Abstract
Background
Radiation in some plant groups has occurred on islands and due to the characteristic rapid pace of phenotypic evolution, standard molecular markers often provide insufficient variation for phylogenetic reconstruction. To resolve relationships within a clade of 21 closely related New Caledonian Diospyros species and evaluate species boundaries we analysed genome-wide DNA variation via amplified fragment length polymorphisms (AFLP).
Results
A neighbour-joining (NJ) dendrogram based on Dice distances shows all species except D. minimifolia, D. parviflora and D. vieillardii to form unique clusters of genetically similar accessions. However, there was little variation between these species clusters, resulting in unresolved species relationships and a star-like general NJ topology. Correspondingly, analyses of molecular variance showed more variation within species than between them. A Bayesian analysis with BEAST produced a similar result. Another Bayesian method, this time a clustering method, Structure, demonstrated the presence of two groups, highly congruent with those observed in a principal coordinate analysis (PCO). Molecular divergence between the two groups is low and does not correspond to any hypothesised taxonomic, ecological or geographical patterns.
Conclusions
We hypothesise that such a pattern could have been produced by rapid and complex evolution involving a widespread progenitor for which an initial split into two groups was followed by subsequent fragmentation into many diverging populations, which was followed by range expansion of then divergent entities. Overall, this process resulted in an opportunistic pattern of phenotypic diversification. The time since divergence was probably insufficient for some species to become genetically well-differentiated, resulting in progenitor/derivative relationships being exhibited in a few cases. In other cases, our analyses may have revealed evidence for the existence of cryptic species, for which more study of morphology and ecology are now required.
Keywords: Cryptic species, Island flora, Morphological diversification, Progenitor/derivative relationships, Species radiation, Woody plants
Background
Island floras are often characterized by high levels of endemism and groups of closely related but morphological and ecological divergent species that are mostly the result of single colonisation events followed by radiation e.g. [1,2]. New Caledonia was cited as one of the 34 biodiversity hotspots recognized by Conservation International [3,4]. Nearly 75% of the native flora is endemic [5], which is the fourth highest for an island [6]. While the continental part of New Caledonia (mainly Grande Terre) was entirely submerged during the Eocene (until 37 mya), a thick layer of heavy-metal-rich oceanic mantle accumulated [7]. Today, around one-third of the main island, Grande Terre, is still overlaid with ultramafic substrates. Generally, Grande Terre is a substrate mosaic [8], which is cited as one reason for the high level of endemism found there e.g. [9]. The climate in New Caledonia ranges from tropical to subtropical, and the main island is split by a mountain range into a humid eastern and a dry western part with prevailing winds and rain coming from the south-east. Taking climatic and geological factors together, Grande Terre has a wide range of environmental diversity. The main vegetation types in New Caledonia are evergreen humid forests, maquis, dry forests, littoral vegetation, and savannah [10].
One plant group that has taken advantage of many available habitats on New Caledonia is Diospyros, which is the largest genus (> 500 species in its broad circumscription [11]) of Ebenaceae, a pantropical family of woody plants. In New Caledonia Diospyros species range from sea level up to ca. 1250 m (the highest point New Caledonia is 1628 m), and species are found in all vegetation types except mangroves, with several species co-occurring in micro-sympatry (Table 1).
Table 1.
Occurrence of Diospyros species in different habitats in New Caledonia
| |
|
Substrate |
||||
|---|---|---|---|---|---|---|
| Limestone | Serpentine | Schist | Ultramafic rock | Volcanic rock | ||
| Vegetation |
Humid mountain forest |
|
|
D. parviflora, D. trisulca |
|
D. flavocarpa, D. labillardierei |
| Humid low land forest |
|
|
|
D. glans, D. pancheri, D. parviflora, D. umbrosa |
D. umbrosa |
|
| Mesophyll forest |
D. minimifolia, D. pustuala, D. tridentata |
|
|
D. erudita |
D. cherrieri, D. erudita, D. minimifolia, D. perplexa, D. pustulata |
|
| Maquis |
|
D. revolutissima |
|
D. vieillardii |
|
|
| Littoral forest | D. calciphila, D. inexplorata | D. impolita | ||||
Habitats are grouped according to vegetation type and substrate. Note that several species are co-occurring and that a few species are found in several habitats.
Diospyros colonised New Caledonia via long-distance dispersal at least four times [12]. In previous studies based on low-copy nuclear and/or multiple plastid markers [12,13], it was possible to resolve phylogenetic relationships for the majority of Diospyros species, except for one group of endemics from New Caledonia. Of the 31 New Caledonian Diospyros species, 24 belong to this clade of closely related endemics. In previous analyses, this strongly supported group is related to species found on islands throughout the Indian and Pacific Oceans as far east as Hawai’i [12,13]. However, due to extremely low levels of sequence divergence, it was not possible to tease apart relationships between these species (they formed a hard polytomy in most individual trees, and there was little informative variation that permitted clustering of pairs or groups of species). Most of these closely related species are morphologically and ecologically clearly differentiated (for examples see [13]), and several species are narrow endemics restricted to small areas.
Amplified fragment length polymorphism (AFLP; [14]) is a fingerprinting technique that has proven to be useful for revealing phylogenetic relationship among closely related taxa (e.g. Hypochaeris, [15]; Lactuca, [16]; Phylica, [17]; Trollius, [18]; Ranunculus alpestris, [19]; Puya, [20,21]; Araucaria, [22]). In contrast to standard phylogenetic markers, AFLP variation is spread across the whole genome, spanning both coding and non-coding DNA regions and may therefore be more representative of overall genetic patterns present as well as being highly informative for phylogenetic analyses at the low phylogenetic level [23,24]. Compared to other fingerprinting techniques AFLP shows increased reproducibility and does not require any prior knowledge of the analysed genomes. However, there are some detrimental issues to consider when working with AFLP data; these include potential non-homology and non-independence of fragments, asymmetry in the probability of loss/gain of fragments, and problems in distinguishing heterozygote from homozygote bands e.g. [23,25]. Despite these difficulties, several authors have used AFLPs to reveal phylogenetic relationships corroborated by analyses of other types of data, especially for species that have diverged recently or radiated within a short period of time e.g. [15,17,23,26].
In this study we focus on this group of closely related species of Diospyros endemic to New Caledonia (Figure 1). Our aim was to clarify species boundaries as well as phylogenetic relationships between these New Caledonian Diospyros species. Integrated in a broader context, the outcome of our research should help us better understand the factors behind and mechanisms of speciation and radiation on islands.
Figure 1.
Bayesian maximum clade credibility tree of New Caledonian Diospyros species based on plastid and nuclear DNA data (taken from Turner et al.[13]). Bold branches are supported (> 70% bootstrap and Bayesian posterior probability). Accessions in blue correspond to the white group found in Structure and PCO, green ones to the grey group (light blue/green accessions included in current data set, dark blue/green accessions failed in current analysis but colour indicates the group to which they most probably belong), accessions in black are not included in the present study.
Results
After excluding 186 replicates the final matrix used for analyses contained 192 individuals and 792 fragments. The AFLP profiles showed good reproducibility with a mean error-rate of 2.4% across all replicated samples. Because the focus of this study was on the phylogenetic relationships between species and species limits rather than intra-specific population genetics, we are presenting and discussing mostly the results of inter-specific relationships. We are presenting here only unrooted trees due to the low resolution of their backbone. We analysed the data using neighbour-joining (NJ) dendrograms and principal coordinate analysis (PCO) with different distance methods, and in both cases the Dice distance gave the highest resolution of relationships between species.
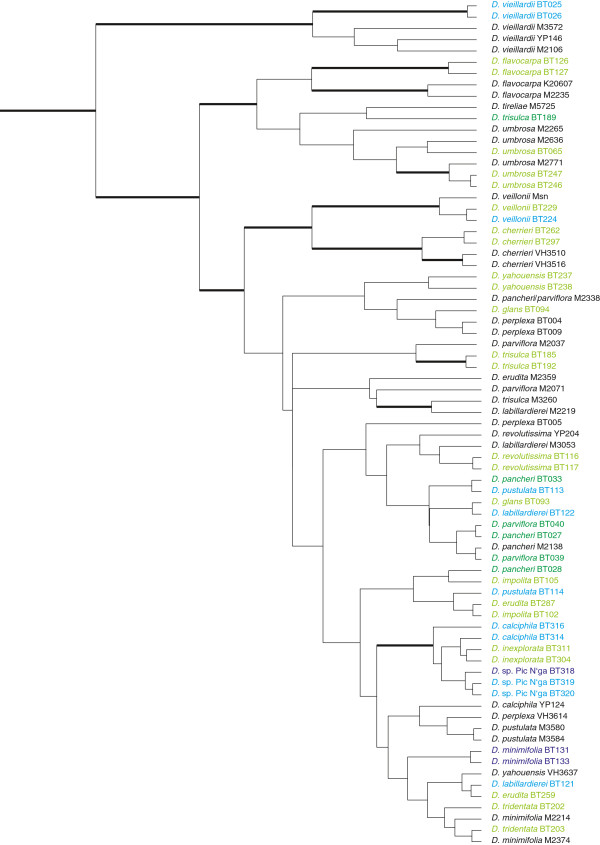
The NJ analysis resulted in a star-like dendrogram with a backbone of short branches lacking bootstrap support greater than 75%. All species except D. minimifolia, D. parviflora and D. vieillardii form single clusters in the NJ tree (Figure 2A). However, only eight (D. calciphila, D. cherrieri, D. inexplorata, D. impolita, D. pustulata, D. trisulca, D. umbrosa and D. yahouensis) of the 21 included species form clusters with bootstrap higher than 80%. The Bayesian inference (BI) produces a similar result. All species except D. labillardierei, D. minimifolia, D. pancheri and D. parviflora form single clusters in the BI tree (Figure 2B). Apart from D. flavocarpa, D. revolutissima, D. tridentata and D. vieillardii, all clustered species have high (> 0.95) posterior probabilities.
Figure 2.
Phylogenetic dendrograms inferred from the data collected in this study. Each species is shown in a different colour. Colours were selected randomly and do not indicate any grouping. A: Neighbour joining dendrogram based on Dice distances. Black dots indicate nodes with > 80% bootstrap support. B: Bayesian maximum clade credibility dendrogram. Black dots indicate nodes with > 0.95 Bayesian posterior probability. Picture credits: D. calciphila: H. Benoît, http://www.endemia.nc; D. cherrieri: C. Chambrey; D. erudita, D. pancheri, D. pustulata, D. umbrosa, D. vieillardii: D. & I. Létocart, http://www.endemia.nc; D. flavocarpa, D. minimifolia, D. revolutissima, D. sp. Pic N’ga: J. Munzinger; D. glans, D. parviflora: J.-L. Ruiz, http://www.endemia.nc; D. impolita: J. Barrault, http://www.endemia.nc; D. labillardierei: B. Turner; D. perplexa, D. yahouensis: V. Hequet; D. veillonii: R. Amice, http://www.endemia.nc.
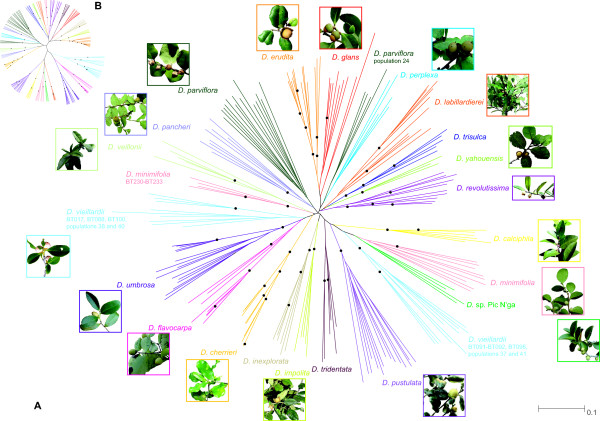
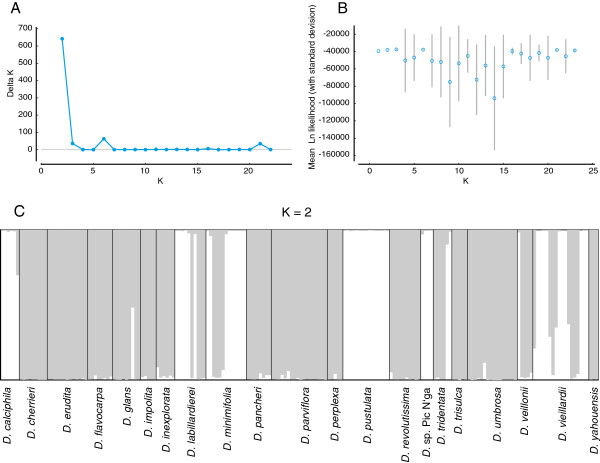
PCO separated accessions into two main groups (hereafter named “white” and “grey”) that can be subdivided into six subgroups (Figure 3). Within the “white” group (defined in the Structure results below) subgroup one includes D. vieillardii (individuals indicated by squares in Figure 3), subgroup two D. calciphila (triangles) and subgroup three the rest of the individuals from this group (circles). In the “grey” group (more extensively described in the Structure results below) subgroup four included D. flavocarpa, D. umbrosa and D. vieillardii (indicated by squares in Figure 3), subgroup five D. erudita and D. glans (triangles) and subgroup six the remaining individuals (circles). A PCO of populations (not shown) based on the pair-wise F ST distances obtained from the AMOVA resulted in similar groups and subgroups of populations as those obtained from the individual-based PCO. Structure analysis gave the highest value of ∆K for K = 2 plus few other suboptimal K values (Figure 4A and B). However the latter contained clusters with negligible membership (“empty” clusters). Both K = 3 and K = 6 resulted in three visible clusters, with one cluster being only found in significantly admixed samples (Additional file 1). Visualisation of K =16 and K = 21 showed two clusters only and both analyses are highly similar to each other (Additional file 1). It has been argued the ad-hoc Evanno method [27] favours by default K =2 over K = 1 when searching for the correct number of clusters [28]. However, PCO separated individuals included in our analyses into two groups as well, and therefore we consider K = 2 as representative for our sample set. For K = 2, the allele-frequency divergence between the two groups was 0.0074. One group (“grey”) includes the majority of accessions (Figure 4C). The other group (“white”) includes D. calciphila, D. labillardierei (population 13 and accession BT179), D. minimifolia (majority of individuals), D. pustulata, D. sp. Pic N’ga, D. tridentata (accessions BT206 and BT207), D. veillonii (accession BT224) and D. vieillardii (population 37 [except accession BT017], population 39 [except accession BT100] and population 41). Seven individuals appear to be admixed (less than 90% identity with one of the groups); most of those are D. vieillardii. Several species (D. labillardierei, D. minimifolia, D. tridentata, D. veillonii and D. vieillardii) and even some populations comprise individuals belonging to each of the two groups.
Figure 3.
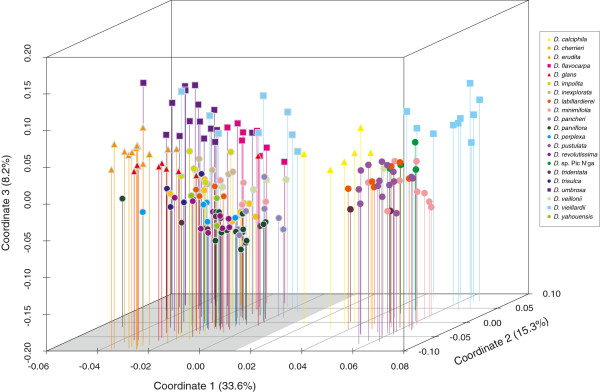
PCO of individual accessions based on Dice distances. Shading of the base-grid of the figure marks the two main groups inferred by Structure analysis – white and grey. Each species is shown in a different colour. Colours were selected randomly and do not indicate any grouping.
Figure 4.
Results of the Structure analysis. A) Delta K values of the K values tested. B) Mean Ln likelihood of K values tested. C) Clustering of K = 2. The two groups are marked in different shades (white and grey).
In order to quantify the amount of genetic variation between species we have performed a non-hierarchical AMOVA with species assigned as “populations”. This analysis showed as little as 30% of the variation to occur among the species. However, in the Structure, PCO, NJ and BI analyses several species seemed to be formed by genetically distinct populations assigned to different clusters and coming in distinct positions in the tree. To avoid mixing up of cryptic variation within a group, we run further AMOVAs with populations assigned as sample localities, despite the relatively low sample size per locality. Results of non-hierarchical AMOVA in this case indicate a higher level of differentiation between populations, resulting in an F ST of 0.38. There was no visible difference in gene diversity between stands of co-occurring species and isolated populations. Several hierarchical AMOVAs (except one based on the Structure results) were not significantly more informative than the non-hierarchical AMOVAs (Table 2). Grouping populations according to geography or ecology, explains a surprisingly low amount of the variation (1.4 – 1.6%). Furthermore, allocating populations to the 21 included species assigns a relatively high percentage of variation at the between-species level (19.4%), but with a highly similar F ST value to the non-hierarchical AMOVA results. When higher-level groupings paralleled the Structure results, we obtained the highest F ST value (0.4), albeit the percentage of variation between the two clusters as defined by Structure was only 9.5%, lower than the percentage of differentiation shown between species. Removing seven admixed samples (less than 90% membership form each of the two groups based on the Structure results) from the AMOVA gave nearly the same results as the analysis including them (Table 2).
Table 2.
Results of different AMOVAs conducted
| |
Percentage of variation |
|
|
||
|---|---|---|---|---|---|
| Analysis | Among groups | Among populations within groups | Within groups | F ST | p value |
| Non-hierarchical |
- |
38.16 |
61.84 |
0.382 |
0.00 |
| Species-wise |
19.43 |
19.15 |
61.42 |
0.386 |
0.00 |
|
Structure |
9.46 |
33.22 |
57.32 |
0.427 |
0.00 |
|
Structure no admixed |
9.93 |
33.39 |
56.68 |
0.433 |
0.00 |
| Geographic |
1.58 |
36.97 |
61.45 |
0.385 |
0.00 |
| Water |
1.37 |
37.20 |
61.43 |
0.386 |
0.00 |
| Soil | 1.54 | 36.92 | 61.54 | 0.385 | 0.00 |
In the non-hierarchical analysis, no grouping was applied. In the species-wise analysis, samples were grouped according to taxonomic features (21 groups corresponding to the 21 species included).
In the Structure analysis, samples were grouped according to the results of Structure analysis (two groups corresponding to the two groups – white and grey – inferred by Structure); in the analysis without admixed samples seven samples with less than 90% identity to one of the two groups were removed. In the geography analysis the samples were grouped according to their origin (three areas – north, middle and south – of New Caledonia). The analysis based on water availability was structured into two groups – humid and dry. In the soil-type based analysis, species were grouped according to the substrate on which they were found (five groups – limestone, serpentine, schist, ultramafic rock and volcanic rock).
The average gene diversity over loci within populations ranged from 0.03 in D. erudita (population 4) to 0.12 in D. parviflora (population 22). Contrary to predictions, the highest number of polymorphic sites, pair-wise differences and average gene diversity were not found in the admixed populations (according to the Structure results) but in D. parviflora (for details see Additional file 2).
Discussion
“Explosive” radiations featuring rapid opportunistic morphological and ecological diversification are phenomena previously reported for some islands (e.g. [29] and references therein). Extreme ancestral bottlenecks, together with on-going hybridization and incomplete lineage sorting, can prevent phylogenetic reconstruction in cases of island radiations if they have been recent and produced many species [30]. However, a good understanding of phylogenetic relationships within radiating groups is key for further evolutionary studies into mechanisms and whether change is adaptive, due to drift in small populations or other phenomena [29].
For the endemic New Caledonian Diospyros species, previous studies, based on multiple plastid [12] and low-copy nuclear [13] markers, showed 21 species to be closely related (Figure 1) and were not able to clearly resolve phylogenetic relationships among them. In the combined data set (plastid and nuclear markers; [13]) only seven of the 21 species included were found to form highly supported groups of accessions from single species. Individuals belonging to each of the remaining 14 species failed to cluster according to their taxonomic circumscription. Dating analysis based on plastid and low-copy nuclear markers showed that the common ancestor of this clade of endemic New Caledonian Diospyros species has arrived in New Caledonia around nine million years ago [13]. Diospyros vieillardii has been shown to be sister to the rest of this endemic clade and separated from the rest of the species around 7.2 million years ago.
Results of the current study using genome-wide AFLP markers reveal that most species form unique groups paralleling recognised species. Around one-third (eight species, NJ dendrogram, Figure 2A) and one-half (11 species, Bayesian tree, Figure 2B) of the species, are genetically distinct with high support (Figure 2). However, the overall AFLP results prove unable to clearly resolve the backbone of trees, similar to previous results obtained from analyses of DNA sequence data [13]. Intra-specific variation was greater (~80%) than that found at inter-specific level (~20%). This low ratio of among- versus within-species divergence in the context of considerable morphological and ecological divergence is indicative of a recent diversification [22]. Such a process can explain why we were able to get clear species boundaries for most species but were unable to clearly resolve phylogenetic relationships between them.
Two species that did not form well-defined clades (D. minimifolia and D. parviflora) were previously considered by White [31] to show variability in leaf morphology that may indicate that they are in fact a collection of several species. For D. minimifolia White [31] mentioned that the type population (close to population 15 of this study) has smaller leaves compared to other populations of this species. In our results this population clusters together with the majority of the D. minimifolia accessions; the population that is separated from the rest (population 16) is from Gaji. According to White [31]D. parviflora is a wide-spread species, showing considerable variability of leaf morphology even within populations, making it impossible to differentiate these into different species. Our results show all accessions of D. parviflora, except those from Plateau de Tango (population 24), to form a group. All included accessions from D. parviflora are from ultramafic localities.
To our surprise, the AFLP results do not show any significant grouping according to ecological (edaphic, climatic, elevational), geographical or morphological factors (Additional file 3). The two weakly differentiated groups revealed by Structure and PCO also do not correspond to any conspicuous phenotypic characteristics. The allele-frequency divergence between the two groups found by Structure is low, which explains why we did not observe the two groups in the Bayesian and NJ tree-building results. Taken together, these results indicate that positive selection has perhaps acted on few genomic regions [32] and has resulted in phenotypic diversification of New Caledonian Diospyros. Variation in copy number of specific genomic regions may be an additional aspect of molecular variation that, although invisible to AFLP markers, could form the basis of adaptation to different environmental conditions [33].
The individuals of D. vieillardii, D. umbrosa and D. flavocarpa form a minimally isolated group (squares in the grey group) in the PCO (Figure 3). Previous phylogenetic analyses (Figure 1) showed these three species to be sister to the rest of the taxa. Due to its morphological and ecological features D. sp. Pic N’ga from Île des Pins could be a hybrid between D. calciphila and D. vieillardii, but D. vieillardii is now not known from this island. In PCO, individuals of this putative species are located between individuals of D. calciphila and D. vieillardii (Figure 3). The split between the two groups observed (Figures 3 and 4) could be relatively old, separating two lineages that developed in isolated regions. For instance, dry periods of the Pleistocene caused aridification in many areas, and some vegetation types persisted only in local refugia e.g. [34-36]. After climatic conditions became more favourable, the two groups probably expanded rapidly into newly suitable habitats where they overlapped; the time scale of these fluctuations (ca. 0.02 – 0.1 myr; [37]) was probably not enough to allow woody species with long generation time such as Diospyros to diverge and become permanently reproductively isolated [22]. There are a few admixed individuals in the Structure analysis (Figure 4), which implies that hybridization might have played a role in evolution of this group.
Accelerated rates of evolution at few genes as a result of positive selection could have resulted in the morphological and ecological diversification apparent today in this group of New Caledonian Diospyros species. Furthermore, in addition to retention of ancestral polymorphisms, frequent gene flow could have acted against genome-wide genetic differentiation between the species. Barriers to gene flow between these species may be highly porous, with only few genes responsible for ecological and morphological adaptations evolving on distinct trajectories under strong selection, which leaves the rest of their genomes open to gene flow [38]. Finding these few genes with AFLP is realistically improbable because they are a miniscule component in comparison the rest of these genomes. In the case of a recent and rapid radiation in plants, it could be argued that the bulk of regions sampled by AFLP have not evolved quickly enough to accumulate substitutions that could indicate species relationships. Our results are similar to those found in various other island genera (e.g. Araucaria in New Caledonia, [22]; Ourisia in New Zealand, [39]).
Diospyros vieillardii, which is sister to the rest of the taxa belonging to this group of New Caledonian endemics [12,13], is confined to ultramafic soils, which supports the hypothesis of this being an exaptation of the progenitor of this New Caledonian Diospyros clade to ultramafic soils when the whole island was still covered by heavy-metal-rich substrates; similar findings have been made in other plant groups in New Caledonia e.g. [9]. Later, erosion reduced the extent of this geological layer to one third of the island [7], and existing species began to move onto other substrates where they subsequently diverged, forming distinct species. Such observations have been made in various other New Caledonian groups (e.g. Araucaria, [22]; Spiraeanthemum, [35]; Codia, [40]). A few studies have examined the adaptive basis and processes involved in rapid radiations in New Caledonia e.g. [41] and Hawai’i (e.g. lobeliads, [42]; silverswords, [43]). Linking ecological parameters and/or phenotypic traits associated with speciation has to be done with caution because range alterations, subsequent evolution, and species extinctions might have erased initial signals found in only a few genes. Therefore, the associations observed today may be misleading, and the specific conditions/traits that were indeed linked to speciation, if any, may no longer be present [44].
Further work involving common garden experiments would provide insights into the effect of environmental conditions on morphological traits and therefore plasticity of genomes of the New Caledonian Diospyros species. Unfortunately, such experiments are time and cost intensive. It is difficult to obtain ripe fruits of all Diospyros species, and in addition it is difficult to germinate and grow them, which is a crucial aspect of conducting such experiments. Reciprocal transplantation of seedlings across environments are of course more easily conducted than common garden experiments, but they are still time consuming and costly; in addition species adapted to one soil type often will not survive when transplanted to other soil types.
Conclusions
Although New Caledonian Diospyros are morphologically and ecologically diverse, they show little genetic divergence (based on DNA sequences and AFLP data). In this case of the endemic clade of New Caledonian Diospyros, AFLP data did not provide enough information to resolve phylogenetic relationships between the species, but it was sensitive enough for testing for the presence of genetic species boundaries. However, the AFLP results exhibit a good correlation with morphology-based species concepts. Further studies of this New Caledonian Diospyros group with deeper sampling of the genome using next generation sequencing methods are needed to get a clearer picture of the processes that formed this group.
Methods
Material
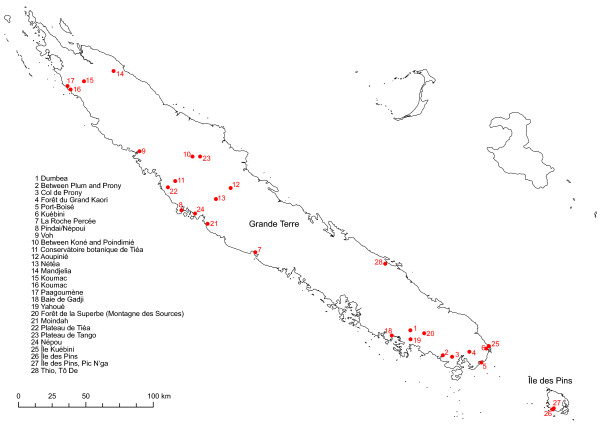
Material from New Caledonian Diospyros species was collected on the main island (Grande Terre) and on a smaller island, Île des Pins. When possible, we collected five individuals per population. Collecting population samples from tropical trees/shrubs is not always easy because the trees can be tall (and leaves therefore out of reach) and individuals are often far from each other. Collecting ten individuals in an area of ten square meters also does not make much sense for a study like this because these individuals are probably offspring from the same mother plant. As the focus of the present study is on the phylogenetic relationships between the species and not on population genetics within species, the authors consider the small size of the samples we collected to be sufficient. For widespread species, we collected populations throughout their range. For distribution of sampling sites, see Figure 5. From samples where fertile material was available, a voucher was made with several duplicates sent to the herbaria at Noumea (NOU), University of Montpellier II (MPU) and the University of Vienna (WU). When sterile, one voucher per population was taken; this was compared to already existing collections in Noumea Herbarium (NOU) from the same location and referred to that species if similar. In total we included in the present study 231 individuals of New Caledonian Diospyros species, which correspond to 20 identified and one unidentified species (due to absence of diagnostic reproductive organs at the time of collection), giving 47 populations in total. Details of the 192 individuals (43 populations) for which we were able to get useable results are given in Table 3. Silica-gel-dried material was used for DNA extraction.
Figure 5.
Map with sampling localities. Dots indicate sampling sites; the numbers associated with each dot refer to the list of sampling sites on this figure. Those numbers are used throughout the present study to characterize sampling sites.
Table 3.
Table of accessions; showing all individuals used in this study
| Taxon | Sample ID | Population | Sampling location | Voucher |
|---|---|---|---|---|
|
D. calciphila F.White |
BT312-BT317 |
1 |
26, littoral forest |
JM6650, JM6653 |
| (MPU, NOU, P) | ||||
|
D. cherrieri F.White |
BT262, BT276-BT278 |
2 |
21, dry forest |
NOU079551, WU062860 |
| NOU054492, NOU054008 | ||||
|
D. cherrieri |
BT293-BT297 |
3 |
24, dry forest |
NOU079547 |
|
D. erudita F.White |
BT259-BT261, BT273-BT275 |
4 |
21, dry forest |
WU062855, NOU079545 |
| NOU079544, WU062870 | ||||
| NOU054010, WU062856 | ||||
| NOU054011, WU062857 | ||||
|
D. erudita |
BT280-BT285, BT287 |
5 |
22, dry forest |
WU062858, Chambrey & Turner 20 (NOU) |
|
D. flavocarpa (Vieill. ex P.Parm.) F.White |
BT126-BT130 |
6 |
10, humid mountain forest |
JM6625 (NOU) |
|
D. flavocarpa |
BT155, BT158-BT159 |
7 |
12, dense humid mountain forest |
JM6632 (NOU) |
|
D. glans F.White |
BT020-BT022 |
8 |
1, forest near river |
NOU053705, NOU030755, WU062846 |
|
D. glans |
BT075 |
9 |
5, dense forest near road |
NOU000819 |
|
D. glans |
BT082, BT084, BT087, BT093-BT094 |
10 |
6, forest near river |
NOU022860 |
|
D. impolita F.White |
BT101-BT105 |
11 |
7, mesophyll forest near beach |
NOU019538 |
|
D. inexplorata F.White |
BT304, BT307-BT311 |
12 |
25, littoral forest |
NOU005818 |
|
D. labillardierei F.White |
BT121-BT125 |
13 |
10, river edge in mountain forest |
JM6624 (NOU) |
|
D. labillardierei |
BT178-BT182 |
14 |
13, river edge |
(NOU031346) |
|
D. minimifolia F.White |
BT134-BT135 |
15 |
11, dry forest |
NOU019556 |
|
D. minimifolia |
BT230-BT234 |
16 |
18, mesophyll forest near beach |
NOU019554 |
|
D. minimifolia |
BT263-BT264, BT266-267, BT269-BT270 |
17 |
21, dry forest |
NOU079549, WU062872 |
| NOU054493 | ||||
|
D. pancheri Kosterm. |
BT029-BT031, BT035 |
18 |
2, forest near road |
JM6619, JM6620 (NOU) |
|
D. pancheri |
BT076-BT079 |
19 |
5, dense forest near road |
|
|
D. parviflora (Schltr.) Bakh. |
BT042 |
20 |
3, wet forest |
|
|
D. parviflora |
BT059, BT062-BT063, BT068 |
21 |
4, wet dense forest |
NOU006656 |
|
D. parviflora |
BT080, BT085, BT089-BT090 |
22 |
6, forest near river |
JM6622 (NOU) |
|
D. parviflora |
BT248-BT250, BT252-BT253 |
23 |
20, humid forest at low elevation |
tree no. 23109 |
|
D. parviflora |
BT289-BT292 |
24 |
23, mountain forest |
NOU079550 |
|
D. perplexa F.White |
BT147-BT151 |
25 |
11, forest near river |
JM6630 (NOU) |
|
D. pustulata F.White |
BT111-BT114 |
26 |
8, dry forest |
|
|
D. pustulata |
BT136-BT140 |
27 |
11, dry forest |
JM6629 (NOU) |
|
D. pustulata |
BT257-BT258, BT265, BT268, BT271-BT272 |
28 |
21, dry forest |
NOU079548, WU062871 |
| NOU053999 | ||||
|
D. revolutissima F.White |
BT116-BT120 |
29 |
9, maquis |
NOU023189 |
|
D. revolutissima |
BT218-BT222 |
30 |
17, maquis |
JM6640 (NOU) |
|
D. tridentata F.White |
BT202-BT207 |
31 |
15, dry forest at low elevation |
JM6639 (NOU) |
|
D. trisulca F.White |
BT185, BT192, BT197, BT199-BT201 |
32 |
14, mountain forest |
NOU031344, JM6637 (NOU) |
|
D. umbrosa F.White |
BT061, BT065-BT066, BT071, BT073 |
33 |
4, wet dense forest |
|
|
D. umbrosa |
BT170-BT171, BT175-BT177 |
34 |
13, dense humid forest |
JM6635 (NOU) |
|
D. umbrosa |
BT246-BT247, BT251, BT254, BT256 |
35 |
20, humid forest at low elevation |
NOU023234 |
|
D. veillonii F.White |
BT224, BT226-BT229 |
36 |
18, mesophyll forest near beach |
NOU019582 |
|
D. vieillardii (Hiern) Kosterm. |
BT017, BT023-BT026 |
37 |
1, forest near river |
JM6618 (NOU) |
|
D. vieillardii |
BT055, BT057-BT058 |
38 |
4, dry open forest |
|
|
D. vieillardii |
BT088, BT091-BT092, BT098, BT100 |
39 |
6, forest near river |
|
|
D. vieillardii |
BT215-BT217 |
40 |
16, maquis |
NOU023242 |
|
D. vieillardii |
BT324-BT325, BT328 |
41 |
28, forest near river |
|
|
D. yahouensis (Schltr.) Kosterm. |
BT237-BT239 |
42 |
19, mesophyll forest |
P00057340 |
| D. sp. Pic N'ga | BT319, BT321-BT323 | 43 | 27, maquis | JM6065 (NOU) |
The numbers of sampling localities are the same as in Figure 2. Voucher-Codes: JMXXXX: collection number J. Munzinger; Tree N° XXX: Tree of New Caledonian Plant Inventory and Permanent Plot Network (NC-PIPPN, [45]); NOUXXXXXXX: Herbarium accession number of Noumea herbarium (NOU); WUXXXXXX: Herbarium accession number of the Herbarium of the University Vienna (WU); P: Herbarium of the Natural History Museum Paris; MPU: Herbarium of the University of Montpellier II.
DNA extraction
For DNA extraction, a modified sorbitol/high-salt CTAB method [46] was used (for details see [13]).
AFLP
Preparation and amplification of fragments followed the protocol of Vos et al. [14] with some modifications. Restriction of genomic DNA with two restriction enzymes and ligation of double-stranded adaptors to the resulting restricted fragments were performed in one step in a thermal cycler (Veriti, AB, Life Technologies; 37°C for 2 h followed by a 30 min hold at 17°C). Reactions comprised 1.1 μL 10x T4 DNA ligase buffer (Promega), 1.1 μL 0.5 M NaCl, 0.55 μL BSA (1 mg/ mL; New England BioLabs), 50 μM MseI adaptors (genXpress), 5 μM EcoRI adaptors (genXpress), 1 U MseI restriction endonuclease (New England BioLabs), 5 U EcoRI restriction endonuclease (New England BioLabs), 1 U T4 DNA ligase (Promega), and 0.5 μg DNA and were made up to a total volume of 11 μL with water. Ligated DNA fragments were diluted 10-fold with sterile water. Preselective amplification reactions contained 1.14 μL 10x RedTaq PCR reaction buffer (Sigma), 0.2 U RedTaq DNA polymerase (Sigma), 0.22 μL dNTPs (10 mM; AB, Life Technologies), 0.58 μL preselective primer pairs (EcoRI-A and MseI-CT, each 5 μM; Sigma), and 2 μL diluted restriction-ligation product, and were brought with water to a total volume of 10 μL. Amplification was carried out in the same machine used for restriction-ligation with the following profile: 2 min at 72°C, 20 cycles of 10 sec denaturing at 94°C, 30 sec annealing at 56°C, 2 min extension at 72°C, and a final extension step for 30 min at 60°C. The preselective PCR products were diluted 10-fold with sterile water. Reactions for selective amplification contained 0.5 μL 10x RedTaq PCR reaction buffer (Sigma), 0.1 U RedTaq DNA polymerase (Sigma), 0.11 μL dNTPs (10 mM; AB, Life Technologies), 0.27 μL MseI-primer (5 μM; Sigma), 0.27 μL EcoRI-primer (1 μM; Sigma), and 1 μL diluted preselective amplification product and were brought to a total volume of 5 μL with water. They were carried out in a GeneAmp PCR System 9700 (AB, Life Technologies) with the following profile: 1 min at 94°C, 9 cycles of 1 sec at 94°C, 30 sec at 65-57°C (reducing the temperature at 1°C per cycle), 2 min at 72°C, 25 cycles of 1 sec at 94°C, 30 sec at 56°C, 2 min at 72°C and a final extension for 30 min at 60°C. The selective PCR products were purified using Sephadex G-50 Superfine (GE Healthcare Bio-Sciences) applied to a MultiScreen-HV 96-Well Plate (Millipore) in three steps of 200 μL each and settled at 750 g (1, 1 and 5 min, respectively). The same speed was used for centrifugation of the samples (5 μL of each selective PCR product), again for 5 min. Two microliters of the eluate were combined with 10 μL HiDi and 0.1 μL GeneScan 500 ROX (AB, Life Technologies) and denatured for 3 min at 95°C before running them on a capillary sequencer (3130xl Genetic Analyzer, AB, Life Technologies).
The selective primer pairs (6Fam-EcoRI-AGC/MseI-CTGA, Vic-EcoRI-ATG/MseI-CTCG and Ned-EcoRI-ATC/MseI-CTGA) were chosen because they generated clear and not too many bands (thus decreasing the risk of fragments co-migrating by chance), with sufficient variability in preliminary tests. Although the genome size of the New Caledonian Diospyros species (1C-value: 1.5 – 2.3 pg; [13]) is smaller than the mean 1C-value of eudicots (2.7 pg, [47]), we found the AFLP profiles generated with Msel primers with four selective bases much clearer than those obtained from primers with just three selective bases.
Reproducibility was checked by repeating ca. 80% of the samples. This high number of repetitions was necessary because of initial difficulties with fragment sizing.
Scoring and phylogenetic analysis
Sizing and scoring of the data was performed with GeneMarker v2.2.0 (SoftGenetics). After pre-analysis using default settings, sizing profiles of all samples were checked and where necessary manually corrected. Most of these corrections concerned one of the following peaks of the size standard: 35 bp, 50 bp and 139 bp. These peaks were often not correctly recognized by the GeneMarker program. High-quality sizing-profiles (score > 90) were obtained for all samples. A panel of scorable fragments was established for each primer combination, and fragments between 65 – 510 bp were scored. The relative fluorescent unit (RFU) threshold was set at 40. Automatic scoring was conducted using Local Southern peak call, peak saturation, base line subtraction, spike removal, pull up correction, and a stutter peak filter of 5% (as described in [48]). The results were exported as presence/absence matrix. The outcome of the automatic scoring was manually checked and corrected for errors. These errors mostly concerned peaks for which shape was atypical. In total 486 samples corresponding to 231 individuals were scored. From 186 individuals replicate samples were performed (between two and five replicates per individual). The differences between the different samples (replicates) were counted and divided by the total number of phenotypic comparisons to get the error rate (calculated according to Bonin et al. [49]). After initial analysis (neighbour-joining, NJ) of the complete data set, replicates of samples and obviously failed samples were excluded from further analyses. As replicated samples of the corresponding individuals mostly clustered together, selection of samples from each individual for further analyses was random and not according to any pattern or protocol. For the final analyses we ended up with 192 individuals.
All three primer-combinations were combined in a single matrix and analysed together. Different distance measures were tested for their power to resolve relationships with our data set. Distance matrixes were calculated in PAUP* v4b10 ([50]; Nei-Li distance) and SplitsTree v4.12.6 ([51]; uncorrected P, Dice, corrected and uncorrected Hamming). Phylogenetic relationships based on previously mentioned distance matrices were reconstructed using SplitsTree v4.12.6 [51] to create unrooted NJ dendrograms. To assess robustness of branches NJ-bootstrap (NJ-BS) analyses were performed using SplitsTree v4.12.6 [51] and PAUP* v4b10 [50]. Bayesian inference (BI) was conducted with BEAST v1.7.5 [52], with two runs each 20 million generations, sampling every 1,000th generation and removal of the first 30% of trees as burn in.
To visualise the pattern of genetic clustering of individuals and populations, we plotted principal coordinate analysis (PCO) using the R-package scatterplot3d [53] based on an individual Dice distance matrix, and respectively, on AMOVA-derived pair-wise F ST distances calculated with Arlequin v3.5.1.2 [54]. To investigate further significant groupings of the included individuals we used the program Structure v2.3.3 [55,56] on the Bioportal computing cluster of the University Oslo [57]. We ran Structure for K = 1–23 with 10 replicates each and a model based on admixture and independent allelic frequencies, without taking into account information regarding sampling localities. Each run had 3 million iterations with 10% additional burn in. The calculation of deltaK (∆K; [27]) and preparation of the input file for Clumpp was done with Harvester [58]. Production of a combined file from the ten replicates of the best K was perfomed using Clumpp v1.1.2 [59] with the full search algorithm. The graphical representation of Structure results was prepared with Distruct v1.1 [60].
Both non-hierarchical and hierarchical analyses of molecular variance (AMOVA) and calculations of population statistics were conducted using Arlequin v3.5.1.2 [54]. For hierarchical AMOVAs groups have been defined based on different possible clusterings (Additional file 4) according to Structure results, taxonomy, distribution patterns and ecological traits.
Availability of supporting data
AFLP presence/absence matrix and phylogenetic analyses are deposited in treeBASE under study 14798 (http://purl.org/phylo/treebase/phylows/study/TB2:S14798).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BT carried out the acquisition and analysis of the data, drafted the manuscript and assisted collecting the plant material. OP helped with data analysis. JM collect and identified the plant material and helped to interpret the results. Previous studies of SD helped to design this project. MWC helped to design this project and gave linguistic support to the manuscript. RS, designed and coordinated the study and helped to draft the manuscript. All authors read, commented, corrected and approved the final manuscript.
Supplementary Material
Structureresults of suboptimal K values (3, 6, 16 and 21) in comparison with K =2. Delta K likelihoods are given for each K.
Table showing the population statistics inferred from non-hierarchical AMOVA based on STRUCTURE results. Populations marked bold differ in this analysis from the general population grouping given in Table 3.
Figure of the neighbour joining dendrogram coloured according to soil type (colour of the branches) and water availability (colour of taxa names). This dendrogram is the same as Figure 3A, but coloured according to ecological features.
Table giving the details of the different AMOVAs conducted. The numbers in the populations column are the same as given in Table 3, respectively, in Additional file 1 for the Structure based AMOVA.
Contributor Information
Barbara Turner, Email: barbara.turner@univie.ac.at.
Ovidiu Paun, Email: ovidiu.paun@univie.ac.at.
Jérôme Munzinger, Email: Jerome.munzinger@ird.fr.
Sutee Duangjai, Email: fforsud@ku.ac.th.
Mark W Chase, Email: m.chase@kew.org.
Rosabelle Samuel, Email: mary.rosabella.samuel@univie.ac.at.
Acknowledgements
This work was funded by a grant from the Austrian Science Fund (FWF, Project-Number: P 22159-B16) awarded to R. Samuel. The authors thank the team of the Department of Systematic and Evolutionary Botany of the University of Vienna as well as the team of the IRD AMAP Noumea for support with this study. Special thank goes to V Klenja for the lab work. Thanks to the following persons for their help with lab work, field work and ideas to improve this work: F Balao Robles, J-P Butin, C Chambrey, G Dagostini, E Grasserbauer, V Hequet, G Kohl, D & I Létocart, W Nigote and H Vandrot. Specimens are deposited in the herbaria of Noumea (NOU), University of Montpellier II (MPU) and the University of Vienna (WU).
References
- Givnish TJ, Millam KC, Mast AR, Paterson TB, Theim TJ, Hipp AL, Henss JM, Smith JF, Wood KR, Sytsma KJ. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae) Proc R Soc B. 2009;13:407–146. doi: 10.1098/rspb.2008.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knope ML, Morden CW, Funk VA, Fukami T. Area and the rapid radiation of Hawaiian Bidens (Asteraceae) J Biogeogr. 2012;13:1206–1216. doi: 10.1111/j.1365-2699.2012.02687.x. [DOI] [Google Scholar]
- Mittermeier RA, Gil PR, Hoffmann M, Pilgrim J, Brooks T, Mittermeier CG, Lamoreux J, da Fonseca GAB. Hotspots revisited: Earth’s biologically richest and most endangered terrestrial ecoregions. Mexico City: CEMEX; 2004. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;13:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Morat P, Jaffré T, Tronchet F, Munzinger J, Pillon Y, Veillon J-M, Chalopin M. Le référentiel taxonomique Florical et les caractéristiques de la flore vasculaire indigène de la Nouvelle-Calédonie. Adansonia. 2012;13:177–219. [Google Scholar]
- Lowry PP. , II. In: Rare, threatened, and endangered floras of Asia and the Pacific rim. Peng CF, Lowry PPII, editor. Taiwan: Institute of Botany, Taipei; 1998. Diversity, endemism and extinction in the flora of New Caledonia: a review; pp. 181–206. [Google Scholar]
- Pelletier B, Payri C, Richer De Forges B. Compendium of marine species from New Caledonia, Documents Scientifiques et Techniques II4. New Caledonia: Institut de Recherche pour le Développement Nouméa; 2006. Geology of the New Caledonia region and its implications for the study of the New Caledonian biodiversity; pp. 17–30. [Google Scholar]
- Maurizot P, Vendé-Leclerc M. New Caledonia geological map, scale 1/500000. Direction de l’Industrie, des Mines et de l’Energie - Service de la Géologie de Nouvelle-Calédonie, Bureau de Recherches Géologiques et Minières. 2009.
- Pillon Y, Munzinger J, Amir H, Lebrun M. Ultramafic soils and species sorting in the flora of New Caledonia. J Ecol. 2010;13:1108–1116. doi: 10.1111/j.1365-2745.2010.01689.x. [DOI] [Google Scholar]
- Jaffré T, Rigault F, Munzinger J. In: Atlas de la Nouvelle-Calédonie. Bonvallot J, Gay J-C, Habert E, editor. Nouméa: IRD Editions; 2012. La végétation; pp. 77–80. [Google Scholar]
- Duangjai S, Wallnöfer B, Samuel R, Munzinger J, Chase MW. Generic delimitation and relationships in Ebenaceae sensu lato: evidence from six plastid DNA regions. Am J Bot. 2006;13:1808–1827. doi: 10.3732/ajb.93.12.1808. [DOI] [PubMed] [Google Scholar]
- Duangjai S, Samuel R, Munzinger J, Forest F, Wallnöfer B, Barfuss MHJ, Fischer G, Chase MW. A multi-locus plastid phylogenetic analysis of the pantropical genus Diospyros (Ebenaceae), with an emphasis on the radiation and biogeographic origins of the New Caledonian endemic species. Mol Phylogenet Evol. 2009;13:602–620. doi: 10.1016/j.ympev.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Turner B, Munzinger J, Duangjai S, Temsch EM, Stockenhuber R, Barfuss MHJ, Chase MW, Samuel R. Molecular phylogenetic of New Caledonian Diospyros (Ebenaceae) using plastid and nuclear markers. Mol Phylogenet Evol. 2013;13:740–763. doi: 10.1016/j.ympev.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;13:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremetsberger K, Stuessy TF, Kadlec G, Urtubey E, Baeza CM, Beck SG, Valdebenito HA, Ruas CF, Matzenbacher NI. AFLP phylogeny of South American species of Hypochaeris (Asteraceae, Lactuceae) Syst Bot. 2006;13:610–626. doi: 10.1600/036364406778388520. [DOI] [Google Scholar]
- Koopman WJM, Zevenbergen MJ, van den Berg RG. Species relationships in Lactuca s.l. (Lactuceae, Asteraceae) inferred from AFLP fingerprints. Am J Bot. 2001;13:1881–1887. doi: 10.2307/3558364. [DOI] [PubMed] [Google Scholar]
- Richardson JE, Fay MF, Cronk QCB, Chase MW. Species delimitation and the origin of populations in island representatives of Phylica (Rhamnaceae) Evolution. 2003;13:816–827. doi: 10.1111/j.0014-3820.2003.tb00293.x. [DOI] [PubMed] [Google Scholar]
- Despré L, Giells L, Redoutet B, Taberlet P. Using AFLP to resolve phylogenetic relationships in a morphologically diversified plant species complex when nuclear and chloroplast sequences fail to reveal variability. Mol Phylogenet Evol. 2003;13:185–196. doi: 10.1016/S1055-7903(02)00445-1. [DOI] [PubMed] [Google Scholar]
- Paun O, Schönswetter P, Winkler M, Tribsch A. IntraBioDiv Consortium. Historical divergence vs. contemporary gene flow: evolutionary history of the calcicole Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Mol Ecol. 2008;13:4263–4275. doi: 10.1111/j.1365-294X.2008.03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte K, Silvestro D, Kiehlmann E, Vesely S, Novoa P, Zizka G. Detection of recent hybridization between sympatric Chilean Puya species (Bromeliaceae) using AFLP markers and reconstruction of complex relationships. Mol Phylogenet Evol. 2010;13:1105–1119. doi: 10.1016/j.ympev.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Jabaily RS, Sytsma KJ. Historical biogeography and life-history evolution of Andean Puya (Bromeliaceae) Bot J Linn Soc. 2012;13:201–224. [Google Scholar]
- Gaudeul M, Rouhan G, Gardner MF, Hollingsworth PM. AFLP markers provide insights into the evolutionary relationships and diversification of New Caledonian Araucaria species (Araucariaceae) Am J Bot. 2012;13:68–81. doi: 10.3732/ajb.1100321. [DOI] [PubMed] [Google Scholar]
- Koopman WJM. Phylogenetic signal in AFLP data sets. Syst Biol. 2005;13:197–217. doi: 10.1080/10635150590924181. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;13:762–768. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meudt HM, Clarke AC. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007;13:106–117. doi: 10.1016/j.tplants.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Bussell JD, Waycott M, Chappill JA. Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspect Plant Ecol, Evol Systematics. 2005;13:3–26. doi: 10.1016/j.ppees.2004.07.001. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol. 2005;13:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y, Glaubitz JC, Matsouka Y, Goddman MM, Sánchez GJ, Doebley J. Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am J Bot. 2008;13:1240–1253. doi: 10.3732/ajb.0800097. [DOI] [PubMed] [Google Scholar]
- Glor RE. Phylogenetic insights on adaptive radiation. Ann Rev Ecol, Evol Systematics. 2010;13:251–270. doi: 10.1146/annurev.ecolsys.39.110707.173447. [DOI] [Google Scholar]
- Lerner HRL, Meyer M, James HF, Hofreiter M, Fleischer RC. Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers. Curr Biol. 2011;13:1838–1844. doi: 10.1016/j.cub.2011.09.039. [DOI] [PubMed] [Google Scholar]
- White F. Flore de la Nouvelle-Calédonie et Dépendances. 19. Ébénacées. Paris: Muséum National d’Histoire Naturelle; 1993. [Google Scholar]
- Kapralov MV, Votintseva AA, Filatov DA. Molecular adaptation during a rapid adaptive radiation. Mol Biol Evol. 2013;13:1051–1059. doi: 10.1093/molbev/mst013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC. Copy number variation and transposable elements feature in recent ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 2010;13:e1000998. doi: 10.1371/journal.pgen.1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintaud J-C, Tanguy J, Puig H. Chorology of New Caledonian palms and possible evidence of Pleistocene rain forest refugia. C R Acad Sci. 2011;13:453–463. doi: 10.1016/s0764-4469(01)01312-9. [DOI] [PubMed] [Google Scholar]
- Pillon Y, Hopkins HC, Munzinger J, Amir H, Chase MW. Cryptic species, gene recombination and hybridization in the genus Spiraeanthemum (Cunoniaceae) from New Caledonia. Bot J Linn Soc. 2009;13:137–152. doi: 10.1111/j.1095-8339.2009.00997.x. [DOI] [Google Scholar]
- Poncet V, Munoz F, Munzinger J, Pillon Y, Gomez C, Couderc M, Tranchant-Dubreuil C, Hamon S, de Kochko A. Phylogeography and niche modelling of the relict plant Amborella trichopoda (Amborellaceae) reveal multiple Pleistocene refugia in New Caledonia. Mol Ecol. 2013. doi:10.1111/mec.12554. [DOI] [PubMed]
- Bennett KD. Milankovitch cycles and their effects on species in ecological and evolutionary time. Paleobiology. 1990;13:11–21. [Google Scholar]
- Kane NC, King MG, Barker MS, Raduski A, Karrenberg S, Yatabe Y, Knapp SJ, Rieseberg LH. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent Helianthus species. Evolution. 2009;13:2061–2075. doi: 10.1111/j.1558-5646.2009.00703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meudt HM, Lockhart PJ, Bryant D. Species delimitation and phylogeny of a New Zealand plant species radiation. BMC Evol Biol. 2009;13:111. doi: 10.1186/1471-2148-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon Y, Munzinger J, Amir H, Hopkins HC, Chase MW. Reticulate evolution on a mosaic of soils: diversification of the New Caledonian endemic genus Codia (Cunoniaceae) Mol Ecol. 2009;13:2263–2275. doi: 10.1111/j.1365-294X.2009.04178.x. [DOI] [PubMed] [Google Scholar]
- Murienne J, Guilbert E, Grandcolas P. Species diversity in the New Caledonian endemic genera Cephalidiosus and Nobarnus (Insecta: Heteroptera: Tingidae), an approach using phylogeny and species distribution modelling. Biol J Linn Soc. 2009;13:177–184. doi: 10.1111/j.1095-8312.2008.01184.x. [DOI] [Google Scholar]
- Givnish TJ, Montgomery RA, Goldstein G. Adaptive radiation of photosynthetic physiology in the Hawaiian lobeliads: light regimes, static light responses, and whole-plant compensation points. Am J Bot. 2004;13:228–246. doi: 10.3732/ajb.91.2.228. [DOI] [PubMed] [Google Scholar]
- Baldwin BG, Sanderson MJ. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proc Natl Acad Sci USA. 1998;13:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough TG. What can phylogenetics tell us about speciation in the Cape flora? Divers Distrib. 2006;13:21–26. doi: 10.1111/j.1366-9516.2006.00208.x. [DOI] [Google Scholar]
- Ibanez T, Munzinger J, Dagostini G, Hequet V, Rigault F, Jaffré T, Birnbaum P. Structural and floristic diversity of mixed tropical rainforest in New Caledonia: New data from the New Caledonian Plant Inventory and Permanent Plot Network (NC-PIPPN) Appl Veg Sci. doi:10.1111/avsc.1270.
- Tel-Zur N, Abbo S, Myslabodski D, Mizrahi Y. Modified CTAB procedure for DNA isolation from epiphytic cacti of genera Hylocereus and Selenicereus (Cactaceae) Plant Mol Biol Report. 1999;13:249–254. doi: 10.1023/A:1007656315275. [DOI] [Google Scholar]
- Bennett MD, Leitch IJ. Angiosperm DNA C-values database (release 8.0, Dec. 2012) http://www.kew.org/cvalues/
- Safer S, Tremetsberger K, Guo Y-P, Kohl G, Samuel MR, Stuessy TF, Stuppner H. Phylogenetic relationships in the genus Leontopodium (Asteraceae: Gnaphalieae) based on AFLP data. Bot J Linn Soc. 2011;13:364–377. doi: 10.1111/j.1095-8339.2011.01117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetic studies. Mol Ecol. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;13:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;13:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligges U, Mächler M. Scatterplot3d - an R package for visualizing multivariate data. J Stat Softw. 2003;13:1–20. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;13:47–50. [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnely P. Inference of population structure using multilocus genotype data. Genetics. 2000;13:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;13:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifeportal. http://www.uio.no/english/services/it/research/hpc/lifeportal/
- Earl DA, Von Holdt BM. Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conserv Genet Resour. 2012;13:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;13:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;13:137–138. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structureresults of suboptimal K values (3, 6, 16 and 21) in comparison with K =2. Delta K likelihoods are given for each K.
Table showing the population statistics inferred from non-hierarchical AMOVA based on STRUCTURE results. Populations marked bold differ in this analysis from the general population grouping given in Table 3.
Figure of the neighbour joining dendrogram coloured according to soil type (colour of the branches) and water availability (colour of taxa names). This dendrogram is the same as Figure 3A, but coloured according to ecological features.
Table giving the details of the different AMOVAs conducted. The numbers in the populations column are the same as given in Table 3, respectively, in Additional file 1 for the Structure based AMOVA.