that oxidized proteins interact with the 20SPT through hydrophobic patches exposed on the protein surface due to partial unfolding upon oxidation [62–64]. It is thought that proteasomal interaction with the hydrophobic protein surface would trigger 20SPT gating [1]. This mechanism is supported by the observation that proteasome opening is triggered by the C-terminal hydrophobic motif HbYX (hydrophobic residue, conserved penultimate tyrosine and a variable C-terminal residue) of three 19S ATPase subunits that interact with the α-subunits of the 20SPT [65]. Notably, peptides containing the motif trigger proteasomal opening independent of 19S unit coupling. Many other reports in the literature show increased peptidolytic proteasomal activity upon incubation with hydrophobic compounds, suggesting that hydrophobic interactions activate the 20SPT. Some structured proteins can be degraded independently by the 20SPT, supposedly through hydrophobic interactions [64,66].
Indeed, there is no extant systematic study reporting the gate conformation of the free 20SPT pool. It is currently assumed that free 20SPT would be in a latent form, which usually means the closed conformation, allowing the entrance of only peptides [67]. The most conclusive data supporting this assumption are related to crystallographic studies of the yeast and mammal 20SPT [68,69]. However, the presence of inhibitors, the yeast growth conditions, and the addition of thiol reductants in the extraction buffers utilized for proteasomal purification make it impossible to infer the conformation of the 20S proteasomal gate in vivo.
A major question to be addressed regarding preferential degradation of oxidized proteins inside cells is whether such proteins are ubiquitinylated and are therefore subject to 26SPT proteolysis. Related to this question is whether proteolysis by free 20SPT, which bypasses ubiquitinylation, is kinetically favored. During oxidative stress, proteins already ubiquitinylated or being ubiquitinylated that are also being oxidized may be reliably degraded after 26SPT recognition. In addition, mild oxidation of specific proteins may augment their ubiquitinylation because oxidation-induced conformational changes might expose specific ubiquitinylation targets, as predicted by the N-end rule [70,71]. The energy and time costs of degrading ubiquitinylated substrates by the 26S proteasome was recently explored [72], showing it to be dependent on protein length and how tightly the protein is folded among other variables involving 26S proteasomal properties and the ubiquitinylation process. By comparing two substrates differing in length and folding, these authors demonstrated that the molar ratio between ATP consumption and protein substrate could vary from 50 to 160. The time course for degradation increased proportionally with increased protein length. According to these data, degradation of ubiquitinylated substrates is a process that consumes large amounts of energy.
The notion that oxidized proteins might promptly interact with the free 20SPT pool is attractive, as the process is most likely kinetically favored. The rate-limiting step of the preferential degradation of oxidized proteins by the 20SPT free pool would be the availability of active 20SPT. Strengthening this hypothesis are the observations that 20SPT uncouples from the 19S regulatory particle during oxidative stress [54,56] and that S-glutathionylation stabilizes the 20SPT active open conformation [30]. Therefore, more accurate approaches are needed to measure the kinetics of degradation of oxidized proteins inside cells.
Final remarks
This review presents data regarding S-glutathionylation as a potential regulatory mechanism for 20SPT [2]. Four main observations supporting this notion were reviewed: (1) modification of specific Cys residues under physiological conditions (high GSH/GSSG ratio); (2) conformational changes of the modified protein; (3) reversibility of the reaction; and (4) concomitant change of the physiological response, e.g., the increased degradation of oxidized proteins. Therefore, we hypothesize that 20SPT S-glutathionylation plays a regulatory role that allows cells to cope with increased oxidative stress (Fig. 4).
There are few studies on proteasomal S-glutathionylation resulting in structural and functional modifications, and thus many questions remain unaddressed. One important question is whether S-glutathionylation would trigger 26S uncoupling per se during oxidative stress. Additionally, two open questions are why and how oxidized proteins interact with 20SPT. Peptide fragments generated by both redox forms of 20SPT are also predicted to differ, as the peptidolytic activities of both forms differ [25,26]. Finally, additional unresolved questions are whether S-glutathionylation is enzymatically regulated and whether oxidized protein degradation through the free S-glutathionylated 20SPT presents a kinetic advantage over degradation via 26SPT.
It is noteworthy that the number of Cys residues in the proteasome core particle evolved from 32 in yeast to 54 in human. No disulfide bond is present in yeast 20SPT but there are two in human 20SPT. As demonstrated previously, the ChT-L activity of the human 20SPT is modulated by both glutathione redox states and at a broad glutathione concentration range [25]. This observation contrasts with yeast 20SPT which is modulated only by the reduced glutathione form. Whether the increased number of Cys residues in the mammal proteasome would be a functional evolutionary advantage is an intriguing open question.
Acknowledgements
I am grateful to the authors for their important contributions to the work described here. The studies by our group cited in the present review were supported by FAPESP (Fundação de Amparo a Pesquisa do Estado de São Paulo), the Instituto Nacional de Ciência, Tecnologia e Inovação de Processos Redox em BioMedicina (Redoxome; CNPq, FAPESP, CAPES), and the Conselho Nacional de Ciência e Tecnologia (CNPq).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Jung T., Grune T. The proteasome and the degradation of oxidized proteins: Part I—Structure of proteasomes. Redox Biol. 2013;1:178–182. doi: 10.1016/j.redox.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mieyal J.J., Chock P.B. Posttranslational modification of cysteine in redox signaling and oxidative stress: focus on s-glutathionylation. Antioxid. Redox Signal. 2012;16:471–475. doi: 10.1089/ars.2011.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziegler D.M. Role of reversible oxidation–reduction of enzyme thiols-disulfides in metabolic regulation. Annu. Rev. Biochem. 1985;54:305–329. doi: 10.1146/annurev.bi.54.070185.001513. [DOI] [PubMed] [Google Scholar]
- 4.Gallogly M.M., Mieyal J.J. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Dalle-Donne I., Rossi R., Colombo G., Giustarini D., Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Hill B.G., Bhatnagar A. Protein S-glutathiolation: redox-sensitive regulation of protein function. J. Mol. Cell. Cardiol. 2012;52:559–567. doi: 10.1016/j.yjmcc.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klatt P., Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur. J. Biochem. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 8.Gutscher M., Sobotta M.C., Wabnitz G.H., Ballikaya S., Meyer A.J., Samstag Y., Dick T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009;284:31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malvezzi A., Higa P.M., T-do Amaral A., Silva G.M., Gozzo F.C., Ferro E.S., Castro L.M., de Rezende L., Monteiro G., Demasi M. The cysteine-rich protein thimet oligopeptidase as a model of the structural requirements for S-glutathiolation and oxidative oligomerization. PLoS One. 2012;7:e39408. doi: 10.1371/journal.pone.0039408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Ruiz A., Cadenas S., Lamas S. Nitric oxide signaling: classical, less classical, and nonclassical mechanisms. Free Radic. Biol. Med. 2011;51:17–29. doi: 10.1016/j.freeradbiomed.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Østergaard H., Tachibana C., Winther J.R. Monitoring disulfide bond formation in the eukaryotic cytosol. J. Cell Biol. 2004;166:337–345. doi: 10.1083/jcb.200402120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan B., Ezeriņa D., Amoako T.N., Riemer J., Seedorf M., Dick T.P. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 2013;9:119–125. doi: 10.1038/nchembio.1142. [DOI] [PubMed] [Google Scholar]
- 13.Le Moan N., Clement G., Le Maout S., Tacnet F., Toledano M.B. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J. Biol. Chem. 2006;81:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 14.Sarkar D.D., Edwards S.K., Mauser J.A., Suarez A.M., Serowoky M.A., Hudok N.L., Hudok P.L., Nuñez M., Weber C.S., Lynch R.M., Miyashita O., Tsao T.S. Increased redox-sensitive green fluorescent protein reduction potential in the endoplasmic reticulum following glutathione-mediated dimerization. Biochemistry. 2013;52:3332–3345. doi: 10.1021/bi400052u. [DOI] [PubMed] [Google Scholar]
- 15.Gupta V., Carroll K.S. Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta doi:pii: S0304-4165(13)00244-4. 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed]
- 16.Pimentel D., Haeussler D.J., Matsui R., Burgoyne J.R., Cohen R.A., Bachschmid M.M. Regulation of cell physiology and pathology by protein S-glutathionylation: lessons learned from the cardiovascular system. Antioxid. Redox Signal. 2012;16:524–542. doi: 10.1089/ars.2011.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winterbourn C.C., Hampton M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Flohé L. The fairytale of the GSSG/GSH redox potential. Biochim. Biophys. Acta. 1830;3139-42:2013. doi: 10.1016/j.bbagen.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Tairum C.A., Jr, de Oliveira MA, Horta BB, Zara FJ, Netto LE. Disulfide biochemistry in 2-cys peroxiredoxin: requirement of Glu50 and Arg146 for the reduction of yeast Tsa1 by thioredoxin. J. Mol. Biol. 2012;424:28–41. doi: 10.1016/j.jmb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Day A.M., Brown J.D., Taylor S.R., Rand J.D., Morgan B.A., Veal E.A. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol. Cell. 2012;45:398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 21.Grek C.L., Zhang J., Manevich Y., Townsend D.M., Tew K.D. Causes and consequences of cysteine s-glutathionylation. J. Biol. Chem. 2013;288:26497–26504. doi: 10.1074/jbc.R113.461368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemp M., Go Y.M., Jones D.P. Non equilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic. Biol. Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Townsend D.M., Manevich Y., He L., Hutchens S., Pazoles C.J., Tew K.D. Novel role for glutathione S-transferase pi. Regulator of protein S-Glutathionylation following oxidative and nitrosative stress. J. Biol. Chem. 2009;284:436–445. doi: 10.1074/jbc.M805586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen E.M., Mieyal J.J. Protein-thiol oxidation and cell death: regulatory role of glutaredoxins. Antioxid. Redox Signal. 2012;17:1748–1763. doi: 10.1089/ars.2012.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demasi M., Shringarpure R., Davies K.J. Glutathiolation of the proteasome is enhanced by proteolytic inhibitors. Arch. Biochem. Biophys. 2001;389:254–263. doi: 10.1006/abbi.2001.2332. [DOI] [PubMed] [Google Scholar]
- 26.Demasi M., Silva G.M., Netto L.E. 20 S proteasome from Saccharomyces cerevisiae is responsive to redox modifications and is S-glutathionylated. J. Biol. Chem. 2003;278:679–685. doi: 10.1074/jbc.M209282200. [DOI] [PubMed] [Google Scholar]
- 27.Dixon D.P., Skipsey M., Grundy N.M., Edwards R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 2005;138:2233–2244. doi: 10.1104/pp.104.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niture S.K., Velu C.S., Bailey N.I., Srivenugopal K.S. S-thiolation mimicry: quantitative and kinetic analysis of redox status of protein cysteines by glutathione-affinity chromatography. Arch. Biochem. Biophys. 2005;444:174–184. doi: 10.1016/j.abb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Zong C., Young G.W., Wang Y., Lu H., Deng N., Drews O., Ping P. Two-dimensional electrophoresis- based characterization of post-translational modifications of mammalian 20S proteasome complexes. Proteomics. 2008;8:5025–5037. doi: 10.1002/pmic.200800387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva G.M., Netto L.E., Simões V., Santos L.F., Gozzo F.C., Demasi M.A., Oliveira C.L., Bicev R.N., Klitzke C.F., Sogayar M.C., Demasi M. Redox control of 20S proteasome gating. Antioxid. Redox Signal. 2012;16:1183–1194. doi: 10.1089/ars.2011.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamieson D.J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 32.Silva G.M., Netto L.E., Discola K.F., Piassa-Filho G.M., Pimenta D.C., Bárcena J.A., Demasi M. Role of glutaredoxin 2 and cytosolic thioredoxins in cysteinyl-based redox modification of the 20S proteasome. FEBS J. 2008;275:2942–2955. doi: 10.1111/j.1742-4658.2008.06441.x. [DOI] [PubMed] [Google Scholar]
- 33.Zmijewski J.W., Banerjee S., Abraham E. S-glutathionylation of the Rpn2 regulatory subunit inhibits 26 S protea- somal function. J. Biol. Chem. 2009;284:22213–22221. doi: 10.1074/jbc.M109.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen K.M., Madsen L., Prag S., Johnsen A.H., Semple C.A., Hendil K.B., Hartmann-Petersen R. Thioredoxin Txnl1/TRP32 is a redox-active cofactor of the 26S proteasome. J. Biol. Chem. 2009;284:15246–15254. doi: 10.1074/jbc.M900016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen K.M., Jensen C., Kriegenburg F., Lauridsen A-MB, Gordon C., Hartmann-Petersen R. Txl1 and Txc1 are co-factors of the 26S proteasome in fission yeast. Antioxid. Redox Signal. 2011;14:1601–1608. doi: 10.1089/ars.2010.3329. [DOI] [PubMed] [Google Scholar]
- 36.Li X., Demartino G.N. Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem. J. 2009;421:397–404. doi: 10.1042/BJ20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bar-Nun S., Glickman M.H. Proteasomal AAA-ATPases: structure and function. Biochim. Biophys. Acta. 1823;67-82:2012. doi: 10.1016/j.bbamcr.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Sledź P., Förster F., Baumeister W.J. Allosteric effects in the regulation of 26S proteasome activities. Mol. Biol. 2013;425:1415–1423. doi: 10.1016/j.jmb.2013.01.036. [DOI] [PubMed] [Google Scholar]
- 39.Stadtmueller B.M., Hill C.P. Proteasome activators. Mol. Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groll M., Bajorek M., Köhler A., Moroder L., Rubin D.M., Huber R., Glickman M.H., Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 41.Kusmierczyk A.R., Kunjappu M.J., Funakoshi M., Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 2008;153:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 42.Bajorek M., Glickman M.H. Keepers at the final gates: regulatory complexes and gating of the proteasome channel. Cell. Mol. Life Sci. 2004;61:1579–1588. doi: 10.1007/s00018-004-4131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibatani T., Carlson E.J., Larabee F., McCormack A.L., Fruh K., Skach W.R. Global organization and function of mammalian cytosolic proteasome pools: implications for PA28 and 19S regulatory complexes. Mol. Biol. Cell. 2006;17:4962–4971. doi: 10.1091/mbc.E06-04-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babbitt S.E., Kiss A., Deffenbaugh A.E., Chang Y.H., Bailly E., Erdjument-Bromage H., Tempst P., Buranda T., Sklar L.A., Baumler J., Gogol E., Skowyra D. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 45.Tanahashi N., Murakami Y., Minami Y., Shimbara N., Hendil K.B., Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 2000;275:14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 46.Hendil K.B., Khan S., Tanaka K. Simultaneous binding of PA28 and PA700 activators to 20S proteasomes. Biochem. J. 1998;332:749–754. doi: 10.1042/bj3320749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Höhn A., König J., Grune T. Protein oxidation in aging and the removal of oxidized proteins. J. Proteomics doi:pii: S1874-3919(13)00021-3. 10.1016/j.jprot.2013.01.004 [DOI] [PubMed]
- 48.Stadtman E.R. Protein oxidation and aging. Free Radic. Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 49.Davies K.J., Goldberg A.L. Proteins damaged by oxygen radicals are rapidly degraded in extracts of red blood cells. J. Biol. Chem. 1987;262:8227–8234. [PubMed] [Google Scholar]
- 50.Davies K.J., Delsignore M.E. Protein damage and degradation by oxygen radicals. III. Modification of secondary and tertiary structure. J. Biol. Chem. 1987;262:9908–9913. [PubMed] [Google Scholar]
- 51.Davies K.J. Protein damage and degradation by oxygen radicals. I. general aspects. J. Biol. Chem. 1987;262:9895–9901. [PubMed] [Google Scholar]
- 52.Inai Y., Nishikimi M. Increased degradation of oxidized proteins in yeast defective in 26 S proteasome assembly. Arch. Biochem. Biophys. 2002;404:279–284. doi: 10.1016/s0003-9861(02)00336-3. [DOI] [PubMed] [Google Scholar]
- 53.Shringarpure R., Grune T., Mehlhase J., Davies K.J. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- 54.Wang X., Yen J., Kaiser P., Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci. Signaling. 2010;3 doi: 10.1126/scisignal.2001232. (ra88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reinheckel T., Ullrich O., Sitte N., Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch. Biochem. Biophys. 2000;377:65–68. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- 56.Grune T., Catalgol B., Licht A., Ermak G., Pickering A.M., Ngo J.K., Davies K.J. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radic. Biol. Med. 2011;51:1355–1364. doi: 10.1016/j.freeradbiomed.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jahngen-Hodge J., Obin M.S., Gong X., Shang F., Nowell T.R., Jr, Gong J., Abasi H., Blumberg J., Taylor A. Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J. Biol. Chem. 1997;272:28218–28226. doi: 10.1074/jbc.272.45.28218. [DOI] [PubMed] [Google Scholar]
- 58.Obin M., Shang F., Gong X., Handelman G., Blumberg J., Taylor A. Redox regulation of ubiquitin-conjugating enzymes: mechanistic insights using the thiol-specific oxidant diamide. FASEB J. 1998;12:561–569. doi: 10.1096/fasebj.12.7.561. [DOI] [PubMed] [Google Scholar]
- 59.da Cunha F.M., Demasi M., Kowaltowski A.J. Aging and calorie restriction modulate yeast redox state, oxidized protein removal, and the ubiquitin-proteasome system. Free Radic. Biol. Med. 2011;51:664–670. doi: 10.1016/j.freeradbiomed.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 60.Reinheckel T., Sitte N., Ullrich O., Kuckelkorn U., Davies K.J., Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem. J. 1998;335:637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kästle M., Reeg S., Rogowska-Wrzesinska A., Grune T. Chaperones, but not oxidized proteins, are ubiquitinated after oxidative stress. Free Radic. Biol. Med. 2012;53:1468–1477. doi: 10.1016/j.freeradbiomed.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 62.Pacifici R.E., Kono Y., Davies K.J. Hydrophobicity as the signal for selective degradation of hydroxyl radical-modified hemoglobin by the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1993;268:15405–15411. [PubMed] [Google Scholar]
- 63.Ferrington D.A., Sun H., Murray K.K., Costa J., Williams T.D., Bigelow D.J., Squier T.C. Selective degradation of oxidized calmodulin by the 20S proteasome. J. Biol. Chem. 2001;276:937–943. doi: 10.1074/jbc.M005356200. [DOI] [PubMed] [Google Scholar]
- 64.Baugh J.M., Viktorova E.G., Pilipenko E.V. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith D.M., Chang S.C., Park S., Finley D., Cheng Y., Goldberg A.L. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's α ring opens the gate for substrate entry. Mol. Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu C.W., Corboy M.J., De Martino G.N., Thomas P.J. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kish-Trier E., Hill C.P. Structural biology of the proteasome. Annu. Rev. Biophys. 2013;42:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H.D., Huber R. Structure of 20S proteasome from yeast at 2.4A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 69.Unno M., Mizushima T., Morimoto Y., Tomisugi Y., Tanaka K., Yasuoka N., Tsukihara T. The structure of the mammalian 20S proteasome at 2.75A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 70.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tasaki T., Sriram S.M., Park K.S., Kwon Y.T. The N-end Rule Pathway. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peth A., Nathan J.A., Godberg A.L. The ATP Costs and Time Required to Degrade Ubiquitinated Proteins by the 26S Proteasome. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.482570. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]


