The architecture of polycation gene carriers has been shown to affect both their transfection efficiency and cytotoxicity. This work reports the synthesis of cyclic polycations and their use for gene transfer to mammalian cells. Cyclic poly((2-dimethylamino) ethylmethacrylate) (pDMAEMA) homopolymers of various molecular weights were synthesized by “intra-chain”click cyclization of α-alkyne-ω-azide heterodifunctional linear precursors prepared by atom transfer radical polymerization (ATRP). Polymers were characterized by size exclusion chromatography and FTIR analyses to confirm efficient cyclization and products with low polydispersity. Cellular uptake, membrane disruption, and nucleic acid delivery efficiency was determined for all polymers. In general, cyclic polymers complexed and delivered nucleic acids with similar efficiencies as their linear counterparts at charge ratios ≤ 5. Notably, cyclic polymers were less cytotoxic than linear polymers due to reduced membrane disruption and are therefore promising alternative structures for biological applications.
Polycations are one of the main classes of materials investigated for nucleic acid delivery.1 When mixed with nucleic acids, polycations complex with their cargo via electrostatic interactions and condense to form nanoparticles called “polyplexes”. The delivery efficiency and biocompatibility of polyplexes are highly dependent on polymer properties such as composition, molecular weight, and architecture.2–4
Perhaps one of the most studied materials for polymeric gene transfer is polyethylenimine (PEI).5 Linear PEI has been reported to be significantly more effective at gene transfer compared to branched PEI,6 possibly due to better nuclear access after cell internalization.7 In contrast, linear poly((2-dimethylamino) ethylmethacrylate) (pDMAEMA) is less effective as a gene carrier compared to both hyperbranched and star pDMAEMA.8–10 A particularly intriguing structure is the knotted polymer.11,12 Synthesized by deactivated atom transfer radical polymerization (DE-ATRP), these polymers have extensive intramolecular cyclization and have been shown to be effective gene transfer agents.
Cyclic polymers, which contain no chain ends, often possess quite different physical properties compared to their linear counterparts.13 However, to date, there are no reported use of cyclic polymers as gene transfer materials, likely in part due to synthetic challenges in their preparation.14 Recently several efficient and scalable synthetic strategies for generating cyclic polymers have been reported.13,14 Among these strategies, the “intra-chain”click cyclization of α-alkyne-ω-azide heterodifunctional linear precursors, first reported by the Grayson group,15 has emerged as a popular approach because it allows for controllable ring size and a rich variety of monomer species that can be applied using atom transfer radical polymerization (ATRP). Inspired by this work, we report here the synthesis of a series of cyclic polycations and further investigate their function asgene transfer agents in comparison to linear analogues. pDMAEMA has been extensively studied as a gene transfer material since its first reported use for this application by Hennink and coworkers.16 pDMAEMA can be also synthesized with low polydispersity by ATRP, and was therefore selected as our model material.17,18
Cyclic pDMAEMAs with degree of polymerizations (DPs) of 25, 50, and 100 were synthesized by combined ATRP and “click” end-to-end coupling under highly dilute conditions as shown in Scheme 1. Detailed synthesis procedures are available in Supplementary Information. Polymers were characterized by GPC (gel permeation chromatography) (Figure S1). Mn and polydispersity (Mw/Mn,, PDI) values were summarized in Table 1. GPC analysis of cyclic p(DMAEMA)50 and p(DMAEMA)100 showed a discernible right-shift towards lower molecular weight due to decreased hydrodynamic volume compared to their linear precursors, confirming efficient intra-molecular cyclization and negligible intermolecularcoupling reaction as reported previously.15,19 Cyclic p(DMAEMA)25 displayed an insignificant shift of GPC elution trace and a slightly increased molecular weight compared to its linear precursor, possibly due to its short chain length. The successful cyclization of all polymers was further confirmed by FT-IR measurements; the characteristic band of azide group centered at ~2100 cm−1 in the linear pDMAEMA-N3 polymers are absent for all the cyclic pDMAEMA polymers (Figure S2). These results indicate that “click” cyclization of α-alkyne-ω-azide heterodifunctional linear polycation is an efficient way to prepare cyclic pDMAEMA polycations.
Scheme 1.
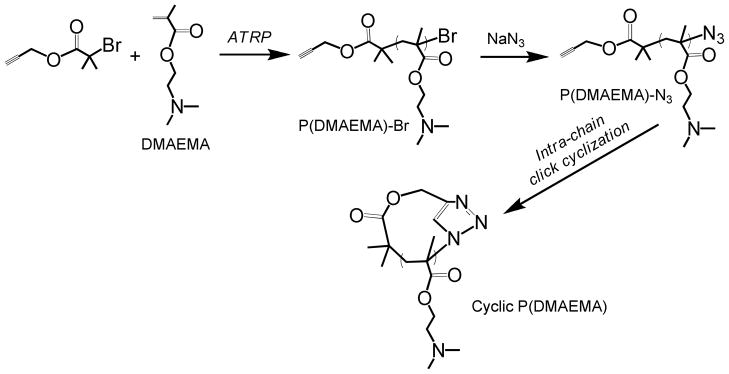
Synthesis of cyclic p(DMAEMA) by integrated ATRP and “intra-chain” click coupling.
Table 1.
Summary of properties of cyclic and linear polymers used in this study.
| Name | Architecture | Degree of Polymerization (DP) | Molecular Weight (Mn, kDa) by GPC | Buffering Capacity | IC50 (μg/mL) to HeLa cells |
|---|---|---|---|---|---|
| L25 | Linear | 25 | 8.5 | 3.38 | 38.1 ± 3.1 |
| C25 | Cyclic | 25 | 10.1 | 2.65 | 244.9 ± 49.5 |
| L50 | Linear | 50 | 13 | 2.27 | 14.6 ± 0.5 |
| C50 | Cyclic | 50 | 11 | 2.24 | 26.0 ± 2.5 |
| L100 | Linear | 100 | 25.9 | 2.07 | 15.6 ± 0.7 |
| C100 | Cyclic | 100 | 23.1 | 2.13 | 23.6 ± 0.6 |
The buffering capacity of pDMAEMA contributes to its effectiveness as a gene delivery agent, likely by improving endosomal release efficiency through the proton sponge effect.5,20 Therefore, we used acid-base titration to determine the buffering capacity (BC), defined as the μmol H+/mg polymer required to decrease the pH value of a 0.2 mg/mL polymer solution from 7.4 to 5.0, of linear vs cyclic pDMAEMA polymers (see Table 1 for BC values and Figure S3 for titration curves). Cyclization did not significantly affect the buffering capacity of the polymers.
The interaction of polymers with plasmid DNA was then assessed by two methods: gel electrophoresis assay (Figure S4) and YOYO-1 condensation assay (Figure S5). All cyclic and linear pDMAEMA polymers efficiently bind DNA by electrophoresis assay, with complete plasmid retention occurring by N/P (DMAEMA monomer to phosphate ratio) of 1.5 for all polymers. The YOYO-1 condensation assay evaluates packaging of plasmid DNA. Plasmid DNA is labeled with the intercalating YOYO-1 dye; DNA condensation results in fluorescence quenching of YOYO-1 due to electronic interactions between nearby dye molecules.21 Surprisingly, YOYO-1 condensation assay revealed that plasmid DNA is not tightly condensed by pDMAEMA polymers added at N/P=5 (<25% quenching for all materials, and almost no fluorescence quenching with linear polymers). However, cyclic polymers do condense plasmid DNA more than their linear analogues.
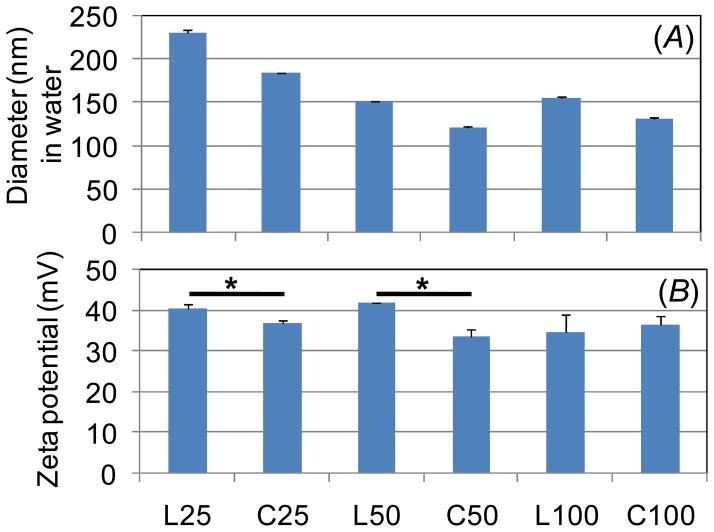
The particle sizes and zeta potentials of the polyplexes (N/P=5) were then measured by dynamic light scattering (DLS) (Figure 1). All polymers formed polyplexes with average hydrodynamic diameter < 250 nm. In addition, the following two trends were observed: (i) particle size decreased with polymer molecular weight and (ii) lower MW cyclic polymers (DP 25 and DP 50) formed smaller polyplexes than their linear counterparts. Higher polycation molecular weight has been correlated with smaller polyplexes for other materials.22,23 The smaller polyplexes formed by cyclic polymers further confirms the YOYO-1 data, suggesting more condensed formation of particles compared to polyplexes formed by linear polymers (Figure S5). Furthermore, zeta potential analysis shows that polyplexes formed by cyclic polymers have lower zeta potential compared to polyplexes formed by linear polymers (Figure 1) which also suggests more efficient plasmid packaging and neutralization of charge by cyclic polymers.
Figure 1.
Hydrodynamic diameter and zeta potential of polyplexes at N/P=5 measured by dynamic light scattering. (* p<0.05)
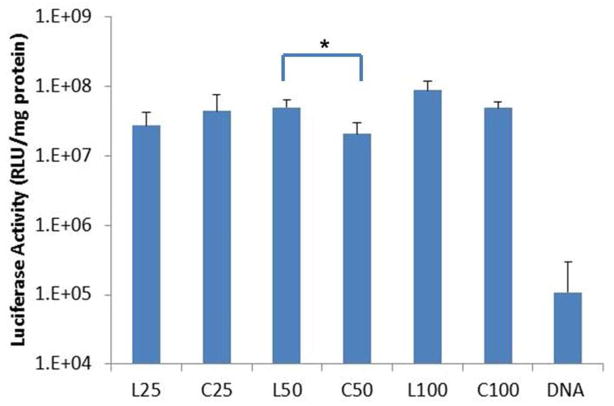
The polyplexes were used to deliver the luciferase reporter gene to HeLa cells at various charge ratios (N/P=2, 5, 8, and 10). In these preliminary studies, increased transfection efficacy was observed with increasing charge ratio, although significant toxicity was observed at N/P=8 and 10 for the higher MW polymers. Therefore, three separate transfections with each polymer tested in triplicate were carried out at N/P=5. Results were reproducible in all three experiments and results from a representative transfection are shown in Figure 2. There is no significant difference in transfection efficiency between linear and cyclic pDMAEMA with DP 25 and 100. However at DP 50, higher transfection efficiency was consistently achieved by linear pDMAEMA compared to cyclic pDMAEMA.
Figure 2.
Transfection efficiency of pDMAEMApolyplexes (N/P=5) to HeLa cells. *p <0.05.
To better understand possible mechanisms for the observed difference of DP 50 polymers, we assessed efficiency of polyplex uptake and also the effect of free polymer on gene transfer. Polyplex uptake was quantified by incubating fluorescently-labeled polyplexes with HeLa cells for 30 min at 37 °C followed by analysis of cells by flow cytometry. No significant difference was observed between polyplexes formed by cyclic vs. linear polymers with DP 25 or 50 and slightly reduced uptake was observed with polyplexes formed by cyclic compared to linear polymers with DP 100 (Figure S6). It should be noted that fluorescence intensity of polyplexes in the OptiMEM media is similar between linear and cyclic polymers (data not shown).
Previous studies have shown that polyplex solutions are comprised of polymer and DNA at low N/P (typically < N/P = 3) and remaining polymer is free in solution.24–26 This excess polymer is critical in facilitating gene transfection efficiency by reducing interaction with cell surface proteins and enhancing endosomal escape; indeed, purified polyplexes demonstrate very low transfection efficiency.24,27 We tested the effect of free cyclic vs. free linear pDMAEMA by transfecting cells complexed with poly-L-lysine (PLL) at N/P=2 and then adding in free cyclic or linear pDMAEMAto a total N/P=5 2hr post transfection. Transfection efficiencies were higher than delivery achieved using PLL polyplexes only but were similar regardless of the specific MW or structure of added polymer (Figure S7).
The mechanism of differential transfection efficiency of pDMAEMA DP 50 polymers therefore remains unclear. One possibility is difference in polyplex stability. Extracellular polyplex stability is needed to protect the nucleic acid from premature degradation while intracellular polyplex unpackaging has been shown to be a limiting factor in gene transfection for larger molecular polycations.28,29 We show in Figures 1 (DLS) and Figure S5 (YOYO-1 condensation) that cyclic polymers package DNA into more condensed particles. The difference in polyplex stability may be more significant between the two DP 50 polymers than the DP 25 or DP 100 polymers.
A second possibility is difference in endosomal release. Wagner and co-workers hypothesized that effective endosomal release can be achieved by combining proton sponge effects and direct membrane destabilization in a single material.32 Hong et al. previously showed that cationic dendrimers interact with mammalian cell membranes resulting in generation of “nanoscale holes”.31 We tested nanohole formation by incubating HeLa cells with polymer in the presence of propidium iodide (PI), a membrane-impermeable dye; nanohole generation results in PI+ cells by dye diffusion into cells. The results, shown in Figure 3, confirm that linear pDMAEMA polymers (DP25 and DP50) interact more with cell membranes than their cyclic counterparts. At DP 100, no difference is observed, possibly due to lower concentrations of endgroups. The increased membrane interaction by linear pDMAEMA compared to cyclic pDMAEMA at DP 50 may therefore contribute to its increased transfection efficiency.
Figure 3.
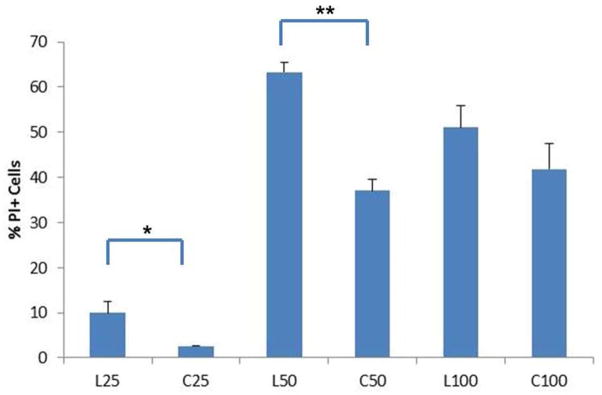
Membrane disruption of HeLa cells induced by pDMAEMA added at equivalent polymer concentrations to N/P=5. Membrane disruption is assessed by cell fluorescence resulting from propidium iodide diffusion through nanoscale holes in the cell membrane. (n=3, *p<0.05, **p<0.001)
Nanohole generation has also been correlated with increased cell toxicity. Therefore, the cytotoxicity of the polymers was tested by determining the IC50 values (concentration for 50% growth inhibition) to HeLa cells by MTS tetrazolium assay (Table 1). The cyclic polymers were significantly less cytotoxic to the cultured cells compared to their linear analogues. The Maynard group investigated the effect of polymer end-group for polymers synthesized by reversible addition-fragmentation chain transfer (RAFT) polymerization and found that trithiocarbonate chain transfer agents (CTAs) produce polymers with lower cytotoxicity compared to dithioester CTAs, possibly due to degradation or reaction of the dithioester chain end with amines or thiols in proteins.30 The polymer end groups of ATRP-synthesized polymers have not been extensively investigated, but these results suggest that end groups in these polymers may contribute to their cytotoxicity.
In summary, this communication reports the synthesis of cyclic, cationic pDMAEMA polymers and their use as gene transfer agents. While better tolerated than PEI, the toxicity of pDMAEMA is still a drawback in its in vivo use.33,34 We demonstrate here that cyclization of pDMAEMA-based polymers reduces cytotoxicity although reduced transfection efficiency is observed at certain MWs. The Szoka and Frechet groups previously reported that cyclic polymers have longer circulation half-lives compared to linear polymers due to reduced renal clearance rates; the increased circulation time can contribute toward better tumor accumulation by passive targeting.35,36 Cyclic polymers are therefore promising materials in gene delivery applications. Future work will include evaluation of cyclic polymers for in vivo gene transfer.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by NIH 1R01NS064404 and NSF 1206426. JZ was supported through the UW Amgen Scholars Program. DSHC is supported by NIH T32 CA138312.
Footnotes
Supporting Information. Experimental information including GPC traces, FT-IR results, buffering curves, electrophoresis images, condensation assay results and additional transfection resultsare included. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pack DW, Hoffman AS, Pun S, Stayton PS. Nature Reviews Drug Discovery. 2005;4:581. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 2.Anderson DG, Akinc A, Hossain N, Langer R. Molecular Therapy. 2005;11:426. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Chu DSH, Schellinger JG, Shi J, Convertine AJ, Stayton PS, Pun SH. Accounts of Chemical Research. 2012;45:1089. doi: 10.1021/ar200242z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer D, Li YX, Ahlemeyer B, Krieglstein J, Kissel T. Biomaterials. 2003;24:1121. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 5.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. Journal of Gene Medicine. 2001;3:362. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 7.Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E. Molecular Therapy. 2002;5:80. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- 8.Newland B, Tai HY, Zheng Y, Velasco D, Di Luca A, Howdle SM, Alexander C, Wang WX, Pandit A. Chemical Communications. 2010;46:4698. doi: 10.1039/c0cc00439a. [DOI] [PubMed] [Google Scholar]
- 9.Xu FJ, Zhang ZX, Ping Y, Li J, Kang ET, Neoh KG. Biomacromolecules. 2009;10:285. doi: 10.1021/bm8010165. [DOI] [PubMed] [Google Scholar]
- 10.Synatschke CV, Schallon A, Jerome V, Freitag R, Muller AHE. Biomacromolecules. 2011;12:4247. doi: 10.1021/bm201111d. [DOI] [PubMed] [Google Scholar]
- 11.Newland B, Zheng Y, Jin Y, Abu-Rub M, Cao HL, Wang WX, Pandit A. Journal of the American Chemical Society. 2012;134:4782. doi: 10.1021/ja2105575. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Cao HL, Newland B, Dong YX, Pandit A, Wang WX. Journal of the American Chemical Society. 2011;133:13130. doi: 10.1021/ja2039425. [DOI] [PubMed] [Google Scholar]
- 13.Jia ZF, Monteiro MJ. Journal of Polymer Science Part a-Polymer Chemistry. 2012;50:2085. [Google Scholar]
- 14.Laurent BA, Grayson SM. Chemical Society Reviews. 2009;38:2202. doi: 10.1039/b809916m. [DOI] [PubMed] [Google Scholar]
- 15.Laurent BA, Grayson SM. Journal of the American Chemical Society. 2006;128:4238. doi: 10.1021/ja0585836. [DOI] [PubMed] [Google Scholar]
- 16.Cherng JY, vandeWetering P, Talsma H, Crommelin DJA, Hennink WE. Pharmaceutical Research. 1996;13:1038. doi: 10.1023/a:1016054623543. [DOI] [PubMed] [Google Scholar]
- 17.Zeng FQ, Shen YQ, Zhu SP, Pelton R. Journal of Polymer Science Part a-Polymer Chemistry. 2000;38:3821. [Google Scholar]
- 18.Zhang X, Xia JH, Matyjaszewski K. Macromolecules. 1998;31:5167. doi: 10.1021/ma980477j. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Ye J, Liu SY. Macromolecules. 2007;40:9103. [Google Scholar]
- 20.Dai FY, Sun P, Liu YJ, Liu WG. Biomaterials. 2010;31:559. doi: 10.1016/j.biomaterials.2009.09.055. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamoorthy G, Duportail G, Mély Y. Biochemistry. 2002;41:15277. doi: 10.1021/bi020440y. [DOI] [PubMed] [Google Scholar]
- 22.Johnson RN, Chu DS, Shi J, Schellinger JG, Carlson PM, Pun SH. Journal of Controlled Release. 2011;155:303. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reschel T, Koňák Čr, Oupický D, Seymour LW, Ulbrich K. Journal of Controlled Release. 2002;81:201. doi: 10.1016/s0168-3659(02)00045-7. [DOI] [PubMed] [Google Scholar]
- 24.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Journal of Gene Medicine. 2004;6:1102. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 25.Clamme JP, Azoulay J, Mely Y. Biophys J. 2003;84:1960. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue YA, Jin F, Deng R, Cai JG, Chen YC, Lin MCM, Kung HF, Wu C. Journal of Controlled Release. 2011;155:67. doi: 10.1016/j.jconrel.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 27.Saul JM, Wang CHK, Ng CP, Pun SH. Adv Mater. 2008;20:19. [Google Scholar]
- 28.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Biotechnology and bioengineering. 2000;67:598. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 29.Strand SP, Lelu S, Reitan NK, de Lange Davies C, Artursson P, Vårum KM. Biomaterials. 2010;31:975. doi: 10.1016/j.biomaterials.2009.09.102. [DOI] [PubMed] [Google Scholar]
- 30.Chang CW, Bays E, Tao L, Alconcel SNS, Maynard HD. Chemical Communications. 2009:3580. doi: 10.1039/b904456f. [DOI] [PubMed] [Google Scholar]
- 31.Hong S, Leroueil PR, Janus EK, Peters JL, Kober M-M, Islam MT, Orr BG, Baker JR, Banaszak Holl MM. Bioconjugate chemistry. 2006;17:728. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 32.Lächelt U, Kos P, Mickler FM, Herrmann A, Salcher EE, Rödl W, Badgujar N, Bräuchle C, Wagner E. Nanomedicine: Nanotechnology, Biology and Medicine. 2013 doi: 10.1016/j.nano.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Cerda-Cristerna BI, Flores H, Pozos-Guillen A, Perez E, Sevrin C, Grandfils C. Journal of Controlled Release. 2011;153:269. doi: 10.1016/j.jconrel.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 34.Dubruel P, Christiaens B, Vanloo B, Bracke K, Rosseneu M, Vandekerckhove J, Schacht E. European Journal of Pharmaceutical Sciences. 2003;18:211. doi: 10.1016/s0928-0987(02)00280-4. [DOI] [PubMed] [Google Scholar]
- 35.Chen B, Jerger K, Frechet JMJ, Szoka FC. Journal of Controlled Release. 2009;140:203. doi: 10.1016/j.jconrel.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasongkla N, Chen B, Macaraeg N, Fox ME, Frechet JMJ, Szoka FC. Journal of the American Chemical Society. 2009;131:3842. doi: 10.1021/ja900062u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.