Summary
Phenotypic high-throughput chemical screens allow for discovery of small molecules that modulate complex phenotypes and provide lead compounds for novel therapies; however, identification of the mechanistically relevant targets remains a major experimental challenge. We report the application of sequential unbiased high-throughput chemical and ultracomplex shRNA screens to identify a novel class of inhibitors that target nicotinamide phosphoribosyl transferase (NAMPT), a rate-limiting enzyme in the biosynthesis of nicotinamide adenine dinucleotide (NAD+), a crucial cofactor in many biochemical processes. The lead compound STF-118804 is a highly specific NAMPT inhibitor, improves survival in an orthotopic xenotransplant model of high-risk acute lymphoblastic leukemia, and targets leukemia stem cells. Tandem high-throughput screening using chemical and ultracomplex shRNA libraries, therefore, provides a rapid chemical genetics approach for seamless progression from small molecule lead identification to target discovery and validation.
INTRODUCTION
Cell-based high-throughput chemical screens are a powerful approach for discovery of small molecules that modulate complex phenotypes, such as viability, and may serve as lead compounds for development of novel therapies. However, subsequent identification of the cognate target or pathway through which the compound acts remains technically challenging, and lack of understanding of the mechanism of action is a roadblock for drug development. Thus, efforts have shifted away from phenotypic screening to in vitro target-based screening approaches. Although the latter are leveraged on an understanding of the mechanism of action, the biological hypothesis is often not confirmed, the target may not be “druggable,” and the discovered molecules may not affect the desired phenotype. Furthermore, despite a major shift to “target-centric” approaches for drug discovery, the FDA over a recent 10 year period (1999–2008) approved more first-in-class new molecular entities (NMEs) that were identified via phenotypic screening (28 NMEs) than target based approaches (18 NMEs) (Swinney and Anthony, 2011). As a consequence, phenotypic screening is experiencing a resurgence in drug discovery despite persistent challenges presented by target identification (Kotz, 2012; Schenone et al., 2013).
Currently, target identification can be accomplished through molecule-target immobilization followed by chemical proteomics (Fleischer et al., 2010; Ong et al., 2009), pattern matching techniques utilizing gene expression profiling (Lamb, 2007; Lamb et al., 2006) and NCI-60 sensitivity (Huang et al., 2005; Paull et al., 1989; Weinstein et al., 1997), or a combination of these techniques (Hahn et al., 2009; Stegmaier et al., 2005). Each of these approaches as currently applied, however, has limitations and technical challenges. Genetic approaches using shRNA screens have been used to understand the genetic pathways involved in mechanisms of action of known chemotherapeutic agents (Brummelkamp et al., 2006; Burgess et al., 2008; Luo et al., 2008; Tsujii et al., 2010), but have yet to be used to identify the target of an unknown agent. Functional genomic approaches based on shRNA screens have been limited by breadth and depth of coverage of available libraries. Recently, this limitation has been addressed by engineering ultracomplex shRNA libraries that target the entire human genome with ~25 shRNAs per gene (Bassik et al., 2013) and contain 1,000s of negative control shRNAs. This allows for RNAi-based, pooled screening of thousands of shRNAs for a specific phenotype that can be monitored by deep sequencing, and significantly reduces both false negative and false positive rates by identifying hit genes based on the comparison between the distribution of phenotypes observed for shRNAs targeting each gene and the distribution of negative control shRNAs. This approach is extremely effective in identifying genes that confer sensitivity or resistance to a drug or toxin using survival-based assays (Bassik et al., 2013) and thus potentially useful in identifying target genes for drugs with an unknown mechanism of action.
There is a critical need for new agents with novel therapeutic targets and improved safety profiles in cancer treatment. This is particularly the case for high-risk and relapsed acute lymphoblastic leukemia (ALL), which is a significant cause of morbidity and mortality in pediatric and adult populations (Pui et al., 2008). Although significant advances have been made in treatment, high-risk ALL continues to pose significant therapeutic challenges. Cytotoxic agents remain the standard of care for acute leukemia, and for decades therapies have relied on similar regimens. Despite numerous efforts to improve treatments with new drug combinations, these approaches have reached a point of diminishing returns since intensified chemotherapies contribute only marginal improvement in outcome and display increased toxicity with long-term sequelae.
We report the use of a chemical genetics approach to identify novel small molecules active in ALL and their targets. We performed sequential unbiased high-throughput phenotypic chemical and ultracomplex, genome-scale shRNA screens to identify a novel class of inhibitors that target NAMPT, a rate-limiting enzyme in the salvage pathway for biosynthesis of NAD+, a crucial cofactor in many biological and biochemical processes.
RESULTS
Identification of novel cytotoxic compounds by high throughput chemical screen
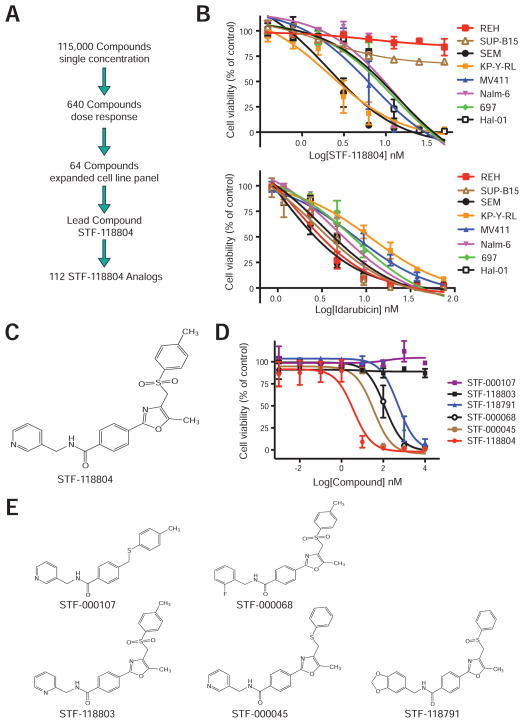
A cell-based phenotypic high-throughput chemical screen (HTS) was conducted using B-ALL cell lines to identify compounds with potential selective anti-leukemic properties (Figure 1A). Screening of 115,000 compounds in HTS format employed a metabolic assay of viability based on redox reduction of resazurin (CellTiter-Blue). Six hundred and forty initial preliminary “hits” were identified that reduced viability of one or both ALL cell lines with high-risk Mixed Lineage Leukemia (MLL) genetic abnormalities (RS411 and HB) by greater than 30%, but did not inhibit the growth of a cell line lacking MLL rearrangement (REH). Compounds that affected all three cell lines, such as the anthracycline idarubicin, were excluded from further analysis. Initial candidates (640) were retested for their dose-dependent toxicity to yield 86 validated compounds with selective properties. The most potent and selective 64 were reordered and tested on an expanded panel of eight human B-ALL cell lines to identify lead compound STF-118804 whose structure (Figure 1C) was verified by NMR analysis and mass spectrometry (data not shown).
Figure 1. Phenotypic chemical HTS screen identifies a class of cytotoxic compounds.
(A) Schematic summary of the experimental approach employed for a phenotypic, cell-based, high-throughput, small molecule screen and subsequent confirmatory analyses that identified lead compound STF-118804, which inhibited human ALL cell lines with nanomolar potency. Structurally related analogs were tested to determine structure-activity-relationship (SAR) of STF-118804.
(B) Human B-ALL cell lines were treated with STF-118804 (upper) or idarubicin (lower) and assessed for viability at day 3 via CellTiter-Blue. Data are from 3 independent experiments performed in triplicate (± SEM).
(C) Chemical structure of STF-118804.
(D) Viability of MV411 cells was quantified after 3-day culture in the presence of the indicated structural analogs of STF-118804. Data represent three independent experiments performed in triplicate (± SEM).
(E) Chemical structures are shown for representative STF-118804 analogs.
STF-118804 reduced the viability of most B-ALL cell lines with high potency demonstrating IC50 values in the low nanomolar range (Table 1). Cell lines with the highest sensitivity (SEM and KP-Y-RL) contained MLL chromosomal translations whereas two non-MLL cell lines (REH and SUP-B15) were resistant (Figure 1B). While the increased sensitivity may not be clinically relevant, it suggested specificity. Idarubicin, which non-specifically intercalates into DNA, affected all cell lines in the low nanomolar range without selectivity (Figure 1B, Table 1). Leukemic samples from 5 pediatric ALL patients were also sensitive to STF-118804 in the low nanomolar range (Table 1). The lead compound displayed 5–10 fold more potency against most leukemias in comparison to cycling human (lineage-negative cord blood) and murine (c-kit+ BM) progenitor cells (Figure S1, Table 1), a therapeutic index considerably larger than that of idarubicin, a drug with known efficacy used in ALL therapy. Thus, high throughput screening identified a promising lead compound with potentially selective anti-leukemic properties.
Table 1.
Inhibitory concentrations for STF-118804 and idarubicin on cell lines, patient samples and normal progenitors
| Cell Type | STF-118804 IC50 (nM) (± 95% confidence interval) |
Idarubicin IC50 (nM) (± 95% confidence interval) |
|---|---|---|
| SEM | 2.7 (2.3–3.2) | 1.0 (0.4–2.4) |
| KP-Y-RL | 2.6 (1.9–3.4) | 11.4 (6.8–19.2) |
| MV411 | 6.0 (5.2–7.0) | 7.6 (3.9–14.9) |
| Nalm6 | 8.8 (8.0–9.8) | 4.6 (2.8–7.5) |
| 697 | 8.6 (7.5–9.8) | 7.6 (4.3–13.4) |
| Hal01 | 9.2 (8.1–10.5) | 3.2 (1.9–5.4) |
| REH | > 10,000 | 1.5 (0.7–5.4) |
| SUP-B15 | > 10,000 | 2.8 (1.0–7.5) |
| MLL-AF4 Patient 1 | 13.9 (6.3–31.1) | ND |
| MLL-AF4 Patient 2 | 32.3 (13.3–78.3) | ND |
| MLL-AF4 Patient 3 | 3.1 (1.1–29) | ND |
| MLL-AF4 Patient 4 | 5.1 (1.3–20.1) | ND |
| TEL-AML Patient 5 | 4.2 (3.1–5.6) | ND |
| Lin− Cord Blood Cells | 40.2 (19.6–82.5) | 11.1 (7.5–16.7) |
| c-kit+ murine BM | 62.3 (52.1–74.5) | 11.5 (9.0–14.6) |
ND, not determined.
Cells were incubated with increasing concentrations of STF-118804 or idarubicin, and viability was assessed using CellTiter-Blue or WST-1 reagent. Assays were performed in triplicate and inhibitory concentrations were calculated using Prism or CalcuSyn software.
Structure activity relationship was assessed for 112 structurally related analogs of STF-118804 in HTS format for their ability to inhibit the in vitro growth of four ALL cell lines. Five representative analogs were individually retested for their toxicity on leukemia cell line MV411 (Figures 1D and 1E, Table S1). These studies identified STF-118804 as a potent representative of a novel class of cytotoxic compounds and defined chemical sub-groups critical for its potency and toxicity.
STF-118804 induces leukemia cell apoptosis without antecedent cell cycle arrest
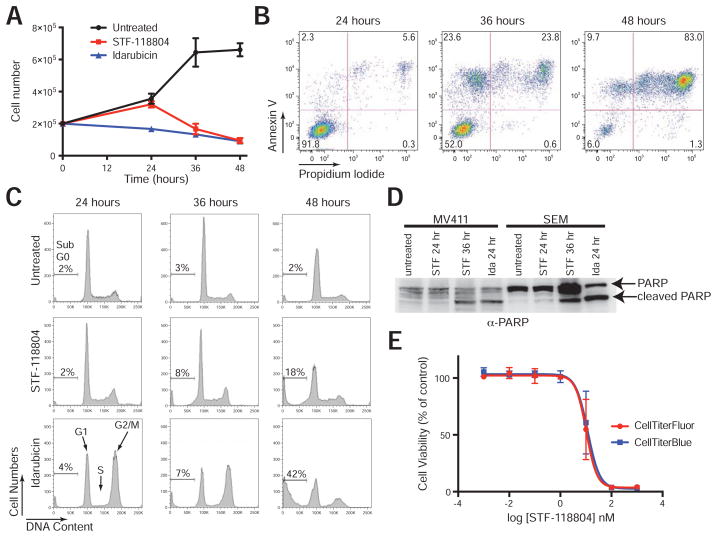
MV411 cells treated with STF-118804 proliferated for 24 hrs comparable with the vehicle control (Figure 2A), but were markedly decreased in number at 36 hrs. This correlated with the onset of apoptosis as quantified by flow cytometry for annexin V positive cells (Figure 2B) and the presence of cleaved PARP at 36 hrs (but not 24 hrs) in western blot analysis (Figure 2D). At 48 hrs, the vast majority of MV411 cells were non-viable as evidenced by propidium iodide staining (Figure 2B).
Figure 2. STF-118804 induces apoptosis without antecedent cell cycle arrest.
(A) Growth of MV411 cells in the absence or presence of STF-11804 (100 nM) or idarubicin (25 nM) was quantified by trypan blue exclusion. Errors bars represent standard error of the mean for three independent experiments.
(B) MV411 cells were treated with STF-118804 (100 nM) for the indicated times and assessed by flow cytometry for annexin V-FITC and propidium iodide staining. Double positive cells correspond to late stages of apoptosis. Data are representative of three independent experiments.
(C) Cell cycle status was assessed by propidium iodide staining of MV411 cells treated or untreated for the indicated times with STF-118804 (100 nM) or idarubicin (25 nM). STF-118804 induced accumulation of a sub-G0 population at 48 hrs without antecedent reduction of the S/G2/M population, in contrast to idarubicin. Data are representative of three independent experiments.
(D) The presence of cleaved PARP (89 kD) was assessed by western blot analysis of lysates from MV411 or SEM leukemia cell lines treated with 100 nM STF-118804 for 24 (STF 24) or 36 (STF 36) hours, or 25 nM idarubicin for 24 hours (Ida 24). Data are representative of two independent experiments.
(E) Dose-response curve of MV411 cells treated with STF-118804 for 72 hours. Cell viability was measured by a resazurin-based assay (CellTiter-Blue) and a live cell protease-based assay (CellTiter-Fluor). Data represent three independent experiments performed in triplicate (± SEM). See also Figure S2.
Cell cycle analysis based on DNA content confirmed that MV411 cells continued to cycle and did not arrest before the onset of apoptosis (appearance of sub-G0 population) at 36 hours of treatment (Figure 2C). This contrasted with idarubicin, which induced rapid cell cycle arrest (24 hrs) characterized by a decreased percentage of cells in G1 and S phases and an accumulation of cells in G2/M phase (Figure 2C). Therefore, STF-118804 displays distinctive cytotoxicity by inducing apoptosis without causing a phase-specific cell cycle arrest.
Assessment of cell viability following treatment with STF-118804 using a live cell protease activity assay (CellTiter-Fluor) gave equivalent results as the resazurin-based assay (CellTiter-Blue) (Figure 2E) confirming that the latter provided a reasonable measure of cell viability in the HTS and validation studies.
In an NCI-60 cell line panel screen, STF-118804 showed potency in various cancer cells including high selectivity for colon and prostate cell lines (Figure S2), suggesting broader applications beyond leukemia. Comparison of the STF-118804 sensitivity profile to known chemotherapeutic agents in the NCI’s database (NCI COMPARE) showed no significant overlap indicating that STF-118804 functions through a unique mode of action.
Functional genomic shRNA screen identifies NAMPT as a candidate target for STF-118804
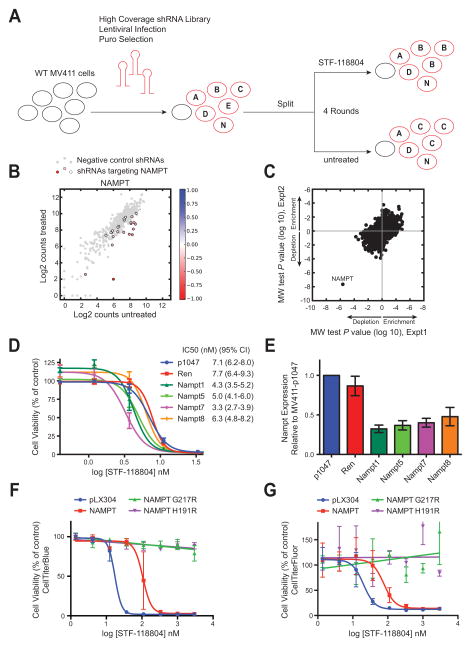
To discover the molecular target of STF-118804, a functional genomic screen was performed to identify shRNAs that conferred sensitivity or resistance to STF-118804 (Figure 3A). MV411 cells were transduced with pooled sublibraries of shRNAs from a high coverage (~25 shRNA per gene) library targeting in total ~9300 human genes and including 500 or more negative control shRNAs per sublibrary (Bassik et al., 2013). Stably transduced cells were subjected to four rounds of treatment with increasing concentrations of STF-118804, or four rounds of mock treatment. Several shRNAs targeting nicotinamide phosphoribosyl transferase (NAMPT) were markedly depleted in the STF-118804-treated population compared to control shRNAs (Figure 3B), indicating that shRNAs against NAMPT conferred sensitivity to STF-118804. NAMPT was the most statistically significant gene to confer sensitivity to STF-118804 in two independent experimental replicates (Figure 3C, Mann Whitney U test). P values for all genes from the two replicate screens are given in Table S2.
Figure 3. Knockdown of NAMPT increases sensitivity to STF-118804.
(A) Scheme is shown for ultracomplex shRNA screen in which wild-type MV411 leukemia cells were transduced with a pooled shRNA lentiviral library and subjected to four rounds of treatment with STF-118804 or four rounds of passage. Frequency of shRNAs in treated and untreated cells was quantified by high-throughput sequencing.
(B) Counts of individual NAMPT shRNAs (colored dots) in STF-118804 treated vs. untreated cells are shown in comparison to distribution of negative control shRNAs (gray dots). Color scale represents enrichment (blue) or depletion (red) of individual shRNAs in treated vs. untreated cells.
(C) Mann Whitney U test P values for enrichment and depletion of STF-118804-treated cells in which different genes are knocked down are shown for two independent experimental replicates.
(D) STF-118804 dose response is shown for MV411 cells expressing empty vector (pMK1047), or shRNAs against Renilla luciferase (Ren), or four different shRNAs against NAMPT. Data are from five independent experiments (± SEM). Respective IC50 and 95% confidence interval values are shown to the right.
(E) Bar graph shows NAMPT transcript levels in cell lines used in panel 3D as measured by qPCR. Data are from four independent experiments (mean ± SEM).
(F) (G) STF-118804 dose-response in HEK293T cells transfected with empty vector (pLX304), or constructs expressing the indicated wt or mutant NAMPT proteins. Cell viability was measured by CellTiter-Blue (F) or CellTiter-Fluor (G). Data are from three independent experiments performed in triplicate (±SEM).
See also Table S2.
To validate the screen results, individual shRNAs against NAMPT were re-tested for their ability to confer sensitivity to STF-118804. MV411 cells stably expressing shRNAs targeting NAMPT exhibited a decrease in STF-118804 IC50 compared to cells expressing empty vector or a control shRNA (Figure 3D). The significant decrease in IC50 corresponded to knock-down of NAMPT expression in the 50–60% range assessed by real-time PCR (Figure 3E), validating the results of the shRNA screen. Although knock-down of NAMPT by itself can reduce NAD+/NADH levels, which could potentially explain decreased signal in a resazurin-based assay, the effect of NAMPT knockdown was dependent on STF-118804 concentration, which, together with the results of the functional genomic screen, strongly implicates NAMPT as the molecular target of STF-118804.
The cytotoxic effects of STF-118804 are exerted through NAMPT inhibition
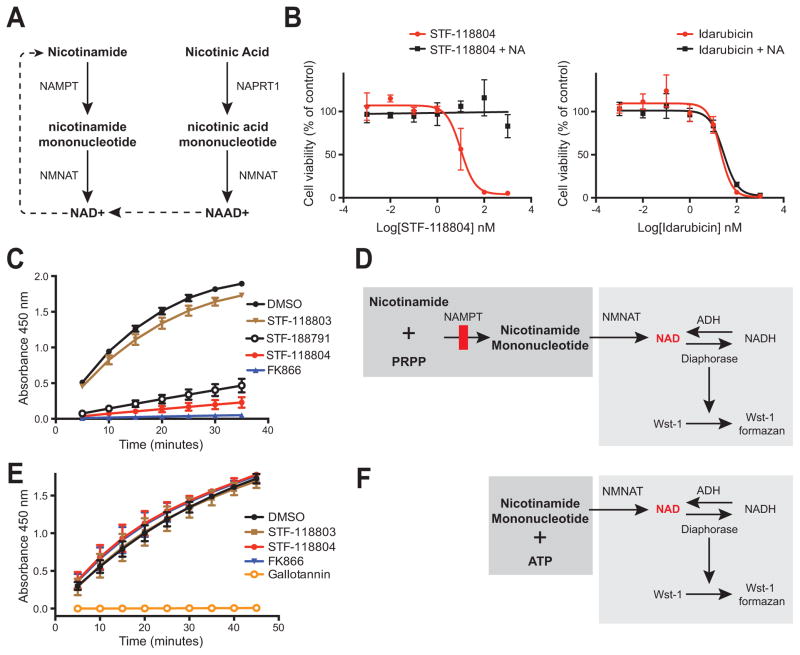
NAMPT is the rate-limiting enzyme in a metabolic salvage pathway that converts nicotinamide to NAD+ (Figure 4A) (Belenky et al., 2007). The Preiss-Handler pathway is an alternative NAD+ biosynthesis pathway that bypasses NAMPT by using nicotinic acid as a substrate to form NAD+ (Figure 4A). Addition of nicotinic acid to in vitro viability assays (measured by live cell protease activity) completely abrogated the toxicity induced in leukemia cells by STF-118804 but not idarubicin (Figure 4B). The observed rescue of viability via the Preiss-Handler pathway suggested that the cytotoxic effects of STF-118804 were due to depletion of NAD+.
Figure 4. STF-118804 specifically inhibits NAMPT enzymatic activity.
(A) The salvage (left side) and Press-Handler (right side) pathways that produce NAD+ from nicotinamide and nicotinic acid, respectively, are shown schematically.
(B) The ability of nicotinic acid (10 μM) to rescue the viability of MV411 cells treated with STF-118804 (left) or idarubicin (right) was assessed at 72 hours of treatment by CellTiter-Fluor. Data represent three independent experiments performed in triplicate (± SEM).
(C) (D) The indicated compounds were tested for inhibition of NAD+ production in a coupled in vitro enzyme assay (shown schematically) in which the enzymes NAMPT and NMNAT produce NAD+, which is indirectly measured via the formation of the colorimetric indicator Wst-1 formazan that is quantified via absorbance at 450 nm. Data represent three independent experiments performed in duplicate (± SEM).
(E) (F) The indicated compounds were assessed for inhibition of NAD+ production by NMNAT, which produces NAD+ from the substrates nicotinamide mononucleotide and ATP (shown schematically). NAD+ production is indirectly measured by formation of NADH, which converts Wst-1 to Wst-1 formazan. STF-118804 did not inhibit NMNAT or other enzymes in the coupled assay including ADH and diaphorase, thus establishing its specificity for NAMPT, whereas gallotannin (10 μM), a known NMNAT inhibitor, prevented production of NAD+. Data represent three independent experiments performed in duplicate (± SEM).
The ability of STF-118804 and analogs to inhibit biosynthesis of NAD+ from nicotinamide was measured using a coupled in vitro enzyme assay containing NAMPT and NMNAT (Figures 4C and 4D). STF-118804 and its less active analog STF-118791 prevented NAD+ production (quantified by Wst-1 formazen synthesis) as did the known NAMPT inhibitor FK866 (all at 10 μM), which was also cytotoxic to MV411 cells (Figure 1D, Table S1). In contrast, the analog STF-118803, which lacked inhibitory activity in cellular assays, did not block NAD+ production in vitro. The observed reduction in NAD+ biosynthesis was caused by specific inhibition of NAMPT in the coupled reaction since none of the analogs or FK866 inhibited production of NAD+ catalyzed by the downstream enzyme NMNAT in a parallel in vitro assay (Figures 4E and 4F). Taken together, the inhibitory activities of STF-118804 and analogs in NAMPT enzymatic assays, which correlate with their SAR in cellular assays, establish that NAMPT is a molecular target.
Over-expression of NAMPT rendered 293T cells more resistant to STF-118804 (Figure 3F and 3G), resulting in a higher IC50 (106 nM, 95% CI 74–151 nM) compared to control cells (17 nM, 95% CI 13–23 nM), further confirming that NAMPT protein levels dictate sensitivity to STF-118804. Previous studies have shown that resistance to known NAMPT inhibitors FK866 and CHS-828/GMX1778 (Olesen et al., 2010; Watson et al., 2009) is conferred by arginine substitutions (G217R or H191R) in the hydrophobic tunnel in which FK866 binds (Khan et al., 2006). The mutant proteins retain enzymatic activity but are not inhibited by FK866 or CHS-828/GMX1778 (Olesen et al., 2010; Watson et al., 2009). Similarly, cells over-expressing G217R or H191R mutants were completely resistant to STF-118804 (Figure 3F and 3G), suggesting that it likely binds in the NAMPT hydrophobic tunnel similar to FK866. These results indicate that STF-118804 cytotoxicity is a result of its ability to inhibit NAMPT, and that STF-118804 does not have significant off-target effects on cell viability.
STF-118804 is highly efficacious in vivo and effectively depletes leukemia initiating cells
The potential efficacy of STF-118804 was assessed in an orthotopic xenograft model of ALL. Sublethally irradiated NSG mice were transplanted with MV411 cells engineered to constitutively express firefly luciferase. Dosing of STF-118804 was initiated two weeks post-transplant when MV411 cells had engrafted and bioluminescent signal was detectable (Figures 5A and 5B). Although STF-118804 plasma levels rapidly decreased after subcutaneous, intra-peritoneal, or oral delivery (Figure S3A), a target plasma concentration of 100 nM was maintained for 22 hours by dosing mice subcutaneously twice a day with a 5.5-hour interval (Figure S3B).
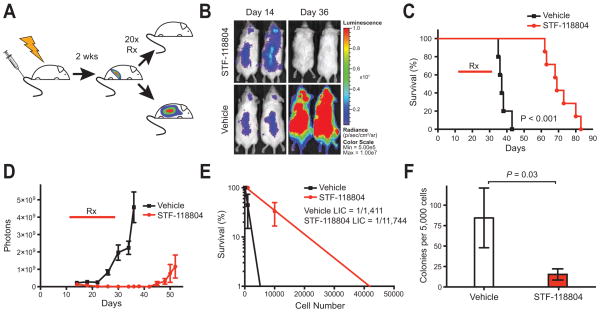
Figure 5. STF-118804 is highly efficacious in an orthotopic xenograft model of ALL.
(A) Sublethally irradiated (2.5 gy) NSG mice were transplanted with MV411 cells (5 × 106) modified to constitutively express firefly luciferase. Two-weeks post transplant mice were randomized into 2 groups, and treated subcutaneously for 20 days with either a split dose of 50 mg/kg STF-118804 (n = 7) or vehicle (n = 5).
(B) Bioluminescent images of representative mice show no detectable tumor at Day 36 of STF-118804 treatment.
(C) Survival curves for mice treated with STF-118804 (days 14–34) indicate survival extension for an average of 34 days longer than vehicle treated mice (P < 0.001, log-rank test).
(D) Bioluminescence quantification shows prolonged suppression of disease for at least 18 days after treatment was discontinued and prevention of relapse in individual mice. Data represent mean numbers of photons from bioluminescence images for STF-118804 treated (n = 7) and vehicle treated (n = 5) mice (± SEM).
(E) Frequencies of leukemia initiating cells (LICs) were quantified at Day 35 by limit-dilution secondary transplantation and Poisson statistics in mice treated with STF-118804 (n = 3) or vehicle (n = 3). Mice treated with STF-118804 showed a LIC frequency of 1/11,744 (1/4,480–1/30,784) whereas mice treated with vehicle showed a LIC frequency of 1/1,411 (1/600–1/3,320). Error bars are standard error of the mean.
(F) Bar graph shows significant reduction (P = 0.03, Student’s t-test) of leukemia colony forming cells in bone marrow of mice treated with STF-118804 (n = 8) versus vehicle (n = 6) at Day 35 (mean ± SEM). Whole bone marrow was plated in human methocult without cytokines, which does not support the growth of endogenous murine progenitors.
See also Figures S3 and S4.
Mice treated with STF-118804 over a 20-day period survived an average of 34 days longer than vehicle-treated animals (Figure 5C). During treatment, bioimaging showed regression of tumor followed by suppression of disease (Figures 5B and 5D). Regrowth was not apparent, as measured by bioluminescence, in individual animals until at least 18 days after treatment was discontinued (Figure 5D). Thus, STF-118804 displayed high efficacy in a xenograft model of ALL.
To assess potential effects on leukemia initiating cells (LICs), transplanted mice were sacrificed after 20 days of treatment and analyzed for levels of residual disease. Whole bone marrow from STF-118804 or vehicle treated mice was transplanted into secondary recipients to determine LIC frequency using Poisson statistics. The LIC frequency in STF-118804 treated mice was significantly lower (~8 fold) than vehicle treated mice showing that STF-118804 was effective in reducing leukemia stem cells (Figure 5E). Additionally, bone marrow from STF-118804 treated mice formed fewer colonies in semi-solid media that supports growth of human cells, but not endogenous murine progenitors (Figure 5F). These results indicate that STF-118804 effectively reduces tumor burden and is also capable of targeting LICs in a xenograft model of high-risk ALL. STF-118804 was tolerated in the efficacious dose range, and the absence of adverse physical or pathological effects indicated that toxicity was not limiting in a 20-day study of mock transplanted mice (Figure S4).
DISCUSSION
We applied a sequential screening approach in a model of high-risk pediatric B-ALL to identify a novel class of NAMPT inhibitors. First, a high-throughput cell-based phenotypic screen identified the highly cytotoxic lead compound STF-118804. Subsequently, an unbiased, ultracomplex, genome-scale shRNA forward genetic screen identified NAMPT as the molecular target of STF-118804. The target was then validated and the lead compound was shown to be efficacious in an orthotopic model. These results demonstrate that sequential high-throughput screening for small molecule identification and shRNA mechanism of action discovery is an effective approach for drug development, combining the advantages of phenotypic screening with the power of functional genomics.
Chemical genetics, or the use of small molecules in shRNA screening, has been used to study the mechanism of action of chemotherapeutic agents with known targets (Brummelkamp et al., 2006; Luo et al., 2008), identify potentially effective drug combinations (Azorsa et al., 2009; Hassane et al., 2010) and understand drug resistance (Vidot et al., 2010). We demonstrate the first use of genome-scale chemical genetics to identify the target of a novel small molecule discovered in an unbiased cell-based phenotypic screen. Other shRNA proof-of-concept studies have used low-coverage libraries with drugs at toxic concentrations, which may bias results by reducing shRNA library complexity. Ultracomplex (~25 shRNAs per gene, 1,000s of negative control shRNAs) shRNA library-based screens provide an advantage over other shRNA approaches or target identification methods since they provide a way to identify genetic hits with high statistical confidence and do not require sophisticated knowledge of chemical SAR, or synthetic chemistry expertise.
NAMPT inhibitors in cancer therapeutics
NAMPT plays a key role in regulating cellular metabolism and represents an attractive therapeutic target in oncology. NAMPT over-expression renders cell lines more resistant to chemotherapeutic agents, while knockdown causes sensitization (Yang et al., 2007). Since cancer cells have altered metabolic needs and higher rates of NAD+ turnover, their reliance on specific metabolic pathways is perturbed (Locasale and Cantley, 2010; Vander Heiden, 2011). Growing evidence indicates metabolic reprogramming is not simply a result of increased proliferation associated with cancer cell growth, but a direct function of oncogenes and tumor suppressors and therefore a driver of oncogenesis (Dang, 2012; DeBerardinis et al., 2008; Ward and Thompson, 2012).
It is unknown why leukemia cells may be particularly susceptible to NAMPT inhibition. One possibility is that NAD+ depletion by NAMPT inhibitors leads to ATP depletion, and cancer cells may be susceptible due to high cell metabolism/turnover. However, this hypothesis is difficult to reconcile with data indicating that normal cord blood cells proliferating at the same rate as leukemia cells were not as susceptible to STF-118804. Various mechanisms may account for the cytotoxic effects of NAMPT inhibition. NAD+ depletion inhibits the NAD+-dependent glyceraldehyde 3-phosphate dehydrogenase, thus inhibiting glycolysis (Tan et al., 2013), which some cancer cells are preferentially dependent on even in aerobic conditions. Childhood B-ALL cells have been shown to undergo a shift in glucose metabolism that may render them more sensitive to NAMPT inhibition (Boag et al., 2006).
Other work suggests that NAD+ depletion might have an impact on the sirtuins, NAD+-dependent protein deacetylases important in cancer (Cea et al., 2011; Imai et al., 2000). SIRT1, a member of the sirtuin family, deacetylates and inactivates p53, which inhibits its tumor suppressor function (Vaziri et al., 2001). SIRT1 is overexpressed in human CML leukemia stem cells (LSCs), and inhibition of SIRT1 increases apoptosis in LSCs and reduces their growth (Li et al., 2012). Deactivation of SIRT1 via NAMPT inhibition may provide a way to target LSCs and is consistent with our observation that STF-118804 is effective against LICs in a xenograft model of high-risk ALL.
STF-118804 represents a novel class of NAMPT inhibitors that may improve upon previously described inhibitors given its substantial pre-clinical efficacy. CB30865 (Fleischer et al., 2010), FK866/APO866/WK175 (Wosikowski et al., 2002), and CHS-828/GMX1778 (Olesen et al., 2008) have been shown to decrease cellular levels of NAD+. They are active against multiple tumor types, including leukemia (Nahimana et al., 2009; Wosikowski et al., 2002). Like STF-118804, they induce apoptosis (Hasmann and Schemainda, 2003) without causing a phase-specific cell cycle arrest (Skelton et al., 1998), and NAMPT knockdown increases sensitivity to CHS-828 while over-expression decreases sensitivity (Watson et al., 2009). However, NAMPT inhibitors that have entered clinical trials to date have yet to show efficacy (Holen et al., 2008; Hovstadius et al., 2002). Their clinical effectiveness may have been limited by poor bioavailability or the selection of an inappropriate indication and/or patient population. Our studies suggest high-risk ALL, colon cancer, and prostate cancer patients may benefit most.
STF-118804 displays a wide therapeutic index, safety and efficacy in vivo, and is the first NAMPT inhibitor with demonstrated efficacy in depleting LICs. Its most potent analogs share a meta-substituted pyridine ring that is also present in other classes of NAMPT inhibitors. However, our SAR studies indicate that other chemical sub-groups can be substituted at this position. Critical structural features that distinguish the STF-118804 inhibitor class include a 5-methyl-oxazole ring sub-structure. Our studies using site-directed mutant NAMPT proteins suggest that STF-118804 binds in a hydrophobic tunnel between the dimerized enzyme subunits similar to FK866 (Khan et al., 2006). No inhibitors have been reported to bind a different surface of NAMPT or within the known tunnel in a different manner.
NAMPT represents a novel metabolic target in ALL. Relapse in ALL is thought to be due to the re-emergence of a leukemia stem cell clone. By targeting bulk leukemia cells as well as leukemia stem cells, STF-118804 has the potential to address the major problem of relapse in high-risk ALL patients. Our studies demonstrate that STF-118804 has high activity in an in vivo xenotransplant model of ALL and is the first NAMPT inhibitor with demonstrated efficacy in depleting LICs. Efficacy and safety in this model provide a rationale for further advancement to lead optimization and clinical testing, and demonstrate that NAD+ biosynthesis inhibitors have the potential to significantly impact high-risk leukemia.
EXPERIMENTAL PROCEDURES
Cell culture
Human leukemia cell lines were obtained from ATCC or DSMZ, with the exception of HB(Smith et al., 1989) and maintained at 37°C and 5% CO2 in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin-glutamine. Cells lines were tested for Mycoplasma by PCR (MD Bioproducts) and results verified by IDEXX RADIL Laboratories. Cells were seeded at a density of 6×105 cells/ml in all cellular assays. Human leukemia cell colony forming assays were performed in semi-solid media (Stem Cell Technologies H4230). Murine progenitors were cultured in M3630 (Stem Cell Technologies) supplemented with 20 ng/ml SCF, 10 ng/ml IL3, 10 ng/ml IL6, 10 ng/ml GM-CSF (Peprotech). Colonies were enumerated at day 7. Cord blood progenitors were enriched from whole blood (collected from newborn placenta with informed consent and Stanford Institutional Review Board approval) by Ficoll separation, and depleted of cells expressing lineage-specific antigens using a lineage depletion antibody cocktail and an AutoMACS magnetic cell separator (Miltenyi Biotech, Auburn, CA). Cord blood progenitors were grown in IMDM (Gibco) containing 20% (v/v) FBS and human cytokines (100 ng/ml TPO, FLT3, SCF and 20 ng/ml IL-6). Primary patient bone marrow ALL samples were obtained at diagnosis after informed consent and IRB approval, and mononuclear cells were enriched by Ficoll-Hypaque (density 1.077 g/mL; GE Healthcare). Cryopreserved cells were thawed, suspended in AIM-V +AlbuMAX media (Invitrogen), 20% (v/v) FBS, and 2 mM L-glutamine, and depleted of cell debris by Ficoll-Hypaque, and suspended in media.
Chemical compounds
Chemical compounds were obtained from ChemDiv, Specs, and Chembridge. STF-118804 was synthesized at ChemDiv. FK866 was acquired from Cayman Chemicals (13287). Gallotannin (16201) and nicotinic acid (72309) were purchased from Sigma Aldrich. Compounds were dissolved in DMSO to stock concentration of 10 mM. For use in cell culture, compounds were diluted to 2 mM in DMSO and serially diluted in media to achieve final concentrations. For use in animals, STF-118804 was formulated in 5% (v/v) DMSO and 20% (w/v) 2-hydroxypropyl-gamma-cyclodextrin (Sigma Aldrich H125).
High-throughput screening
High-throughput screening was performed at the Stanford High-Throughput Biosciences Center using libraries from ChemDiv, Specs, and Chembridge. Human leukemia cell lines (HB, RS411, and REH) were plated in clear bottom/black walled 384-well microplates (E&K Scientific) using a Matrix Wellmate (Thermo Scientific). Compounds were added using a robotic pin tool in an automated incubator, and plates were held at 37°C/5% CO2 for 48 hours. Cell viability was assessed 4 hours after addition of CellTiter-Blue reagent (Promega G8081) in an AnalystGT (Molecular Devices) plate reader at an excitation of 555 nm and emission of 590 nm.
Cell viability assays
Human cell lines or lineage negative cord blood cells were seeded into 96-well plates (6×105 cell/ml). Compounds were added in increasing concentrations and cells were incubated at 37°C/5% CO2 for 72 hours. To detect viability, CellTiter-Blue reagent was added at 1:10 dilution, and plates were incubated for 4 hours at 37°/5% CO2 prior to reading on a Flexstation II 384 (Molecular Devices) or a Synergy H1 (BioTek) at an excitation of 555 nm and emission detection of 590 nm. Cell viability was also measured by CellTiter-Fluor (Promega G6082). The cell-permeable fluorogenic peptide substrate glycylphenylalanyl-aminofluorocoumarin (GF-AFC) reagent was added at 1:2 dilution, and plates were incubated for 30 minutes at 37°/5% CO2 prior to reading on a Synergy H1 at an excitation of 380 nm and emission detection of 505 nm. Cord blood cells were enumerated on a hemocytometer and cell viability was assessed by trypan blue exclusion dye. All inhibitory concentrations (IC50) were calculated using Prism (Graphpad) software.
Primary patient samples were plated in 96-well plates and treated with increasing concentrations of STF-118804 for 48 hours at 37°C in 5% CO2. WST-1 reagent (Roche Diagnostics) was added to the culture medium (1:10 dilution) and absorbance was measured at 450 nm using a Bio-Rad model 680 microplate reader. All assays were performed in triplicate. Inhibitory concentrations (IC50) were calculated using CalcuSyn version 2.0 software (Biosoft).
Apoptosis assays
Apoptosis was assessed by flow cytometry using an annexin V-FITC detection kit (BD Biosciences 556420) per manufacturer’s directions. Cells were collected, washed in PBS, and resuspended in binding buffer (0.1 M Hepes pH 7.4, 1.4 M NaCl, 25 mM CaCl2). Annexin V-FITC and propidium iodide were added and cells were incubated for 5 minutes at room temperature. Binding buffer was added and cells were analyzed using a FACScan Analyzer (custom Stanford and Cytek), and data were analyzed using FlowJo v9.4.9.
Cell cycle analysis
Cellular DNA content was analyzed via propidium iodide staining. Cells were collected by centrifugation, resuspended in PBS, and fixed by addition of cold (−20°C) 100% ethanol to a final concentration of 70% (v/v). Cells were washed and resuspended in propidium iodide staining solution [50 μg/ml propidium iodide (Sigma P41070), 3.8 mM sodium citrate, and 0.02 mg/ml RNase] and incubated at 4°C in the dark. Samples were analyzed using a FACScan Analyzer and data were analyzed using FlowJo v9.4.9.
Western blot analysis
Pelleted cells were resuspended in SDS lysis buffer (50 mM Tris HCl pH 6.8, 2% (w/v) SDS, and 10% (v/v) glycerol) prior to electrophoresis in SDS-PAGE, electrophoretic transfer, and immunoblotting using a PARP-specific antibody (Cell Signaling Technology 95425) and rabbit anti-goat HRP secondary antibody (Invitrogen 811620).
STF-118804 sensitivity/resistance screen
Four 55,000-element shRNA lentivirus sub-libraries targeting a total of ~9300 human genes (~25 shRNAs per gene) and containing 500 or more negative control shRNAs were used to transduce MV411 cells in four pools. After puromycin selection and expansion, MV411 cells stably expressing a sub-library of pooled shRNAs were split and subjected to four rounds of treatment with STF-118804 or mock-treatment. For each round, cells were treated with STF-118804 for 48 hours and then allowed to recover in the absence of compound for 48 hours. Increasing concentrations of STF-118804 were used for later rounds of treatment (range 6–12 nM). A minimum of 5–6 × 107 cells per treatment condition was maintained at all times to ensure ~1000× coverage of the 55,000-element sub-library. Genomic DNA was purified from STF-118804 treated and untreated cells, shRNA sequencing libraries (Supplemental Experimental Procedures) were prepared as described (Bassik et al., 2013) and frequencies of shRNA-encoding cassettes were determined by next-generation sequencing (Illumina HiSeq). The STF-118804 resistance ρ conferred by an individual shRNA was calculated as described (Bassik et al., 2013). The set of ρ values of all shRNAs for a given gene were compared to the set of ρ for the negative control shRNAs, and the significance by Mann-Whitney U test for enrichment (resistance) or depletion (sensitivity) was calculated. The STF-118804 resistance screen was carried out in two independent replicates. To calculate P values combining data from both experimental replicates, shRNA phenotypes (STF-118804 resistance ρ) from the replicates were averaged before calculating a P value for each gene (Table S2).
NAMPT shRNA knockdown and overexpression
Individual shRNA sequences (Supplemental Experimental Procedures) were cloned into the BstXI site of the pMK1047 lentiviral vector for stable transduction of MV411 cells, which were tested for STF-118804 dose response. NAMPT expression in transduced cells was quantified by real-time PCR (primers given in Supplemental Experimental Procedures). Relative expression was calculated using the comparative Ct method with ACTB as a reference gene.
HEK293T cells were transiently transfected with pLX304 (Addgene) encoding wt NAMPT (CCSB-Broad Lentiviral Expression Library, Thermo Scientific) or mutant derivatives using TurboFect (Thermo Scientific). pLX304-NAMPT (G217R) and pLX304-NAMPT (H191R) were created by standard site-directed mutagenesis. 24 hours after transient transfection, cells were seeded in 96-well plates at 10,000 cells/well. Cells were treated with STF-118804 for 72 hours to measure dose-response using CellTiter-Blue or CellTiter-Fluor.
Enzyme assays
The enzymatic activities of NAMPT (CY-1251) and NMNAT (CY-1252) were measured using in vitro kits (MBL International) employing the 2-Step Method per manufacturer’s instructions. Compounds, NAMPT and/or NMNAT enzymes, and their substrates were mixed and incubated at 30°C for 1 hour. Reagents for the indicator reaction (Wst-1) were then added and absorbance was read at 450 nm every 5 minutes at 30 C on a Tecan Infinite M100 multimode plate reader (Tecan).
Animal studies
Male NSG mice (NOD.Cg-PrkdcscidIL2rgtm1Wjl/SzJ) 6–8 weeks of age purchased from the Jackson Laboratory (Bar Harbor, ME) were sublethally irradiated (2.5 gy) and injected via tail vein with MV411 cells (5×106) that were modified to stably express firefly luciferase via lentivirus infection [Cignal Lenti Positive Control (luc) SA Biosciences (CLS-PCL-8)]. Two weeks post-transplant, mice were randomized to be treated by subcutaneous injections of STF-118804 (25 mg/kg) or vehicle (20% (w/v) 2-hydroxypropyl-gamma-cyclodextrin/ 5% (v/v) DMSO) twice daily for 20 consecutive days. Analysis was performed in a non-blinded fashion and statistical methods were not used to estimate sample size. For imaging, mice were injected intra-peritoneally with 150 mg/kg D-luciferan firefly potassium salt (Xenogen XR-1001) and anesthetized with 3% isoflurane. Mice were imaged using an IVIS 200 spectrum imaging system (PerkinElmer), and images were analyzed via Living Image v4.3.1 software (Caliper LifeSciences). All data collected from mice were included for analysis. All experiments on mice in this study were performed with the approval of and in accordance with the Stanford University Administrative Panel on Laboratory Animal Care.
Limiting dilution analysis
Bone marrow was collected after animals had undergone 20 days of treatment (35 days post-transplant) with vehicle or STF-118804. Femurs and tibias were flushed with cold PBS/10% (v/v) FBS, and recovered cells were washed, counted, resuspended in saline and held at 4°C. Serially diluted bone marrow cells were injected into the tail vein of sub-lethally irradiated NSG recipient mice (n = 3 for each dilution).
NCI-60 cell panel
Sensitivity of the NCI-60 cancer cell line panel to STF-118804 was performed at the NCI using published methods (http://dtp.nci.nih.gov/branches/btb/ivclsp.html).
Pharmacokinetics
Blood was collected over a 24-hour period by retro-orbital bleeding of mice that had been treated with STF-118804. Plasma was collected and stored at −80 C. Plasma concentrations of STF-118804 were quantified by mass spectrometry.
Toxicity studies
NSG mice (n = 3 in each group) were injected with saline, STF-118804, or vehicle subcutaneously twice a day for 10 days. Blood was collected via retro-orbital bleeding on the first and last days of dosing for pharmacokinetic analyses. Complete necropsies were performed 1 day after the last dose. Bone and tissues were fixed in Cal Ex II (Fischer Scientific) or 10% (v/v) buffered neutral formalin, respectively, routinely processed for histology, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (Histo-Tech Laboratory) for histological analysis. Blood chemistry analysis was performed in the Stanford Veterinary Service Center’s clinical pathology laboratory.
Supplementary Material
Highlights.
Sequential phenotypic and functional genomic screens enable drug target discovery
Structurally novel class of NAMPT inhibitors display wide therapeutic index
High efficacy of NAMPT inhibition in xenograft model of acute lymphoblastic leukemia
SIGNIFICANCE.
Identifying the target and molecular mechanism of action of small molecules is a critical challenge in drug discovery. By using a strategy of sequential unbiased phenotypic small molecule screens and ultracomplex shRNA library screens, we identified a novel class of highly specific NAMPT inhibitors active in extending survival and depleting leukemia-initiating cells in acute lymphoblastic leukemia. As a proof of concept, these studies establish a chemical genetics approach to drug discovery based on combined use of cell-based chemical and functional genomic screens to identify a phenotypically active class of compounds and subsequently define molecular target and mechanism of action.
Acknowledgments
We thank K. Grimes, D. Mochly-Rosen, and advisors of the SPARK Translational Research Program at Stanford for their support and guidance. We acknowledge the advice and services of the Shared FACS Facility, Transgenic Research Center, Small Animal Imaging Facility, and High-Throughput Bioscience Center. We thank L. Alexandrova and the Stanford Mass Spectrometry Facility for pharmacokinetic analysis, S. Lynch for NMR analysis, and N. Cyr for graphical assistance. These studies were supported by funds from the NIH (CTSA awards UL1-RR02744 and UL1-TR000093; NIH Tumor Biology grant T32-009151; 1U01CA168370-01, J.S.W. and M.T.M.; RO1 GM80783, M.T.M.), the Lucile Packard Foundation for Children’s Health, the SPARK Translational Research Program at Stanford, the Leukemia and Lymphoma Society, the St. Baldrick’s Foundation (M.C.W.), a Howard Hughes Collaborative Initiative Award (J.S.W.), and the UCSF Program for Breakthrough Biomedical Research (J.S.W. and M.T.M.).
C.J.M., M.C.W., M.C.B., M.L.C. designed the study; C.J.M., M.C.W., A.J.D., M.I., O.P., R.R. performed experiments; C.J.M., M.C.W., M.C.B., M.K., M.L.C. analyzed data; D.E.S., P.B. designed experiments and analyzed data; D.M.B. reviewed murine pathology; M.C.B., M.K., M.T.M., J.S.W. provided reagents and technical advice; C.J.M., M.C.W., M.L.C. wrote the manuscript with input from all authors.
Footnotes
Supplemental Information includes four figures, two tables, and Supplemental Experimental Procedures and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azorsa DO, Gonzales IM, Basu GD, Choudhary A, Arora S, Bisanz KM, Kiefer JA, Henderson MC, Trent JM, Von Hoff DD, et al. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. Journal of Translational Medicine. 2009;7:43. doi: 10.1186/1479-5876-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A systematic Mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends in Biochemical Sciences. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Boag JM, Beesley AH, Firth MJ, Freitas JR, Ford J, Hoffmann K, Cummings AJ, de Klerk NH, Kees UR. Altered glucose metabolism in childhood pre-B acute lymphoblastic leukaemia. Leukemia. 2006;20:1731–1737. doi: 10.1038/sj.leu.2404365. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, Hannon GJ, Lowe SW, Hemann MT. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci U S A. 2008;105:9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cea M, Soncini D, Fruscione F, Raffaghello L, Garuti A, Emionite L, Moran E, Magnone M, Zoppoli G, Reverberi D, et al. Synergistic interactions between HDAC and sirtuin inhibitors in human leukemia cells. PLoS One. 2011;6:e22739. doi: 10.1371/journal.pone.0022739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Fleischer TC, Murphy BR, Flick JS, Terry-Lorenzo RT, Gao ZH, Davis T, McKinnon R, Ostanin K, Willardsen JA, Boniface JJ. Chemical proteomics identifies Nampt as the target of CB30865, an orphan cytotoxic compound. Chem Biol. 2010;17:659–664. doi: 10.1016/j.chembiol.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Hahn CK, Berchuck JE, Ross KN, Kakoza RM, Clauser K, Schinzel AC, Ross L, Galinsky I, Davis TN, Silver SJ, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009;16:281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- Hassane DC, Sen S, Minhajuddin M, Rossi RM, Corbett CA, Balys M, Wei L, Crooks PA, Guzman ML, Jordan CT. Chemical genomic screening reveals synergism between parthenolide and inhibitors of the PI-3 kinase and mTOR pathways. Blood. 2010;116:5983–5990. doi: 10.1182/blood-2010-04-278044. [DOI] [PubMed] [Google Scholar]
- Holen K, Saltz LB, Hollywood E, Burk K, Hanauske AR. The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Invest New Drugs. 2008;26:45–51. doi: 10.1007/s10637-007-9083-2. [DOI] [PubMed] [Google Scholar]
- Hovstadius P, Larsson R, Jonsson E, Skov T, Kissmeyer AM, Krasilnikoff K, Bergh J, Karlsson MO, Lonnebo A, Ahlgren J. A Phase I study of CHS 828 in patients with solid tumor malignancy. Clin Cancer Res. 2002;8:2843–2850. [PubMed] [Google Scholar]
- Huang R, Wallqvist A, Thanki N, Covell DG. Linking pathway gene expressions to the growth inhibition response from the National Cancer Institute’s anticancer screen and drug mechanism of action. The Pharmacogenomics Journal. 2005;5:381–399. doi: 10.1038/sj.tpj.6500331. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Khan JA, Tao X, Tong L. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat Struct Mol Biol. 2006;13:582–588. doi: 10.1038/nsmb1105. [DOI] [PubMed] [Google Scholar]
- Kotz J. Phenotypic screening, take two. SciBX: Science-Business eXchange. 2012;5:1–3. [Google Scholar]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nature Reviews Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford E, Peck D, Modell J, Blat I, Wrobel M, Lerner J, Brunet J, Subramanian A, Ross K, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and diseases. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Li L, Wang L, Wang Z, Ho Y, McDonald T, Holyoake TL, Chen W, Bhatia R. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Cantley LC. Altered metabolism in cancer. BMC Biology. 2010;8:88. doi: 10.1186/1741-7007-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahimana A, Attinger A, Aubry D, Greaney P, Ireson C, Thougaard AV, Tjornelund J, Dawson KM, Dupuis M, Duchosal MA. The NAD biosynthesis inhibitor APO866 has potent antitumor activity against hematologic malignancies. Blood. 2009;113:3276–3286. doi: 10.1182/blood-2008-08-173369. [DOI] [PubMed] [Google Scholar]
- Olesen UH, Christensen MK, Bjorkling F, Jaattela M, Jensen PB, Sehested M, Nielsen SJ. Anticancer agent CHS-828 inhibits cellular synthesis of NAD. Biochem Biophys Res Commun. 2008;367:799–804. doi: 10.1016/j.bbrc.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Olesen UH, Petersen JG, Garten A, Kiess W, Yoshino J, Imai S, Christensen MK, Fristrup P, Thougaard AV, Bjorkling F, et al. Target enzyme mutations are the molecular basis for resistance towards pharmacological inhibition of nicotinamide phosphoribosyltransferase. BMC Cancer. 2010;10:677. doi: 10.1186/1471-2407-10-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, Plowman J, Boyd MR. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. Journal of the National Cancer Institute. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- Schenone M, Dancik V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton LA, Ormerod MG, Titley JC, Jackman AL. Cell cycle effects of CB30865, a lipophilic quinazoline-based analogue of the antifolate thymidylate synthase inhibitor ICI 198583 with an undefined mechanism of action. Cytometry. 1998;33:56–66. doi: 10.1002/(sici)1097-0320(19980901)33:1<56::aid-cyto7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Smith SD, McFall P, Morgan R, Link M, Hecht F, Cleary M, Sklar J. Long-term growth of malignant thymocytes in vitro. Blood. 1989;73:2182–2187. [PubMed] [Google Scholar]
- Stegmaier K, Corsello SM, Ross KN, Wong JS, Deangelo DJ, Golub TR. Gefitinib induces myeloid differentiation of acute myeloid leukemia. Blood. 2005;106:2841–2848. doi: 10.1182/blood-2005-02-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney DC, Anthony J. How were new medicines discovered? Nature Reviews Drug Discovery. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Tan B, Young DA, Lu ZH, Wang T, Meier TI, Shepard RL, Roth K, Zhai Y, Huss K, Kuo MS, et al. Pharmacological inhibition of nicotinamide phosphoribosyltransferase (NAMPT), an enzyme essential for NAD+ biosynthesis, in human cancer cells: metabolic basis and potential clinical implications. J Biol Chem. 2013;288:3500–3511. doi: 10.1074/jbc.M112.394510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujii H, Eguchi Y, Chenchik A, Mizutani T, Yamada K, Tsujimoto Y. Screening of cell death genes with a mammalian genome-wide RNAi library. J Biochem. 2010;148:157–170. doi: 10.1093/jb/mvq042. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nature Reviews Drug Discovery. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vidot S, Witham J, Agarwal R, Greenhough S, Bamrah HS, Tigyi GJ, Kaye SB, Richardson A. Autotaxin delays apoptosis induced by carboplatin in ovarian cancer cells. Cellular Signalling. 2010;22:926–935. doi: 10.1016/j.cellsig.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M, Roulston A, Belec L, Billot X, Marcellus R, Bedard D, Bernier C, Branchaud S, Chan H, Dairi K, et al. The small molecule GMX1778 is a potent inhibitor of NAD+ biosynthesis: strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1-deficient tumors. Mol Cell Biol. 2009;29:5872–5888. doi: 10.1128/MCB.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Myers TG, O’Connor PM, Friend SH, Fornace AJ, Jr, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- Wosikowski K, Mattern K, Schemainda I, Hasmann M, Rattel B, Loser R. WK175, a novel antitumor agent, decreases the intracellular nicotinamide adenine dinucleotide concentration and induces the apoptotic cascade in human leukemia cells. Cancer Res. 2002;62:1057–1062. [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.