SUMMARY
Sensory dendrites depend on cues from their environment to pattern their growth and direct them toward their correct target tissues. Yet, little is known about dendrite-substrate interactions during dendrite morphogenesis. Here, we describe MNR-1/menorin, which is part of the conserved Fam151 family of proteins and is expressed in the skin to control the elaboration of “menorah”-like dendrites of mechanosensory neurons in Caenorhabditis elegans. We provide biochemical and genetic evidence that MNR-1 acts as a contact-dependent or short-range cue in concert with the neural cell adhesion molecule SAX-7/L1CAM in the skin and through the neuronal leucine-rich repeat transmembrane receptor DMA-1 on sensory dendrites. Our data describe an unknown pathway that provides spatial information from the skin substrate to pattern sensory dendrite development nonautonomously.
INTRODUCTION
Neurons receive and process information through often elaborately branched dendritic arbors. Such arbors exist in both the central and peripheral nervous systems, and the molecular mechanisms that govern their development appear to be generally conserved (Parrish et al., 2007; Jan and Jan, 2010). The formation of dendritic arbors is crucial for the ability of neurons to integrate information and sample the environment appropriately (Hall and Treinin, 2011). These arbors may vary greatly in shape and complexity, reflecting the different types of input they receive. Accordingly, loss of dendritic complexity and structure has been linked to a range of neurological conditions, including autism spectrum disorders, schizophrenia, and Alzheimer’s disease (Kaufmann and Moser, 2000; Kulkarni and Firestein, 2012).
Our understanding of dendrite morphogenesis in the sensory system has advanced significantly through the use of model organisms (reviewed in Jan and Jan, 2010). For instance, in Drosophila, the larval sensory dendrite arborization (da) neurons are subdivided into four groups based on progressive arbor complexity. Several different classes of molecules, including transcription factors, cytoskeletal proteins, components of the secretory pathway, microtubule-based motor proteins, and signal transduction molecules, have been shown to orchestrate the elaboration of the arbors (Corty et al., 2009; Jan and Jan, 2010). Two recent reports have revealed a role for basement membrane-neuron interactions mediated by neuronal integrins as a mechanism to confine da neuron arbors to a two-dimensional plane and ensure dendritic self-avoidance (Han et al., 2012; Kim et al., 2012). These observations are consistent with earlier findings in vertebrate models (Moresco et al., 2005; Marrs et al., 2006), thereby highlighting the conservation of molecular mechanisms that regulate dendrite arborization.
In Caenorhabditis elegans, the mechanosensory PVD neurons display stereotypic dendritic arbors that form during postembryonic development (Halevi et al., 2002; Tsalik et al., 2003). On each side of the animal, a primary dendrite grows anteriorly and posteriorly from the lateral PVD cell body and repeatedly branches off perpendicular secondary branches. These branches, in turn, bifurcate where the muscle quadrants meet the lateral hypodermal ridges (skin) (Figures 1A, 1B, and 1D). The tertiary branches grow parallel to the primary branches but avoid overlap with adjacent tertiary neurites, and each branch forms several perpendicular quaternary branches. The resulting structures resemble candelabra and have hence been named menorahs (Figures 1A, 1B, and 1D; Oren-Suissa et al., 2010). PVD dendrite formation is perturbed in mutants of the vesicular transport machinery, such as myosin and dynein (Aguirre-Chen et al., 2011), demonstrating that mechanisms of dendrite patterning are conserved in PVD neurons. Other studies have implicated additional genes in the regulation of PVD morphogenesis, such as the fusogen eff-1 (Oren-Suissa et al., 2010) and transcription factors (e.g., unc-86/BRN3a and mec-3/Lhx-5; Smith et al., 2010, Smith et al., 2013), and suggested a role for the UNC-6/netrin pathway in PVD dendrite self-avoidance (Smith et al., 2012). Lastly, the leucine-rich repeat (LRR) transmembrane receptor dma-1 has been shown to act in PVD dendrites to promote PVD branching (Liu and Shen, 2012).
Figure 1. MNR-1 Is a Conserved Protein that Is Required for Development, but Not Maintenance, of Dendritic Arbors.
(A) Schematic of the PVD neuron in its tissue context. Primary (1°), secondary (2°), tertiary (3°), and quaternary (4°) dendritic branches are indicated. hyp, hypodermis (skin).
(B) Lateral view of an adult wild-type animal. PVD sensory neurons are visualized by a fluorescent reporter (wdIs52). Anterior is to the left, scale bars indicate 50 µm, and an arrowhead denotes the PVD axon that travels toward the ventral nerve cord in all panels.
(C) Lateral view of an adult mnr-1(dz175) mutant animal.
(D and E) Details of (B) and (C) as indicated.
(F and G) Dorsal views of adult wild-type and mnr-1(dz175) mutant animals. FLP sensory neurons are visualized by a fluorescent reporter (muIs32). Arrows indicate defective dendrites.
(H) Timeline of RNAi gene knockdown initiated at different developmental stages.
(I) Lateral images of adult animals after RNAi against the genes indicated, starting at the larval L4 stage.
(J) Quantification of defective animals after RNAi gene knockdown. Data are represented as mean ± SE of proportion. Images for RNAi throughout life and starting at the young adult stage are provided in Figure S2.
(K) Schematic protein structures of MNR-1 in different species. Accession numbers and mutant alleles are indicated in parentheses. The Pfam_B73514 domain may be a nematode-specific protein domain. Signal peptides were predicted using SignalP and transmembrane helices were predicted using TMHHM (http://www.cbs.dtu.dk). Irrespective of the position or probability (shade of gray) of a transmembrane (TM) domain, all members of the Fam151 family with aDUF2181 are predicted to be luminal/extracellular. Most proteins are predictions only and may not be complete. Numbers in parentheses in DUF2181 indicate percent sequence similarity in pairwise comparisons with the MNR-1 DUF2181. The sequence of the DUF2181 shows distant similarity to glycerophosphoryl diester phosphodiesterases (data not shown).
(L) Protein alignments showing the position of the missense mutations dz180 and dz188.
(M) Phylogenetic tree based on DUF2181 sequences from different species using Multalin (http://multalin.toulouse.inra.fr).
See also Figures S1, S2, S3, and S4.
Although a variety of neuron-intrinsic factors that regulate sensory dendrite morphogenesis have been identified, less progress has been made in identifying extraneuronal factors that provide substrate-derived information to orchestrate the growth and branching of dendrites. The best-known examples of target-derived/extrinsic cues that regulate dendritic arbors are neurotrophins. Dendrite arborization of pyramidal neurons is controlled by neurotrophins that are expressed in different cortical layers of the brain (McAllister et al., 1997). Ablation of ectoderm in the chicken wing resulted in defects in the ramification patterns of sensory arbors, suggesting a role for skin-derived cues (Martin et al., 1989; Honig et al., 2005). In zebrafish, extracellular heparan sulfates are required for the correct development of sensory arbors of Rohon-Beard somatosensory neurons (Wang et al., 2012). Together, these findings indicate that the innervation of the skin by somatosensory neurons is governed by target-derived molecules; however, a bona fide skin-derived signaling system that controls arbor formation of somatosensory dendrites has not been identified.
In this study, we report the identification of a factor, which we name MNR-1 (for menorin), that is required for the stereotypic branching pattern of PVD somatosensory arbors in C. elegans. MNR-1 is a member of the conserved, previously unstudied Fam151 family of proteins. We show that MNR-1 is produced in the skin and acts as a contact-dependent or short-range cue to control PVD dendritic branching. Furthermore, we provide genetic and biochemical evidence that MNR-1 acts together with the cell adhesion molecule (CAM) SAX-7/L1CAM in the skin and the LRR transmembrane receptor DMA-1 on PVD. This work reveals a molecular program that provides information from the surrounding skin to shape the development and patterning of sensory dendritic arbors.
RESULTS
MNR-1 Is Required for Development, but Not Maintenance, of PVD Dendrites
To understand dendrite arborization, we conducted a genetic screen for mutants with defects in PVD somatosensory neurons in C. elegans. One class of mutants represented by four recessive alleles affected a previously unstudied gene, W01F3.1 (Figure S1 available online; Table S1; Experimental Procedures). Because of the fully penetrant phenotype characterized by disorganization of the PVD dendritic “menorahs,” we named the gene mnr-1/menorin. The mnr-1 mutants were characterized by disoriented growth of all higher-order PVD branches (secondary to quaternary), with many instances of crossovers, looping, and loss of orthogonality (Figures 1B–1E). Moreover, tiling of menorahs across the primary branch was severely impaired in these mutants, as was self-avoidance of sister branches (Figure 1). In contrast, the axon of PVD did not exhibit obvious guidance defects and we did not detect defects in the viability, fertility, or locomotion of mutant animals (data not shown). Similar defects in dendrite arborization were seen in the two FLP neurons that cover the head region of the worm with a similarly structured mechanosensory arbor, including tangled higher-order branches and loss of characteristic orthogonal dendrites (Figures 1F and 1G). In contrast, a survey of other neuronal classes (branched and unbranched) in C. elegans showed no major defects in mnr-1 mutants, ruling out a global function in nervous-system patterning for mnr-1 (data not shown). Of note, the commissures of D-type motor neurons, about half of which fasciculate with secondary PVD branches (Smith et al., 2010), seemed to be unaffected in mnr-1 mutants (data not shown). Taken together, the defects in mnr-1 mutants appear to be specific for PVD and FLP dendrites and are not generally observed in other neurons, including those that share the same molecular environment as PVD dendrites.
To determine when MNR-1 function is required for PVD dendrite formation, we conducted a series of RNAi experiments starting at different developmental time points. Constitutive knockdown of mnr-1 or mec-3, a LIM homeobox transcription factor with an early role in PVD differentiation (Way and Chalfie, 1989; Smith et al., 2010), resulted in reduced arbor complexity in PVD (Figures 1H–1J, S2A, and S2B). Intriguingly, RNAi beginning at the early L4 larval stage against mnr-1, but not mec-3, resulted in terminal branching defects in PVD (Figures 1H–1J), indicating a role during later larval stages for mnr-1, but not mec-3. In contrast, RNAi against either mnr-1 or mec-3 starting merely 12 hr later at the onset of adulthood failed to result in defects in PVD dendrite structure (Figures 1J, S2C, and S2D). These results show a requirement for MNR-1 during development until arbor stabilization occurs at the onset of adulthood, but suggest that MNR-1 may play a lesser role in PVD dendrite maintenance.
MNR-1 Is a Member of the Conserved Fam151 Family of Proteins
The MNR-1 protein is predicted to contain a signal peptide followed by a domain of unknown function number 2181 (DUF2181) and a nematode-specific domain (Figure 1K). Proteins that contain a DUF2181 define the family of proteins with sequence similarity 151 (Fam151). The DUF2181 is an ancient protein domain that can be identified in simple eukaryotes, such as the recently sequenced choanoflagellate Salpingoeca (Figures 1K–1M, S3, and S4; Ruiz-Trillo et al., 2007). Most Fam151 proteins, including MNR-1, are predicted to contain transmembrane domains at either the N terminus or C terminus. The genomes of nematodes, the choanoflagellate Salpingoeca, and D. melanogaster encode a protein with a single DUF2181, as do lampreys and Ciona, a tunicate. In contrast, vertebrate genomes generally harbor two genes encoding this domain, which have been designated Fam151a and Fam151b (Figures 1K–1M, S3, and S4). The Fam151b family encodes proteins with a single DUF2181, whereas the Fam151a family encodes proteins with two DUF2181s (Figure 1K). The MNR-1 DUF2181 is more closely related to proteins with a single DUF2181 (the Fam151b family) and the N-terminal DUF2181 of proteins with two DUF2181s (the Fam151a family) (Figures 1M, S3, and S4). Thus, the Fam151b form may represent the ancestral form, with gene duplications occurring in the clade leading to jawed vertebrates.
Two of the four mnr-1 mutant alleles identified in our screen represented premature stop codons (Figures 1K–1M). Most of our studies were conducted with an early nonsense allele, mnr-1(dz175), which results in a truncation of the protein after 208 amino acids within the DUF2181 and is likely a complete loss-of-function allele (Figures 1K–1M, and S3). The two remaining alleles encode missense mutations that alter highly conserved residues in the DUF2181 (Figures 1K and 1L). Together, these findings suggest that the DUF2181 is important for MNR-1 function.
MNR-1 Function Promotes Higher-Order Branch Formation
To understand the function of MNR-1, we examined mnr-1 mutants at the late L3 larval stage. We first quantified the numbers of secondary, tertiary, quaternary, and ectopic tertiary branches, as well as their average lengths (Figures 2A–2F). We found strongly decreased numbers of tertiary and quaternary branches, which on average were shorter than the corresponding wild-type branches. On the other hand, we observed an increased number of secondary and ectopic tertiary branches that were shorter than normal secondary and tertiary branches in wild-type animals.
Figure 2. mnr-1 Functions to Establish Stable Tertiary and Quaternary Dendritic Branches.
(A and B) Lateral views of a wild-type (A) and a mnr-1 mutant animal (B) at the late L3 larval stage. Anterior is to the left in all panels and ventral is down. White arrowheads indicate tertiary branches in all panels.
(C–F) Quantification of secondary, tertiary, quaternary, and ectopic tertiary branch numbers and average branch length. Data are represented as mean ± SEM. Statistical comparisons were performed using one-sided ANOVA with the Tukey correction. Statistical significance is indicated (****p < 0.0005). n = 18 wild-type control animals (870 dendritic branches); n = 22 mnr-1(dz175) mutant animals (999 dendritic branches).
(G) Time-lapse still images of an mnr-1(dz175) mutant animal. Green arrowhead: ectopic tertiary branch; white arrowhead: fused/overlapping tertiary branch; yellow arrowhead: retracting tertiary branch. The times after mounting of early L3 animals are shown, and scale bars denote 5 mm in (G)–(I).
(H) Time-lapse still images of a wild-type animal at the times indicated. White arrowheads indicate tertiary branches and red arrowheads indicate quaternary branches.
(I) High-magnification time-lapse still images of an mnr-1(dz175) mutant animal. A green arrowhead indicates an ectopic tertiary branch.
(J and K) Quantification of secondary (J) and ectopic tertiary (K) branches from time-lapse movies for the genotypes indicated. Data are represented as mean ± SEM. Statistical comparisons were performed using Student’s t test (statistical significance is indicated). n = 4 wild-type control animals (264 dendrites); n = 4 mnr-1(dz175) mutant animals (508 dendrites). For sample movies, see Movies S1 and S2.
To better understand this phenotype, we conducted time-lapse experiments (Figures 2G–2K; Movies S1 and S2). We found that secondary branches in mnr-1 mutant animals failed to result consistently in tertiary branches at expected branching points. For example, one branch over the course of 7 hr failed to form stable tertiary branches (Figure 2G, asterisk). This was not due to an inherent inability to form branches because the same secondary branch formed ectopic tertiary branches in rapid succession that retracted again later (Figures 2G and 2I, green arrowheads). Tertiary branches in mnr-1 mutants that formed at presumably the appropriate place sometimes appeared to fuse with neighboring tertiary branches or at least failed to avoid them. Although some of those tertiary branches persisted over the imaging period (white arrowhead), others separated and retracted again (yellow arrowhead). In contrast, wild-type animals formed stable tertiary branches within 25 min (Figure 2G, from 2:05 hr to 2:30 hr, white arrowhead) as well as subsequent quaternary branches (red arrowheads). The increased number of secondary and ectopic tertiary branches in mnr-1 mutants could be the result of faster growth or slower retraction of branches (or both). We quantified growth and retraction rates for secondary and ectopic tertiary branches and found neither changed significantly compared with the rates for wild-type animals (Figures 2J and 2K). We conclude that the function of MNR-1 promotes and stabilizes tertiary and quaternary branch formation.
MNR-1 Functions in a Pathway with SAX-7/L1CAM and the LRR-Transmembrane Receptor DMA-1 to Pattern PVD Dendrites
In a separate screen for genes involved in neuronal branching, we isolated dz156, a nonsense allele of SAX-7/ L1CAM (C.A.D.-B. and H.E.B., unpublished data). The sax-7 locus in C. elegans encodes a transmembrane CAM of the immunoglobulin (Ig) superfamily (Figure 3A) that is homologous to L1CAM. Mutations in L1CAM result in CRASH syndrome in humans (reviewed in Chen and Zhou, 2010). SAX-7 exists in at least two different extracellular isoforms (short [S] and long [L]) with four or six extracellular Ig domains (Figure 3A). We found that sax-7 mutant animals exhibited strong defects in PVD arbor formation. This phenotype was shared by an additional sax-7 null allele (nj48), but not by an allele that removes only the long isoform SAX-7L (nj53; Figures 3A–3C and S5A–S5C). We also did not observe this phenotype in animals that carry a predicted null allele of lad-2, a paralog of sax-7 in C. elegans (Figure S5D; Wang et al., 2008). Thus, the short (but not the long) isoform of SAX-7 is necessary for correct patterning of PVD dendrites in C. elegans.
Figure 3. MNR-1, SAX-7/L1CAM, and DMA-1 Act Genetically in the Same Pathway.
(A) Schematic of the SAX-7/L1CAM with alleles indicated. Arrowheads show proteolytic cleavage sites resulting in C-terminal fragment 1 (CFT1) and CFT2 (Pocock etal., 2008). ANK, ankyrin-binding domain; Ig, immunoglobulindomain; FER, conserved FERM domain;FN(III), fibronectin domain III; PDZ, PDZ domain; TM, transmembrane domain.
(B and C) Lateral images of sax-7 mutant alleles. Anterior is to the left, an arrowhead indicates the PVD axon, and scale bar = 50 µm in all micrographs.
(D) Schematic of the DMA-1 protein with domains indicated. The tm5159 allele is a 672 bp deletion that results in a frameshift in DMA-1 after 100 amino acids.
(E) Lateral image of a dma-1(tm5159) mutant L4 animal.
(F–I) Quantification of branch numbers in mnr-1(dz175), sax-7(dz156), and dma-1(tm5159) single- and double-mutant animals. Data are represented as mean ± SEM. Statistical comparisons were performed using one-sided ANOVA with the Tukey correction and statistical significanceis indicated (*p<0.05, ****p < 0.0005; ns: not significant [p >0.05]). n= 18wild-type controls (870 dendritic branches); n = 22 mnr-1(dz175) mutantanimals (999 dendriticbranches); n= 25sax-7(dz156) mutant animals (898 dendritic branches), n = 26 dma-1(tm5159) mutant animals (431 dendritic branches), n = 33 sax-7(dz156); mnr-1(dz175) mutant animals (1,323 dendritic branches), n = 26 dma-1(tm5159); mnr-1(dz175) mutant animals (325 dendritic branches), n = 19 dma-1(tm5159); sax-7(dz156) mutant animals (312 dendritic branches). Data for wild-type control and mnr-1 animals in (F)–(K) are identical to those in Figure 2 and are shown for comparison only.
(J and K) Quantification of growth and retraction speed in secondary (J) and ectopic tertiary (K) branches from time-lapse movies for the genotypes indicated. Data are represented as mean ± SEM and statistical significance is indicated. n = 4 dma-1(tm5159) mutant animals (248 dendrites); n = 4 sax-7(dz156) mutant animals (319 dendrites). Data for wild-type controls are identical to those in Figures 2J and 2K and are shown for comparisononly. For sample movies, see Movies S1, S2, S3, and S4.
See also Figure S5.
The defects in mnr-1 and sax-7/L1CAM mutants were reminiscent of mutants in the LRR transmembrane receptor DMA-1, which is expressed and acts in PVD neurons (Liu and Shen, 2012; Figures 3D and 3E). This prompted us to quantify and compare PVD dendrite phenotypes in single and double mutants of mnr-1, sax-7, and dma-1 (Figures 3F–3I and S5E–S5H). In the sax-7 mutants, as in the mnr-1 mutants, we found a reduced number of shorter quaternary and tertiary branches and an increased number of secondary and ectopic tertiaries. The branching phenotypes were not further enhanced in a sax-7; mnr-1 double mutant, establishing that sax-7 and mnr-1 act genetically in the same pathway in this process (Figures 3F–3I and S5E–S5H). Time-lapse analyses of sax-7 and dma-1 mutant animals showed that just as in mnr-1 mutants, the different numbers of branches were not a reflection of changes in the speed of neurite growth or retraction (Figures 3J and 3K; Movies S3 and S4). However, in dma-1 null mutant animals, the numbers of secondary, tertiary, and ectopic tertiary branches were decreased compared with the sax-7; mnr-1 double mutant, showing that dma-1 has sax-7- and mnr-1-independent functions. Moreover, branch length was reduced in dma-1 mutants compared with wild-type animals (Figures 3F–3I and S5E–S5H). Overall, dma-1 appeared to be epistatic in double mutants with sax-7 and mnr-1 with regard to both branch number and length, which is consistent with the interpretation that most, if not all, of sax-7 and mnr-1’s function in the formation of higher-order branches requires dma-1.
MNR-1 and SAX-7 Function in the Skin to Pattern PVD Dendrites
PVD menorahs grow out from the primary dendrite toward the dorsal and ventral hypodermal ridges along the body, first over the lateral hypodermal ridge and later sandwiched between the dorsal or ventral body wall muscles and a thin sheet of syncytial hypodermis (Figure 1A; Smith et al., 2010; Oren-Suissa et al., 2010). DMA-1 was previously shown to be expressed and act in PVD neurons (Liu and Shen, 2012). We thus sought to determine where MNR-1 and SAX-7 act to pattern PVD dendrites.
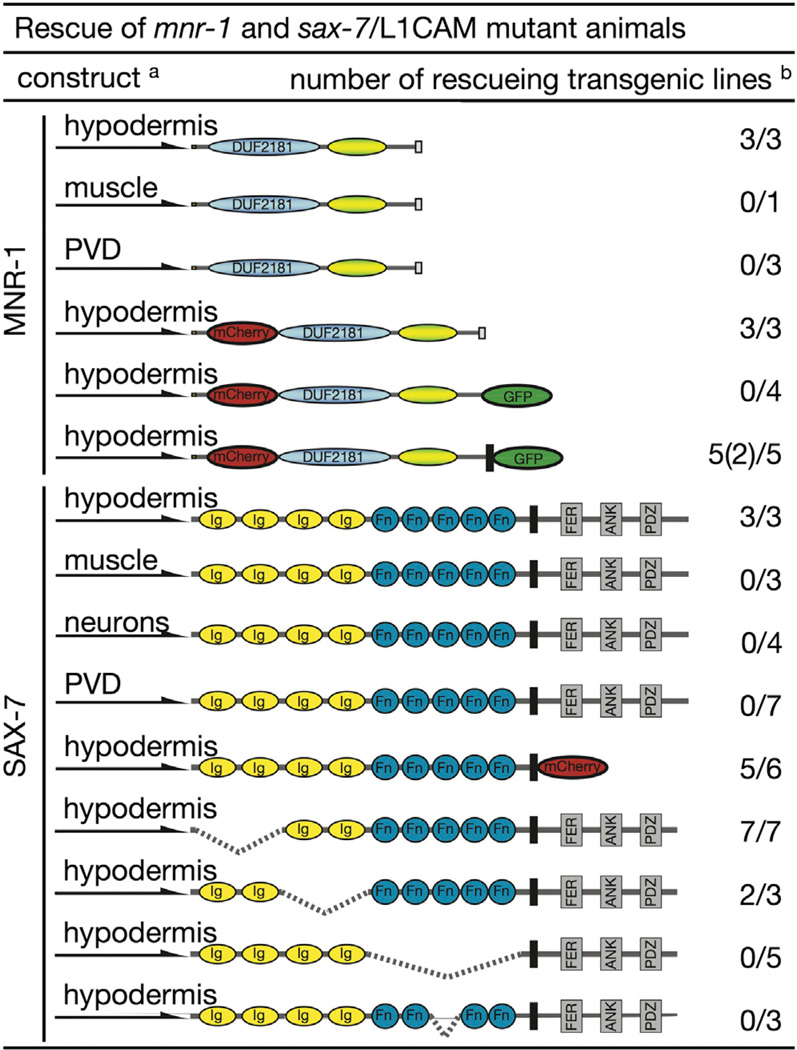
A fosmid-based transcriptional GFP reporter for MNR-1 showed clear GFP expression in the ventral, dorsal, and lateral hypodermal ridges starting at the L3 larval stage (Figures 4A, 4B, S6A, and S6B). In accordance with this pattern, transgenic expression of a mnr-1 cDNA in the hypodermis, but not in muscle or PVD, restored PVD arbor formation in mnr-1(dz175) mutants, including aspects of secondary, tertiary, and quaternary dendrite formation (Figures 4C–4E and 5). This suggested that MNR-1 functions from the skin to pattern PVD dendritic arbors.
Figure 4. Expression Patterns of MNR-1 and SAX-7.
(A) Schematic of the mnr-1∷GFP reporter construct, which should be governed by most, if not all, regulatory elements of the mnr-1 locus (Tursun et al., 2009).
(B) Image of an adult animal, showing expression of the mnr-1∷GFP reporter in the lateral and ventral hypodermal ridges, but not in seam cells. At earlier larval stages, some expression is also seen in a limited number of neurons and the anterior pharyngeal bulb (Figures S6A and S6B). dhr, dorsal hypodermal ridge; lhr, lateral hypodermal ridge; sc, seam cells; vhr, ventral hypodermal ridge; an asterisk indicates the vulva.
(C–E) Images of an mnr-1(dz175) mutant animal transgenically rescued by the hypodermically driven mCherry∷MNR-1 fusion. The fusion is localized throughout the hypodermis in a punctate pattern (D and E). Similar punctate patterns are seen when mCherry∷MNR-1 is expressed in muscle or when a SAX-7∷mCherry fusion or mCherry alone is expressed in the hypodermis (data not shown). An arrow points to coelomocytes and an arrowhead points to the PVD axon. Anterior is to the right and ventral is down. Scale bar indicates 50 µm.
(F) Magnification of coelomocytes, showing the vesicular red fluorescent staining of the mCherry∷MNR-1 fusion (arrow). Scale bar indicates 10 µm.
(G) Optical section of a nonpermeabilized HEK293 cell transiently transfected with mCherry∷MNR-1. The red channel shows fluorescence of the mCherry∷MNR-1 construct, and the green channel shows localization of extracellular mCherry∷MNR-1 as determined by an α-mCherry monoclonal antibody. Partial overlap likely indicates that a part of the fusion is localized intracellularly to the secretory pathway. Similar results were obtained with COS-7 cells (data not shown). DIC, differential interference contrast. Scale bar indicates 20 µm.
(H and I) Ventral images of adult animals of the genotypes indicated, stained with a monoclonal α-SAX-7 antibody. Arrowheads indicate sublateral lines stained with the α-SAX-7 antibody. Scale bar indicates 50 µm.
See also Figure S6.
Figure 5. Summary of mnr-1 and sax-7 Rescue Experiments.
Shown are the constructs, promoters used, and the numberofrescuing lines out of the total number of lines (partial rescuing lines in parentheses). aDeletions are indicated as dashed lines. Color coding and abbreviations as in Figures 2 and 3. bRescue was defined as restoration of secondary, tertiary, and quaternary branches, i.e., the establishment of menorah-like structures. Generally, rescue of sax-7 mutants resulted in more wild-type-like menorahs when compared with rescue of mnr-1 mutants with the mnr-1 cDNA. Partial rescue was defined as lines in which <50% of the animals showed restoration of PVD-like branching patterns. Transgenic animals were always compared with nontransgenic siblings and the number of transgenic animals scored was n = 30–100 per transgenic line.
To determine where within the hypodermis the MNR-1 protein is localized, we expressed a rescuing mCherry∷MNR-1 fusion protein under control of a hypodermal promoter in mnr-1(dz175) null mutants (Figures 4C–4E and 5). We found that rescued tertiary dendrites grew precisely at the boundary of the lateral hypodermal ridge (Figures 4C–4E). The mCherry-tagged MNR-1 was distributed evenly within the hypodermal ridges in a punctate pattern that appeared to be independent of SAX-7 and DMA-1 (Figures 4E, S6C, and S6D). Interestingly, the tagged protein also labeled vesicles within the coelomocytes, a set of scavenger cells that are known to take up and degrade secreted proteins from the extracellular space (Figure 4F; Fares and Greenwald, 2001). Thus, the mCherry∷MNR-1 fusion was extracellular and either diffusible under the experimental conditions or cleaved from the membrane. To provide further support for these findings, we transiently expressed the functional mCherry∷MNR-1 fusion in human embryonic kidney 293 (HEK293) cells and immunohistochemically stained transfected cells with an α-mCherry antibody without prior cell permeabilization as previously described (Fujisawa et al., 2007). We observed staining of the cell perimeter of nonpermeabilized cells expressing mCherry∷MNR-1, but not in nontransfected cells or with an unrelated primary antibody (Figures 4G and S6E), suggesting that the mCherry∷MNR-1 fusion is localized to the outer surface of transfected cells. Moreover, a C-terminal heterologous transmembrane domain in place of the endogenous transmembrane domain of MNR-1 resulted in transgenic rescue of mnr-1 mutants, whereas removal of the transmembrane domain failed to produce significant rescue (Figure 5). Taken together, our results suggest that a membrane-bound form of MNR-1 acts nonautonomously from the skin to shape PVD sensory dendrites.
To determine where SAX-7 acts, we immunohistochemically stained wild-type worms with an antibody specific for SAX-7 (Hadwiger et al., 2010). We found that SAX-7 localized to both the ventral and dorsal nerve cords and thin lines in lateral positions to the nerve cords (Figures 4H and 4I). This staining may be associated with the nervous system (e.g., the sublateral nerve cords) or the hypodermal ridges, and appeared to be independent of either MNR-1 or DMA-1 (Figure S6F). Transgenic rescue experiments showed that expression of the short isoform SAX-7S from a hypodermal promoter rescued PVD dendrite defects in sax-7 mutants (Figure 5). In contrast, expression of SAX-7S from a muscle or panneuronal or PVD-specific promoter failed to rescue the sax-7 mutant phenotype (Figure 5). Moreover, SAX-7S function appeared to be independent of the first or second pair of Ig domains as well as the intracellular domain but seemed to be dependent on the fibronectin domains, specifically the third fibronectin domain (Figure 5). We conclude that SAX-7 acts from the skin to pattern PVD branches and requires at least the third fibronectin domain, but not the intracellular domain, for this function.
MNR-1 Acts as a Short-Range or Contact-Dependent Guidance Cue
Muscle-specific expression of MNR-1 in mnr-1(dz175) mutants did not restore the stereotypical PVD menorahs but instead resulted in a distinct phenotype. In these animals, secondary branches grew aberrant tertiary branches together with highly disorganized quaternary neurites, often in a zigzag pattern but sometimes in parallel to the primary branch, rather than forming the wild-type pattern of orthogonal tertiary and quaternary branches (Figures 6A and 6B). The resulting tangled structures did not resemble menorahs but rather were reminiscent of baobab trees. To further characterize this phenotype, we conducted time-lapse imaging experiments. Surprisingly, expression of MNR-1 in muscle rescued aspects of the mnr-1 mutant phenotype in that it restored the formation of stable tertiary branches and suppressed both excessive branching of secondary and ectopic tertiary branches (Figures 6A, 6B, 6I, and 6J; Movie S5). These findings suggest that MNR-1 can act as a contact-dependent (or short-range) branching cue.
Figure 6. MNR-1 Acts Instructively with SAX-7/L1CAM through the LRR Transmembrane Receptor DMA-1.
(A and B) Lateral image of an adult mnr-1(dz175) mutant animal expressing mnr-1 ectopically in muscle (A) together with a schematic (B). Arrowheads indicate baobab-like branching pattern in all panels, indicated in red in (B). Anterior is to the left and a scale bar indicates 50 µm in all micrographs.
(C–H) Lateral images of different adult animals. The mnr-1(gof) gain-of-function strain is an integrated transgene (dzIs43) that expresses a wild-type mnr-1 cDNA from a myo-3 muscle-specific heterologous promoter (mnr-1(gof): Is[myo-3prom∷mnr-1]). Figure 6G is identical to Figure 3E and shown for comparison only.
(I and J) Quantification of secondary (I) and ectopic tertiary (J) branches from time-lapse movies for the genotypes indicated. Data are represented as mean ± SEM and statistical significance is indicated. n = 4 mnr-1(gof) mutant animals (260 dendrites). For sample movies, see Movies S1, S2, and S5. Data for wild-type control and mnr-1 mutant animals are identical to those in Figures 2J and 2K and shown for comparison only.
(K) Quantification of branching patterns in animals of the genotypes indicated.
(L) Quantification of transgenic rescue with heterologous promoters driving a sax-7S wild-type cDNA in mnr-1(gof); sax-7(dz156) mutant animals as indicated. Nontransgenic siblings showed no effect (data not shown). n = 23–58 for each transgenic line.
(M–P) Images of sax-7; mnr-1 double-mutant animals expressing a sax-7S and mnr-1 cDNA alone or in combination from a seam-cell-specific (grd-10prom) promoter as indicated. A seam-cell-specific tagRFP served as a dominant injection marker. Arrows indicate dendrites being attracted to the seam cells only in animals expressing both SAX-7S and MNR-1. Similar results were obtained with three independent transgenic lines each.
See also Figure S7.
mnr-1 Acts Instructively through sax-7 and dma-1 to Pattern PVD Dendrites
To determine whether mnr-1 acts permissively or instructively, we ectopically expressed MNR-1 in muscle in an otherwise wild-type background. Ectopic expression of MNR-1 resulted in a gain-of-function phenotype with a mix of menorah- and baobab-like dendritic arbors (Figures 6C and 6D), showing that PVD dendrite branches responded to endogenous hypodermis-derived and ectopic muscle-derived MNR-1 in distinct ways. Both normal and aberrant dendritic arbors were dependent on sax-7 and dma-1 (Figures 6C–6H and 6K), demonstrating that MNR-1 plays an instructive branch-promoting role in coordinating the growth of tertiary and quaternary PVD branches and requires SAX-7 and DMA-1 for this process.
Our experiments suggest that MNR-1 and SAX-7 are necessary in the hypodermis to control development of PVD menorahs. To determine whether coexpression of SAX-7 and MNR-1 in other cells is sufficient to induce branched PVD dendrites, we exploited the mnr-1 gain-of-function phenotype. Using hypodermal- and muscle-specific promoters, we transgenically expressed SAX-7 in the mnr-1(gof); sax-7(null) strain, in which MNR-1 is ectopically expressed in the muscle in addition to its endogenous hypodermal expression. We found that expression of SAX-7 in the hypodermis resulted in a mix of menorah and baobab structures, i.e., it restored the mnr-1(gof) phenotype or, in other words, rescued the sax-7 mutant phenotype. In contrast, expression of SAX-7 in muscle failed to restore either baobab- or menorah-like branching patterns. These results show that SAX-7 and MNR-1 are not sufficient in muscle to attract and shape tertiary and quaternary dendrites, but that MNR-1 from the muscle can act in trans with SAX-7 in the hypodermis. In contrast to the situation in muscle, combined expression of MNR-1 and SAX-7 in seam cells (another hypodermal tissue) in sax-7; mnr-1 double null mutants was sufficient to attract and induce branches on this hypodermal tissue (Figures 6M–6P). Yet, when individually expressed in seam cells, MNR-1 could attract branches to the seam cells only when endogenous SAX-7 was present (Figure S7). We conclude that MNR-1’s function in the seam cells is also SAX-7 dependent and that additional cell-specific factors that act in concert with SAX-7 and MNR-1 must exist in the hypodermis for normal patterning of sensory dendrites.
MNR-1 Physically Interacts with SAX-7
To determine how MNR-1, SAX-7, and DMA-1 interact,we transfected one set of HEK293 cells with tagged versions of MNR-1 and SAX-7 alone or in combination, and another set with a tagged version of DMA-1. Differently transfected cells were cocultured in a 1:1 ratio (MNR-1/SAX-7:DMA-1) for another day, lysed, and subjected to coimmunoprecipitation experiments. We found that MNR-1 could be precipitated with an antibody against SAX-7 and vice versa, suggesting that SAX-7 and MNR-1 are part of a complex (Figure 7A). This interaction appeared stronger in the presence of DMA-1, raising the possibility of a tripartite complex. Moreover, MNR-1 appeared to precipitate only full-length SAX-7S, and not the shorter, proteolytically generated forms that comprise the transmembrane and intracellular parts of SAX-7 (Figure S7F; Pocock et al., 2008). Collectively, these findings suggest that MNR-1 forms a complex with SAX-7 and (possibly) DMA-1, and that the extracellular domains of SAX-7 are important for the formation of this complex (Figure 7B).
Figure 7. MNR-1 Forms a Complex with SAX-7.
(A) Input (total cell lysates) and immunoprecipitates (IP) from cocultured transfected cells and detected with antibodies in western blots (WB) as indicated. Similar amounts of input were used in each lane. A control IP for mCherry∷MNR-1 is shown in Figure S7E. When mCherry alone was transfected as a control, SAX-7S was not precipitated (data not shown). Note that mCherry∷MNR-1 and SAX-7∷V5 are functional in vivo (Figures 4 and 5; data not shown). kDa, kilodalton in all panels.
(B) Schematic of cellular interactions. Length of proteins is drawn to scale. The position of immuno tags is indicated.
DISCUSSION
We have shown here that MNR-1/menorin plays a role in patterning the dendrites, but not the axons, of PVD somatosensory neurons in C. elegans. MNR-1/menorin is an extracellular protein that forms a complex with the Ig fibronectin III domain containing SAX-7/L1CAM, and acts as a contact-dependent, short-range cue nonautonomously from the skin through the LRR-containing transmembrane receptor DMA-1 on PVD neurons.
MNR-1/menorin and SAX-7/L1CAM Pattern PVD from the Skin through DMA-1
L1CAM has previously been shown to be required for dendrite development (Demyanenko et al., 1999; Yamamoto et al., 2006). It is known to function through both homophilic and heterophilic interactions with other proteins in a variety of signaling pathways, such as semaphorin, integrin, and fibroblast growth factor receptor signaling pathways (Schmid and Maness, 2008). We find that both SAX-7 and MNR-1 act in the hypodermis to pattern PVD dendrites. SAX-7 is localized to a sublateral line that could be adjacent to where tertiary branches form, whereas MNR-1 does not display comparable subcellular localization (Dong et al., 2013 [this issue of Cell]). Thus, SAX-7 could function as a scaffolding protein that provides spatial specificity, with binding specificity being provided by a cofactor. MNR-1 could be such a cofactor and provide specificity by allowing formation of a tripartite complex with the DMA-1 receptor on PVD. Several observations argue in favor of this model. First, SAX-7 binds MNR-1 in vitro and this interaction may be stronger in the presence of DMA-1. Second, mnr-1 and sax-7 act genetically in the same pathway. Third, the dma-1 mutation is epistatic to mnr-1 and sax-7. Lastly, wherever it was tested by gain-of-function experiments in vivo, mnr-1 required both sax-7 and dma-1 to exert its function (Figures 6 and S7). An important goal for the future will be to determine how SAX-7 is spatially localized to a distinct line and whether such localization is mediated by factors in the hypodermis or elsewhere.
We found that MNR-1 and SAX-7 act instructively on PVD branches when coexpressed in cis in hypodermal tissues, but not in muscle (Figure 6). The inability of MNR-1 and SAX-7 to function in cis in muscle may indicate either additional permissive factors in the hypodermis or nonpermissive factors in muscle. Intriguingly, the muscle gain-of-function analysis showed that, in certain experimentally induced cellular contexts, MNR-1 could also function in trans to SAX-7, albeit possibly to a lesser degree. For example, some aspects of dendrite arborization could be restored when MNR-1 was expressed in muscle and SAX-7 was expressed in the hypodermis, such as suppression of supernumerary secondary and ectopic tertiary branches in baobab-like dendrites (Figures 6A, 6B, 6I, and 6J). However, only coexpression in the hypodermis resulted in the formation of correct tertiary and quaternary branches, i.e., menorah-like structures. We propose that additional factors in the hypodermis are required for correct PVD patterning.
Stepwise Control of PVD Menorah Formation
The development of a PVD menorah is a highly stereotyped process that occurs in discrete steps (Smith et al., 2010). Our experiments showed reduced tertiary and quaternary branching and increased secondary and ectopic tertiary in mnr-1 and sax-7 mutants (Figure 2). One possible explanation for these seemingly contrary findings is that different cofactors at successive developmental stages account for distinct functions of mnr-1 and sax-7 during the steps of arbor development. Alternatively, the primary function of MNR-1 and SAX-7 could be to establish a stable tertiary branch. This could allow the formation of higher-order branches and in turn signal to suppress further branching in lower-order dendrites. The latter scenario is consistent with the following observations. First, the intracellular domain of SAX-7 is dispensable for SAX-7 function in PVD development (Figure 5), raising the possibility that signaling downstream of DMA-1 in PVD may be required. Second, expression of MNR-1 in muscle of mnr-1 mutant animals suppresses supernumerary secondary and tertiary branching. In other words, the formation of stable tertiary dendrites (even as part of disorganized baobab-like dendritic trees) is sufficient to suppress extra secondary branching. Third, it is known that, in wild-type animals, the dynamic initiation of secondary branches is suppressed in the vicinity of secondary branches that have formed stable tertiary branches (Smith et al., 2010). A feedback mechanism in PVD neurons that begins with the formation of stable tertiary branches could provide an explanation for our observations.
The DUF2181 Menorin Domain
The conserved DUF2181 is an ancient protein domain of about 250 amino acids that is already present in several protists, including the choanoflagellate Salpingoeca (Figure 1; Ruiz-Trillo et al., 2007). Choanoflagellates are colony-forming unicellular eukaryotes that serve as models for the evolution of multicellularity. Several classes of genes that are important for intercellular communication and are considered hallmarks of metazoa (e.g., genes encoding CAMs such as cadherins) first appear in this group of organisms (Abedin and King, 2008). It is thus plausible that the DUF2181 also first arose in these organisms, perhaps as an important component of cell-cell communication. Elaborate branching patterns such as those observed in the PVD dendrites arguably require sophisticated cellular interactions. In this context, it is interesting to note that the peripheral endings of low-threshold mechanosensory receptors with their orthogonal lanceolate endings in the hairy skin of mice (Li et al., 2011) display a striking similarity to the menorah-like dendritic arbors of PVD. Given the intriguing role that MNR-1 and SAX-7/L1CAM play in establishing the stereotypical branching patterns of PVD somatosensory neurons, future studies will have to establish whether similar functions of Fam151 proteins are conserved in other organisms.
EXPERIMENTAL PROCEDURES
Strains and Genetics
N2 Bristol was used as the wild-type strain and worms were grown under standard conditions at 20°C. The mutant alleles used in this work were mnr-1(dz175), mnr-1(dz179), mnr-1(dz180), mnr-1(dz188), sax-7(nj48), sax-7(nj53), sax-7(dz156), lad-2(tm3056), and dma-1(tm5159). Transgenesis was performed using standard procedures. For details and a complete list of strains, see the Extended Experimental Procedures.
Identification of mnr-1 and Molecular Cloning
In a mutant screen for PVD defects (using either ynIs30 or otIs138 as PVD reporters), we isolated three alleles of mnr-1 (dz175, dz179, and dz180). An additional allele (dz188) was identified by complementation tests. Using a one-step SNP mapping and whole-genome sequencing method (Doitsidou et al., 2010), we identified the causative molecular lesion in two mutant alleles with identical phenotypes, dz175 and dz179. Both dz175 and dz179 carried different early stop codons in the same gene, mnr-1 (W01F3.1), within the mapped region. Two additional alleles, dz180 and dz188, failed to complement dz175 for the PVD phenotype and contained missense mutations in mnr-1. All plasmid constructs and molecular biology were performed according to standard procedures. For details, see Figure S1 and the Extended Experimental Procedures.
RNAi
For constitutive RNAi, L4 hermaphrodites were grown for 1 to 2 days and allowed to lay a brood, which was analyzed after 2 to 3 days as young adults. For late larval RNAi, L3 hermaphrodites were placed on RNAi-seeded plates and analyzed 2 to 3 days later as adults. To exclude maintenance defects, 1-day-old adults were placed on RNAi-seeded plates and scored 2 days later for PVD defects. For details, see the Extended Experimental Procedures.
Imaging and Quantification of Branching
For quantification of branching, synchronized starved L1s were transferred to seeded plates and allowed to grow for 30 hr. At 30 hr (corresponding to mid- to late L3) larvae were immobilized in 1–5 mM levamisole (Sigma). Z stacks and maximum-intensity projections were produced using a Zeiss Axioimager Z1 Apotome. All branches within 100 µm of the primary branch anterior to the cell body were traced, measured, and classified into primary, secondary, tertiary, quaternary, and ectopic tertiary branches using the NeuronJ plugin of the ImageJ 1.46r software. Statistical comparisons were conducted using one-sided ANOVA with the Tukey correction (Prism [GraphPad Software]).
Time-lapse imaging of animals at the L3 stage (by gonadal development) was conducted using an inverted Nikon TE2000-S microscope equipped with a Perkin-Elmer UltraVIEW spinning disk unit. Volocity software (version 6.2.1) was used to collect the raw files, and processing and video editing were carried out using the ImageJ 1.46r software. Statistical comparisons were made using Student’s t test (Prism [GraphPad Software]). For details, see the Extended Experimental Procedures.
Immunohistochemistry
Worms were freeze-fractured and fixed by methanol/acetone (Duerr, 2006). Immunodetection was accomplished with the use of a primary monoclonal α-SAX-7 antibody (1:500, Developmental Studies Hybridoma Bank[DSHB]; Hadwiger et al., 2010) followed by a secondary antibody (1:500, Alexa Fluor 555 donkey a-mouse IgG (H+L), A-31570; Life Technologies).
Cell Culture Experiments with MNR-1, SAX-7, and DMA-1
For cellular localization studies of MNR-1, HEK293T cells were transfected with pcDNA3.1∷mCherry∷mnr-1 using Lipofectamine 2000 (Life Technologies). After 24 hr, the cells were fixed, either permeabilized or not (Fujisawa et al., 2007), stained with an α-mCherry primary antibody (1:500, ab125096; Abcam) and developed with a secondary Alexa Fluor 488 goat α-mouse IgG (1:500, A-11001; Life Technologies).
For coimmunoprecipitation experiments, HEK293T cells were transfected separately with pcDNA3.1∷3XHA∷dma-1 or the ligand vectors pcDNA3.1∷ mCherry∷mnr-1 and pcDNA3.1∷sax-7S∷V5 (a gift from Dr. O. Hobert) alone or in combination. Twenty-four hours posttransfection, the cells were trypsinized, mixed in a 1:1 ratio, and cocultured for another day. Protein lysates were subjected to immunoprecipitation with the following antibodies: rat monoclonal α-HA (Roche), mouse monoclonal α-V5 (Life Technologies), and mouse monoclonal α-mCherry (Abcam). Western blots were probed using the same antibodies, except for mCherry∷MNR-1, which was identified using a rabbit polyclonal α-mCherry.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kang Shen (Stanford University) for communicating results prior to publication; N. Baker, S. Emmons, A. Melé ndez, R. Townley, and members of the Bülow laboratory for discussions and comments on the manuscript; Ji Sze, O. Hobert, and J. Culotti for DNA clones; the Caenorhabditis Genetics Center,L.Chen, and S. Mitani for strains; C. Rubin for the α-mCherry antibody; M. Akabas for the use of his cell culture facility; B. Calder and A. Fiser for advice on bioinformatic analyses; M. Lázaro-Peña for injections; S. Bhatt and B. Gonzalez for constructing strains; and C. Crocker for artwork. This work was funded in part by the NIH (T32GM007288 and F31HD066967 to C.A.D.-B., T32GM007491 and F31 NS076243 to M.A., T32GM07491 to E.T., 5R01HD055380 and R21NS081505 to H.E.B., and P30HD071593 and P30CA013330 to Albert Einstein College of Medicine). N.R. is the recipient of a Fulbright fellowship. H.E.B. is an Alfred P. Sloan fellow.
Footnotes
SUPPLEMENTAL INFORMATIONM
Supplemental Information includes Extended Experimental Procedures, seven figures, one table, and five movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.08.058.
REFERENCES
- Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–948. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- Aguirre-Chen C, Bülow HE, Kaprielian Z. C. elegans bicd-1, homolog of the Drosophila dynein accessory factor Bicaudal D, regulates the branching of PVD sensory neuron dendrites. Development. 2011;138:507–518. doi: 10.1242/dev.060939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhou S. “CRASH”ing with the worm: insights into L1CAM functions and mechanisms. Dev. Dyn. 2010;239:1490–1501. doi: 10.1002/dvdy.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development. 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko GP, Tsai AY, Maness PF. Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. J. Neurosci. 1999;19:4907–4920. doi: 10.1523/JNEUROSCI.19-12-04907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Poole RJ, Sarin S, Bigelow H, Hobert O. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE. 2010;5:e15435. doi: 10.1371/journal.pone.0015435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Liu OW, Howell AS, Shen K. An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell. 2013;155(his issue):296–307. doi: 10.1016/j.cell.2013.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr JS. Immunohistochemistry. WormBook. 2006;19:1–61. doi: 10.1895/wormbook.1.105.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H, Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa K, Wrana JL, Culotti JG. The slit receptor EVA-1 coactivates a SAX-3/Robo mediated guidance signal in C. elegans. Science. 2007;317:1934–1938. doi: 10.1126/science.1144874. [DOI] [PubMed] [Google Scholar]
- Hadwiger G, Dour S, Arur S, Fox P, Nonet ML. A monoclonal antibody toolkit for C. elegans. PLoS ONE. 2010;5:e10161. doi: 10.1371/journal.pone.0010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevi S, McKay J, Palfreyman M, Yassin L, Eshel M, Jorgensen E, Treinin M. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. EMBO J. 2002;21:1012–1020. doi: 10.1093/emboj/21.5.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Treinin M. How does morphology relate to function in sensory arbors? Trends Neurosci. 2011;34:443–451. doi: 10.1016/j.tins.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Wang D, Soba P, Zhu S, Lin X, Jan LY, Jan YN. Integrins regulate repulsion-mediated dendritic patterning of Drosophila sensory neurons by restricting dendrites in a 2D space. Neuron. 2012;73:64–78. doi: 10.1016/j.neuron.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig MG, Camilli SJ, Surineni KM, Knight BK, Hardin HM. The contributions of BMP4, positive guidance cues, and repulsive molecules to cutaneous nerve formation in the chick hindlimb. Dev. Biol. 2005;282:257–273. doi: 10.1016/j.ydbio.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat. Rev.Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb. Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kim ME, Shrestha BR, Blazeski R, Mason CA, Grueber WB. Integrins establish dendrite-substrate relationships that promote dendritic self-avoidance and patterning in Drosophila sensory neurons. Neuron. 2012;73:79–91. doi: 10.1016/j.neuron.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sun L, Gabel CV, Fang-Yen C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS ONE. 2013;8:e53419. doi: 10.1371/journal.pone.0053419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni VA, Firestein BL. The dendritic tree and brain disorders. Mol. Cell. Neurosci. 2012;50:10–20. doi: 10.1016/j.mcn.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, Jankowski MP, Luo W, Heintz N, Koerber HR, et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Shen K. The transmembrane LRR protein DMA-1 promotes dendrite branching and growth in C. elegans. Nat. Neurosci. 2012;15:57–63. doi: 10.1038/nn.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs GS, Honda T, Fuller L, Thangavel R, Balsamo J, Lilien J, Dailey ME, Arregui C. Dendritic arbors of developing retinal ganglion cells are stabilized by beta 1-integrins. Mol. Cell. Neurosci. 2006;32:230–241. doi: 10.1016/j.mcn.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Martin P, Khan A, Lewis J. Cutaneous nerves of the embryonic chick wing do not develop in regions denuded of ectoderm. Development. 1989;106:335–346. doi: 10.1242/dev.106.2.335. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Moresco EM, Donaldson S, Williamson A, Koleske AJ. Integrin-mediated dendrite branch maintenance requires Abelson (Abl) family kinases. J. Neurosci. 2005;25:6105–6118. doi: 10.1523/JNEUROSCI.1432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren-Suissa M, Hall DH, Treinin M, Shemer G, Podbilewicz B. The fusogen EFF-1 controls sculpting of mechanosensory dendrites. Science. 2010;328:1285–1288. doi: 10.1126/science.1189095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu. Rev. Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Pocock R, Bénard CY, Shapiro L, Hobert O. Functional dissection of the C. elegans cell adhesion molecule SAX-7, a homologue of human L1. Mol. Cell. Neurosci. 2008;37:56–68. doi: 10.1016/j.mcn.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Burger G, Holland PWH, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 2007;23:113–118. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Maness PF. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr. Opin. Neurobiol. 2008;18:245–250. doi: 10.1016/j.conb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Watson JD, Spencer WC, O’Brien T, Cha B, Albeg A, Treinin M, Miller DM., 3rd. Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans. Dev. Biol. 2010;345:18–33. doi: 10.1016/j.ydbio.2010.05.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Watson JD, VanHoven MK, Colón-Ramos DA, Miller DM., 3rd. Netrin (UNC-6) mediates dendritic self-avoidance. Nat. Neurosci. 2012;15:731–737. doi: 10.1038/nn.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, O’Brien T, Chatzigeorgiou M, Spencer WC, Feingold-Link E, Husson SJ, Hori S, Mitani S, Gottschalk A, Schafer WR, et al. Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron. 2013;79:266–280. doi: 10.1016/j.neuron.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsalik EL, Niacaris T, Wenick AS, Pau K, Avery L, Hobert O. LIM homeobox gene-dependent expression of biogenic amine receptors in restricted regions of the C. elegans nervous system. Dev. Biol. 2003;263:81–102. doi: 10.1016/s0012-1606(03)00447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang W, Cheever T, Schwarz V, Opperman K, Hutter H, Koepp D, Chen L. The C. elegans L1CAM homologue LAD-2 functions as a coreceptor in MAB-20/Sema2 mediated axon guidance. J. Cell Biol. 2008;180:233–246. doi: 10.1083/jcb.200704178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wolfson SN, Gharib A, Sagasti A. LAR receptor tyrosine phosphatases and HSPGs guide peripheral sensory axons to the skin. Curr. Biol. 2012;22:373–382. doi: 10.1016/j.cub.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JC, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes. Dev. 1989;3(12A):1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Ueda R, Takahashi K, Saigo K, Uemura T. Control of axonal sprouting and dendrite branching by the Nrg-Ank complex at the neuron-glia interface. Curr. Biol. 2006;16:1678–1683. doi: 10.1016/j.cub.2006.06.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.