1. Introduction
Diverse metal ions participate in multiple roles in protein structure and function. In addition to their role in catalysis, electron transfer, and ligand binding, metal ions are important for the structural integrity of many proteins. Physiological responses are known to ensure adequate cellular levels of essential metal ions, but less is known about both the process of protein metalation and metal ion availability within cellular compartments such as the mitochondrion.
In general, metalation reactions are expected to occur during protein biosynthesis or shortly thereafter, since many metal cofactors are important determinants of protein stability. On cytoplasmic ribosomes, protein biosynthesis and chain elongation are coupled to chaperone-mediated protein folding for many proteins.1 Nascent polypeptides are protected within the ribosome and can only fold as they emerge from the exit tunnel. The molecular chaperones bind to nascent polypeptides and actively guide the polypeptides in folding through cycles of binding and release. Some proteins achieve a native conformer before final release from the ribosome.2,3 Metalloproteins are likely metalated at this stage. Biological metalation is dependent on both the availability of adequate bioavailable pools of metal ion and selectivity mechanisms to ensure metalation with the appropriate metal ion.
Specific metalation is a significant issue, as proteins have only limited discrimination between binding various metal ions. Limited control over metalation is imposed by diverse preferences in coordination geometry and hardness/softness of ligand donor atoms between multiple metal ions. Moreover, for metal ions such as Zn(II), these imposed limits are more flexible. Zn(II) is able to coordinate with a variety of donor ligands due to its borderline hardness, and conversion between coordination numbers is possible due to a lack of any ligand field stabilization effects for Zn(II). These effects have the potential to raise both the difficulty in precise Zn site metalation and the misincorporation of Zn into other metalloproteins.
Metalloproteins are compartmentalized within cells. Metalation of metalloproteins within mitochondria is expected to occur after protein import into the organelle. The vast majority of mitochondrial proteins, if not all, are imported as unfolded proteins, and folding occurs within the organelle. Iron, zinc, copper, and manganese ions are cofactors in metalloenzymes and metalloproteins within mitochondria. Thus, bioavailable pools of these metal ions must exist within the mitochondria for efficient metalation reactions.
In this review, we will discuss the various paradigms that ensure specificity in metalation reactions during metalloprotein biosynthesis and the available literature on bioavailable metal ion pools in cells, with particular focus on the mitochondrial organelle. Perturbation of metal pools through disease or environmental factors can lead to detrimental mismetalation, underlining the importance of maintaining metal specificity. Thus, regulation of both metalation specificity and metal ion accessibility must exist.
2. Metalation of Proteins
One strategy to achieve precise metalation is to take advantage of compartment specific differential metal ion availability. Recently, such compartment specific metalation was elegantly shown in the cyanobacterium Synechocystis.4 Two structurally related proteins, MncA and CucA, within the periplasm were found to be the predominant manganese-and copper-containing proteins, respectively. Yet both proteins prefer to bind Cu(II), as evidenced by in vitro metalation of recombinant proteins. However, precise metalation is achieved by differential compartmentalization during the metalation reactions, despite the similar ligand set and metal preference. The activity of the mangano-protein, MncA, is dependent on incorporation of Mn(II) within the cytoplasm, where copper and zinc are highly restricted. Metalated MncA is then secreted into the periplasm as a holoenzyme via the Tat pathway. In contrast, metalation of the copper protein, CucA, occurs within the periplasm after export of the nascent chain via the Sec system.
A second strategy for metalation is through directed distribution. One well-studied pathway of metal ion insertion in eukaryotic cells is the metallochaperone-mediated pathway in the cytoplasm. Metallochaperones mediate copper ion shuttling to sites of copper utilization.5 Protein-mediated copper targeting imparts metal ion specificity to metalation reaction. Atx1 and Ccs1 are two notable examples of metallochaperones in yeast. The human orthologs are designated ATOX1 and CCS. Atx1 shuttles Cu(I) to the Ccc2 P-type ATPase transporter localized in post-Golgi vesicles in yeast.6 Ccs1 provides Cu(I) for activation of Sod1.7 Ccs1 also catalyzes the formation of an essential disulfide on newly synthesized Sod1 in addition to inserting Cu(I).8 However, a Ccs1-bypass mechanism for Cu-metalation of Sod1 exists in mammals, nematodes, and insects. In mammalian cells lacking Ccs1, partially active Sod1 is present.9 This is expanded in the nematode C. elegans, as no obvious homologue of CCS exists, yet Sod1 is active and oxidized.10 A similar situation exists in flies, as flies lacking Ccs1 show a less severe phenotype than flies lacking Sod1.11 Cu(I) delivery from Ccs1 and Atx1 to target proteins occurs via ligand exchange reactions within heterodimeric complexes.7 The requirement for specific protein/protein interaction imparts selectivity in metal transfer reactions. Thus, Sod1 cannot be copper metalated by Atx1 and Ccc2 cannot receive Cu(I) through Ccs1.
Metallochaperones for zinc and manganese are not known, but one candidate metallochaperone for Fe(II) was recently discovered.12 Fe-loading of ferritin in the cytoplasm of animal cells is facilitated by the poly(rC)-binding protein-1 (PCBP1). PCBP1 binds ferritin and is competent to stimulate Fe-loading of ferritin in vitro.12
3. Mitochondrial Organization
3.1. Mitochondrial Ultrastructure
The mitochondrion is a highly dynamic subcellular compartment like other organelles. Yet, structurally the mitochondrion is extremely complex and contains not only its own genome but also several subcompartments. This complexity is compounded given that a dynamic double membrane exists largely as a tubular reticulum that continually undergoesfusion and fission events encloses mitochondria.13–15 In combination, these factors place enormous demands on metal distribution and bioavailability in addition to protein import. As such, we will briefly outline mitochondrial structure and protein import in order to place the maturation of the numerous apo-metalloenzymes and potential sources of bioavailable metal in context.
The first of the two membranes composing mitochondria is the porous outer membrane (OM). The channels within the OM are approximately 25 Å and permit diffusion of small molecules. In contrast, the inner membrane (IM) is a barrier to free ion diffusion and contains several cation and metabolite transporters. Structurally, the IM exists as segments in close juxtaposition to the OM as well as segments that invaginate into the inner compartment called the matrix. The invaginations, termed cristae, are folded into lamellae, or tubular structures that increase the surface area of the IM.16 The IM appears to be dynamic in that it is predominantly cristae tubules in the mitochondria of cells with high energy demand.17 The IM adjacent to the OM is designated as the inner boundary membrane (IBM). The mean distance across the OM and IBM is 20 nm, although the distance narrows to only about 14 nm at junction points. Three aqueous compartments therefore exist within mitochondria. The first volume between the OM and the IBM is called the inter-membrane space (IMS) and is interrupted by junction points in which the IM and OM are in contact. The IMS is in partial equilibrium with a second volume enclosed by the cristae membrane created by an invagination of the IBM. Near the connection of the IBM and cristae, ringlike constrictions of ~28 Å form that limit diffusion between the IMS and cristae lumen.14 Lastly, the volume enclosed within the IM is the matrix compartment where the mitochondrial genome resides.
3.2. Mitochondrial Genome
The mitochondrial DNA is not uniformly distributed in the tubular network of the organelle, rather multiple copies of the genome localize in discrete zones called nucleoids. An average nucleoid in mammalian cells contains 5–7 genomes, and these structures are stabilized by an array of DNA-binding proteins that are distinct from the typical histones organizers of the nuclear DNA.18 The mitochondrial genome encodes a limited subset of mitochondrial proteins with an average number of protein products being 10–20 in various eukaryotes. The mitochondrial-encoded polypeptides are typically membrane subunits of the oxidative phosphorylation pathway (OXPHOS) consisting of complexes I—V. The human mitochondrion encodes 13 OXPHOS subunits, whereas the Plasmodium mitochondrial genome encodes only 3 core subunits (Cob, Cox1, Cox3) of complexes III and IV.
3.3. Mitochondrial Proteome
In addition to mitochondrial encoded polypeptides, proteomic analysis has revealed upward of 1000 distinct proteins in yeast to mammalian cells. 19–21 Analysis of the proteome in a suite of diverse mouse tissues reveals changes in both abundance and composition, demonstrating that tissuespecific expression of certain mitochondrial proteins exists.21 Since the mitochondrial genome encodes a very small subset of the organellar proteome, the majority of mitochondrial proteins are therefore nuclear-encoded and targeted to the organelle. Furthermore, approximately half of mitochondrial proteins are translated on ribosomes associated with the OM of the organelle through cis-acting signals in the transcripts.22 Protein import into the mitochondrion occurs in both cotranslational and post-translational processes.23,24 Cotranslational import occurs at sites on the OM that are in contact with the IM.24,25 Post-translational import of most mitochondrial proteins is achieved through N-terminal mitochondrial targeting sequences, although internal target sequences are known.
3.4. Mitochondrial Protein Import
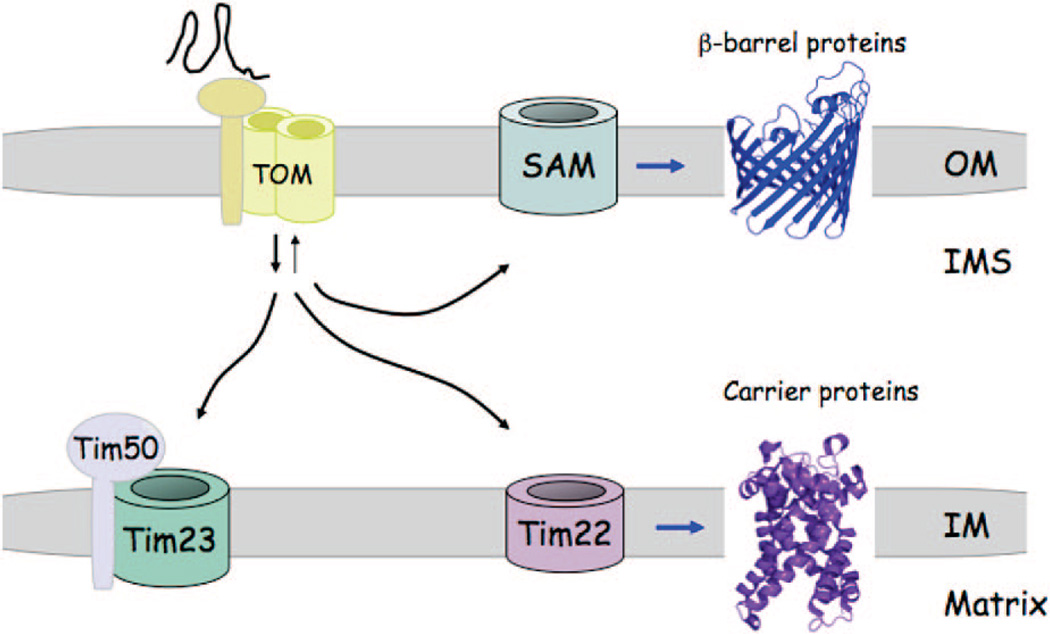
The translocation of preproteins into mitochondria occurs through import channels formed in the OM and IM (Figure 1) The OM channel consists of several subunits forming distinct receptor and insertion complexes, both of which together form the TOM (translocase of the outer membrane) complex.26 The yeast receptor complex (Tom20, Tom22, and Tom70) functions in specific recognition of proteins with targeting sequences. The insertion complex is composed of four associated subunits with a central hydrophilic pore formed by Tom40.27 The pore diameter is only 20–25 Å, which is too small to accommodate fully folded proteins. Thus, the preproteins are extruded through the channel as unfolded apo-polypeptides.
Figure 1.
Model for mitochondrial protein import. Proteins are imported into mitochondria in the unfolded state through the TOM complex in the outer membrane. Receptors in the TOM complex faciliate recognition of proteins with mitochondrial targeting sequences. Proteins with N-terminal targeting sequences are routed directly to the TIM23 complex in the inner membrane by the Tim50 protein that also gates the TIM23 complex. A subset of polytopic helical IM proteins are routed to the TIM22 complex for membrane insertion. Proteins within the OM consisting of β-barrel structures are routed to the SAM complex for folding and OM insertion.
Multiple sorting steps occur upon transit through the TOM complex.26–28 β-Barrel proteins destined for the OM progress to the sorting and assembly machinery (SAM) complex in the OM for folding and membrane insertion (Figure 1). Soluble chaperone complexes, e.g. the Tim9/Tim10 hexamer, escort OM preproteins to the SAM complex. In contrast, polytopic IM proteins with internal target sequences proceed to a translocase of the IM called the TIM22 complex for membrane insertion while also being guided by Tim9/Tim10. One common class of proteins inserted into the IM by the TIM22 complex is the mitochondrial carrier proteins that form helical bundle structures. The majority of mitochondrial proteins contain an N-terminal positively charged presequence that directs the molecule to a second translocase in the IM called the TIM23 complex for either extrusion into the matrix or IM insertion via Oxa1. The TIM23 channel is gated in order to maintain membrane potential and again accommodates only unfolded proteins through a limited diameter of 13–24 Å. Import through the TIM23 complex occurs in conjunction with the TOM complex such that proteins being extruded through the TOM complex are engaged by Tim50, a component of the TIM23 complex. The interaction of preproteins with Tim50 opens the TIM23 gated pore. An ATP-driven motor, contained within the matrix Hsp70 protein, then mediates import through the TIM23 complex. Translocation is followed by proteolytic cleavage of the precursor signal sequence by the mitochondrial processing protease (MPP) and ATP-dependent protein folding mediated by Hsp70 and Tim44.
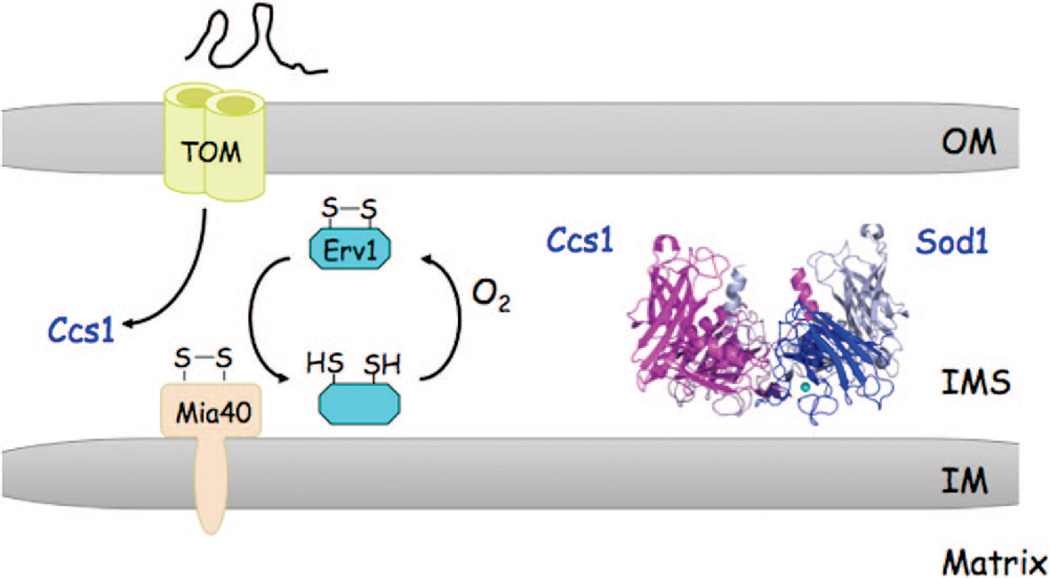
Numerous proteins localized within the IMS require only translocation across the outer membrane via the TOM complex. Three different classes of IMS proteins are known. One class of proteins contains a bipartite targeting sequence consisting of a mitochondrial import sequence followed by a hydrophobic signal that arrests translocation through the TIM23 channel, leading to lateral transfer within the IM. Subsequent proteolytic cleavage of the IM tether leads to soluble IMS localization. A second class of IMS proteins consists of low molecular weight cysteine-rich polypeptides that exist within the IMS as disulfide-bonded conformers. An example of this protein class is the small Tim9/Tim10 complex that chaperones polypeptides to either the SAM or TIM22 complexes for insertion into the OM or IM, respectively (Figure 2). These Cys-rich proteins are captured in the IMS after extrusion through the TOM channel by the MIA import system consisting of Mia40 and Erv1.29–31 Disulfide interchange between oxidized Mia40 and reduced thiolates on the imported protein generate an intermolecular disulfide that traps the molecules within the IMS. The sulfhydryl oxidase, Erv1, then forms a ternary complex with Mia40 and the imported protein to effectively transfer the disulfide(s) to the imported protein, thereby releasing it from Mia40.32 Failure to generate the disulfides in the imported proteins results in mitochondrial depletion via diffusion back through the TOM complex. A third class of proteins require the presence of affinity sites on IM anchored proteins for retention within the IMS.27
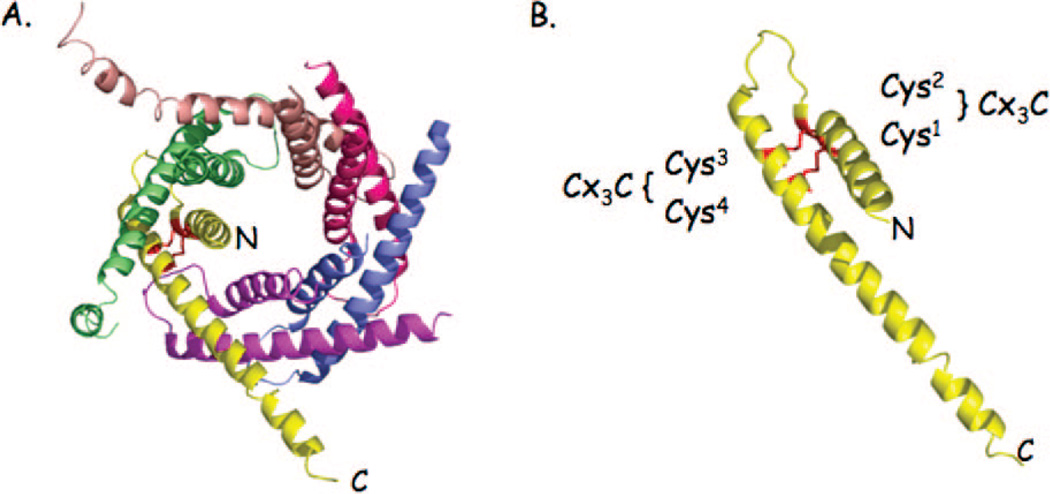
Figure 2.
Ribbon diagram of the Tim9/Timl0 complex. (A) The Tim9/Timl0 hexameric complex existing within the IMS mediates transfer of hydrophobic polypeptides to either the SAM or TIM22 complex for membrane insertion. The N- and C-termini of one Tim9 monomer in the complex are shown for orientation. Tim9 or Timl0 subunits consist of twin Cys-X3-Cys sequence motifs that form two intrachain disulfides per subunit, stabilizing the hexameric complex (panel B).
4. Protein Folding and Maturation within Mitochondria
As mentioned, the vast majority of mitochondrial proteins are imported into the organelle and multiple translocation events require proteins to be in an unfolded state. Protein folding occurs either within the membrane or within one of the soluble subcompartments of the organelle. Protein homeostasis requires rapid and precise folding to minimize the levels of unfolded or misfolded conformers that are prone to aggregation. A misfolded conformer of one protein can induce off-pathway folding reactions of other proteins, leading to detrimental aggregates. The presence of protein chaperones within the soluble matrix compartment is one deterrent to such deleterious events. In addition, a myriad of proteases exist within the mitochondria that may minimize accumulation of misfolded proteins.
A host of mitochondrial proteins require a metal ion cofactor for function and/or stability. Zinc, iron, copper, and manganese ions are commonly required for normal mitochondrial physiology. The folding of metalloproteins is dependent on the availability and the selective insertion of the appropriate metal ion. Mis-metalation by a non-native metal ion may yield either an inactive protein or a misfolded state prone to aggregation. The question arises whether quality control pathways exist to ensure proper metalation within the various mitochondrial compartments.
5. Distribution of Metalloproteins within Mitochondria
Metalloproteins are distributed throughout the mitochondria and, as such, detail the demands for metal distribution within a highly compartmentalized organelle. The bioavailable metal ion pools that must exist to metalate the imported nascent chains and specific metalation reactions will be discussed after a survey of the metalloproteome of mitochondrion.
5.1. Iron Metalloproteome
One of the most abundant mitochondrial metal ions is iron. Iron is present in polynuclear sulfur-bridged iron–sulfur (Fe/S) centers, binuclear oxo-bridged complexes, and hemo-proteins. Outside of heme synthesis, the major proteinacious demand on mitochondrial iron is the OXPHOS respiratory chain residing predominantly in the cristae membrane as well as in the IBM. Fe/S clusters exist within the respiratory complexes I, II, and III. Complex I in mammalian cells itself requires 8 distinct Fe/S centers within the 45-subunit complex.33 In addition, hemoproteins are found in each of the electron transfer complexes II–IV of the respiratory chain as well.
Within the matrix, Fe/S centers of different nuclearities are present in a myriad of proteins in addition to those associated with iron sulfur cluster (ISC) machinery, such as the Fe/S scaffold proteins Isu1, Isu2, and Nfu1.34 These proteins include aconitase, homoaconitase, biotin synthase, lipoate synthase, and ferredoxin.34 Binuclear oxo-bridged iron proteins include mitochondrial ferritin within the matrix of mammalian cells, the IM-associated Coq7 hydroxylase involved in ubiquinone formation, and an alternative oxidase found in the respiratory chain of plants and certain fungi. 35–37 Iron proteins within the IMS include the heme-containing cytochrome c peroxidase, the cytochrome b2 oxidoreductase, and the Fe/S Rieske protein of the respiratory complex III exposed to the IMS.
5.2. Zinc Metalloproteome
Zinc is equally abundant to iron within yeast mitochondria. The zinc metalloproteome includes a series of zinc metalloproteinases distributed throughout mitochondria (Figure 3). These proteases in yeast include the iAAA, mAAA, Oct, the MPP protease complex, Atp23, and Oma1. While numerous metalloproteinases can be activated in vitro by diverse divalent cations, Zn(II) appears to be physiological metal ion bound. Moreover, these Zn-dependent proteases all share a common essential HEXXH or HXXEH metal binding motif.
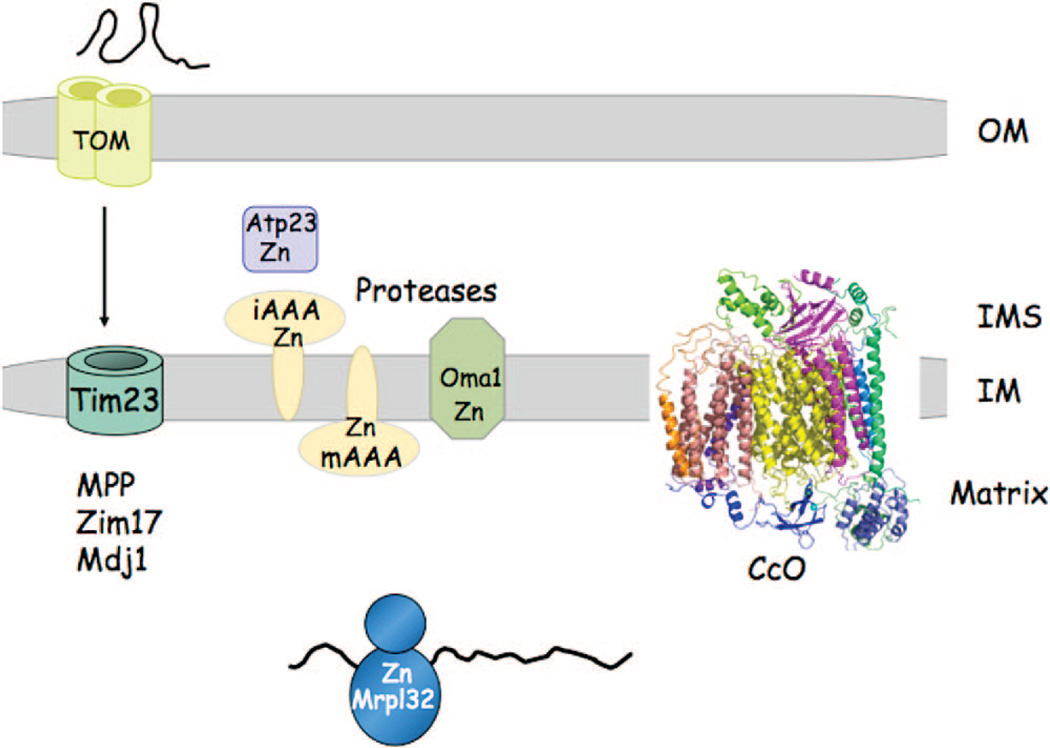
Figure 3.
Zinc proteome of yeast mitochondria. A subset of the Zn(II) metalloproteins of yeast mitochondria are shown. A myriad of IM-associated metalloproteinases use Zn(II) as a catalytic cofactor. These include the mAAA, iAAA, Omal proteases of the IM, and the IMS protease Atp23. The mitochondrial processing protease (MPP) that functions in cleavage of the N-terminal mitochondrial targeting sequence consists of two subunits, Masl and Mas2, that are metal-dependent, most likely with Zn(II).44 Another component of the protein import machinery is the Zn-requiring Ziml7, which functions with the Hsp70 protein Sscl to facilitate protein import.48 The DnaJ protein Mdj 1 contains a Cys-rich motif consistent with Mdj 1 being a Zn-dependent enzyme. One subunit, Mrpl32, of the mitoribosome is a candidate Zn(II) stabilized protein. Cytochrome oxidase contains an essential Zn-containing subunit, Cox4 in yeast.170
The iAAA protease is composed of possibly three or more Yme1 subunits, each binding Zn(II). The proteolytic active site of iAAA faces the IMS compartment and is essential for respiratory growth at elevated temperature. In addition to Yme1 within the IMS, Zn(II) appears important in the protein import. Protein import into the IMS through the Mia40 pathway may also be facilitated by Zn(II) binding to maintain protein thiolates in an import competent state.38,39 Hot13 was shown to interact with Mia40 and enhance its oxidation by Erv1.40 It was proposed that Hot13 serves as a Zn(II) buffer in the IMS to facilitate removal of Zn(II) from imported proteins. The fate of the Hot13-bound Zn(II) is not clear.
The remaining proteases are active within the matrix beyond the solute barrier imposed by the inner membrane. The mAAA protease is composed of Yta10 and Yta12 multimers, each requiring Zn(II) for activity. The mAAA protease is required for respiratory competency through both the proteolytic activation of the mito-ribsome via Mrpl32 activation and proteolysis of nonassembled inner membrane proteins.41,42 Oct1 and MPP sequentially process mitochondrial-targeting sequences as preproteins are extruded through the TIM23 import complex. MPP consists of two subunits encoded by MAS1 and MAS2 in yeast. Both subunits have proteolytic activity dependent on metal ions.43 Zn(II) appears to be physiological metal ion bound, although Mn(II) or Co(II) can support partial proteolytic activity.44 Atp23 is required for the coordinate processing and assembly of the F1F0-ATP synthase.45,46 Oma1 (for overlapping activity of mAAA) also functions in the degradation of misfolded proteins within the inner membrane.47
Zn(II) is a cofactor for several other enzymes in the mitochondrial matrix in addition to the proteases detailed above. For instance, Zn(II) is a cofactor of Zim17 that associates with Ssc1 to facilitate protein import as well as protein folding within the matrix.48 A DnaJ component of Sscl, Mdj1 also contains a Cys-rich motif consistent with Mdj1 being a Zn-dependent enzyme. Mdj1 accounts for 1% of the mitochondrial proteins,49 so Mdj1 is likely an abundant mitochondrial Zn-protein. Matrix Zn-metalloenzymes include Adh3, Adh4, and Leu9 in yeast. Zn(II) is likely associated with the yeast mitoribosome, as Mrpl32 has two essential Cys-X2-Cys motifs that are likely donor atoms for the candidate Zn(II) site. Whereas the cytoplasmic ribosomes have multiple Zn-containing subunits, Zn(II) in the mitoribosome appears restricted to Mrpl32. The Cox4 subunit of cytochrome c oxidase (CcO) is a Zn-requiring molecule with Zn(II) binding contributing to the stability of Cox4. In addition, the Coq4 protein contains a conserved HDXXH motif commonly found in catalytic Zn sites. Coq4 is a key component of the protein complex involved in coenzyme Q biogenesis.171
5.3. Copper Metalloproteome
The copper metalloproteome of mitochondria is dominated by cytochrome c oxidase (CcO), the terminal electron acceptor in the respiratory chain. It is predominantly localized in the cristae membrane, and in yeast, it is largely found in a supercomplex with the bc1 complex.50,51 Two mitochondrial-encoded subunits of CcO, Cox1 and Cox2, have copper centers forming the CuB and CuA sites in CcO, respectively. A series of low abundance proteins within the IMS bind Cu(I) ions transiently for the subsequent metalation of CcO during its biogenesis. These Cu-binding proteins include the soluble protein Cox17, which is the Cu(I) donor to two IM-tethered proteins, Sco1 and Cox11. In turn, Sco1 and Cox11 then donate Cu(I) to Cox2 and Coxl, respectively. In addition to CcO, approximately 1–5% of total cellular Cu,ZnSod1 localizes within the IMS. Retention of Sod1 within the IMS is dependent on its cognate Cu(I)-binding metallochaperone Ccs1.52,53 Import of both Ccs1 and Cox17 occurs through the TOM complex in a Mia40-dependent manner. Both metallochaperones are likely copper metalated within the IMS for activation of their downstream targets, assuming they are imported in an unfolded state, as are most proteins entering the TOM channel.
5.4. Manganese Metalloproteome
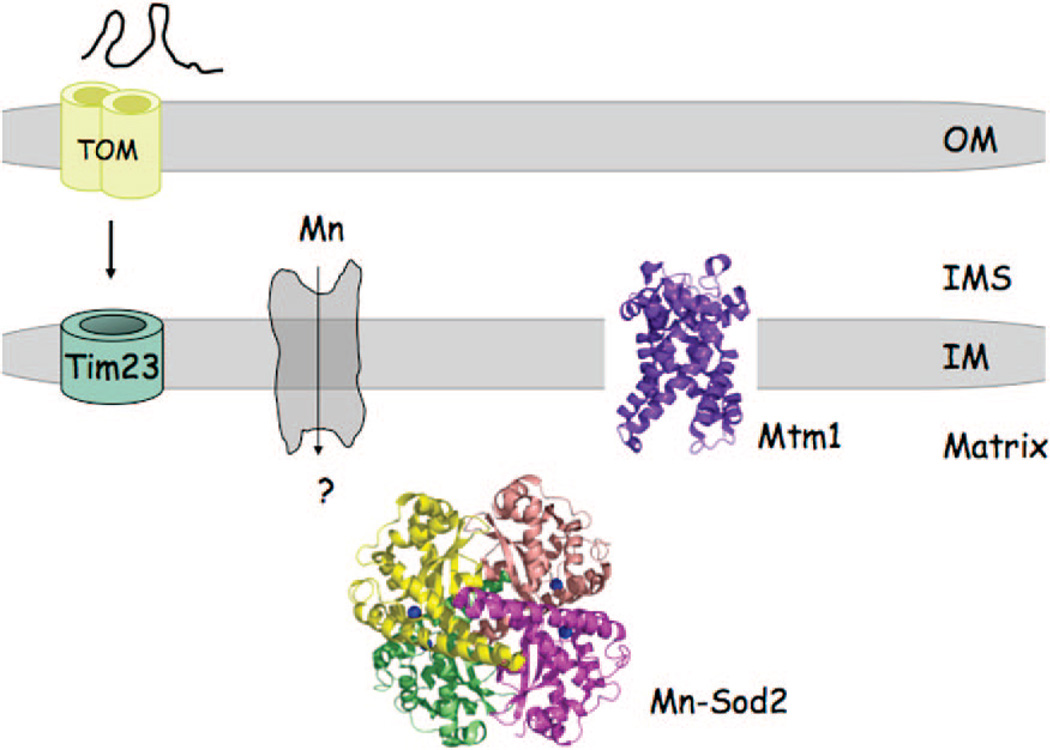
The only definitive mangano-protein in the mitochondria is MnSod2.54 MnSod2 protects against oxidative damage by scavenging superoxide anions within the matrix, just as the Cu,ZnSod1 protects against superoxide anions within the IMS. Sod2 is imported into the matrix as an apoprotein, and its metalation is coupled to the folding reaction because Mn(II) cannot be loaded into a folded apo-Sod2 in vivo.55
6. Metal Ion Transport in Mitochondria
Based on the known metalloproteome of mitochondria, metal distribution throughout the organelle is essential. Specifically, Fe(II) and Zn(II) are required throughout mitochondria, whereas Mn(II) is needed within the matrix, and copper is needed within the IMS. However, how mitochondria acquire, distribute, and maintain adequate metal levels is only partially understood. While several families of metal ion transporters are known in many cellular compartments, only a paucity of information is available on mitochondrial metal ion permeases. From these limited examples, and yet to be described functions from among transporter families, mitochondria must obtain homeostatic quantities of metal. As such, we will outline both known mitochondrial transporters and other transporters known to facilitate intracellular metal distribution.
6.1. Iron Transport
Iron transporters are known for many cellular compartments including mitochondria. From among a family of 35 similar mitochondrial carrier family (MCF or SLC25) transporters identified in yeast,58 Fe(II) import into the matrix involves at least two redundant members termed Mrs3 and Mrs4.56,57 Cells lacking the Mrs3/Mrs4 transporters have reduced mitochondrial iron as well as impaired Fe/S and hemoprotein biosynthesis.57,59,60 Mrs3/4 orthologs in vertebrates are the SLC25 carriers designated mitoferrin (Mfrn1 and Mfrn2).61 Mfrn1 is highly expressed in the hematopoietic tissues of zebrafish and mouse. Missense mutations in zebrafish Mfrn1 result in an erythroid developmental arrest arising from defective heme formation due to impaired mitochondrial iron uptake.61 Likewise, erythroblasts generated from murine embryonic stem cells defective for Mfrn1 show an impaired incorporation of radio-iron into heme, a step that occurs within the mitochondrial matrix. Mouse and zebrafish contain Mfn2, but it is nonredundant with Mfn1.61 Two other mitochondrial IM cation diffusion facilitators, Mmt1 and Mmt2, are also implicated in iron homeostasis, but the mechanism is unclear.62
The form of iron utilized by these MCF carriers remains undetermined. In a computational study of MCF proteins based on the structure of the ADP/ATP carrier (Figure 1), key contact residues were identified in MCFs that appear to recognize and distinguish substrates. Based on these analyses, Mrs3/Mrs4 are predicted to transport a complex distinct from a keto or amino acid.63 Thus, mitochondrial iron transport likely occurs as a chelate rather than ionic Fe(II). The pathway of Fe delivery from the plasma membrane transporters to mitochondria is unclear. Moreover, such a potential chelate could facilitate Fe delivery from the plasma membrane, while preventing detrimental Fenton chemistry.
6.2. Zinc Transport
Cellular zinc acquisition is achieved through a conserved Zrt-, Irt-like protein (ZIP or SLC39) family of transporters.64 ZIP transporters in yeast (Zrt1, Zrt2, and Zrt3 shown in Figure 4) and mammals predominately translocate Zn(II) from the extracellular space or organelles into the cytoplasm with rare exception (i.e., Yke4). A second class of eukaryotic Zn(II) transporters is the cation diffusion facilitator (CDF, ZnT, or SLC30) family that translocate Zn(II) from the cytoplasm either into organelles or out of the cell with rare exception in yeast and mammal.64’65 Both families of transporters are polytopic intrinsic membrane proteins and with respect to CDFs use gradients of other ions, e.g. H+ or K+, to drive Zn(II) transport.64 Fourteen candidate ZIP and ten distinct ZnT transporters are known in the human,66 whereas yeast possesses five ZIP as well as five CDF transporters characterized to date.
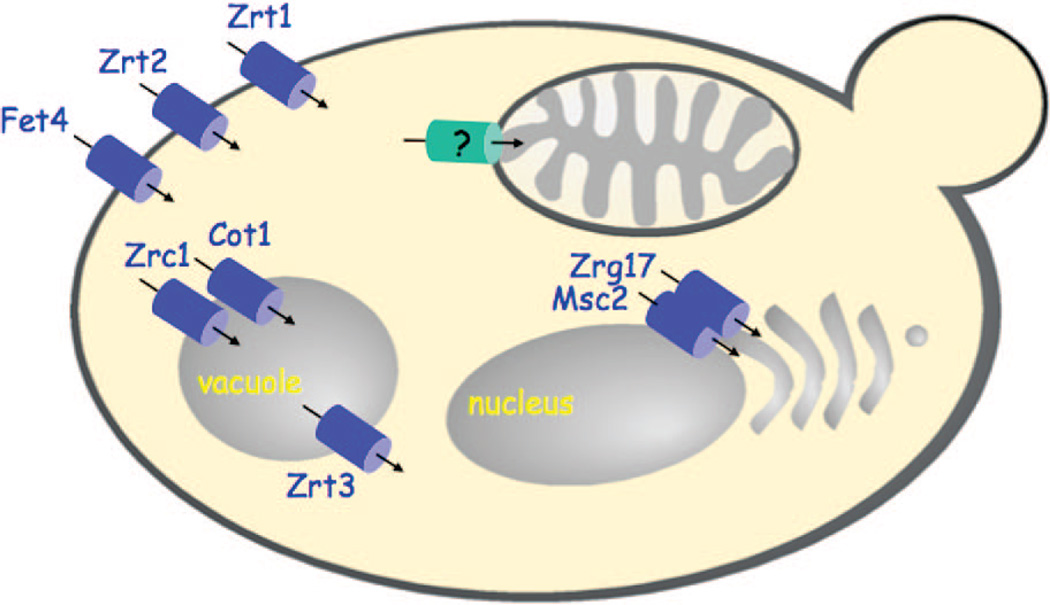
Figure 4.
Fungal Zn(II) transporters. Zn(II) transporters are known for the import of Zn(II) into yeast and the distribution of Zn(II) to the ER/Golgi compartments and vacuole. No mitochondrial Zn(II) transporter has been identified to date.
Zinc is important within the secretory pathway for metalation of both secreted metalloproteins and resident proteins involved in protein folding and post-translational modification. Zn(II) can be supplied to the Golgi apparatus either through vesicular trafficking from the endoplasmic reticulum (ER) or through Golgi-specific transporters. In yeast, two CDF transporters, Msc2 and Zrg17, form a heteromeric complex that mediates ER Zn(II) uptake (Figure 4).64 In vertebrates, CDF transporters including the ZnT5/ZnT6 pair, ZnT7, and ZnT8 are Golgi-specific transporters providing Zn(II) directly to the secretory pathway. Among these CDFs, certain transporters exhibit tissue specific function. For example, ZnT8 is the key transporter for the Zn(II) accumulation in pancreatic β cells for insulin secretion.67 Likewise, ZnT2 and ZnT4 are key Zn(II) transporters in mammary glands providing Zn(II) to milk.68,69 As with many transporters, the localization of ZnT transporters is not static. Differential splicing results in localization of ZnT5 in the Golgi or more disperse distribution including the plasma membrane.65
Two CDF transporters, Zrc1 and Cot1, mediate Zn(II) uptake into the vacuole in yeast (Figure 4). Vacuolar Zn(II) uptake serves as a storage pool for Zn(II) as well as a mechanism to minimize Zn toxicity in the cytoplasm.70 Zinc stored within the vacuole can be subsequently exported via the ZIP transporter Zrt3.71
As mentioned, mitochondria require significant levels of zinc, yet no ZnT or ZIP mitochondrial transporter has yet to be isolated. From observations that Zn(II) impairs mitochondrial respiratory function, the calcium uniporter blocker ruthenium red was found to block the deleterious Zn effects on neural mitochondria.72–75 Ruthenium red was found to inhibit uptake of Zn(II) into the mitochondrial matrix, suggesting that the calcium uniporter may mediate Zn(II) transport across the IM.76 Zn(II) uptake is also impaired when the membrane potential is depleted, suggesting that uptake is an active process.76 Although the actual calcium importer remains unknown, the uncoupling proteins 2 and 3 (UCP2 and UCP3) are important for Ca(II) import.77
6.3. Copper Transport
Copper transporters exist to translocate Cu(I) from the cytoplasm, but their localization is restricted to the trans-Golgi network and internal vesicles. These transporters identified to date are all members of the P-type ATPase family. In yeast the Ccc2 transporter is restricted to the TGN, whereas the mammalian P-type ATPases (ATP7A and ATP7B) traffic from the TGN to cytoplasmic vesicles in response to high cellular Cu concentrations where they efflux Cu(I). These transporters are not responsible for Cu(I) translocation to the mitochondrion.
6.4. Manganese Transport
As discussed, mitochondria also require Mn(II) to support Sod2 activity within the matrix (Figure 5). While the identity of the mitochondrial Mn(II) importer remains elusive, several other transporters are required to maintain active metalated Sod2 through as of yet unresolved means. Mitochondrial manganese levels in yeast require the presence of a Nramp transporter Smf2, which localizes to internal vesicles. Nramp (natural resistance-associated macrophage protein or SLC11) transporters are a family of conserved metal ion transporters.78 Although Nramp transporters are known for manganese, iron, cadmium, and copper, Smf2 has selectivity for Mn(II). Yeast cells lacking Smf2 contain the Sod2 polypeptide within the mitochondrial matrix, but the enzyme is inactive due to manganese deficiency.54 Smf2 may mobilize Mn(II) pools from Golgi-like vesicles, and this Mn(II) is somehow delivered to the mitochondrion as well as the secretory pathway.79 The process may involve vesicular fusion or specific Mn(II) chaperones. In addition to Smf2, the yeast phosphate transporter Pho84 appears to also function in low affinity manganese uptake, which appears to be biologically active, as demonstrated through downstream activation of Sod2.80
Figure 5.
Manganese proteome of yeast mitochondria. The Mn(II)-dependent Sod2 exists with the matrix as a soluble protein. Sod2 is imported into the matrix as an unfolded protein and metalled within the matrix during the folding reaction. The identity of the Mn(II) transporter in the IM is unknown, but the mitochondrial carrier protein Mtm1 influences the metalation of Sod2. In the absence of Mtm1, Sod2 is mismetalated by Fe(II).98
In addition to Smf2 and Pho84, yeast cells lacking the MCF transporter Mtm1 also lack active Sod2.81 However, in contrast to the case of smf2 mutants, mitochondrial manganese levels are normal; therefore, Mtm1 cannot be the Mn(II) permease. A third potential route for mitochondrial Mn(II) uptake is through the Mg(II) channel Mrs2. Mrs2 is a member of the ubiquitous CorA superfamily. However, detailed patch clamp analysis has shown that this channel discriminates Mg(II) from Mn(II).82
One major unanswered question is how metal ions such as Zn(II), Cu(I), and Mn(II) are translocated into the mitochondrion. Genetic and high throughput deletion screens have failed to identify the mode of transport to date.
7. Bioavailable Metal Ion Pools
7.1. Labile Metal Pools within the Cytoplasm
Much has been written about the lack of free or labile copper or zinc pools in the cytoplasm of S. cerevisiae or E. coli.83,84 Using the E. coli Zn-responsive transcription factors Zur and ZntR, the steady-state labile Zn(II) concentration in E. coli was predicted to be substantially less than a single aqueo Zn(II) ion per cell.84 This work was confirmed by transcript profiling in E. coli cells cultured in minimal LB medium.85 Therefore, cytoplasmic Zn(II) levels must be sequestered in a bound, less available state. In cells propagated in rich medium, expression levels of target genes regulated by both factors are off, making assessment of the cytoplasmic Zn(II) levels more complex.86–87 The fact that Zur activation requires the addition of a Zn(II) chelator to the growth medium suggests that cytoplasmic Zn(II) levels must be significantly elevated over that in cells cultured in LB medium. Thus, bioavailable Zn(II) within the cytoplasm is dependent on the growth conditions and may undergo oscillations due to expression changes in Zn(II) transporters.64 Oscillations in yeast transcriptional networks controlling expression of metal transporters are well-established.88 In yeast exhibiting a regeneration time of 8 h, respiratory oscillations were observed with cycle times of approximately 40 min.88 The myriad of cytoplasmic metalloproteins in eukaryotes implies that bioavailable metal pools must exist, perhaps a pool associated with the ribosome for facile metalation of newly synthesized polypeptides emerging from the ribosome. One question is whether oscillations occur within labile metal ion pools to accommodate variable expression of metalloproteins.
7.2. Mitochondrial Iron
Eukaryotic cells have a pool of accessible iron, designated the labile iron pool (LIP) that has been defined as a low molecular weight, weakly chelated iron pool that constitutes a low fraction of the total cellular iron.89,90 The LIP represents a minor fraction of the total cellular iron, but the redox state of the iron remains unclear. The presence of a LIP has been observed within mitochondria. As evidence, mitochondrial-specific Fe(II)-dependent fluorophores reveal 16 µM of accessible Fe within the matrix of hepatic mitochondria.91’93 Although the ligand for this LIP is not known, it was estimated that the Fe(II) is bound with apparent stability constants (log Kapp) of between 9 and 14, as determined with chelating fluors of varying affinities.91 A variety of ligands including citrate, carboxylates, nucleotides, inositol phosphates, and phospholipids have been implicated as stabilizing the LIP pools existing within the cytoplasm and mitochondria.90,92
In the absence of a known ligand, the exact biological role of the LIP remains elusive. Nonetheless, several lines of evidence point to the importance of the LIP. First and foremost, increasing evidence exists for a role of mitochondrial LIP in the pathogenesis of human diseases such as Friedreich’s ataxia, Parkinson’s disease, and X-linked sideroblastic anemia.89,93 Bioavailable Fe(II) is also known to be an important catalyst for the initiation and propagation of reactive oxygen species (ROS) and is implicated in the pathogenesis of diverse neuronal disorders. The link between neuronal injury resulting from Fe-dependent mitochondrial ROS production was recently shown through pharmalogically induced excitotoxicity and the protective effects of the mitochondrial Fe-chelator N,N<’-bis(2-hydroxybenzyl)ethyl-enediamine-N,N<’-diacetic acid.94 The mitochondrial LIP lias also been implicated as the proximate iron donor for heme synthesis,95 although a more recent study found that heme iron was directly routed to the ferrochelatase after import.96
A mitochondrial LIP accumulates in cells impaired in the biogenesis of Fe/S clusters within the matrix. RNA silencing of the mitochondrial inner membrane transporter, ABCB7, which contributes to Fe/S biogenesis in the cytoplasm, leads to a marked iron deposition in the mitochondria of hepato-cytes.97 Yeast cells devoid or depleted of Atm1, Erv1, Yah1, Grx5, or Ssq1 show a significant rise in the LIP, as assessed by the chromogenic chelator bathophenanthroline sulfonate (BPS). Expansion of the matrix LIP in ssq1Δ, grx5Δ, ggc1Δ, or mtm1Δ cells results in inactivation of Mn-Sod2 within the matrix due to iron mismetalation.98 The bioavailability of mitochondrial iron in ggc1Δ, mtm1Δ, ssq1Δ, or grx5Δ cells appears to be controlled by the yeast frataxin homologue, Yfh1, as Fe-metalation of Mn-Sod2 in the matrix is Yfh1-dependent.98 While matrix iron also expands within yfh11Δ mitochondria, it forms insoluble Fe(III) phosphate particles,99 thereby making it inaccessible for Sod2 mismetalation.
The function of frataxin has also been characterized in mammalian systems, and whether loss of frataxin alters mitochondrial iron is the subject of current debate.100 Frataxin is a ubiquitous mitochondrial iron-binding protein involved in the biosynthesis of Fe/S clusters and heme. Its deficiency in humans causes Friedreich’s ataxia, a severe neurodegenerative disease. Attenuation of frataxin in HeLa cells compromised Fe/S cluster biogenesis without altering mitochondrial iron levels. In a similar study, fibroblasts from Friedreich’s ataxia patients deficient in frataxin do not contain elevated LIP, as assessed by a mitochondrial-specific Fedependent fluorophore. However, an accumulation of iron is observed within cardiac mitochondria of late stage frataxin null mice.101
The mitochondrial labile iron pool can also be buffered by mitochondrial ferritin, which forms a 24 subunit oligomer capable of sequestering Fe(III) in animals.102 Expression of mito-ferritin in HeLa cells led to a diminution in the mitochondrial LIP resulting in an attenuation in ROS production. However, mito-ferritin is not ubiquitous in animal tissues. Expression of mito-ferritin is restricted to certain organs, including testis, heart, pancreas, and kidney. In tissues not expressing mito-ferritin, e.g. liver, frataxin may be a key factor modulating the LIP.
The LIP is also affected in cells depleted for the essential ferredoxin, Yah1. Cells depleted of Yah1 show a significant oxygen-dependent expansion of the matrix LIP consisting of 2–4 nm Fe(III) phosphate nanoparticles.103 Likewise, mitochondria isolated from Yah1 depleted cells cultured aerobically but subsequently treated with reductant showed a marked reduction in the LIP, suggesting that matrix ferric phosphate nanoparticle formation was dependent on ferric iron and the process is reversible.103 Since ferric phosphate nanoparticles do not accumulate in the mitochondria of aerobically grown wild-type yeast, one scenario is that wild-type cells contain a ligand capable of maintaining ferric ions in both a soluble and redox inactive state.
7.3. Mitochondrial Zinc
Mammalian cells have well-described pools of labile Zn(II) in vesicular compartments, including an as yet ill-defined structure termed the zincosome.64,104 Labile Zn(II) pools have been described in the brain, pancreas, prostate, and retina. These advances have been made possible by the development of Zn-specific fluorophores. The correlation of the cellular fluorescence with labile Zn(II) pools is complicated by caveats such as the binding affinity of the fluorophore, discrimination among diverse metal ions, and stabilities of fluorphores, including a tendency to compartmentalize.105 A key caveat is the Zn(II) binding affinity. Since labile Zn(II) pools may be as low as nanomolar, a fluor with sensitivity in the nanomolar concentration range is preferred, assuming its presence within the cell does not perturb the normal Zn(II) distribution.
One well-defined labile Zn(II) pool resides in presynaptic boutons in mossy fiber neurons within the hippocampus.106 These presynaptic vesicles are also enriched with glutamate as a neurotransmitter. The vesicles accumulate Zn(II) up to 0.3 mM through the ZnT-3 Zn(II) transporter. Ablation of ZnT-3 attenuates the labile vesicular pool of Zn(H).106–108 Stimulation of the mossy fiber neurons results in release of Zn(II) and glutamate into the synaptic cleft and subsequent accumulation in the postsynaptic neuron.109 Synaptic cleft Zn(II) is a modulator of glutamate receptors, and postsynaptic neuronal Zn(II) may have a second messenger effect.106
Zn(II) is also enriched in secretory granules in specific cells. For example, insulin granules within pancreatic β cells are enriched with Zn(II) to stabilize the proinsulin hexamer facilitating conversion to insulin.110 Islet-specific expression of ZnT-8 is responsible for the Zn(II) accumulation within the proinsulin secretory vesicles, as overexpression of ZnT-8 results in Zn(II) accumulation.67 Zn(II) is also enriched in both mast cell and intestinal paneth cell granules.111,112 In each case a specific ZnT Zn(II) transporter is responsible for generating the labile Zn(II) pool. The ZnT-4 transporter involved in mast cell Zn granules is also important for mammary gland Zn(II) secretion into milk.68 Lastly, prostate epithelial cells accumulate high levels of Zn(II) and citrate for secretion into seminal fluid. Zn(II) accumulation in this case appears to be dependent on hZIP1, as Zn(II) accumulates in response to hZIP1 overexpression and decreases upon suppression of hZIP1.172,173 Zn(II) ions may be important to maintain sperm in a quiescent state prior to ejaculation.113 Zn(II) levels in seminal plasma approach 2 mM.114,115 A significant fraction of Zn(II) within prostate cells localizes to mitochondria.117,118 Accumulation of Zn(II) in this case has been shown to inhibit mitochondrial aconitase activity. Inhibition of mitochondrial aconitase decreases citrate utilization for ATP production, thereby making it available for secretion into seminal fluid.172
Labile Zn(II) pools have also been observed within mitochondria. A labile mitochondrial Zn(II) pool distinct from protein bound Zn(II) was identified using the mitochondrial specific Zn-responsive fluorophore RhodZin-3 in neuronal cells.116,119 The labile pool was shown to be dependent on the IM membrane potential and to be mobilized upon mitochondrial depolarization. Moreover, modulation of mitochondrial Zn was shown to effect both mitochondrial respiration and ROS production.116 We have identified a labile Zn(II) pool largely within the matrix of yeast mitochondria that is responsive to the exogenous Zn(II) concentration. The mitochondrial Zn(II) pool appears nonprotein-acious, is accessible to RhodZin-3, and fractionates as a low molecular weight cationic complex (Atkinson and Winge, unpublished observation).
7.4. Mitochondrial Copper
Mitochondria in yeast and human cells contain a labile copper pool that is used in the metalation of cytochrome oxidase (CcO) and Sod1. The labile copper pool is a low mass ligand complex that resides within the matrix compartment. Although the structure of the ligand has yet to be elucidated, it stably binds Cu(I) in an anionic complex that can be depleted by the targeted expression of two heterologous Cu(I) binding molecules, human Sod1 or yeast Crs5 metallothionein, within the matrix.120 The functional consequence of attenuating the CuL pool is apparent in Cu-limited cells, where the presence of either competitor molecule within the matrix diminishes both CcO and resident Sod1 activity and, as a result, impairs either respiratory dependent or hyperoxic growth, respectively. Supplementation of the cultures with exogenous Cu salts causes a significant expansion in the matrix CuL pool and reverses these observed phenotypes, arguing that it is the source of copper used in the metalation of CcO as well as Sod1 within the IMS.
We postulate that Cu(I) binding to the ligand within the cytosol triggers the translocation of the anionic CuL complex to the mitochondrion. The facile expansion of the matrix CuL pool upon supplementation of cells with exogenous copper is consistent with the CuL translocation model. Yeast cells depleted of Cup1, which serves to buffer against changes in cytosolic copper levels, exhibit an expansion of the matrix CuL pool over wild-type yeast which is dramatically enhanced upon growth in copper salts. Thus, trafficking of copper to the mitochondria is more facile in cells unable to buffer copper in the cytosol, suggesting some competition between the ligand and other cytosolic fates for copper.
The CuL complex translocation model predicts transport occurs both into and out of the matrix. Therefore, either two transporters or a single bidirectional transporter exist within the IM. We postulate that once the CuL diffuses through channels in the OM, a transporter functions to move it from the IMS into the matrix. Its regulated release by a transporter would then result in the channeling of either Cu(I) or the CuL complex from the matrix back to the IMS for subsequent use by Cox17 and Ccs1 in downstream mitochondrial metalation reactions (discussed in detail below). The protein-mediated regulation of transporter activity is supported by the observation that the deletion of either Coa1 or Shy1, two CcO assembly factors with roles in Cox1 maturation, attenuates matrix copper levels in yeast.121 The regulation of the export step by a subset of proteins that participate either in CcO assembly or Sod1 activation would effectively limit the amount of free copper within IMS and may in fact have been the driving force that necessitated the existence of the Cox17 copper chaperone pathway in the IMS.
Labile copper pools have been reported in mammalian cells using Cu-responsive fluorophores. Mouse fibroblast 3T3 cells incubated with a Cu(I) sensor show a perinuclear staining pattern that colocalizes with Golgi and mitochondria.122 Another Cu(I) specific fluor developed was shown to detect Cu(I) in HEK293 cells incubated with copper salts.123 A labile copper pool is also postulated to exist in early hematopoietic progenitor cells in human umbilical cord blood cultures. As evidence, treatment of cultures with the copper chelator tetraethylenepentamine (TEPA) results in preferential expansion of early hematopoietic progenitor cells without cell differentiation. 124–126 TEPA appeared to diminish a labile copper pool within the cells. One potential implication of this work is that chelator treatment of stem cells may enhance long-term ex vivo expansion for therapeutic applications. An outstanding question is whether TEPA treatment depletes either the Golgi or mitochondrial labile copper pools. If TEPA treatment attenuates the mitochondrial matrix copper pool, cytochrome oxidase biogenesis may be impaired, which may diminish ATP production. Stem cell proliferation without differentiation may occur if the cells partially switch to glycolysis for ATP generation. Otto Warburg suggested that such a reprogramming of cellular energy metabolism may contribute to tumor proliferation.127
The Golgi labile copper pool may arise from the presence of the Cu(I) efflux transporters ATP7A and ATP7B in mammalian cells. These ATP-driven Cu(I) pumps translocate Cu(I) to the TGN lumen for binding to secretory copper metalloproteins. Epidermal melanocytes synthesize melanin within secretory vesicles called melanosomes. ATP7A translocates Cu(I) into melanosomes for activation of tyrosinase.128 In yeast the Ccc2 P-type ATPase transports Cu(I) into the TGN for the Cu-activation of Fet3, a component of the high affinity iron uptake system.129
7.5. Mitochondrial Manganese
No information exists on labile pools of Mn(II) within the mitochondrial matrix.
8. Protein Metalation within the Mitochondrion
8.1. Iron Metalation Reactions
Formation of heme and Fe/S centers occurs within the mitochondrial matrix. Heme synthesis is achieved by ferrochelatase associated with the inner leaflet of the IM, placing it within the matrix. The ferrochelatase reaction inserts Fe(II) into protoporphyrin IX to generate heme. The source of Fe(II) for this reaction is likely from Mrs3/Mrs4 transporters and not a matrix store.96 Although ferrochelatase can utilize Zn(II), little Zn-protoporphyrin forms in wild-type cells. This specificity may arise from a coupling of heme formation by ferrochelatase with the iron transporters Mrs3/Mrs4.
The iron sulfur cluster (ISC) machinery mediates formation of Fe/S clusters within the matrix and is composed of at least 10 proteins.34 The initial step is the assembly of an Fe/S cluster on the Isu1 scaffold protein, or the redundant homologue Isu2. The sulfide ions for the cluster are provided by the Nfs1 cysteine desulfurase in conjunction with Isd11. A catalytic cysteinyl residue in Nfs1 reacts with the sulfur atom of a cysteine substrate, forming a persulfide intermediate that is subsequently transferred to Isu1 via Isd11.130 The source of iron for Fe/S biogenesis is less clear. However, the yeast frataxin homologue, Yfh1, is believed to be an immediate Fe(II) donor to Isu1.131–134 Yfh1 is capable of transferring Fe(II) to Isu1 for Fe/S cluster formation in vitro.136,141 Moreover, the interaction between Yfh1 and Isu1 is Fe-dependent, as mutations that attenuate Fe(II) binding impair the interaction.134,140 Monomeric yeast Yfh1 binds two Fe(II) ions,133,135 whereas the Drosophila Yfh1 binds a single Fe(II) ion with a Kd of 6 µM.136 Yfh1 is capable of undergoing an iron-mediated oligomerization,137,138 yet oligomerization is not critical for Yfh1 function, as mutants that fail to undergo Fe-induced oligomerization are functional in vivo.139 After assembly, Fe/S clusters formed on Isu1 are transferred to acceptor proteins (listed in section 5.1) in a step requiring additional ISC assembly components.34
8.2. Zinc Metalation Reactions
No information exists on specific Zn(II) metalation reactions within the mitochondrion.
8.3. Copper Metalation Reactions
8.3.1. Sod1 Copper Ion Metalation
Copper activation of Sod1 and metalation of CcO during its biosynthesis occur within the IMS and are mediated by metallochaperones Ccs1 and Cox17, respectively. Cu(I) transfer is likely mediated by ligand exchange reactions in transient heterodimeric protein interactions, as with similar reactions occurring in the cytoplasm. A portion of the cytoplasmic Ccs1 is imported into the IMS via Mia40 and appears to be trapped within the IMS upon oxidative folding (Figure 6). The presence of Ccs1 within the IMS effectively traps Sod1 within the IMS, presumably through Ccs1-activation of Sod1. One model for trapping is that the disulfide in Ccs1 generated through import is transferred to Sod1.53,142 The activity of Sod1 is dependent on formation of the disulfide bond.8 The resulting reduced Ccs1 is poised for Cu(I) binding and subsequent Cu(I) transfer to Sod1. The source of Cu(I) for Sod1 activation appears to be derived from the aforementioned matrix copper-ligand complex. Thus, copper metalation of Sod1 necessitates transport of Cu(I) from the matrix to the IMS. Activated Sod1 contains both Zn(II) and Cu(I). No information is available on the route of Zn(II) binding to Sod1 or the Zn(II) source.
Figure 6.
Mitochondrial Sod1 metalation. The metalation of Sod1 within the IMS is mediated by Ccs1. Ccs1 is imported by the Mia40/ Erv1 pathway through disulfide-capture and oxidative folding. A reactive disulfide in Mia40 generated by Erv1 forms an intermolecular disulfide with the imported protein, e.g. Ccs1, and release from Mia40 occurs through disulfide interchange, leading to transfer of the disulfide to the imported protein. Ccs1 has Cys residues in the N- and C-terminal domains that may participate in this oxidative folding. These are the two domains that bind Cu(I) for transfer to Sod1. Thus, it is not clear how a disulfide in Ccs1 is reduced to permit Cu(I) metalation of imported Sod1. In the absence of Ccs1, Sod1 is not retained within the IMS.53,142
8.3.2. Cytochrome Oxidase Copper Ion Metalation
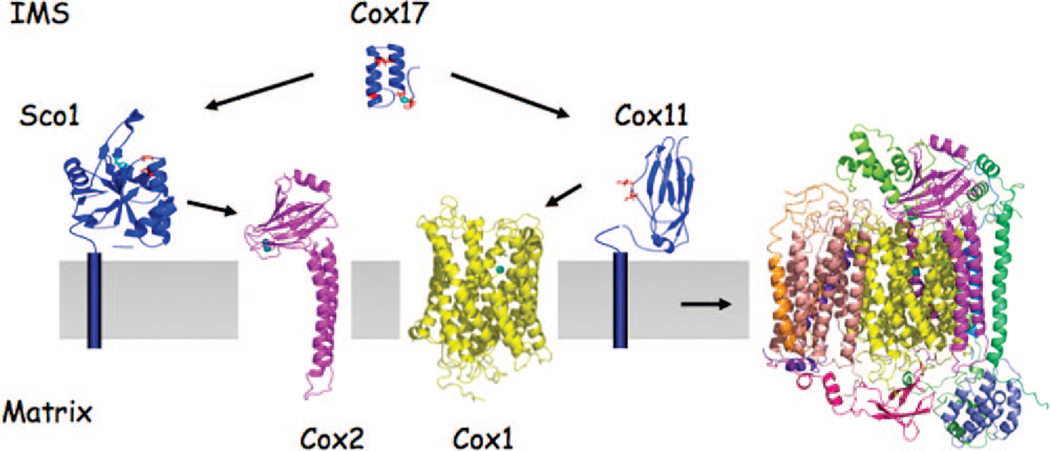
In the copper metalation of CcO, Cox17 transfers Cu(I) to two IM-tethered proteins Sco1 and Cox11 for subsequent transfer to Cox2 and Cox1, respectively (Figure 7). Cu(I) coordination in Cox17, Ccs1, and Cox11 is through thiolate ligation, whereas both thiolate and histidyl ligands participate in Cu(I) coordination within Sco1. This is significant, as the IMS has a more oxidizing redox potential than the cytoplasm.143 Furthermore, numerous thiol-rich proteins are stabilized with disulfide bonds.144 How certain Cys residues form specific disulfide bridges while other Cys residues are maintained in a reduced state to facilitate metal binding is a significant issue. Little information exists on how the redox state of proteins within the IMS is maintained.
Figure 7.
Cu-metalation of CcO. During biogenesis, CcO metalation is mediated by chaperone proteins. Cox17 is the Cu(I) donor to both Scol and Cox11 for subsequent transfer to Cox2 and Cox1, respectively. Cox1 and Cox2 are synthesized on mitoribosomes and assembled into the mature CcO complex in a stepwise process. Biogenesis commences with the maturation of Cox1 and heme a and CuB site formation followed by addition of Cox2 with the CuA site. Several of the small peripheral subunits are added at a late stage of assembly.
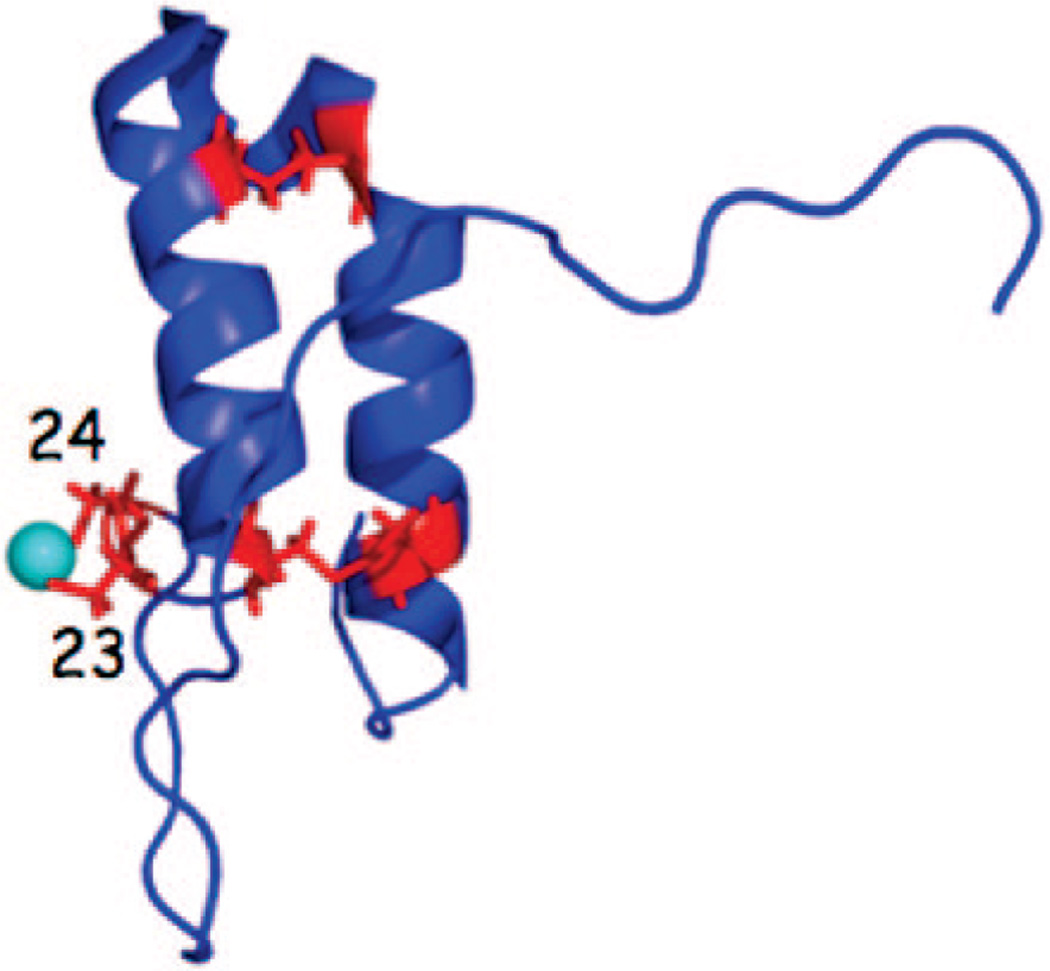
Coxl7 and two other key IMS-localized CcO assembly factors (Cox19 and Cox23) contain a conserved twin Cys-X9-Cys sequence motif. The four cysteines within the Cox17 Cys-X9-Cys motifs are present in two disulfides stabilized within a helical hairpin conformation. Two additional vicinal cysteinyl residues outside the twin Cys-X9-Cys motifs form a bent, two coordinate Cu(I) complex (Figure 8).145 With the six total Cys residues, the apo-molecule is capable of forming three disulfides, yet the dual disulfide molecule is the likely species within the IMS to facilitate both Cu(I) binding and a stable redox state within the IMS. The later point is supported, as the redox potential of the IMS was recently shown to be approximately −255 mV with some uncertainty due to the lack of precise information on the pH of the compartment.143 A redox potential of −255 mV would support the double disulfide conformer, as the midpoint redox potential of the triple disulfide form to the double disulfide form is −197 mV,146 whereas the double disulfide configuration to the fully reduced state has a midpoint potential of −340 mV.147 Furthermore, neither Cu(I) coordination nor Cox17 function are dependent on the presence of two disulfide cross-links, as revealed through mutational analysis.148
Figure 8.
Structure of the Cu(I) conformer of Coxl7. The four conserved Cys residues of the twin Cys-X9-Cys motif are shown in the disulfide linkage in red. Two additional vicinal cysteinyl residues (Cys23 and Cys24) outside the twin Cys-X9-Cys motifs form a bent, two coordinate Cu(I) complex.145 The redox potential of the IMS likely supports the double disulfide conformer with the vicinal Cys residues reduced to coordinate Cu(I).146,147
Sco1 is tethered to the IM by a single transmembrane helix with the C-terminal globular domain protruding into the IMS (Figure 7).149,150 Human cells have a second Sco protein, Sco2, which is also important in CcO biogenesis.151 A single Cu(I) binding site exists within the globular domain of Sco1 and Sco2, consisting of two cysteinyl residues within a Cys-X3-Cys motif and a conserved histidyl residue. The Cu(I) ion coordinated by Sco1 and Sco2 is solvent-exposed and poised for ligand exchange transfer reactions. The structures of the metal-free Sco1 and Cu1Scol complex are similar other than significant rearrangements restricted to loop 8.152 The movement of this loop orients the Cu(I) binding His residue in the proper orientation for Cu binding.152 The structural dynamics of loop 8 suggest that it may be an important interface for a functional interaction. Conserved residues at the base of the loop were shown to be important for a function with Cox2.153 Cu(I) binding to Cox17, Sco1, and Sco2 may be expected to be dependent on the Cu(I)-binding cysteines being maintained in the reduced state during the Cu(I) transfer reactions. However, recent evidence suggests human Cu-Cox17 is capable of transferring Cu(I) as well as two electrons to human oxidized Sco1, enabling metalation of Sco1.154 Thus, Sco1 may exist within the IMS in an oxidized resting state.
The Cu ions bound by Sco1 are likely then transferred to Cox2 to form the binuclear CuA in Cox2 (Figure 7). The CuA site in Cox2 is formed within a ten-stranded β-barrel and two cysteine residues within a Cys-X3-Cys motif bridging the two Cu ions.155 The domain of Cox2 containing the CuA site protrudes into the IMS with the CuA site 8 Å above the membrane surface.156 It is not clear whether Cu-Sco1 is capable of transferring Cu(I) and electron equivalents to Cox2 with oxidized cysteines or whether a redox system exists to maintain the Cox2 cysteines in the thiolate state. However, Sco1 has a thioredoxin-like fold, so Sco1 may have a redox function with the CuA site cysteines.157,158 Alternatively, Sco1 may function as a redox switch, in which oxidation of the two Cys residues in the Cu(I) binding Cys-X3-Cys motif may faciliate Cu(I) transfer to Cox2.152 The Cys residues within the Cys-X3-Cys motif in Cox2 may be involved in ligand exchange reactions to move Cu(I) from Sco1 to Cox2. An initial Cu(I) coordination in Cox2 may be a distorted two-coordinate complex involving the two Cys residues prior to rearrangements to form the final coordination sites.
Like Sco1 and Sco2, Cox11 is tethered to the IM by a single transmembrane helix with a C-terminal globular domain protruding into the IMS (Figure 7). This domain binds Cu(I) through at least two thiolate residues.159,160 Substitution of three cysteines within the globular domain attenuates Cu(I) binding and abrogates in vivo function. A specific role for Cox11 in the biogenesis of the CuB site was demonstrated in Rhodobacter sphaeroides. CcO isolated from R. sphaeroides cox11Δ cells lacked CuB but contained both hemes in Cox1. Cu(I) transfer from Cox11 to Cox1 to form the CuB site likely occurs concurrently with the insertion of heme a3 in newly synthesized Cox1. No information exists on whether Cu-Cox17 is capable of transferring Cu(I) and electron equivalents to oxidized Cox11. The Cox1 CuB site consists of three histidyl residues, so redox issues are relevant.
The presence of the copper-ligand complex in the matrix and copper metalation in the IMS necessitates translocation of Cu(I) to the IMS. The IM transporter has not been identified to date; however, some clues about this process have emerged. Multiple screens have been conducted to identify yeast deletion strains that have altered mitochondrial copper pools. Two deletion strains identified to date are genes that encode the CcO assembly factors Coa1 and Shy1. These two proteins have essential roles in Coxl maturation. Cells lacking either Coa1 or Shy1 have diminished matrix copper levels. These observations may suggest that the Cu(I) translocation to the IMS is coupled to Cox1 maturation. Coa1 and Shy1 may have a secondary role in modulating the transport process. The translocated Cu(I) is likely presented to Cox17 for subsequent IMS delivery to Sco1 and Cox11. This proposed coupling of Cu(I) translocation and Cox1 maturation may restrict the amount of Cu(I) transported to minimize any Cu-catalyzed ROS production in the IMS.
8.4. Manganese Metalation Reactions
Manganese activation of Sod2 within the matrix occurs during the folding step of the Sod2 polypeptide, as evidenced by the fact that prefolded Sod2 polypeptides cannot be activated with manganese.55 Likewise, relocalization of Sod2 within the yeast cytoplasm leads to impaired manganese activation. As mentioned, Mn-activation of Sod2 requires the function of both the Smf2 Mn(II) transporter, which localizes to internal vesicles, and the Mtm1 mitochondrial carrier protein.54,81 Cells lacking Smf2 are deficient in mitochondrial Mn(II), whereas cells lacking Mtm1 contain normal manganese levels. Thus, Mtm1 is not a Mn(II) transporter. It remains unclear how Smf2 contributes Mn(II) to the mitochondrion.
The lack of Sod2 dismutase activity in mtm1Δ yeast cells is due to an accumulation of Fe(II) within the mitochondrion.98 The accumulation of Fe(II) in mtm1Δ mitochondria leads to the mismetalation of Sod2 by Fe(II) during the folding reaction.98 The reason for the iron accumulation in mtm1Δ mitochondria remains unclear, but the absence of Mtm1 may lead to the accumulation of a metabolite capable of binding Fe(II) within the matrix. The Fe(II) mismetalation of Sod2 is dependent on Yfh1, suggesting that Yfh1 maintains Fe(II) in the bioavailable state. In wild-type yeast the matrix Fe(II) pools are maintained at sufficiently low levels to allow for Mn(II) activation of Sod2. Conditions that lead to an expansion of the matrix Fe(II) pool result in a Fe(II)/Mn(II) competition for availability to Sod2.
Mis-metalation of Sod is also known in E. coli cells that contain two related Sod proteins, MnSod and FeSod. Each protein can bind the other metal ion, but the resulting protein is catalytically inactive due to an aberrant redox potential.161–163 MnSod isolated from E. coli contains a mixture of bound Mn(II) and Fe(II).161,162 Mn(II) metalation of the FeSod has not been reported in E. coli. An interesting unresolved question is how E. coli controls the Fe(II) and Mn(II) pools, permitting proper metalation of FeSod and MnSod.
Proper metal ion metalation reactions within the mitochondrial matrix are therefore dependent on the control of bioavailable pools of metal ions. Mismetalation reactions may be possible with proteins binding Fe(II) or Zn(II), since both metal ions are abundant in mitochondria. For instance, the bacterial IscU Fe/S scaffold protein was isolated with a bound Zn(II) rather than a Fe/S cluster.164 It is unclear whether mitochondrial Isu1 is capable of Zn(II) binding and if so whether the Zn(II) conformer precludes Fe/S cluster assembly. Alternatively, Zn(II) binding could be transient and, as with the Zn(II) associated with Mia40 and Hot13, serve to maintain reduced metal ligands.
9. Role of Mitochondria in Controlling Cellular Metal Pools
The mitochondrion modulates iron homeostasis in yeast. Cells defective in Fe/S cluster biogenesis within the mito-chondrial matrix exhibit constitutive activity of the iron– metalloregulatory transcription factor Aft1.165 Aft1 is normally active only under conditions of iron deficiency in yeast. Thus, the activation of Aft1 in cells impaired in mitochondrial Fe/S cluster biogenesis mimics an iron deficient state despite normal iron levels.166 Iron sensing by Aft1 requires proper mitochondrial Fe/S cluster biosynthesis, as well as the export of an ill-defined intermediate through the Atm1 IM exporter.167 The signal for iron-inhibition of Aft1 activity appears to correlate with the substrate exported by Atm1. This substrate is then subsequently used by the cytoplasmic Fe/S biogenesis machinery to mature Fe/S clusters in cytoplasmic and nuclear Fe/S proteins.
In human cells the CcO assembly factors Sco1 and Sco2 have a role in cellular copper homeostasis.168 Fibroblast cell lines from patients with mutations in Sco1 or Sco2 exhibit significant reductions in total cellular copper content due to enhanced export. A similar reduction is also present in fibroblasts of patients with mutations in Cox15. Interestingly, the copper-deficiency can be reversed by overexpression of Sco2. The signaling pathway by which mutant CcO assembly factors modulate cellular copper levels has not yet been resolved.
Genome wide screens are being conducted to assess the impact of a particular gene deletion on the metal quota of cells. One high throughput study of deletion strains in S. cerevisiae revealed a set of mitochondrial gene products that when deleted caused perturbations in the cellular metal quota.169 Cellular Zn and Cu but not Fe levels were increased in cells depleted of two mito-ribosomal subunits. Cells depleted of assembly factors for the bc1 respiratory complex showed an attenuation in cellular Cu and Fe levels. Clearly, numerous interconnections exist between cellular compartments that result in modulation of the metal quota or ionome of a cell.
10. Major Future Challenges
Major gaps in our knowledge of metal ion homeostasis exist in the identity of various metal ion transporters in mitochondria. It is unclear what homeostatic mechanisms exist to regulate the bioavailable pools of metal ions within compartments such as the mitochondria. The importance of metalation reactions in diverse compartments is suggestive that regulatory steps may be significant. An important future line of research will be to evaluate whether cells have any quality control mechanisms to ensure proper metalation. If mismetalation induces a non-native conformation, perhaps the Hsp70 protein chaperone machinery will facilitate refolding and proper metalation. Another major challenge is to assess how cells integrate information on metal ions in various compartments to ensure homeostasis and maintenance of the metal ion quota of cells.
Biographies

Aaron Atkinson is a postdoctoral fellow with Dr. Winge’s group at the University of Utah in the Department of Hematology and Biochemistry. He received his Ph.D. degree in Biology from Dartmouth College in 2007 under the guidance of Dr. Mary Lou Guerinot. While there, his research focused on metal selectivity of Zrtlrt-like transporters and metal distribution in Arabidopsis. He received his B.S. from the University of Utah, where he worked with both Dr. Gary Drews and Dr. Leslie Sieburth. Currently, he is studying mitochondrial zinc homeostasis as it relates to both isolated Zn-dependent mutants in yeast and characterization of a labile cationic zinc complex within mitochondria.

Dennis Winge is Professor of Internal Medicine and Biochemistry at the University of Utah. He received his Ph.D. degree in Biochemistry from Duke University in 1975 under the guidance of Dr. K. V. Rajagopalan and his B.S. from Concordia College, Moorhead MN, in 1969. He completed postdoctoral fellowships at the University of Geneva and Duke University prior to accepting the position at the University of Utah in 1979. For several years, his research focused on a series of metal-responsive transcriptional activators in yeast with the goal of elucidating the mechanism of metal specificity in response. The activators included the Cu-responsive Ace1 and Mac1 factors, the Zn-responsive Zap1 protein, and the Fe-responsive Aft1 and Aft2 proteins. In recent years, his research has focused more on mitochondrial metal ion homeostasis with the goal of understanding metalation reactions with the mitochondrion. Presently, he is studying the bioavailability of copper and zinc ions in mitochondria and the assembly of the redox cofactor sites in cytochrome oxidase.
References
- 1.Frydman J. Annu. Rev. Biochem. 2001;70:603. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Netzer WJ, Hartl FU. Nature. 1997;388:343. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 3.Young JC, Agashe VR, Siegers K, Hartl FU. Nat. Rev. Mol. Cell Biol. 2004;5:781. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 4.Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, Cheek TR, Gray J, Banfleld MJ, Dennison C, Robinson NJ. Nature. 2008;455:1138. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 5.Huffman DL, O’Halloran TV. Annu. Rev. Biochem. 2001;70:677. doi: 10.1146/annurev.biochem.70.1.677. [DOI] [PubMed] [Google Scholar]
- 6.Lin S-J, Pufahl RA, Dancis A, O’Halloran TV, Culotta VC. J. Biol. Chem. 1997;272:9215. [PubMed] [Google Scholar]
- 7.O’Halloran TV, Culotta VC. J. Biol. Chem. 2000;275:25057. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa Y, Torres AS, O’Halloran TV. EMBO J. 2004;23:2872. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong PC, Waggoner D, Subramaniam JR, Tessarollo L, Bartnikas TB, Culotta VC, Price DL, Rothsten J, Gitlin JD. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2886. doi: 10.1073/pnas.040461197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen LT, Culotta VC. J. Biol. Chem. 2005;280:41373. doi: 10.1074/jbc.M509142200. [DOI] [PubMed] [Google Scholar]
- 11.Kirby K, Jensen LT, Binnington J, Hilliker AJ, Ulloa J, Culotta VC, Phillips JP. J. Biol. Chem. 2008;283:35393. doi: 10.1074/jbc.M807131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H, Bencze KZ, Stemmler TL, Philpott CC. Science. 2008;320:1207. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermann GJ, Shaw JM. Annu. Rev. Cell Dev. Biol. 1998;14:265. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- 14.Frey TG, Mannella CA. Trends Biochem. Sci. 2000;25:319. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 15.Hoppins S, Nunnari J. Biochim. Biophys. Acta. 2009;1793:20. doi: 10.1016/j.bbamcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Zick M, Rabl R, Reichert AS. Biochim. Biophys. Acta. 2009;1793:5. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Vogel F, Bornhovd C, Neupert W, Reichert AS. J. Cell Biol. 2006;175:237. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogenhagen DF, Rousseau D, Burke S. J. Biol. Chem. 2008;283:3665. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 19.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13207. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohlmeier S, Kastaniotis AJ, Hiltunen JK, Bergmann U. J. Biol. Chem. 3956;279 doi: 10.1074/jbc.M310160200. [DOI] [PubMed] [Google Scholar]
- 21.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. Cell. 2008;134:112. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. PLoS One. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay A, Ni L, Weiner H. Biochem. J. 2004;382:385. doi: 10.1042/BJ20040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia M, Darzacq X, Delaveau T, Jourdren L, Singer RH, Jacq C. Mol. Biol. Cell. 2007;18:362. doi: 10.1091/mbc.E06-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellems RE, Allison VF, Butow RA. J. Cell Biol. 1975;65:1. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohnert M, Pfanner N, van der Laan M. FEBS Lett. 2007;581:2802. doi: 10.1016/j.febslet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Neupert W, Herrmann JM. Annu. Rev. Biochem. 2007;76:723. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 28.Mokranjac D, Neupert W. Biochim. Biophys. Acta. 2008;1777:758. doi: 10.1016/j.bbabio.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 29.Herrmann JM, Kohl R. J. Cell Biol. 2007;176:559. doi: 10.1083/jcb.200611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. J. Cell Biol. 2007;179:389. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sideris DP, Tokatlidis K. Mol. Microbiol. 2007;65:1360. doi: 10.1111/j.1365-2958.2007.05880.x. [DOI] [PubMed] [Google Scholar]
- 32.Stojanovski D, Milenkovic D, Muller JM, Gabriel K, Schulze-Specking A, Baker MJ, Ryan MT, Guiard B, Pfanner N, Chacinska A. J. Cell Biol. 2008;183:195. doi: 10.1083/jcb.200804095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bych K, Kerscher S, Netz DJ, Pierik AJ, Zwicker K, Huynen MA, Lill R, Brandt U, Balk J. EMBO J. 2008:27, 1736. doi: 10.1038/emboj.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lill R, Muhlenhoff U. Annu. Rev. Biochem. 2008;77:669. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 35.Levi S, Corsi B, Bosisio M, Invernizzi R, Volz A, Sanford D, Arosio P, Drysdale J. J. Biol. Chem. 2001;276:24437. doi: 10.1074/jbc.C100141200. [DOI] [PubMed] [Google Scholar]
- 36.Stenmark P, Grunler J, Mattsson J, Sindelar PJ, Nordlund P, Berthold DA. J. Biol. Chem. 2001;276:33297. doi: 10.1074/jbc.C100346200. [DOI] [PubMed] [Google Scholar]
- 37.Maxwell DP, Wang Y, Mcintosh L. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8271. doi: 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu H, Woodburn J. J. Mol. Biol. 2005;353:897. doi: 10.1016/j.jmb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Rissler M, Wiedemann N, Pfannschmidt S, Gabriel K, Guiard B, Pfanner N, Chacinska A. J. Mol. Biol. 2005;353:485. doi: 10.1016/j.jmb.2005.08.051. [DOI] [PubMed] [Google Scholar]
- 40.Mesecke N, Bihlmaier K, Grumbt B, Longen S, Terziyska N, Hell K, Herrmann JM. EMBO Rep. 2008;9:1107. doi: 10.1038/embor.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arlt H, Steglich G, Perryman R, Guiard B, Neupert W, Langer T. EMBO J. 1998;17:4837. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. Cell. 2005;123:277. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Gakh O, Cavadini P, Isaya G. Biochim. Biophys. Acta. 2002;1592:63. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 44.Luciano P, Tokatlidis K, Chambre I, Germanique JC, Geli V. J. Mol. Biol. 1998;280:193. doi: 10.1006/jmbi.1998.1858. [DOI] [PubMed] [Google Scholar]
- 45.Zeng X, Neupert W, Tzagoloff A. Mol. Biol. Cell. 2007;18:617. doi: 10.1091/mbc.E06-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osman C, Wilmes C, Tatsuta T, Langer T. Mol. Biol. Cell. 2007;18:627. doi: 10.1091/mbc.E06-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaser M, Kambacheld M, Kisters-Woike B, Langer T. J. Biol. Chem. 2003;278:46414. doi: 10.1074/jbc.M305584200. [DOI] [PubMed] [Google Scholar]
- 48.Burri L, Vascotto K, Fredersdorf S, Tiedt R, Hall MN, Lithgow T. J. Biol. Chem. 2004;279:50243. doi: 10.1074/jbc.M409194200. [DOI] [PubMed] [Google Scholar]
- 49.Voisine C, Cheng YC, Ohlson M, Schilke B, Hoff K, Beinert H, Marszalek J, Craig EA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1483. doi: 10.1073/pnas.98.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bianchi C, Genova ML, Parenti Castelli G, Lenaz G. J. Biol. Chem. 2004;279:36562. doi: 10.1074/jbc.M405135200. [DOI] [PubMed] [Google Scholar]
- 51.Heinemeyer J, Braun HP, Boekema EJ, Kouril R. J. Biol. Chem. 2007;282:12240. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 52.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. J. Biol. Chem. 2001;276:38084. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 53.Kawamata H, Manfredi G. Hum. Mol. Genet. 2008;17:3303. doi: 10.1093/hmg/ddn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luk E, Culotta VC. J. Biol. Chem. 2001;276:47556. doi: 10.1074/jbc.M108923200. [DOI] [PubMed] [Google Scholar]
- 55.Luk E, Yang M, Jensen LT, Bourbonnais Y, Culotta VC. J. Biol. Chem. 2005;280:22715. doi: 10.1074/jbc.M504257200. [DOI] [PubMed] [Google Scholar]
- 56.Foury F, Roganti T. J. Biol. Chem. 2002;277:24475. doi: 10.1074/jbc.M111789200. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Lyver ER, Knight SAB, Pain D, Lesuisse E, Dancis A. J. Biol. Chem. 2006281:22493. doi: 10.1074/jbc.M604246200. [DOI] [PubMed] [Google Scholar]
- 58.Roussel D, Harding M, Runswick MJ, Walker JE, Brand MD. J. Bioenerg. Biomembr. 2002;34:165. doi: 10.1023/a:1016027302232. [DOI] [PubMed] [Google Scholar]
- 59.Muhlenhoff U, Stadler JA, Richhardt N, Seubert A, Eickhorst T, Schweyen RJ, Lill R, Wiesenberger G. J. Biol. Chem. 2003;278:40612. doi: 10.1074/jbc.M307847200. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Lyver ER, Knight SAB, Lesuisse E, Dancis A. J. Biol. Chem. 2005;280:19794. doi: 10.1074/jbc.M500397200. [DOI] [PubMed] [Google Scholar]
- 61.Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, Trede NS, Barut BA, Zhou Y, Minet E, Donovan A, Brownlie A, Balzan R, Weiss MJ, Peters LL, Kaplan J, Zon LI, Paw BH. Nature. 2006;440:96. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 62.Li L, Kaplan J. J. Biol. Chem. 1997;272:28485. doi: 10.1074/jbc.272.45.28485. [DOI] [PubMed] [Google Scholar]
- 63.Robinson AJ, Kunji ERS. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2617. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eide D. J. Biochim. Biophys. Acta. 2006;1763:711. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 65.Jackson KA, Helston RM, McKay JA, O’Neill ED, Mathers JC, Ford D. J. Biol. Chem. 2007;282:10423. doi: 10.1074/jbc.M610535200. [DOI] [PubMed] [Google Scholar]
- 66.Cousins RJ, Liuzzi JP, Lichten LA. J. Biol. Chem. 2006;281:24085. doi: 10.1074/jbc.R600011200. [DOI] [PubMed] [Google Scholar]
- 67.Chimienti F, Devergnas S, Pattou F, Schuit F, Garcia-Cuenca R, Vandewalle B, Kerr-Conte J, Van Lommel L, Grunwald D, Favier A, Seve M. J. Cell Sci. 2006;119:4199. doi: 10.1242/jcs.03164. [DOI] [PubMed] [Google Scholar]
- 68.Huang L, Gitschier J. Nat. Genet. 1997;17:292. doi: 10.1038/ng1197-292. [DOI] [PubMed] [Google Scholar]
- 69.Chowanadisai W, Lonnerdal B, Kelleher SL. J. Biol. Chem. 2006;281:39699. doi: 10.1074/jbc.M605821200. [DOI] [PubMed] [Google Scholar]
- 70.Simm C, Lahner B, Salt D, LeFurgey A, Ingram P, Yandell B, Eide DJ. Eukaryotic Cell. 2007;6:1166. doi: 10.1128/EC.00077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacDiarmid CW, Gaither LA, Eide D. EMBO J. 2000;19:2845. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saris NE, Niva K. FEBS Lett. 1994;356:195. doi: 10.1016/0014-5793(94)01256-3. [DOI] [PubMed] [Google Scholar]
- 73.Sensi SL, Ton-That D, Weiss JH. Neurobiol. Dis. 2002;10:100. doi: 10.1006/nbdi.2002.0493. [DOI] [PubMed] [Google Scholar]
- 74.Dineley KE, Richards LL, Votyakova TV, Reynolds I. J. Mitochondrion. 2005;5:55. doi: 10.1016/j.mito.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 75.Sensi SL, Ton-That D, Sullivan PG, Jonas EA, Gee KR, Kaczmarek LK, Weiss JH. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6157. doi: 10.1073/pnas.1031598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malaiyandi LM, Vergun O, Dineley KE, Reynolds IJ. J. Neurochem. 2005;93:1242. doi: 10.1111/j.1471-4159.2005.03116.x. [DOI] [PubMed] [Google Scholar]
- 77.Trenker M, Malli R, Fertschai I, Levak-Frank S, Graier WF. Nat. Cell Biol. 2007;9:445. doi: 10.1038/ncb1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Culotta VC, Yang M, Hall MD. Eukaryot. Cell. 2005;4:1159. doi: 10.1128/EC.4.7.1159-1165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Culotta VC, Yang M, O’Halloran TV. Biochim. Biophys. Acta. 2006;1763:747. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jensen LT, Ajua-Alemanji M, Culotta VC. J. Biol. Chem. 2003;278:42036. doi: 10.1074/jbc.M307413200. [DOI] [PubMed] [Google Scholar]
- 81.Luk E, Carroll M, Baker M, Culotta VC. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10353. doi: 10.1073/pnas.1632471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schindl R, Weghuber J, Romanin C, Schweyen RJ. Biophys. J. 2007;93:3872. doi: 10.1529/biophysj.107.112318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rae RD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Science. 1999;284:805. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 84.Outten CE, O’Halloran TV. Science. 2001;292:2488. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 85.Yamamoto K, Ishihama A. J. Bacterial. 2005;187:6333. doi: 10.1128/JB.187.18.6333-6340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Binet MR, Poole RK. FEBS Lett. 2000;473:67. doi: 10.1016/s0014-5793(00)01509-x. [DOI] [PubMed] [Google Scholar]
- 87.Hantke K. J. Mol. Microbiol. Biotechnol. 2002;4:217. [PubMed] [Google Scholar]
- 88.Klevecz RR, Bolen J, Forrest G, Murray DB. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1200. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Levi S, Rovida E. Biochim. Biophys. Acta. in press doi: 10.1016/j.bbagen.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 90.Veiga N, Torres J, Mansell D, Freeman S, Dominguez S, Barker CJ, Diaz A, Kremer C. J. Biol. Inorg. Chem. 2008;14:51. doi: 10.1007/s00775-008-0423-2. [DOI] [PubMed] [Google Scholar]
- 91.Rauen U, Springer A, Weisheit D, Petrat F, Korth HG, de Groot H, Sustmann R. ChemBioChem. 2007;8:341. doi: 10.1002/cbic.200600311. [DOI] [PubMed] [Google Scholar]
- 92.Kruszewski M. Acta Biochim. Pol. 2004;51:471. [PubMed] [Google Scholar]
- 93.Petrat F, Weisheit D, Lensen M, de Groot H, Sustmann R, Rauen U. Biochem. J. 2002;362:137. doi: 10.1042/0264-6021:3620137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liang LP, Jarrett SG, Patel M. J. Neurosci. 2008;28:11550. doi: 10.1523/JNEUROSCI.3016-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tangeras A. Biochim. Biophys. Acta. 1985;843:199. doi: 10.1016/0304-4165(85)90140-0. [DOI] [PubMed] [Google Scholar]
- 96.Lange H, Kispal G, Lill R. J. Biol. Chem. 1999;274:18989. doi: 10.1074/jbc.274.27.18989. [DOI] [PubMed] [Google Scholar]
- 97.Cavadini P, Biasiotto G, Poli M, Levi S, Verardi R, Zanella I, Derosas M, Ingrassia R, Corrado M, Arosio P. Blood. 2007;109:3552. doi: 10.1182/blood-2006-08-041632. [DOI] [PubMed] [Google Scholar]
- 98.Yang M, Cobine PA, Molik S, Naranuntarat A, Lill R, Winge DR, Culotta VC. EMBO J. 2006;25:1775. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lesuisse E, Santos R, Matzanke BF, Knight SAB, Camadro J-M, Dancis A. Hum. Mol. Genet. 2003;12:879. doi: 10.1093/hmg/ddg096. [DOI] [PubMed] [Google Scholar]
- 100.Sturm B, Bistrich U, Schranzhofer M, Sarsero JP, Rauen U, Scheiber-Mojdehkar B, de Groot H, Ioannou P, Petrat F. J. Biol. Chem. 2005;280:6701. doi: 10.1074/jbc.M408717200. [DOI] [PubMed] [Google Scholar]
- 101.Puccio H, Simon D, Cossee M, Criqui-Filipe P, Tiziano F, Melki J, Hindelang C, Matyas R, Rustin P, Koenig M. Nat. Genet. 2001;27:181. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 102.Campanella A, Rovelli E, Santambrogio P, Cozzi A, Taroni F, Levi S. Hum. Mol. Genet. 2009;8:1. doi: 10.1093/hmg/ddn308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mao R, Martinho M, Morales JG, Kim H, Ellis EA, Lill R, Hendrich MP, Munck E, Lindahl PA. Biochemistry. 2008;47:9888. doi: 10.1021/bi801047q. [DOI] [PubMed] [Google Scholar]
- 104.Haase H, Beyersmann D. Biochem. Biophys. Res. Commun. 2002;296:923. doi: 10.1016/s0006-291x(02)02017-x. [DOI] [PubMed] [Google Scholar]
- 105.Thompson RB, Peterson D, Mahoney W, Cramer M, Maliwal BP, Suh SW, Frederickson C, Fierke C, Herman P. J. Neurosci. Methods. 2002;118:63. doi: 10.1016/s0165-0270(02)00144-9. [DOI] [PubMed] [Google Scholar]
- 106.Frederickson CJ, Suh SW, Silva D, Frederickson CJ, Thompson RB. J. Nutr. 2000;130:1471S. doi: 10.1093/jn/130.5.1471S. [DOI] [PubMed] [Google Scholar]
- 107.Palmiter RD, Cole TB, Quaife CJ, Findley SD. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14934. doi: 10.1073/pnas.93.25.14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Linkous DH, Flinn JM, Koh JY, Lanzirotti A, Bertsch PM, Jones BF, Giblin LJ, Frederickson CJ. J. Histochem. Cytochem. 2008;56:3. doi: 10.1369/jhc.6A7035.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Y, Hough CJ, Suh SW, Sarvey JM, Frederickson CJ. J. Neurophysiol. 2001;86:2597. doi: 10.1152/jn.2001.86.5.2597. [DOI] [PubMed] [Google Scholar]
- 110.Dodson G, Steiner D. Curr. Opin. Struct. Biol. 1998;8:189. doi: 10.1016/s0959-440x(98)80037-7. [DOI] [PubMed] [Google Scholar]
- 111.Ho LH, Ruffin RE, Murgia C, Li L, Krilis SA, Zalewski PD. J. Immunol. 2004;172:7750. doi: 10.4049/jimmunol.172.12.7750. [DOI] [PubMed] [Google Scholar]
- 112.Giblin LJ, Chang CJ, Bentley AF, Frederickson C, Lippard SJ, Frederickson CJ. J. Histochem. Cytochem. 2006;54:311. doi: 10.1369/jhc.5A6724.2005. [DOI] [PubMed] [Google Scholar]
- 113.Yoshida K, Kawano N, Yoshiike M, Yoshida M, Iwamoto T, Morisawa M. Mol. Hum. Reprod. 2008;14:151. doi: 10.1093/molehr/gan003. [DOI] [PubMed] [Google Scholar]
- 114.Arver S. Acta Physiol. Scand. 1982;116:67. doi: 10.1111/j.1748-1716.1982.tb10600.x. [DOI] [PubMed] [Google Scholar]
- 115.Arver S, Sjoberg HE. Acta Physiol. Scand. 1982;116:159. doi: 10.1111/j.1748-1716.1982.tb07125.x. [DOI] [PubMed] [Google Scholar]
- 116.Sensi SL, Ton-That D, Sullivan PG, Jonas EA, Gee KR, Kaczmarek LK, Weiss JH. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6157. doi: 10.1073/pnas.1031598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Costello LC, Liu Y, Franklin RB, Kennedy MC. J. Biol. Chem. 1997;272:28875. doi: 10.1074/jbc.272.46.28875. [DOI] [PubMed] [Google Scholar]
- 118.Liu Y, Franklin RB, Costello LC. Prostate. 1997;30:26. doi: 10.1002/(sici)1097-0045(19970101)30:1<26::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 119.Sensi SL, Ton-That D, Weiss JH, Rothe A, Gee KR. Cell Calcium. 2003;34:281. doi: 10.1016/s0143-4160(03)00122-2. [DOI] [PubMed] [Google Scholar]
- 120.Cobine PA, Pierrel F, Bestwick ML, Winge DR. J. Biol. Chem. 2006;281:36552. doi: 10.1074/jbc.M606839200. [DOI] [PubMed] [Google Scholar]
- 121.Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR. EMBO J. 2007;26:4335. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang L, McRae R, Henary MM, Patel R, Lai B, Vogt S, Fahrni C. J. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11179. doi: 10.1073/pnas.0406547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zeng L, Mller EW, Pralle A, Isacoff EY, Chang CJ. J. Am. Chem. Soc. 2006;128:10. doi: 10.1021/ja055064u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peled T, Glukhman E, Hasson N, Adi S, Assor H, Yudin D, Landor C, Mandel J, Landau E, Prus E, Nagler A, FIbach E. Exp. Hematol. 2005;33:1092. doi: 10.1016/j.exphem.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 125.Peled T, Landau E, Mandel J, Glukhman E, Goudsmid NR, Nagler A, FIbach E. Exp. Hematol. 2004;32:547. doi: 10.1016/j.exphem.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 126.Prus E, Peled T, Fibach E. Leuk. Lymphoma. 2004;45:583. doi: 10.1080/10428190310001598035. [DOI] [PubMed] [Google Scholar]
- 127.Warburg O. Science. 1956;124:269. [PubMed] [Google Scholar]
- 128.Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Nature. 2008;454:1142. doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.De Silva DM, Askwith CC, Eide D, Kaplan J. J. Biol. Chem. 1995;270:1098. doi: 10.1074/jbc.270.3.1098. [DOI] [PubMed] [Google Scholar]
- 130.Wiedemann N, Urzica E, Guiard B, Muller H, Lohaus C, Meyer HE, Ryan MT, Meisinger C, Muhlenhoff U, Lill R, Pfanner N. EMBO J. 2006;25:184. doi: 10.1038/sj.emboj.7600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R. Hum. Mol. Genet. 2002;11:2025. doi: 10.1093/hmg/11.17.2025. [DOI] [PubMed] [Google Scholar]
- 132.Stehling O, Elsasser HP, Bruckel B, Muhlenhoff U, Lill R. Hum. Mol. Genet. 2004;13:3007. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 133.Cook JD, Bencze KZ, Jankovic AD, Crater AK, Busch CN, Bradley PB, Stemmler AJ, Spaller MR, Stemmler TL. Biochemistry. 2006;45:7767. doi: 10.1021/bi060424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang T, Craig EA. J. Biol. Chem. 2008;283:12674. doi: 10.1074/jbc.M800399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bencze KZ, Kondapalli KC, Cook JD, McMahon S, Mllan-Pacheco C, Pastor N, Stemmler TL. Crit. Rev. Biochem. Mol. Biol. 2006;41:269. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kondapalli KC, Kok NM, Dancis A, Stemmler TL. Biochemistry. 2008;47:6917. doi: 10.1021/bi800366d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gakh O, Adamec J, Gacy AM, Twesten RD, Owen WG, Isaya G. Biochemistry. 2002;41:6798. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- 138.Karlberg T, Schagerlof U, Gakh O, Park S, Ryde U, Lindahl M, Leath K, Garman E, Isaya G, Al-Karadaghi S. Structure. 2006;14:1535. doi: 10.1016/j.str.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 139.Aloria K, Schilke B, Andrew A, Craig EA. EMBO Rep. 2004;5:1096. doi: 10.1038/sj.embor.7400272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gerber J, Muhlenhoff U, Lill R. EMBO Rep. 2003;4:906. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yoon T, Cowan JA. J. Am. Chem. Soc. 2003;125:6078. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- 142.Reddehase S, Grumbt B, Neupert W, Hell K. J. Mol. Biol. 2008;385:331. doi: 10.1016/j.jmb.2008.10.088. [DOI] [PubMed] [Google Scholar]
- 143.Hu J, Dong L, Outten CE. J. Biol. Chem. 2008;283:29126. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Khalimonchuk O, Winge DR. Biochim. Biophys. Acta. 2008;1783:618. doi: 10.1016/j.bbamcr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Band L, Bertini I, Ciofi-Baffoni S, Janicka A, Martinelli M, Kozlowski H, Palumaa P. J. Biol. Chem. 2007;283:7912. doi: 10.1074/jbc.M708016200. [DOI] [PubMed] [Google Scholar]
- 146.Palumaa P, Kangur L, Voronova A, Sillard R. Biochem. J. 2004;382:307. doi: 10.1042/BJ20040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Voronova A, Meyer-Klaucke W, Meyer T, Rompel A, Krebs B, Kazantseva J, Sillard R, Palumaa P. Biochem. J. 2007;408:139. doi: 10.1042/BJ20070804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Heaton D, Nittis T, Srinivasan C, Winge DR. J. Biol. Chem. 2000;275:37582. doi: 10.1074/jbc.M006639200. [DOI] [PubMed] [Google Scholar]
- 149.Rentzsch N, Krummeck-WeiB G, Hofer A, Bartuschka A, Ostermann K, Rodel G. Curr. Genet. 1999;35:103. doi: 10.1007/s002940050438. [DOI] [PubMed] [Google Scholar]
- 150.Nittis T, George GN, Winge DR. J. Biol. Chem. 2001;276:42520. doi: 10.1074/jbc.M107077200. [DOI] [PubMed] [Google Scholar]
- 151.Leary SC, Kaufman BA, Pellechia G, Gguercin G-H, Mattman A, Jaksch M, Shoubridge EA. Hum. Mol. Genet. 2004;13:1839. doi: 10.1093/hmg/ddh197. [DOI] [PubMed] [Google Scholar]
- 152.Banci L, Bertini I, Calderone V, Ciofi-Baffoni S, Mangani S, Marinelli M, Palumma P, Wang S. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8595. doi: 10.1073/pnas.0601375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rigby K, Cobine PA, Khalimonchuk O, Winge DR. J. Biol. Chem. 2008;283:15015. doi: 10.1074/jbc.M710072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Banci L, Bertini I, Ciofi-Baffoni S, Hadjiloi T, Martinelli M, Palumaa P. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6803. doi: 10.1073/pnas.0800019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Hakashima R, Yaono R, Yoshikawa S. Science. 1995;269:1069. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 156.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguichi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 157.Williams JC, Sue C, Banting GS, Yang H, Glerum DM, Hendrickson WA, Schon EA. J. Biol. Chem. 2005;280:15202. doi: 10.1074/jbc.M410705200. [DOI] [PubMed] [Google Scholar]
- 158.Abajian C, Rosenzweig AC. J. Biol. Inorg. Chem. 2006;11:459. doi: 10.1007/s00775-006-0096-7. [DOI] [PubMed] [Google Scholar]
- 159.Carr HS, George GN, Winge DR. J. Biol. Chem. 2002;277:31237. doi: 10.1074/jbc.M204854200. [DOI] [PubMed] [Google Scholar]
- 160.Khalimonchuk O, Rodel G. Mitochondrion. 2005;5:363. doi: 10.1016/j.mito.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 161.Beyer WF, Jr., Fridovich I. J. Biol. Chem. 1991;266:303. [PubMed] [Google Scholar]
- 162.Privalle CT, Fridovich I. J. Biol. Chem. 1992;267:9140. [PubMed] [Google Scholar]
- 163.Vance CK, Mller AF. Biochemistry. 1998;37:5518. doi: 10.1021/bi972580r. [DOI] [PubMed] [Google Scholar]
- 164.Ramelot TA, Cort JR, Goldsmith-Fischman S, Kornhaber GJ, Xiao R, Shastry R, Acton TB, Honig B, Montelione GT, Kennedy MA. J. Mol. Biol. 2004;344:567. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 165.Chen OS, Hemenway S, Kaplan J. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12321. doi: 10.1073/pnas.192449599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen OS, Crisp RJ, Valachovic M, Bard M, Winge DR, Kaplan J. J. Biol. Chem. 2004;279:29513. doi: 10.1074/jbc.M403209200. [DOI] [PubMed] [Google Scholar]
- 167.Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR. J. Biol. Chem. 2005;280:10135. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- 168.Leary SC, Cobine PA, Kaufman BA, Guercin GH, Mattman A, Palaty J, Lockitch G, Winge DR, Rustin P, Horvath R, Shoubridge EA. Cell Metab. 2007;5:9. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 169.Eide DJ, Clark S, Nair TM, Gehl M, Gribskov M, Guerinot ML, Harper JF. Genome Biol. 2005;6:R77. doi: 10.1186/gb-2005-6-9-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Coyne HJ, 3rd., Ciofi-Baffoni S, Banci L, Bertini I, Zhang L, George GN, Winge DR. J. Biol. Chem. 2007;282:8926. doi: 10.1074/jbc.M610303200. [DOI] [PubMed] [Google Scholar]
- 171.Marbois B, Gin P, Gulmezian M, Clarke CF. Biochim. Biophys. Acta. 2009;1791:69. doi: 10.1016/j.bbalip.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Singh KK, Desouki MM, Franklin RB, Costello LC. Mol. Cancer. 2006;5:14. doi: 10.1186/1476-4598-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Franklin RB, Ma J, Zou J, Guan Z, Kukoyi BI, Feng P, Costello LC. J. Inorg. Biochem. 2003;96:435. doi: 10.1016/s0162-0134(03)00249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]