Abstract
Incidental findings have long posed challenges for healthcare providers, but the scope and scale of these challenges have increased with the introduction of new technologies. This article assesses the impact of incidental findings on the introduction of prospective pharmacogenomic testing into clinical use. Focusing on the challenges of the incidentalome, the large set of incidental findings potentially generated through genotyping, the paper argues that provisional approaches to managing incidental findings may be implemented if necessary to allow benefits of pharmacogenomic testing to be realized in the clinical setting. In the longer term, approaches to returning incidental findings may need to focus on limiting the number of incidental findings to a number that can be addressed by patients and providers.
Keywords: clinical pharmacogenomics, incidental findings, incidentalome, translational genomics
Unexpected findings are frequently generated incidentally when medical tests are performed. For example, hyperkalemia is a common incidental finding when a basic metabolic panel is drawn [1] , as is thrombocytopenia when complete blood counts are performed [2]. These two examples of incidental findings are particularly common because of technical limitations, but any laboratory abnormality that is unexpected will often prove spurious. This is because the specificity of virtually every laboratory test is less than 100%, and false-positive results are more common among those patients in whom no reason for an abnormality has been identified [3]. Because of this well-known problem, medical schools have long taught that laboratory tests and other diagnostic evaluations should be used parsimoniously. This approach helps minimize the number of incidental findings that are generated that will later prove to be factitious [3].
As medical technologies have become more sensitive and more comprehensive, however, the challenges posed by incidental findings, and the ability of providers to prevent them, have evolved. MRI, for example, is so sensitive that it frequently reveals unexpected abnormalities [4]. The nature of the technology, however, makes it difficult for providers to minimize the discovery of these incidental findings. This is because MRI generates detailed images of cross-sections of the body; images are not easily limited to a single organ of concern.
The problem of unavoidable incidental findings also raises significant challenges for those working to implement multiplex genotyping into clinical care using such technologies as SNP chips and next-generation sequencers. While genetic tests have been in clinical use for decades and have raised their own challenges related to incidental findings, multiplex genetic technologies are only now beginning to be utilized for clinical care. In 2006, Kohane et al. coined the term ‘incidentalome’ to draw attention to the significant number of incidental findings that could be generated through the use of these newer technologies [5].
One helpful observation made by Kohane et al. is that a new dimension is introduced to the challenge of incidental findings when we move from one genetic test to technologies that perform multiple genetic tests in parallel. As none of the individual tests can boast a specificity of 100%, each additional test increases the likelihood of obtaining a false-positive result. Furthermore, since a confirmatory test is the only way to distinguish between a false-positive and a true-positive result, the incidentalome could lead to a large number of expensive or risky follow-up evaluations [5] .
When they introduced the concept of the incidentalome, Kohane et al. anticipated a time in the future when multiplex genotyping technologies would be introduced into clinical care. That time has now come. Programs at institutions such as Vanderbilt University Medical Center and St Jude’s Children’s Research Hospital have begun to perform pharmacogene-focused genetic tests on patients in order to inform prescribing practices [6,7]. Indeed, pharmacogenomic applications are at the vanguard of the application of genomic testing to clinical care. In this paper, we analyze the impact of the incidentalome on the use of multiplex genetic technologies for clinical pharmacogenomics, and propose practical solutions for addressing incidental findings in the early days of this work.
Incidental findings from pharmacogenetic tests
Given the significant pleiotropy exhibited in the human genome, virtually any genetic test used for pharmacogenetic purposes is likely to generate at least one incidental finding. For example, we recently examined publications that reference at least one of 34 genes included in Illumina’s VeraCode ADME Core Panel, a SNP-based panel designed to target variations relevant to pharmacogenomics. We demonstrated that in addition to pharmacogenomic associations, at least one incidental genotype–phenotype association could be identified for 26 of these 34 genes [8].
However, when we consider single gene tests that have been part of the standard of care for a number of years, we find no evidence that incidental findings have posed significant barriers to the implementation of such tests for pharmacogenetic applications. Two features have tended to minimize the impact of incidental findings. First, even when genetic variants with pharmacogenetic relevance are known to be pleiotropic, the number and strength of incidental findings associated with a single gene are usually modest. Take for example the major histocompatibility complex allele, HLA-B*5701. Testing for this variant prior to initiation of abacavir has been recommended by the US FDA since 2008 [101] and has been hailed as a pharmacogenetics success story [9]. Potential incidental findings for this variant include an elevated risk for Behçet’s syndrome [10] and protection against progression of HIV infection [11] . However, these associations are very preliminary. Given the strength of their association with the HL A-B*5701 variant and their limited replication record, they have not been considered important incidental findings. Given current evidence, providers considering the use of abacavir for a patient with HIV infection need not worry about incidental findings when ordering a genetic test to identify the HLA-B*5701 variant.
Indeed, our own research on pharmacogenes demonstrates that the majority of genotype–phenotype associations have not been adequately replicated, and not all associations are strong enough to reach clinical relevance [8]. We defined a genotype–phenotype association as potentially clinical relevant if evidence for an association had been found in at least two studies, and at least one of those studies estimated the odds ratio for the association to be ≥2.0 or ≤0.5. Furthermore, we excluded a genotype–phenotype association if at least one study examining this correlation had failed to find an association. Using these strict criteria for potential clinical relevance, we found that only eight of the target pharmacogenes were likely to generate one or more relevant incidental findings.
A second factor that has minimized the adverse effect of incidental findings on clinical pharmacogenetics has been the very focused settings where pharmacogenetic testing is usually applied. Abacavir, for example, is usually prescribed by providers who specialize in the care of patients with HIV. According to the FDA, only one other antiviral used to treat HIV has a pharmacogenetic indication reported in its drug label – maraviroc [102] . For the most part, then, we may expect that the specialist physicians who are already using pharmacogenetic tests have not been faced with managing multiple test results at the same time. When incidental findings have been generated, it is likely that this has occurred in settings where specialists were already in frequent contact with their chronically ill patients. Given these factors, we may expect that any discussions about incidental findings generated by focused single-gene tests have been handled relatively easily.
The incidentalome in pharmacogenomic testing
Despite this past experience, there is reason to believe that challenges related to incidental findings will have a larger impact on future attempts to translate pharmacogenetic science into the clinic. A number of institutions have recently initiated projects focused on introducing multiplex genetic tests into primary care settings for pharmacogenomic applications [6,7,12] . These clinical pharmacogenomic projects are designed to generate genotyping results prospectively, that is, before a provider is considering a medication with a pharmacogenomic indication in the care of the patient. This approach is attractive for a number of reasons. First, medications are occasionally needed urgently, but genotyping can take time. Prospective testing can make it possible for genotyping information to be available as soon as the need for a medication is recognized. Second, multiplex genetic testing technologies, from SNP-based to next-generation sequencing technologies, are becoming more economical. As these costs approach that of a single gene test, the economic incentive to use a single gene test rather than a multiplex technology decreases. Also, given that pharmacogenomic indications are emerging for such commonly used medications as clopidogrel, warfarin and the statin class of lipid-lowering agents, the chances continue to increase that multiple single gene tests could be required over the course of a patient’s medical care. Prospective testing using a multiplex genotyping technology eliminates the need for such repeated testing, and thus potentially improves the economic advantage of multiplex testing even further.
Although the technology used for pharmacogenomic testing is likely to change, the healthcare model for performing this testing is likely to remain the same. Direct-to-consumer genetic testing has received a great deal of attention, and companies providing this service do report pharmacogenomic variants [103] . However, medicines are prescribed only by properly licensed healthcare providers. For this reason, we anticipate that the vast majority of pharmacogenomic testing will continue to take place in traditional healthcare settings.
The transition to prospective, multiplex pharmacogenetic testing in healthcare settings could, however, have a significant effect on the factors that have previously minimized the challenge of managing incidental findings. Whereas the single gene tests already in use for pharmacogenetic applications appear not to generate a significant number of incidental genotype–phenotype associations with sufficient strength to support clinical relevance, this is likely to change as multiplex genetic tests are put into use. This is not only because many pharmacogenes are inherently pleiotropic, but also because many of the genotyping technologies proposed for use, including whole-exome sequencing (WES) and whole-genome sequencing (WGS), will generate data related to genes with no pharmacogenetic relevance. This challenge is analogous to the results generated by MRI, where images are generated of an entire region of the body rather than the specific organ of interest.
In addition, prospective pharmacogenomic testing is designed to gather information relevant to a broad set of pharmaceuticals that might be used in the future, rather than those being considered for the treatment of a single condition. For this reason, the responsibility for ordering and following up on prospective pharmacogenomic tests will almost certainly fall to generalist physicians. That these generalists will also be the physicians following up on incidental findings makes a certain amount of sense, given that incidental findings generated are likely to be diverse as well. There are limits, however, to the ability of generalist physicians to incorporate a broad set of new considerations into routine clinical care. In the next section, we will consider the implications of the scale of the incidentalome on the time and attention of primary care providers and their patients.
Practical limitations of time & attention
Any effort to incorporate pharmacogenomic testing, including the management of incidental findings, into primary care practice will depend on a pragmatic account of the context of primary care medicine. Consider, for example, that preventive healthcare interventions prescribed by current guidelines already require a significant amount of time from clinicians and patients. A study from 2003 demonstrated that a primary care physician with a patient panel of 2500 patients would need to dedicate 7.4 h per working day just to address recommended preventive care interventions [13] . This conclusion is striking, since primary care providers must address not only preventive care, but also manage existing health problems.
This study, combined with a great deal of clinical experience, indicates that clinicians cannot address every preventive health issue without significant changes to their practice, such as significantly decreasing their patient panel. For this reason, some have proposed that time and attention during the clinical encounter should be focused on only the most important preventive health issues [14] . In fact, one hoped-for benefit from genomic medicine is that this type of information could help triage which health issues are potentially of greatest importance for each individual patient [15] . In order for genomic medicine to deliver on this vision of medical care focused on each patient’s individual risks, however, the same principle will need to be applied to incidental findings. That is, little benefit can be realized if genomic medicine allows providers to focus on a few especially important preventive health issues, but then requires them to address each incidental finding, no matter its importance, with patients. The potential for genomic medicine, and in particular pharmacogenomic medicine, to deliver on its promise depends on developing effective ways to filter and manage incidental findings.
This insight, among others, has motivated the development of schema for classifying findings generated by multiplex genetic tests [16,17]. An adapted version of a schema proposed by Rudnik-Schöneborn et al. is provided in Table 1 [17] . This classification system is organized primarily around the criteria of validity and ‘actionability’. Category 1 results are scientifically and clinically valid results that may be used to inform specific treatments or clinical interventions for a patient’s own health. Category 2 results are those that meet the same standards for validity, but are ‘actionable’ primarily in the setting of reproductive planning. Results in Category 3 are those with strong validity that do not support specific clinical interventions, primarily because the relevant conditions are not considered preventable. Category 4 includes all results with inadequate validity for use in clinical settings [17] . The schema proposed by Berg et al. is similar in many respects, and places additional focus on the degree to which specific results can be expected to bring about psychological distress in patients [16].
Table 1.
Schema for categorizing incidental genetic findings.
| Category | Description |
|---|---|
| 1 | High utility (i.e., result indicating elevated risk for a potentially treatable disorder) |
|
|
| 2 | Possible benefit (i.e., carrier status for a heritable condition) |
|
|
| 3 | Questionable benefit (i.e., result indicating elevated risk for a disorder whose risk is not considered modifiable) |
|
|
| 4 | Low benefit (i.e., result indicating a genetic variant only weakly associated with disease risk) |
Adapted with permission from [17]. Translated into English by the authors.
This type of schema will be central to any effort to manage incidental findings. However, we anticipate that organizing results in this way alone will not be adequate for clinical efforts to return and utilize incidental genetic findings. To see why this is the case, let us consider briefly how a patient and primary care provider might address an incidental genetic finding during a health maintenance visit. A patient might carry a risk allele that confers a significantly increased risk for developing coronary artery disease. The provider could return this result to the patient by explaining how the patient’s risk compares with that of the general population. The patient and provider could then discuss the clinical and personal meaning of the result. Finally, they could work together to develop a plan to mitigate this risk. Optimistically, a patient and provider could address ten such results in a health maintenance visit, assuming that these discussions could in some ways focus the recommended preventive health interventions that are recommended by such bodies as the US Preventive Services Task Force (USPSTF).
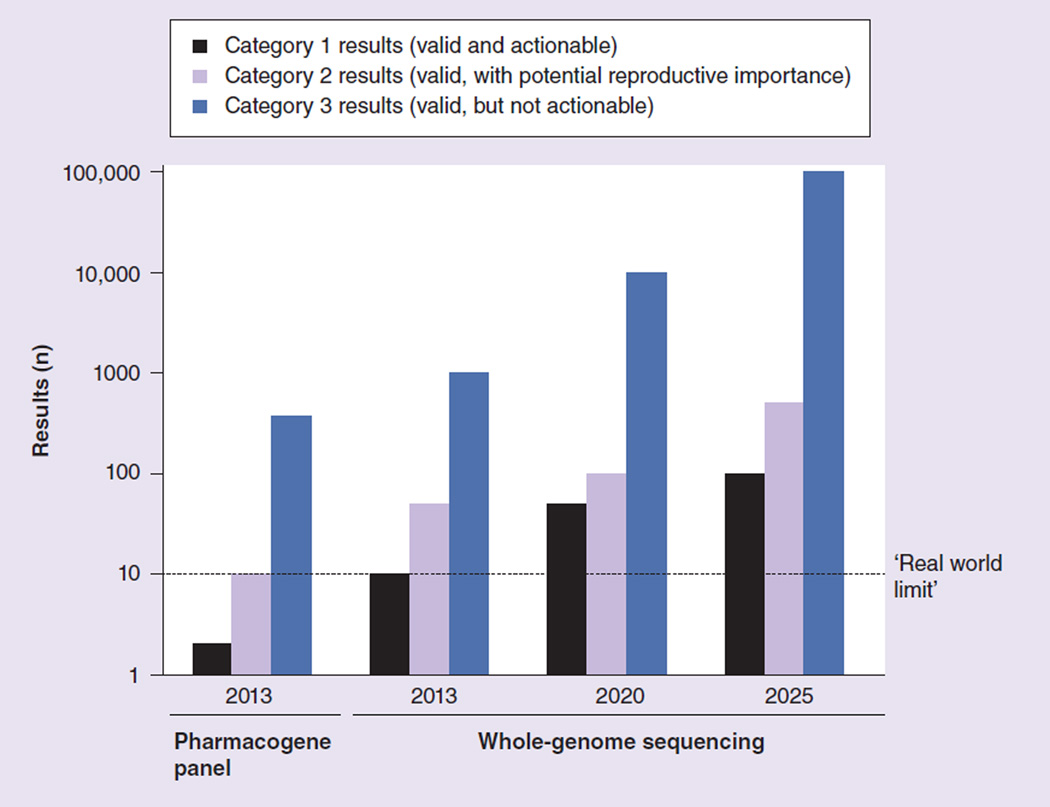
In light of this estimate, we can consider how category-only approaches might affect the ability of providers to address all valid incidental findings generated through pharmacogenomic testing. According to our recent study, when a patient undergoes testing for variants in 34 pharmacogenes, the number of clinically actionable incidental findings (i.e., those in Category 1 in the Rudnik-Schöneborn et al. schema or bin 1 in the Berg et al. schema) are likely to fall well below the real-world limitation of ten results. However, as the genotyping technology used for pharmacogenomic applications progresses from panels of pharmacogenes to WES or WGS, and research identifying genotype–phenotype correlations continues, it is likely – even inevitable – that the number of results that are both valid and actionable will grow to exceed reasonable expectations for what a patient and provider might be able to address together (Figure 1).
Figure 1.
Numbers of incidental findings generated by clinical genotyping technologies, projected speculatively through 2025.
Once the number of validated, clinically useful incidental findings exceeds a practical limit, only the highest priority results should be returned. For example, patients and their primary care providers may be provided with a ‘top ten list’ of the most important incidental findings from Category 1. They may then consider each of these in turn and develop an action plan. The effective management of incidental findings requires not only categorization of results, but also prioritization.
With these observations in mind, we can see that approaches for managing incidental findings from clinical pharmacogenomic testing will need to develop over time. We will later explore what sorts of categorization and prioritization tools might be needed to help primary care providers effectively manage the incidentalome in the era of clinical WES and WGS. But for now we can observe simply that such tools are not available. In the next section we will consider how clinical pharmacogenomics can proceed now, even when the incidentalome remains an unsolved challenge.
The tail wagging the dog
The FDA has already recommended, and in some cases required, the use of a pharmacogenetic test prior to the use of certain medications [102] . In addition, a growing number of these pharmaceuticals are those used widely by adult patients, many of whom take more than one medication with a pharmacogenetic indication. We have arguably already reached the point where the potential benefits of prospective pharmacogenomic testing have begun to outweigh the risks.
Given that effective policies and tools for managing incidental findings have not yet been developed, however, one might also reasonably conclude that the risks they pose outweigh the benefits of prospective pharmacogenomic testing. If this interpretation causes the majority of institutions to delay introducing prospective pharmacogenomic testing while they await clearer guidance on how incidental findings should be managed, then the prediction that the incidentalome poses a barrier to genomic medicine will have proven prescient [5] . If the benefits conferred by pharmacogenomic results are significant, however, then delaying the implementation of such testing because of incidental findings is tantamount to allowing ‘the tail to wag the dog’.
An alternative solution, albeit an imperfect one, is to provisionally introduce prospective pharmacogenomic testing before definitive policies and procedures for managing incidental findings have been developed. This interim form of pharmacogenomic testing would certainly fall short of the grand predictions that have been proposed for genomic medicine. However, such an approach would at least make it possible to deliver the limited benefits of pharmacogenomics that could be made available today. Let us now turn to identifying policies appropriate to such early pharmacogenomic projects, taking our lead from pilot programs already introduced by pioneering institutions.
First, our earlier discussion serves to demonstrate that WES and WGS technologies are not appropriate for early pharmacogenomic programs. Given that financial resources for these types of projects are necessarily limited, the cost of next-generation sequencing technologies alone likely rules out their use in all settings except well-funded research. But more to the point of our analysis, WES and WGS technologies simply generate too many incidental findings. Although to our knowledge no one has generated a comprehensive accounting of the incidental findings potentially generated by WES and WGS, some estimate that these technologies will generate 100 incidental findings per person [18]. Some of these may fall into Categories 1 and 2. However, the majority of these findings are likely to fall into Category 3 and provide no actionable or immediately relevant information.
At this time, WES and WGS increase the burdens and risks of clinical pharmacogenomic testing while providing no marginal benefit over simpler technologies. It is unclear whether this balance will change in the future. For now, though, panels limited to pharmacogenes, such as the VeraCode ADME Core Panel adopted by Vanderbilt’s PREDICT project [7], provide a much more favorable balance of risks and benefits on the whole. The time-tested commitment to parsimony in ordering diagnostic evaluations continues to provide helpful guidance, even in the era of clinical pharmacogenomics.
Another element of an interim approach to managing results from prospective pharmacogenomic testing is that every clinical use, whether it be a pharmacogenomic application or the return of an incidental finding, should be reviewed by an institutional committee. A number of institutions have addressed this challenge by recruiting area experts to serve on a subcommittee of the hospital Pharmacy and Therapeutics Committee [7]. One benefit of such an approach is that a subcommittee can ensure that incidental findings meet appropriate standards of validity and utility before they are reported to patients and providers [17,19] . This task may one day be accomplished by informatics tools integrated into electronic medical records, most likely adapted from algorithms designed for managing incidental findings in the research setting [20–22]. For now it is an effort best undertaken by groups of stakeholders with appropriate expertise. An additional benefit of this approach is that the incidentalome can be addressed in manageable pieces. Institutional committees reviewing results can take the total number of results being returned into account, and implement prioritization as necessary.
A final, but indispensable, element of any clinical pharmacogenomic project is an appropriate informed consent process. For projects designed as research studies, relevant regulations related to informed consent for research participation will apply. However, for projects designed only to provide clinical testing for pharmacogenomic applications that have already been validated (and thus will not be utilizing either WES or WGS), a written consent process may be neither necessary nor preferable. Healthcare providers frequently obtain verbal or implicit consent for routine health interventions, such as laboratory tests or simple imaging studies. This same approach may work well for focused pharmacogenomic tests. One exception worth noting, however, is that regulations in a number of jurisdictions require written informed consent whenever a genetic test is performed in a healthcare context. These include, for example, a federal law in Germany [104] and state-level laws in a number of US states [105] .
When verbal consent is used for pharmacogenomic testing, the conversation between the patient and provider should simply cover:
Why the testing is being proposed
The types of incidental findings that may be produced (using the categories discussed above as an outline)
How incidental findings in each category will be addressed
How patients will be informed of changes to how their results will be used
Pamphlets or other informational documents might facilitate this discussion and help patients later recall what was discussed.
There is currently significant debate about the role patient preferences should play when Category 1 results are identified [23,24]. That this debate remains unresolved serves to emphasize the value of interim approaches that are characterized by local communication and the involvement of multiple stakeholders. For our part, we believe that patient preferences should be elicited in the informed consent process and respected whenever possible.
As clinical genotyping programs evolve, however, new policies will become necessary. The approaches used for identifying and delivering incidental findings will need to be revised, and with these changes will come new requirements for appropriate informed consent. In the next section, we will turn our attention to the clinical pharmacogenomics of the future. We will develop a more detailed picture of the policies that will be needed as clinical pharmacogenomic projects expand to more comprehensive genotyping technologies and more expansive clinical goals.
Future perspective
As we argued earlier, informatics tools used to analyze and deliver genomic results will need to be designed both to categorize results and also prioritize results within categories. This is easier said than done, of course. Algorithms for categorizing results will depend on tools capable of linking genetic variants with specific phenotypes, and then classifying such associations according to quality of evidence and clinical utility. Groups such as the Human Genome Epidemiology (HuGE) network and the Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative are currently working on performing these types of analyses [25,106,107]. The capability to automate such a process, however, remains nascent.
The development of tools capable of prioritizing Category 1 results will be even more challenging. Prioritization schema should ideally be personalized to each individual patient’s situation. From the genotype side, incidental findings indicating higher absolute risks for the development of diseases should be prioritized higher, with diagnostic genetic results ranked highest of all. On the phenotype side, age and gender at a minimum should be taken into account in any algorithm used to prioritize results. More sophisticated systems might be able to consider a patient’s personal medical history and even personal preferences in selecting results with the highest priority.
Our suggestion of a ‘top ten list’ for the highest priority results from Category 1 is just one idea. We believe there will need to be a great deal of careful exploration of other options. The overall aim of any prioritization schema will be to ensure that those results that can provide the most benefit are provided adequate time and attention by both patients and providers, for it is only through proper time and attention that any benefit can be realized from a genetic result.
Some results in Category 1 could have a net negative utility. That is, adding one additional result to those that must be addressed may distract patients and providers from focusing on more important results, thus decreasing the overall utility of these results. Such low-utility results will need to be provided through alternative mechanisms. For example, some findings may only prove important when certain clinical events take place; these can be returned on a just-in-time basis. Other results may be delivered through patient portals or at the time of a future health maintenance visit.
Algorithms for generating results from Category 2 will only be applied to raw genotyping data when patients affirmatively choose to receive them because they are addressing issues related to reproduction. Although patients and providers will need to develop approaches that make sense in their contexts, we anticipate that most will prefer to address these findings during clinic visits dedicated specifically to reproductive planning. For this reason, we expect that in most cases discussions about Category 2 results will not compete with routine health maintenance.
Analyses used to generate results from Category 3 should only be applied once an adult patient has been asked to consider the risks and benefits of receiving this type of result and has affirmatively agreed to receive them. Like results from Category 2, we do not anticipate that discussions related to these results will take place during routine health visits. Instead, we envision opportunities for patients to obtain and consider these results through consultation with genetic counselors or other specialists.
As we work our way through low-utility Category 1, Category 2 and Category 3 results, we begin to see that a number of practical factors could limit the extended benefits hoped for when expanding pharmacogenomic testing from focused panels of pharmacogenes to WES or WGS technologies. In ideal conditions, patients will have longitudinal relationships with primary care providers, have adequate access to specialists including genetic counselors, and will themselves be capable of attending to a large set of health concerns and health guidance. We know, however, that these ideal conditions are rarely achieved. The data may be virtually unlimited, but resources are not.
We should be clear, then, that the approach we are proposing will not guarantee, under real-world conditions, that all potential results generated through WES or WGS would be addressed in a conversation between a patient and a qualified provider. We are aware of the liability concerns that are raised by this approach [26]. Although a complete analysis of relevant liability risks are beyond the scope of this paper, we can make a few brief observations here. First, one of the key arguments of this paper is that the goal of prioritization is to improve the quality of care, not to decrease it. Adequately addressing the most important results is a superior approach compared with inadequately addressing a great number of results. It is for this reason that prioritization is already an important tool in clinical medicine [14] .
Second, informed consent can be used to notify patients how incidental findings will be managed and filtered, including information about how results will be prioritized. The precise informed consent process used will depend on the overall complexity of the program. Verbal consent is likely to suffice for targeted testing of pharmacogenes, while a more formal accounting of risks and benefits would be appropriate for programs involving WES or WGS.
Third, we believe a majority of patients will be content to receive only the highest priority results as a part of their preventive healthcare. Empirical findings from the setting of return of research results indicate that many research subjects want to know ‘everything’ they can be told from their genome [27–29]. Despite this perspective, patients may accept a more focused approach if their personal concerns are taken into account in prioritization algorithms, and if other opportunities are provided to review lower priority results, such as through patient portals or clinic visits with genetic counselors. This hypothesis about patient preferences can and should be tested empirically.
In the final analysis, though, it seems that a broad discussion on the legal obligations of healthcare providers in the era of genomic medicine remains a top priority. Two concerns in particular will need to be addressed. First, if technologies are used that produce more results than can reasonably be addressed by clinicians and patients, are approaches that focus only on higher priority Category 1 results acceptable? In other words, what responsibility do healthcare providers have to view and address all results generated?
Second, what responsibility do providers have to follow-up on results over time? Genotyping performed in 2013 or 2014 will immediately generate a small number of incidental findings. Over time, however, it will support analyses that can reveal ever increasing numbers of incidental findings. Assuming that informatics tools capable of categorizing and prioritizing results can be developed, it is reasonable to assume that they could also be used to apply these algorithms iteratively over time. With this capability will come the need to develop practice guidelines on how providers should respond to new, high- priority results that could be generated for patients no longer under their care. A reasonable duty to rescue could apply in certain cases [30], but a great deal of work is still needed to identify the boundaries and practical limitations of this duty. Going further, any guidance that is developed will not provide once-and-for-all answers, but will instead require ongoing assessment and evaluation.
Conclusion
Given the growing evidence supporting the utility of pharmacogenomic testing, the justification for implementing prospective pharmacogenomic testing in routine clinical care continues to increase. However, it is possible that the challenge of managing and addressing the incidentalome could further delay the clinical implementation of this testing. This is especially true given that optimal informatics tools for categorizing and prioritizing results have not yet been developed. A provisional approach to managing incidental findings should be implemented so that the benefits of pharmacogenomic testing may be realized in the clinical setting. Developing approaches for managing incidental findings in greater numbers will need to focus on balancing the number of incidental findings returned with the practical ability of patients and providers to address them, and developing informed consent procedures appropriate to the numbers and types of results generated. The development of a clearer account of the legal obligations of healthcare providers in the age of genomic medicine remains a top priority.
Executive summary.
Background
The incidentalome is the potentially large set of incidental findings that can be generated through multiplex genotyping technologies.
A new dimension to the challenge of incidental findings is introduced as we move from clinical pharmacogenomic testing with a single-gene assay to technologies that perform multiple genetic tests in parallel.
Incidental findings from pharmacogenetic tests
Two key changes that will take place in this transition to multiplex genotyping technologies are a greatly increased number of results to address and manage and a transition from management of results in specialized medical care settings to primary care settings.
Preventive health measures must necessarily focus on a set of the most important and highest utility interventions. This principle extends to the use of incidental genetic findings from pharmacogenomic testing.
Schema for categorizing incidental findings are an important element of approaches to managing large numbers of results. Categories also facilitate discussions with patients about what types of incidental findings can be expected as a part of informed consent to clinical pharmacogenetic testing.
‘The tail wagging the dog’
If significant clinical benefits from pharmacogenomics can be realized, then implementation should not be delayed in order to allow time for definitive methods for managing incidental findings to be developed. Provisional methods for managing incidental findings can be implemented now.
Appropriate informed consent is an indispensable part of any clinical pharmacogenomic effort, including those that utilize both provisional and more comprehensive approaches to incidental findings.
Future perspective
Appropriate methods for categorizing and prioritizing incidental findings will need to be developed before whole-exome sequencing or whole-genome sequencing are implemented in routine clinical care for pharmacogenomic purposes.
The development of a clearer account of the legal obligations of healthcare providers in the age of genomic medicine remains a top priority.
Acknowledgements
The authors would like to acknowledge the contributions of EW Clayton (Vanderbilt University) and M Rothstein (University of Louisville) for formative conversations on the issues addressed in this paper. The authors would also like to thank S Rudnik-Schöneborn (University of Aachen, Germany) for her contributions, including her permission to reproduce the content in Table 1.
This work was funded in part by the Vanderbilt Genome-Electronic Records Project, NIH/NHGRI grant 1U01HG006378-01 and the Vanderbilt Institute for Clinical and Translational Research (VICTR), NCATS/NIH grant UL1TR000011. This work is also part of the research project Greifswald Approach to Individualized Medicine (GANI_MED). The GANI_MED consortium is funded by the Federal Ministry of Education and Research and the Ministry of Cultural Affairs of the Federal State of Mecklenburg-West Pomerania (support codes: 03IS2061A & 03IS2061E).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
of interest
of considerable interest
- 1.Owens H, Siparsky G, Bajaj L, Hampers LC. Correction of factitious hyperkalemia in hemolyzed specimens. Am. J. Emerg. Med. 2005;23(7):872–875. doi: 10.1016/j.ajem.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Zandecki M, Genevieve F, Gerard J, Godon A. Spurious counts and spurious results on haematology analysers: a review. Part I: platelets. Int. J. Lab. Hematol. 2007;29(1):4–20. doi: 10.1111/j.1365-2257.2006.00870.x. [DOI] [PubMed] [Google Scholar]
- 3. Jackson BR. The dangers of false-positive and false-negative test results: false-positive results as a function of pretest probability. Clin. Lab. Med. 2008;28(2):305–319. doi: 10.1016/j.cll.2007.12.009. □Clear and helpful account of the problem of false-positive results in the use (and overuse) of clinical diagnostic tests
- 4.Vernooij MW, Ikram MA, Tanghe HL, et al. Incidental findings on brain MRI in the general population. N. Engl. J. Med. 2007;357(18):1821–1828. doi: 10.1056/NEJMoa070972. [DOI] [PubMed] [Google Scholar]
- 5. Kohane IS, Masys DR, Altman RB. The incidentalome: a threat to genomic medicine. JAMA. 2006;296(2):212–215. doi: 10.1001/jama.296.2.212. □□In this paper the authors coin the term ‘incidentalome’ and introduce some of the challenges that are developed in greater detail in the current paper
- 6. Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin. Pharmacol. Ther. 2012;92(5):563–566. doi: 10.1038/clpt.2012.140. □Along with reference [7] addresses in some detail the practical decisions made by institutions working to implement prospective pharmacogenomic testing into clinical care
- 7. Pulley J, Denny J, Peterson J, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther. 2012;92(1):87–95. doi: 10.1038/clpt.2011.371. □Along with reference [6] addresses in some detail the practical decisions made by institutions working to implement prospective pharmacogenomic testing into clinical care.
- 8.Westbrook MJ, Wright MF, van Driest SL, et al. Mapping the incidentalome: estimating incidental findings generated through clinical pharmacogenomics testing. Genet. Med. 2013;15(5):325–331. doi: 10.1038/gim.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlos R, Phillips EJ. Individualization of antiretroviral therapy. Pharmacogenomics Pers. Med. 2012;5:1–17. doi: 10.2147/PGPM.S15303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piga M, Mathieu A. Genetic susceptibility to Behcet’s disease: role of genes belonging to the MHC region. Rheumatology (Oxford) 2011;50(2):299–310. doi: 10.1093/rheumatology/keq331. [DOI] [PubMed] [Google Scholar]
- 11.Salgado M, Simon A, Sanz-Minguela B, et al. An additive effect of protective host genetic factors correlates with HIV nonprogression status. J. Acquir. Immune Defic. Syndr. 2011;56(4):300–305. doi: 10.1097/QAI.0b013e3182036f14. [DOI] [PubMed] [Google Scholar]
- 12.O’Donnell PH, Bush A, Spitz J, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin. Pharmacol. Ther. 2012;92(4):446–449. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yarnall KSH, Pollak KI, Østbye T, Krause KM, Michener JL. Primary care: is there enough time for prevention? Am. J. Public Health. 2003;93(4):635–641. doi: 10.2105/ajph.93.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maciosek MV, Coffield AB, Edwards NM, Flottemesch TJ, Goodman MJ, Solberg LI. Priorities among effective clinical preventive services. Am. J. Prev. Med. 2006;31(1):52–61. doi: 10.1016/j.amepre.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl. Res. 2009;154(6):277–287. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 16. Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet. Med. 2011;13(6):499–504. doi: 10.1097/GIM.0b013e318220aaba. □One of two articles that discuss schema for organizing the results generated by multiplex genetic tests
- 17. Rudnik-Schöneborn S, Langanke M, Erdmann P, Robienski J. [Ethical and legal aspects of handling incidental findings from genetics: challenges and solutions] Ethik in der Medizin. 2013 Epub ahead of print. □One of two articles that discuss schema for organizing the results generated by multiplex genetic tests
- 18.Ormond KE, Wheeler MT, Hudgins L, et al. Challenges in the clinical application of whole-genome sequencing. Lancet. 2010;375(9727):1749–751. doi: 10.1016/S0140-6736(10)60599-5. [DOI] [PubMed] [Google Scholar]
- 19.Langanke M, Brothers KB, Erdmann P, et al. Comparing different scientific approaches to personalized medicine: research ethics and privacy protection. Pers. Med. 2011;8(4):437–444. doi: 10.2217/pme.11.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegenscheid K, Seipel R, Schmidt C, et al. Potentially relevant incidental findings on research whole-body MRI in the general adult population: frequencies and management. Eur. Radiol. 2013;23(3):816–826. doi: 10.1007/s00330-012-2636-6. [DOI] [PubMed] [Google Scholar]
- 21.Langanke M, Erdmann P. [MRI as a scientific examination and the problem of communicating incidental findings, ethical challenges regarding research subjects] In: Theiβen H, Langanke M, editors. Tragfähige Rede von Gott. Festgabe für Heinrich Assel zum 50 Geburtstag am 9. Februar 2011. Germany: Hamburg; 2011. pp. 197–240. [Google Scholar]
- 22.Erdmann P, Langanke M, Assel H. Proceedings of the Societas Ethica Annual Conference, The Quest for Perfection. The Future of Medicine/Medicine of the Future. Lugano, Switzerland: Linköping University Electronic Press; 2011. Incidental findings – understanding of risks by research subjects and implications for research ethics; pp. 15–38. [Google Scholar]
- 23.Allyse M, Michie M. Not-so-incidental findings: the ACMG recommendations on the reporting of incidental findings in clinical whole genome and whole exome sequencing. Trends Biotechnol. 2013 doi: 10.1016/j.tibtech.2013.04.006. pii, S0167-7799(13)00092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green R, Berg J, Grody W, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoury MJ. Human Genome Epidemiology (HuGE): translating advances in human genetics into population-based data for medicine and public health. Genet. Med. 1999;1:71–73. doi: 10.1097/00125817-199903000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Marchant GE, Campos-Outcalt DE, Lindor RA. Physician liability: the next big thing for personalized medicine? Pers. Med. 2011;8(4):457–467. doi: 10.2217/pme.11.33. [DOI] [PubMed] [Google Scholar]
- 27.Halverson CME, Ross LF. Attitudes of African–American parents about biobank participation and return of results for themselves and their children. J. Med. Ethics. 2012;38(9):561–566. doi: 10.1136/medethics-2012-100600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy J, Scott J, Kaufman D, Geller G, Leroy L, Hudson K. Public expectations for return of results from large-cohort genetic research. Am. J. Bioethics. 2008;8(11):36–43. doi: 10.1080/15265160802513093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz-Canela M, Valle-Mansilla JI, Sulmasy DP. What research participants want to know about genetic research results: the impact of “genetic exceptionalism”. J. Empir. Res. Hum. Res. Ethics. 2011;6(3):39–46. doi: 10.1525/jer.2011.6.3.39. [DOI] [PubMed] [Google Scholar]
- 30.Beskow LM, Burke W. Offering individual genetic research results: context matters. Sci. Transl. Med. 2010;2(38) doi: 10.1126/scitranslmed.3000952. 38cm20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.Information for Healthcare Professionals: Abacavir (marketed as Ziagen) and Abacavir-Containing Medications. 2008 www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm123927.htm.
- 102. Table of pharmacogenomic biomarkers in drug labels. 2013 www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm. □□A constantly updated list of pharmaceuticals whose US FDA-approved labeling contains information about pharmacogenomic indications.
- 103.Genetic testing for carrier status, disease risk and drug response. 2013 www.23andme.com/health/drugs.
- 104.Law on Genetic Examinations of Human Subjects (Gendiagnostikgesetz: GenDG) German: 2009. www.gesetze-im-internet.de/bundesrecht/gendg/gesamt.pdf. [Google Scholar]
- 105.Genetic information: legal issues relating to discrimination and privacy. 2008 http://assets.opencrs.com/rpts/RL30006_20080310.pdf.
- 106.Human Genome Epidemiology Network (HuGE Net) 2011 www.cdc.gov/genomics/hugenet/default.htm.
- 107.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) 2012 doi: 10.1097/GIM.0b013e318184137c. www.egappreviews.org. [DOI] [PMC free article] [PubMed]