Abstract
Mammalian mitochondria contain about 1100 proteins, nearly 300 of which are uncharacterized. Given the well-established role of mitochondrial defects in human disease, functional characterization of these proteins may shed new light on disease mechanisms. Starting with yeast as a model system, we investigated an uncharacterized but highly conserved mitochondrial protein (named here Sdh5). Both yeast and human Sdh5 interact with the catalytic subunit of the succinate dehydrogenase (SDH) complex, a component of both the electron transport chain and the tricarboxylic acid cycle. Sdh5 is required for SDH-dependent respiration and for Sdh1 flavination (incorporation of the flavin adenine dinucleotide cofactor). Germline loss-of-function mutations in the human SDH5 gene, located on chromosome 11q13.1, segregate with disease in a family with hereditary paraganglioma, a neuroendocrine tumor previously linked to mutations in genes encoding SDH subunits. Thus, a mitochondrial proteomics analysis in yeast has led to the discovery of a human tumor susceptibility gene.
Mitochondrial defects have been implicated in a variety of human disorders, including cancer (1, 2). Nearly one-third of the mammalian mitochondrial proteome is currently uncharacterized. Many of these uncharacterized proteins are evolutionarily conserved, a strong indication that they perform fundamentally important cellular functions (3). We initiated a project to determine the function of one of these proteins, named here Sdh5, using yeast as the primary model system. The Sdh5 protein family is highly conserved in eukaryotes and in some prokaryotic species, including Rickettsia, which is related to the bacterium that became the ancestral mitochondrion (fig. S1) (4, 5).
The mitochondrial localization of yeast Sdh5 (originally named EMI5/YOL071W) was suggested by proteomics studies (6), and we confirmed this by fluorescence microscopy of a strain expressing Sdh5–green fluorescent protein and subcellular fractionation of a strain expressing Sdh5-GFP (green fluorescent protein) (figs. S2 and S3). Both forms of tagged Sdh5 were expressed from the native SDH5 promoter and were fully functional. We further showed that Sdh5 resides in the mitochondrial matrix (Fig. 1A) and is predominantly soluble (Fig. 1B).
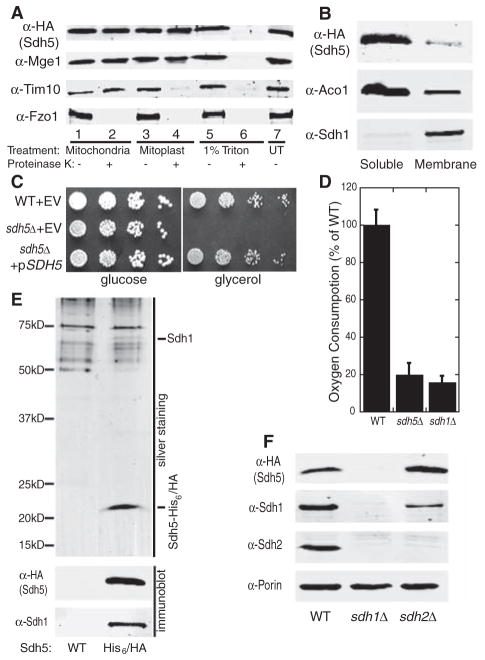
Fig. 1.
Sdh5 is required for respiration and interacts with Sdh1. (A) Mitochondria, mitoplasts generated by hypotonic swelling, and 1% Triton X-100–solubilized mitochondria from a strain expressing Sdh5-HA were treated with (+) or without (−) proteinase K and analyzed by immunoblotting with an untreated mitochondria control (UT). Mge1, Tim10, and Fzo1 are matrix, intermembrane space, and outer membrane proteins, respectively. (B) Soluble and membrane fractions of purified mitochondria (4) as in (A) were immunoblotted. Aco1, soluble matrix protein; Sdh1, membrane-associated matrix protein. (C) Serial dilutions of WT and sdh5Δ strains containing empty vector (EV) or a plasmid expressing Sdh5-HA were spotted on glucose or glycerol medium and grown at 30°C for 2 or 3 days, respectively. (D) Oxygen consumption in WT, sdh5Δ, and sdh1Δ strains grown to mid-log phase in raffinose media (±SD, n = 3 biological replicates). Similar results were obtained in glucose medium. (E) Tandem purification eluates (4) from a WT strain and a strain expressing Sdh5-His6/HA were resolved with SDS-PAGE and visualized by silver staining (top panel) or immunoblot with antibodies to Sdh1 and HA (lower panels). (F) Immunoblot of purified mitochondria from WT, sdh1Δ, or sdh2Δ strains expressing Sdh5-HA. Porin, mitochondrial loading control.
Respiratory-deficient mutants of S. cerevisiae are viable on fermentable carbon sources such as glucose, but are inviable on nonfermentable carbon sources such as glycerol. We found that a yeast strain with a deletion of SDH5 (sdh5Δ) grew normally on glucose medium but failed to grow on glycerol medium, a phenotype that was rescued by an SDH5-expressing plasmid (Fig. 1C). The sdh5Δ strain also showed impaired oxygen consumption, similar to the respiratory-deficient sdh1Δ strain (7) (Fig. 1D), as well as the respiration-related phenotypes of H2O2 hypersensitivity and decreased chronological life-span (figs. S4 and S5). The sdh5Δ respiratory deficiency was not due to defective mitochondrial DNA (mtDNA), because the sdh5Δ strain complemented the glycerol growth defect of a rho0 (mtDNA-deficient) strain in mating tests (fig. S6). Thus, the sdh5Δ strain is respiratory-deficient, despite having a functional mitochondrial genome.
Silver staining of the final eluates from tandem affinity purification of Sdh5-His6/HA revealed two proteins specific for tagged Sdh5 as compared to the wild type, including Sdh5 itself at ~22 kD (Fig. 1E). The second migrated at ~70 kD and was identified by mass spectrometry as Sdh1, the catalytic subunit of the succinate dehydrogenase (SDH) complex. The presence of both Sdh5 and Sdh1 in the final eluate was confirmed by immunoblot (Fig. 1E). The SDH complex is a component of both the tricarboxylic acid (TCA) cycle and the electron transport chain (ETC). In the latter, the SDH complex is known as complex II. It is a highly conserved heterotetramer, with Sdh1 and Sdh2 as catalytic subunits anchored to the mitochondrial inner membrane by Sdh3 and Sdh4 (fig. S12) (8). Both sdh1Δ and sdh5Δ mutants were respiratory-deficient and failed to grow on glycerol medium, but grew weakly with ethanol as the carbon source (Fig. 1D and fig. S7) and exhibited acetate hyperexcretion, a phenotype shared by only four other TCA cycle mutants (9). The importance of the Sdh1–Sdh5 interaction was confirmed by the observation that Sdh5 (like Sdh2) is completely degraded in the sdh1Δ strain (Fig. 1F). In contrast, loss of SDH2 led to an increase in the Sdh5 protein level (Fig. 1F), presumably due to enhanced Sdh1/Sdh5 complex formation in the absence of Sdh2, the major Sdh1 partner.
The Sdh1-Sdh5 interaction is likely to be functionally important because the sdh5Δ mutant lacks SDH activity (Fig. 2A), as previously observed for the sdh1Δ mutant (10). The activity of malate dehydrogenase, another TCA cycle enzyme, was not affected by SDH5 deletion (Fig. 2A). Because SDH is complex II in the ETC, we performed in-gel activity staining of ETC complexes after separation by blue native–polyacrylamide gel electrophoresis (BN-PAGE). As shown in Fig. 2B, complex II/SDH activity was absent in the sdh5Δ mutant, whereas the activities of complexes IV and V were normal. We then examined ETC complex assembly and stability by Coomassie blue staining after BN-PAGE and found that complex II/SDH was specifically absent in sdh5Δ mitochondria (Fig. 2C). The loss of complex II/SDH in the sdh5Δ mutant was confirmed by anti-Myc immunoblot after BN-PAGE of wild-type (WT) and sdh5Δ strains expressing Sdh3-Myc (Fig. 2D). Sdh5 is not a stable component of complex II/SDH because it migrates in a ~90-kD SDS-sensitive complex during BN-PAGE (Fig. 2E), which is distinct from complex II (~200 kD). This ~90-kD complex is likely to be the Sdh5-Sdh1 (70-kD) heterodimer.
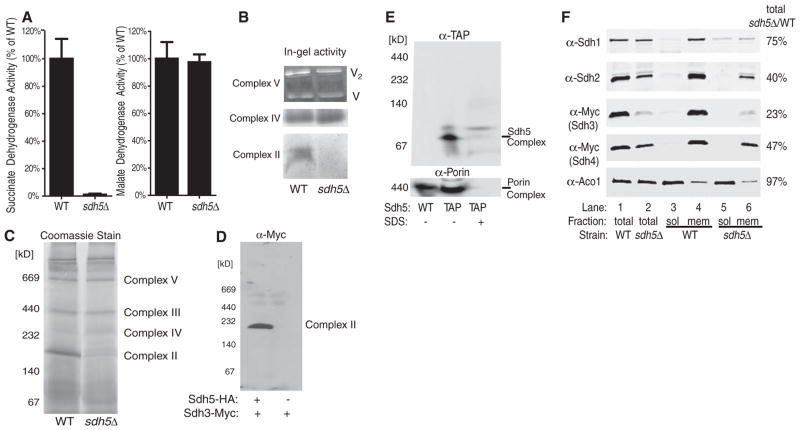
Fig. 2.
Sdh5 is required for SDH activity and stability. (A) SDH and malate dehydrogenase activity (4) from WT and sdh5Δ mitochondria, normalized to total protein (±SD, n = 3 biological replicates). (B) In-gel activity assay of ETC complexes after BN-PAGE of mitochondrial membranes from WT and sdh5Δ strains. V2, complex V dimer. (C) Coomassiestained BN-PAGE of mitochondrial membranes from WT and sdh5Δ strains. (D) Immunoblot of BN-PAGE–separated complex II/SDH using an antibody to Myc to show Myc-tagged Sdh3 in WT and sdh5Δ mitochondria. (E) Immunoblot of BN-PAGE–separated WT and Sdh5-TAP mitochondria using an antibody to TAP, without and with 1% SDS pretreatment. Porin is an ~440-kD control. (F) Immunoblot of mitochondria from WT and sdh5Δ strains in which Sdh3 or Sdh4 was Myc-tagged, separated into soluble (sol) and membrane (mem) fractions (4), or unfractionated (total). Aco1, soluble matrix protein. The indicated percentage is the amount remaining in sdh5Δ mitochondria relative to WT mitochondria.
The levels of all four SDH subunits were significantly decreased in the sdh5Δ mutant, probably because of degradation in the absence of a stable SDH complex (Fig. 2F, lane 1 versus lane 2). The residual Sdh1 level was higher than that of the other subunits, but much of it was in the soluble fraction, unassociated with the SDH complex (Fig. 2F). These data suggest that the SDH complex assembles in the absence of Sdh5, but the complex is nonfunctional and Sdh1 is not stably bound. As a result, the unstable complex is more susceptible to degradation and is disrupted by detergent extraction during the BN-PAGE procedure (the fractionation shown in Fig. 2F was detergent-free).
Multiple cofactors are required for activity of the SDH complex, including the flavin adenine dinucleotide (FAD) in Sdh1 (11). In other ETC complexes, cofactors are important for complex assembly and stability in addition to enzymatic activity (12). The strongly fluorescent FAD is covalently attached to Sdh1 and can be detected by in-gel fluorometry after SDS-PAGE (13). Deletion of SDH5 caused a complete loss of FAD cofactor attachment (flavination) of Sdh1, although Sdh1 was still present (Fig. 3A). The flavination of two other mitochondrial flavo-proteins, visualized in the same gel, was unaffected by either SDH5 or SDH1 deletion (Fig. 3A).
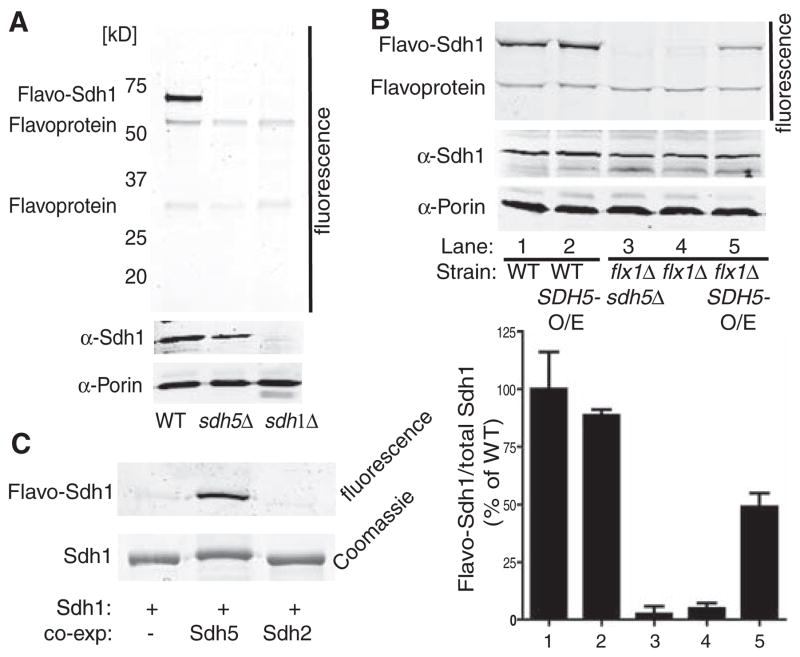
Fig. 3.
Sdh5 is necessary and sufficient for Sdh1 flavination. (A) WT, sdh5Δ, and sdh1Δ mitochondria were resolved by SDS-PAGE and imaged (4) for covalent FAD (top panel) or immunoblotted (lower panels). (B) Fluorescence gel image (top panel) and immunoblot (lower panels) as in (A), with whole-cell extract from WT or flx1Δ sdh5Δ strains containing EV, CEN plasmid SDH5 (flx1Δ: ~1 copy per cell), or 2μ plasmid SDH5 (O/E: ~10 copies per cell). The bar graph shows normalized FAD fluorescence (±SD, n = 3 biological replicates) (bottom panel). (C) His-tagged yeast Sdh1 was expressed alone or with Sdh5 or Sdh2 in E. coli, purified, and analyzed for FAD fluorescence as in (A) and by Coomassie blue staining.
Sdh5 overexpression in WT cells did not increase Sdh1 flavination (Fig. 3B, lane 1 versus lane 2), possibly due to flavination that was already stoichiometric. A significant effect of Sdh5 over-expression might require a state of reduced Sdh1 flavination as was observed in a strain with decreased mitochondrial FAD caused by deletion of the mitochondrial FAD transporter Flx1 (13, 14). The flx1Δ mutant displayed almost no Sdh1 flavination, but Sdh5 overexpression restored flavination to about 50% of WT levels (Fig. 3B, lanes 4 and 5). As shown in Fig. 3C, FAD incorporation was almost undetectable in Escherichia coli–expressed Sdh1 but was increased dramatically when Sdh5, but not Sdh2, was coexpressed. These data demonstrate that Sdh5 is both necessary and sufficient for Sdh1 flavination.
The amino acid sequence of yeast Sdh5 is 44% identical (from residues 33 to 158 of 163) to its human ortholog C11orf79, which we rename here hSDH5 (fig. S1). We hypothesized that hSDH5 is required for flavination of SDHA (human Sdh1) and thus for SDH activity. Previous genetic studies have shown that mutations causing loss of SDH activity are responsible for hereditary forms of a rare neuroendocrine tumor called paraganglioma (PGL). Of the four familial PGL syndromes (PGL1 to PGL4), PGL1, PGL3, and PGL4 have been associated with mutations in SDHD, SDHC, and SDHB, respectively (15). The gene for PGL2 (Online Mendelian Inheritance in Man number %601650) has not been identified, but it maps to chromosome 11q13.1, the genomic locus of the hSDH5 gene (16).
We sequenced hSDH5 in three affected individuals from different branches of the previously described Dutch PGL2 lineage (17). In all three individuals, we found a single nucleotide c.232G>A change in exon 2 (fig. S8), which causes a Gly78 → Arg78 (G78R) mutation in the most conserved region of the protein (fig. S1). None of 400 unaffected control individuals carried the c.232G>A mutation. Within the affected lineage, the mutation was found to cosegregate with the disease haplotype in all 45 individuals who inherited this haplotype, and was not detected in 44 unaffected members without this haplotype. Thirty-three individuals with the mutation have developed the disease, but not seven individuals (2009 median age = 74 years) who inherited the mutation from their mother (Fig. 4A). This suggests an SDHD-like parent-of-origin-specific inheritance pattern. Only five individuals (2009 median age = 42 years) with a paternal mutation have not developed overt PGL. Because penetrance of the disease increases with age (17), these individuals may develop tumors in the future, or tumors may already be present but undetected.
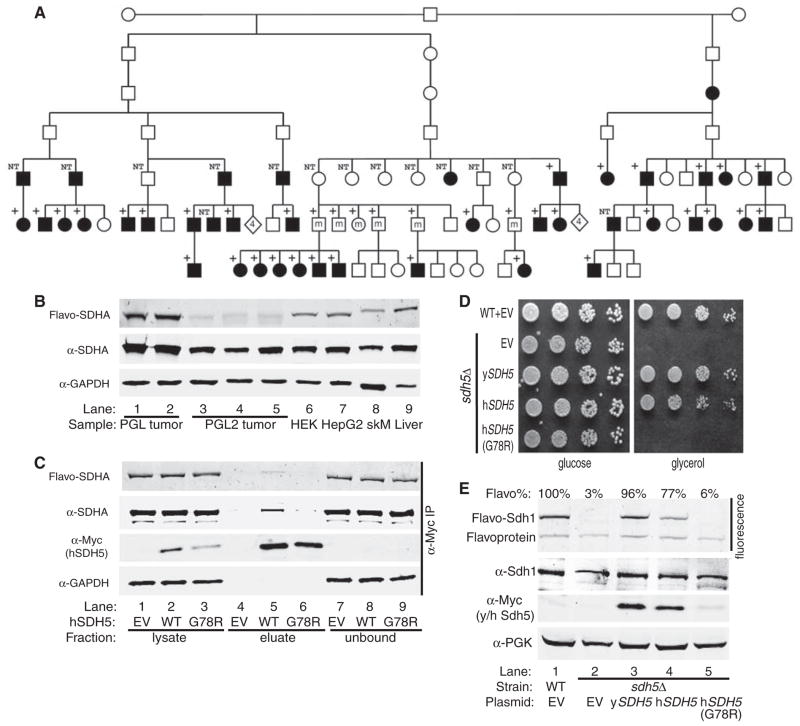
Fig. 4.
A human SDH5 loss-of-function mutation in PGL2. (A) The heterozygous c.232G>A mutation segregates with disease in the PGL2 lineage (17). Black symbols, affected persons; white symbols, unaffected persons; +, heterozygous mutation; NT, not tested. Diamonds with the number 4 represent four unaffected individuals. Individuals who are not affected because they carry the mutation on their maternal chromosome 11 are marked by an m. One healthy maternal mutation carrier and five non-affected paternal mutation carriers are not shown in the pedigree for privacy reasons. (B) Fluorescence gel image (top panel) and immunoblotting (lower panels) of samples from human tumors, cell lines, and mouse tissues. Lanes 1 and 2, sporadic PGL tumors; lanes 3 to 5, PGL2 tumors (hSDH5 G78R); lanes 6 and 7, HEK293 and HepG2 human cell lines; lanes 8 and 9, normal mouse skeletal muscle (skM) and liver. (C) Lysate from HEK293 cells containing EV or expressing WT or G78R hSDH5-Myc were immunoprecipitated with agarose beads conjugated with antibody to Myc. Lysate, eluate, and unbound fraction were FAD-imaged (top panel) and immunoblotted (lower three panels). (D) Serial dilutions of WT and sdh5Δ strains containing EV or plasmids expressing yeast Sdh5-Myc, WT human SDH5-Myc, or G78R hSDH5-Myc were spotted on glucose or glycerol medium and grown at 30°C for 2 or 3 days, respectively. (E) Fluorescence gel image (top panel) and immunoblotting of whole-cell extract from the five strains in (D) (lower panels). PGK, loading control. FAD fluorescence was normalized to Sdh1 protein level and expressed as a percentage relative to WT (Flavo%, ±SD, n = 3 biological replicates).
SDHA flavination was decreased by ~95% in tumors from three patients with PGL2 (hSDH5 G78R) in comparison with control tumors from two sporadic PGL patients or with cultured human embryonic kidney and hepatoma cells and mouse skeletal muscle and liver tissue (Fig. 4B). Mitochondrial localization of hSDH5 was not compromised by this mutation (fig. S9). As observed in yeast, SDHA coimmunoprecipitated with WT hSDH5-Myc. The G78R hSDH5 mutation, however, dramatically impaired the interaction of hSDH5 with SDHA and slightly decreased the level of hSDH5 protein (Fig. 4C). We conclude that the G78R mutation destabilizes hSDH5 and impairs its interaction with SDHA.
When expressed in yeast from the yeast SDH5 promoter, WT hSDH5 complemented the sdh5Δ glycerol growth defect, but the G78R mutant had no effect (Fig. 4D). Overexpression of any of the individual SDH subunits or the proposed SDH chaperone TcM62 (18) failed to complement the sdh5Δ mutant phenotype (fig. S10). Further, an extensive high-copy suppressor screen of the sdh5Δ glycerol growth defect using a yeast genomic library recovered many independent isolates of SDH5 but failed to identify any other suppressing genes. Therefore, the specific rescue of the sdh5Δ glycerol growth defect by native-level expression of hSDH5 is strong evidence of the conservation of Sdh5 function from yeast to humans. Furthermore, expression of hSDH5 in an sdh5Δ mutant strain increased the flavination of yeast Sdh1 to 77% of the WT level, but the G78R mutant had no effect (Fig. 4E). The lack of activity of the G78R hSDH5 mutant in yeast is likely due to destabilization of the protein (Fig. 4E).
The mechanism of covalent FAD insertion into proteins is controversial. Although other covalent cofactors are inserted by specialized enzymes (19), an autocatalytic mechanism has been proposed for flavination (20). However, covalent FAD attachment to Sdh1 was shown to require at least one additional protein (21), which we hypothesize to be Sdh5. Whether Sdh5 participates in the chemistry of FAD attachment (enzymatic function) or simply maintains Sdh1 in a conformation that is susceptible to autocatalytic FAD attachment (chaperone function) is a question that remains to be addressed.
The identification of hSDH5 as the PGL2 gene has potential clinical implications. Approximately 70% of familial cases of head and neck PGL are due to germline mutations in SDHB, SDHC, or SDHD (22), and 10% of sporadic PGL cases are associated with mutations in SDHB or SDHD (23, 24). Mutations in SDHB, SDHC, and SDHD have also been detected in gastrointestinal stromal tumors (25), pheochromocytomas (24), renal cell carcinoma (26), and other diseases (27). Conceivably, hSDH5 mutations may underlie disease development in patients who have tested negative for mutations in SDHB, SDHC, and SDHD. The inclusion of hSDH5 will allow more comprehensive genetic testing, which is suggested for the clinical management of PGL, even for sporadic cases (28). The same is also true of the SDHAF1 gene, which encodes a protein involved in SDH assembly and which was recently identified as a causative factor in infantile leukoencephalopathy (29). In summary, starting with a previously uncharacterized mitochondrial protein in yeast, we have shown through biochemical and genetic analyses that the protein plays a critical role in the biogenesis and function of a respiratory complex and that its mutational inactivation confers tumor susceptibility in humans.
Supplementary Material
Footnotes
References and Notes
- 1.Wallace DC. Annu Rev Genet. 2005;39:359. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin MT, Beal MF. Nature. 2006;443:787. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 3.Pagliarini DJ, et al. Cell. 2008;134:112. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.See supporting material on Science Online.
- 5.Andersson SG, et al. Nature. 1998;396:133. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 6.Sickmann A, et al. Proc Natl Acad Sci USA. 2003;100:13207. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman KB, Solomon SD, Boeke JD. Gene. 1992;118:131. doi: 10.1016/0378-1119(92)90260-v. [DOI] [PubMed] [Google Scholar]
- 8.Sun F, et al. Cell. 2005;121:1043. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Romano JD, Kolter R. J Bacteriol. 2005;187:940. doi: 10.1128/JB.187.3.940-948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson KM, von Kieckebusch-Guck A, Lemire BD. J Biol Chem. 1991;266:21347. [PubMed] [Google Scholar]
- 11.Lemire BD, Oyedotun KS. Biochim Biophys Acta. 2002;1553:102. doi: 10.1016/s0005-2728(01)00229-8. [DOI] [PubMed] [Google Scholar]
- 12.Pierrel F, et al. EMBO J. 2007;26:4335. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bafunno V, et al. J Biol Chem. 2004;279:95. doi: 10.1074/jbc.M308230200. [DOI] [PubMed] [Google Scholar]
- 14.Tzagoloff A, Jang J, Glerum DM, Wu M. J Biol Chem. 1996;271:7392. doi: 10.1074/jbc.271.13.7392. [DOI] [PubMed] [Google Scholar]
- 15.Baysal BE. J Med Genet. 2008;45:689. doi: 10.1136/jmg.2008.058560. [DOI] [PubMed] [Google Scholar]
- 16.Mariman EC, van Beersum SE, Cremers CW, Struycken PM, Ropers HH. Hum Genet. 1995;95:56. doi: 10.1007/BF00225075. [DOI] [PubMed] [Google Scholar]
- 17.van Baars F, Cremers C, van den Broek P, Geerts S, Veldman J. Hum Genet. 1982;60:305. doi: 10.1007/BF00569208. [DOI] [PubMed] [Google Scholar]
- 18.Dibrov E, Fu S, Lemire BD. J Biol Chem. 1998;273:32042. doi: 10.1074/jbc.273.48.32042. [DOI] [PubMed] [Google Scholar]
- 19.Dumont ME, Ernst JF, Hampsey DM, Sherman F. EMBO J. 1987;6:235. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuts DP, Scrutton NS, McIntire WS, Fraaije MW. FEBS J. 2009;276:3405. doi: 10.1111/j.1742-4658.2009.07053.x. [DOI] [PubMed] [Google Scholar]
- 21.Robinson KM, Lemire BD. J Biol Chem. 1996;271:4061. doi: 10.1074/jbc.271.8.4061. [DOI] [PubMed] [Google Scholar]
- 22.Baysal BE, et al. J Med Genet. 2002;39:178. doi: 10.1136/jmg.39.3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiavi F, et al. JAMA. 2005;294:2057. doi: 10.1001/jama.294.16.2057. [DOI] [PubMed] [Google Scholar]
- 24.Timmers H, Gimenez-Roqueplo AP, Mannelli M, Pacak K. Endocr Relat Cancer. 2009;16:391. doi: 10.1677/ERC-08-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McWhinney SR, Pasini B, Stratakis CA. N Engl J Med. 2007;357:1054. doi: 10.1056/NEJMc071191. [DOI] [PubMed] [Google Scholar]
- 26.Ricketts C, et al. J Natl Cancer Inst. 2008;100:1260. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- 27.Birch-Machin MA, Taylor RW, Cochran B, Ackrell BA, Turnbull DM. Ann Neurol. 2000;48:330. [PubMed] [Google Scholar]
- 28.Cascon A, et al. Horm Metab Res. 2009 Apr 2; (e-pub ahead of print) [Google Scholar]
- 29.Ghezzi D, et al. Nat Genet. 2009 May 24; (e-pub ahead of print) [Google Scholar]
- 30.We thank members of the Rutter laboratory for helpful discussions; the J. Shaw and J. Kaplan laboratories for antibodies and technical assistance; J.-M. Heo, T. Smith, E. Taylor, E. van Wijk, S. van der Velde-Visser, and C. Beumer for technical assistance; B. Lemire for the antibodies to Sdh1 and Sdh2; C. Outten for the antiserum to Tim10; and J. Lindsley for critical reading of the manuscript. This work was funded by NIH grants DK071962 and GM087346 (to J.R.), by the Dutch Cancer Society (grant UL 2002-2723), and by the European Union 6th Framework Program, Project No. 518200 (to P.D.). J.R. and H.H. plan to file a patent related to genetic testing of the hSDH5 gene.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.