Abstract
Among the mammalian genes encoding DNA ligases (LIG), the LIG3 gene is unique in that it encodes multiple DNA ligase polypeptides with different cellular functions. Notably, this nuclear gene encodes the only mitochondrial DNA ligase and so is essential for this organelle. In the nucleus, there is significant functional redundancy between DNA ligase IIIα and DNA ligase I in excision repair. In addition, DNA ligase IIIα is essential for DNA replication in the absence of the replicative DNA ligase, DNA ligase I. DNA ligase IIIα is a component of an alternative non-homologous end joining (NHEJ) pathway for DNA double-strand break (DSB) repair that is more active when the major DNA ligase IV-dependent pathway is defective. Unlike its other nuclear functions, the role of DNA ligase IIIα in alternative NHEJ is independent of its nuclear partner protein, X-ray repair cross-complementing protein 1 (XRCC1). DNA ligase IIIα is frequently overexpressed in cancer cells, acting as a biomarker for increased dependence upon alternative NHEJ for DSB repair and it is a promising novel therapeutic target.
Introduction
DNA ligases play an essential role in maintaining genomic integrity by joining breaks in the phosphodiester backbone of DNA that occur during replication and recombination, and as a consequence of DNA damage and its repair. Three human genes, LIG1, LIG3 and LIG4 encode ATP-dependent DNA ligases. These enzymes have related catalytic regions that catalyze the same three-step ligation reaction but different flanking domains that mediate protein:protein interactions with different partners (Ellenberger and Tomkinson, 2008).
While almost all eukaryotes have homologs of the LIG1 and LIG4 genes, the LIG3 gene is less widely distributed. Initially, it was thought that the LIG3 gene was restricted to vertebrates but, with the sequencing of more genomes, it has now been found in about 30% of eukaryotes, including members of 4 of the 6 ancestral eukaryotic groups (Simsek and Jasin, 2011). This distribution suggests that the LIG3 gene arose relatively early during the evolution of eukaryotes but was not always retained. As eukaryotes became more complex, it was presumably advantageous to have multiple LIG genes that encoded a broader repertoire of DNA ligases to participate in the increasing number of specialized DNA transactions, including immunoglobulin gene rearrangements in immune cells, meiosis and germ cell development, the use of poly (ADP-ribose) to signal DNA damage and the different DNA repair pathways in proliferating and terminally differentiated cells. Notably, the DNA ligases encoded by vertebrate LIG3 genes have acquired several conserved accessory domains that flank the core catalytic region during evolution (Simsek and Jasin, 2011). As discussed below, these domains play critical roles in dictating the multiple cellular functions of the DNA ligases encoded by the LIG3 gene in vertebrate DNA metabolism.
The absence of the LIG3 gene in yeasts has prevented the use of genetically tractable lower eukaryotes, such as S. cerevisiae and S. pombe, as models to gain insights into the cellular functions of and interplay between the DNA ligases encoded by the LIG1, LIG3 and LIG4 genes in higher eukaryotes. Based on the efforts of many laboratories, there is an emerging picture of functional redundancy among the DNA ligases in mammalian cells that is complicated by the LIG3 gene encoding multiple DNA ligase polypeptides. In this review, we focus on the structure and function of the DNA ligases encoded by the mammalian LIG3 gene.
LIG3 gene
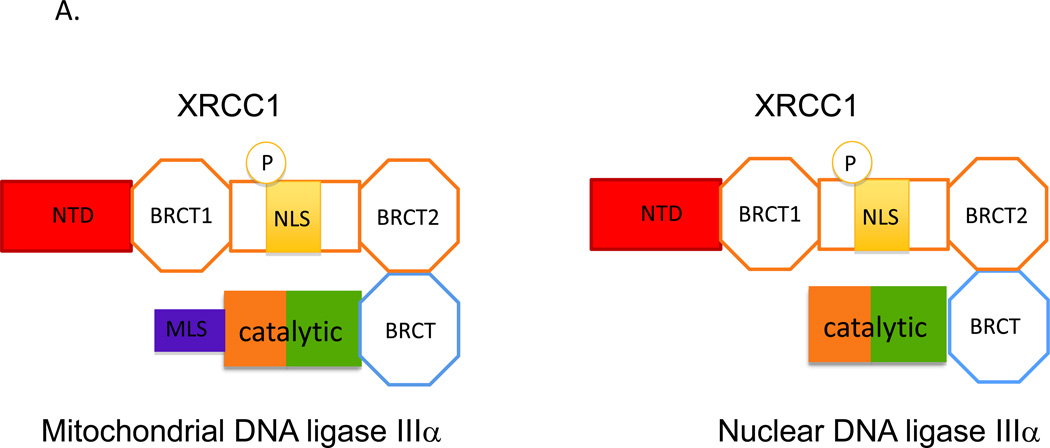
The human LIG3 gene is located on human chromosome 17 at q11.2–q12 (Chen et al., 1995; Wei et al., 1995). Unlike the LIG1 and LIG4 genes, the LIG3 gene encodes three or possibly four different DNA ligase polypeptides (Fig. 1). Mitochondrial and nuclear versions of DNA ligase IIIα are generated in all cells by alternative translation initiation (Lakshmipathy and Campbell, 1999). The DNA ligase IIIα mRNA open reading frame encodes an N-terminal mitochondrial leader sequence (MLS) that is cleaved off during entry into mitochondria (Fig. 2). Thus, translation initiation at the first ATG of the full-length open reading frame generates mitochondrial DNA ligase IIIα whereas translation initiation at an internal ATG adjacent to a Kozak consensus sequence generates nuclear DNA ligase IIIα (Chen et al., 1995; Wei et al., 1995; Lakshmipathy and Campbell, 1999). Since there is no obvious NLS (NLS) within the DNA ligase IIIα polypeptide, it has been suggested, as shown in Figure 2, that nuclear localization is dependent upon complex formation with a partner protein XRCC1 that does have a NLS (Caldecott, 2003; Parsons et al., 2010). The interaction of DNA ligase IIIα with XRCC1 and other partner proteins is described below. Prior to the cloning of the human LIG genes, biochemical studies had identified a 70 kDa DNA ligase in addition to a 125 kDa DNA ligase I and a 100 kDa DNA ligase III that was designated DNA ligase II (Soderhall and Lindahl, 1975; Tomkinson et al., 1991). Amino acid sequencing of peptides from purified DNA ligase II revealed that this polypeptide was encoded by the LIG3 gene and is most likely generated by proteolysis of DNA ligase IIIα during purification (Wang et al., 1994; Chen et al., 1995; Husain et al., 1995). Thus, the confusing nomenclature of the mammalian DNA ligases can be attributed to a purification artifact during attempts to purify and characterize these enzymes.
Figure 1. DNA ligases encoded by the LIG3 gene.
The four DNA ligase polypeptides encoded by the mammalain LIG3 gene are shown schematically. The positions of the zinc finger (ZnF, red) and the catalyic region, which contains the DNA binding domain (DBD, light brown), nucleotidyl transferase domain (NTase, light green) and oligonucleotide/oligosaccharide-fold binding domain (OBD, dark green), are indicated. Mitochondrial and nuclear versions of DNA ligase IIIα are generated by alternative translation initiation. The position of the mitochondrial leader sequence (MLS, purple) is indicated. An alterative splicing event that occurs in male germ cells replaces the C-terminal BRCT domain (BRCT, blue) of DNA ligase IIIα with short amino acid sequence that functions as a NLS (NLS, grey) in DNA ligase IIIβ.
Figure 2. Mitochondrial and nuclear versions of DNA ligase IIIα.
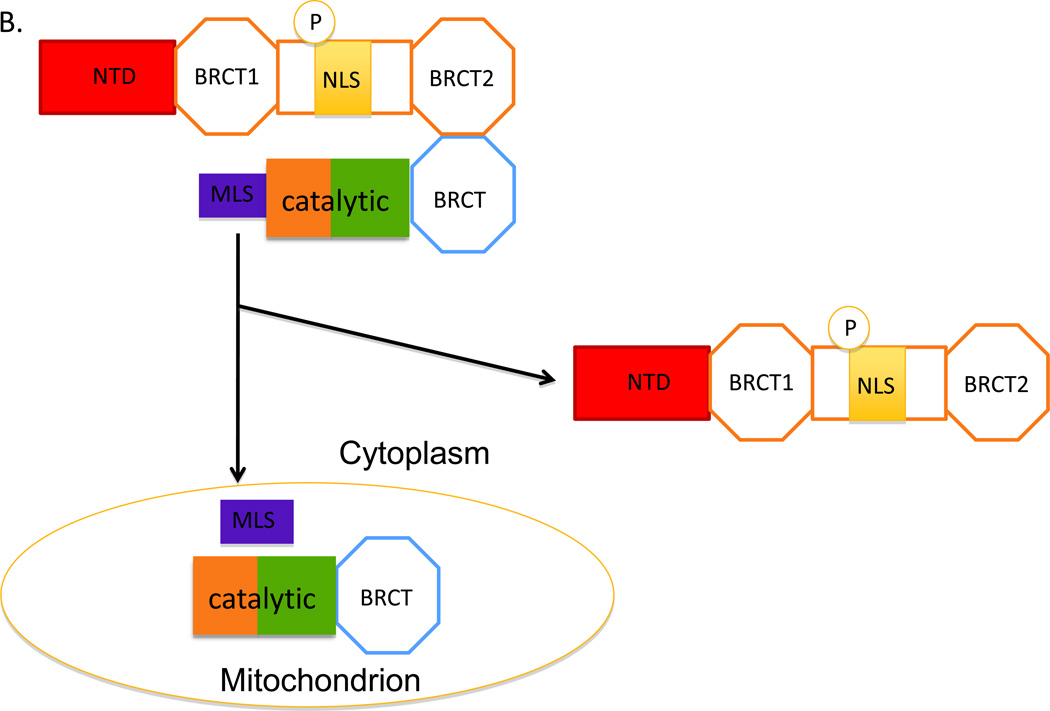
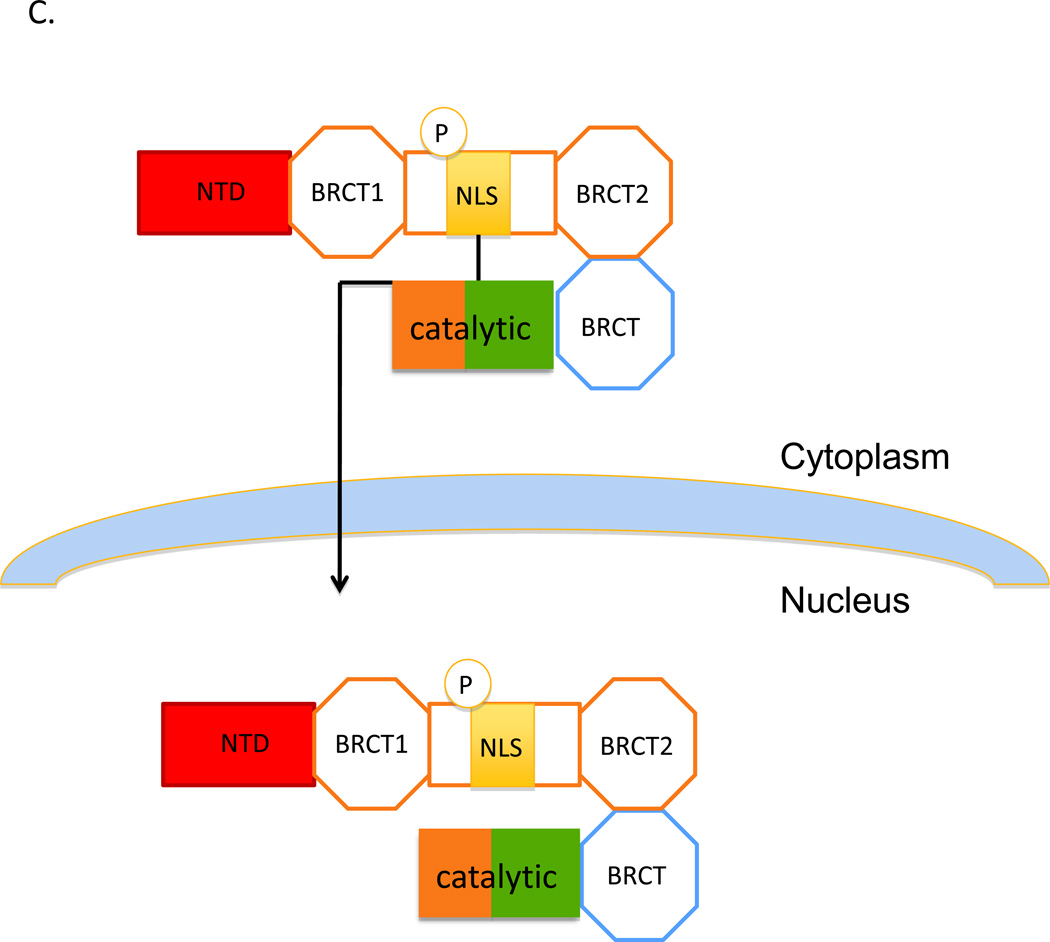
A. Two polypeptides are generated from DNA ligase IIIα mRNA by alternative translation. Mitochondrial DNA ligase IIIα has an additional N-terminal mitochondrial leader sequence (MLS, purple) in a addition to the common catalytic region (brown/green) and C-terminal BRCT domain (BRCT). Since this domain mediates the interaction with the C-terminal BRCT2 domain of XRCC1, it is assumed that both the mitochondrial and nuclear versions of DNA ligase IIIα will form complexes with XRCC1, which also contains an N-terminal domain (NTD, red), a second BRCT domain (BRCT1) and a nuclear localization domain (NLS, yellow) whose activity appears to be dependent upon phosphorylation (P) of adjacent residues. B. The complex containing mitochondrial DNA ligase IIIα is targed to the mitochondria by the MLS. It is assumed that the activity of the MLS is much greater than the activity of XRCC1 NLS. The MLS initiates passage of the DNA ligase IIIα polypeptide through the mitochondrial membrane and is then removed by proteolysis. Unfolding of the DNA ligase IIIα polypeptide as it passes through the mitochondrial membrane disrupts the interaction with XRCC1, leaving free XRCC1 in the cytoplasm. C. The transport of DNA ligase IIIα lacking the MLS into the nucleus is mediated by the NLS of XRCC1 which binds to the nuclear pore complex, resulting in passage of the XRCC1 into the nucleus.
The LIG3 gene is ubiquitously expressed at low levels in all human tissues and cells except for the testes where expression levels are about 10-fold higher (Chen et al., 1995; Wei et al., 1995). Further analysis revealed that the elevated levels of expression occur in primary spermatocytes undergoing recombination prior to the first meiotic division and that a distinct mRNA species, DNA ligase IIIβ, is generated by an alternative splicing mechanism that has only been detected in male germ cells (Mackey et al., 1997). The alternative splicing, which replaces the exon encoding the C-terminal 77 amino acids of DNA ligase IIIα with a novel 17- to 18-amino acid sequence (Fig. 1), begins in early pachytene spermatocytes with DNA ligase IIIβ mRNA detectable throughout pachytene and in round spermatids (Mackey et al., 1997). Since the 5’ end of DNA ligase IIIβ mRNA is identical to that of DNA ligase IIIα mRNA, this transcript is also capable of encoding nuclear and mitochondrial versions of DNA ligase IIIβ by alternative translation (Fig. 1). It should be noted that DNA ligase IIIβ mRNA has not been detected in the ovary (Mackey et al., 1997). This may reflect differences between oogenesis and spermatogenesis. Furthermore, organisms such as S. cerevisiae, that lack a LIG3 homolog undergo meiosis. Thus, although the expression pattern of DNA ligase IIIβ during spermatogenesis suggests that this enzyme participates in the completion of homologous recombination events that occur during meiotic prophase, this has not yet been definitively demonstrated. Conceivably, DNA ligase IIIβ may be involved in maintaining genomic integrity when histones are replaced with protamines late in the haploid phase of spermatogenesis.
DNA ligase polypeptides encoded by the LIG3 gene
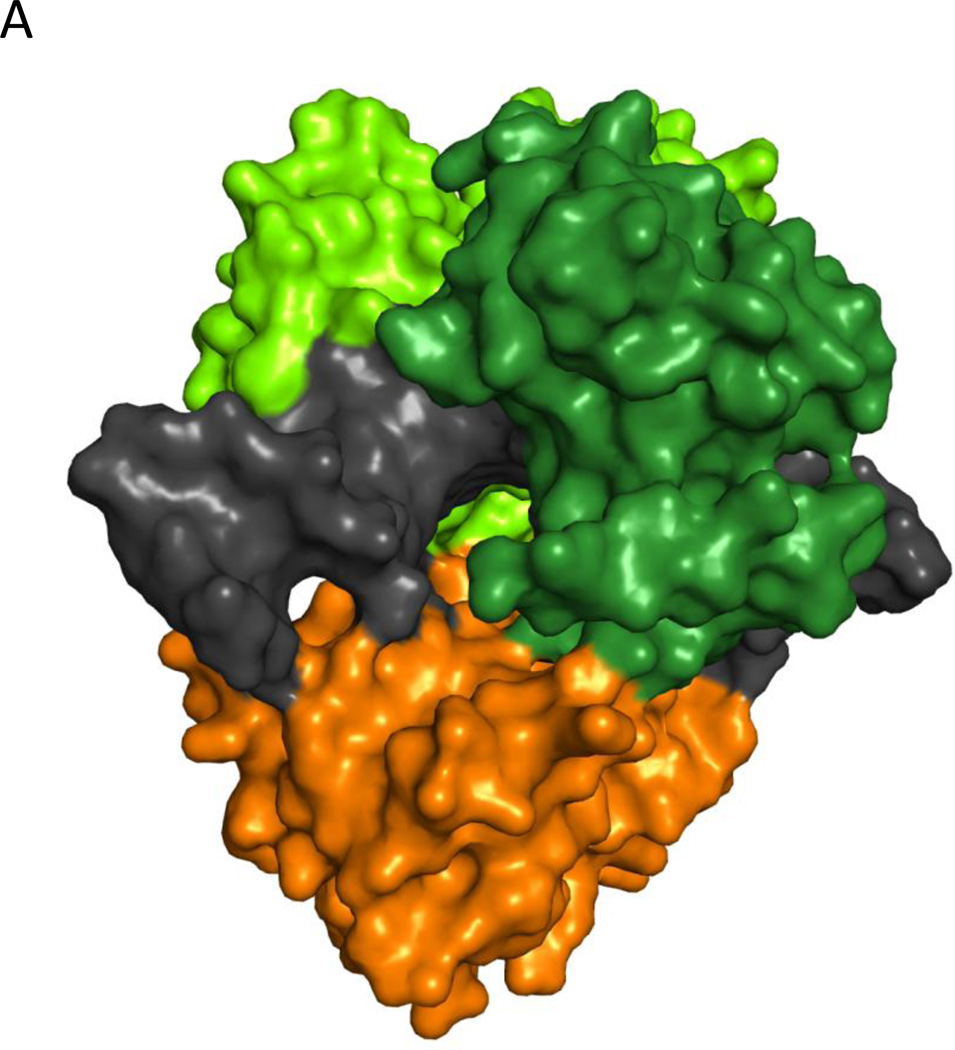
The related catalytic regions of the human DNA ligases contain three domains, a DNA binding domain (DBD), a nucleotidyl transferase domain (NTase) and an oligonucleotide/oligosaccharide-fold binding domain (OBD) (Ellenberger and Tomkinson, 2008). Similar to DNA ligase I, the DNA ligase III polypeptide adopts a flexible, extended conformation in the absence of DNA (Cotner-Gohara et al., 2010). When it engages a DNA nick, the domains of the catalytic region contact and encircle the nicked DNA in a compact, closed clamp structure shown in Figure 3A. (Cotner-Gohara et al., 2010). The protein architecture and conformation of the catalytic domains in this structure are remarkably similar to those of the DNA ligase I catalytic domains bound to nicked DNA despite these polypeptides only sharing 21% amino acid identity (Cotner-Gohara et al., 2010).
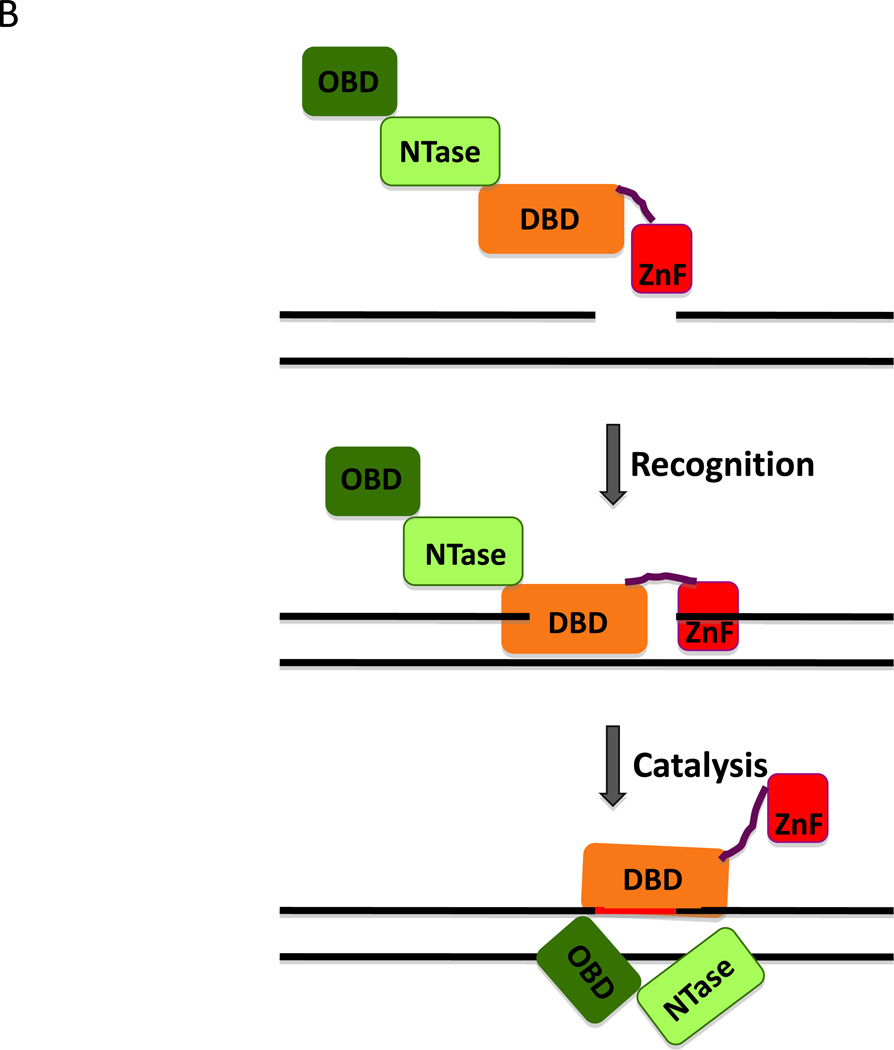
Figure 3. Interaction of DNA ligase III with nicked DNA.
A. Space filling model showing the DNA binding (DBD, light brown), nucleotidyl transferase (NTase, light green) and oligonucleotide/oligosaccharide-fold binding domains (OBD, dark green) of DNA ligase III enaging a short oligonucleotide (Black) containing a non-ligatable nick (Cotner-Gohara et al., 2010). The image was made using PyMol (http://www.pymol.org). B. A fragment of DNA ligase III encompassing the zinc finger (ZnF, red) and the catalytic region (DNA binding domain, DBD; nucleotidyl transferase domain, NTase; oligonucleotide/oligosaccharide-fold binding domain, OBD) and a nicked DNA duplex are shown schematically (upper panel). The single strand interruption is initially recognized by a combination of the ZnF and the DBD (middle panel). If the nick is ligatable, the ZnF is displaced by a combination of the NTase and OBD. The DBD, NTase and OBD encirle and ligate the nicked DNA.
A unique feature of the DNA ligases encoded by the LIG3 gene compared with the other human DNA ligases is an N-terminal zinc finger (ZnF) (Fig. 1). This ZnF is structurally related to the pair of ZnFs at the N-terminus of poly (ADP-ribose polymerase 1 (PARP1) that facilitate binding of PARP1 to DNA breaks and other abnormal DNA structures (Mackey et al., 1999). Although the DNA ligase III ZnF also binds to DNA breaks, it is not required for nick ligation but does enable the enzyme to efficiently join nicked DNA at high salt concentrations (Mackey et al., 1999). Interestingly, the DNA ligase III ZnF and DBD cooperate to form a nick-binding module with the NTase and OBD forming a second nick-binding module (Cotner-Gohara et al., 2008). These two modules have different DNA binding properties with ZnF/DBD module being more tolerant of different nick structures, including gaps, whereas the NTase/OBD module preferentially binds to ligatable nicks (Cotner-Gohara et al., 2008). Based on these properties and biophysical studies, a jackknife model for the ligation of DNA nicks by DNA ligase III has been proposed (Cotner-Gohara et al., 2008; Cotner-Gohara et al., 2010). In this model, the ZnF/DBD module acts as a nick sensor that initially engages DNA breaks with the DNA ligase III polypeptide in an extended conformation (Fig. 3B). If the nick is ligatable, the DNA ligase III polypeptide undergoes a conformational change with NTase/OBD displacing the ZnF/DBD module and then forming the compact, closed clamp structure with the DBD around the DNA nick (Fig. 3B).
Among the human DNA ligases, DNA ligase III has the most robust intermolecular DNA joining activity (Chen et al., 2000). This activity is not only dependent upon the ZnF but also involves key residues within the DBD (Cotner-Gohara et al., 2008; Cotner-Gohara et al., 2010). It has been proposed that the NTase/OBD and ZnF/DBD modules each engage a DNA end (Cotner-Gohara et al., 2010), enabling DNA ligase III to juxtapose and ligate the DNA ends. Alternatively, it is possible that intermolecular ligation involves an interaction in trans between the ZnF and DBD of two DNA ligase III molecules, each bound to a DNA end.
At the C-terminus of DNA ligase IIIα is a breast cancer susceptibility protein (BRCA) 1-related C-terminal (BRCT) domain (Bork et al., 1997). This domain, which is about 100 amino acids long, has been found in many proteins involved in DNA repair and the DNA damage response, and is often involved in protein:protein interactions (Bork et al., 1997). In the case of nuclear DNA ligase IIIα, the BRCT domain interacts with the DNA repair protein, X-ray repair cross-complementing protein 1 (XRCC1) (Fig. 2A), an interaction that is described in more detail below. As a result of the alternative splicing event, the majority of the BRCT domain is missing from DNA ligase IIIβ, which does not interact with XRCC1. The short additional amino acid sequence at the C-terminus of DNA ligase IIIβ acts as a NLS (Fig. 1).
Protein partners of DNA ligase III
XRCC1
XRCC1 was the first DNA ligase III-interacting protein identified (Caldecott et al., 1994). These proteins form a stable, constitutive complex in the nucleus (Caldecott et al., 1995). The human XRCC1 gene was cloned based on its ability to complement the DNA damage sensitive phenotypes of a mutant Chinese Hamster Ovary cell line, EM9, that is sensitive to DNA alkylating agents and ionizing radiation, and has an increased frequency of spontaneous sister chromatid exchanges (Thompson et al., 1990). XRCC1 contains two BRCT domains, one of which, the C-terminal BRCT2 domain, interacts with the C-terminal BRCT of DNA ligase IIIα forming a heterodimer (Fig. 2A). The BRCT2 domain of XRCC1 and the BRCT domain of DNA ligase IIIα are also capable of forming homodimers. Recent structural studies have shown that residues adjacent to the XRCC1 BRCT2 domain also contribute to the interface with the DNA ligase IIIα BRCT domain, thus providing a mechanism by which formation of the DNA ligase IIIα:XRCC1 heterodimer is favored over formation of homodimers (Cuneo et al., 2011).
Since the steady state levels of DNA ligase IIIα are reduced in xrcc1 cell lines and these levels are restored to normal levels by expression of wild type XRCC1 but not a mutant version with amino acid changes in the BRCT2 domain that disrupt the interaction with DNA ligase IIIα (Caldecott et al., 1995; Taylor et al., 2000a), it appears that XRCC1 is required for the stability of nuclear DNA ligase IIIα. Moreover, since XRCC1 has a NLS (Masson et al., 1998), it is possible that DNA ligase IIIα, which lacks a NLS, must interact with XRCC1 in the cytoplasm for transport into the nucleus. Recently, it has been shown that phosphorylation of XRCC1 by a cytoplasmic form of casein kinase 2 is required for nuclear accumulation of XRCC1 and the stabilization of nuclear DNA ligase IIIα (Parsons et al., 2010). Based on these studies, it has been suggested that XRCC1 and DNA ligase IIIα form a complex in the cytoplasm that is protected from ubiquitin-mediated by phosphorylation of XRCC1 and targeted to the nucleus via the NLS of XRCC1 (Fig. 2C). It is likely that the DNA ligase IIIα polypeptide with the N-terminal MLS will also interact with XRCC1 in the cytoplasm (Fig. 2A). We envision that the activity of the DNA ligase IIIα MLS predominates over that of the XRCC1 NLS, resulting in targeting of the complex to mitochondria. Then we predict that the MSL-dependent folding of DNA ligase IIIα as it passes through the mitochondrial membrane disrupts the interaction with XRCC1 (Fig. 2B).
Although XRCC1 has no known catalytic activity, it does interact with a large number of DNA repair proteins (Caldecott, 2003; Ellenberger and Tomkinson, 2008). This has led to the suggestion that XRCC1 is a scaffold protein that co-ordinates the activities of DNA repair enzymes, enhancing the efficiency of DNA repair. In support of this idea, XRCC1 and DNA ligase IIIα have been identified as subunits of different multiprotein complexes, including complexes that catalyze the repair of oxidized base lesions (Luo et al., 2004; Das et al., 2006).
PARP1
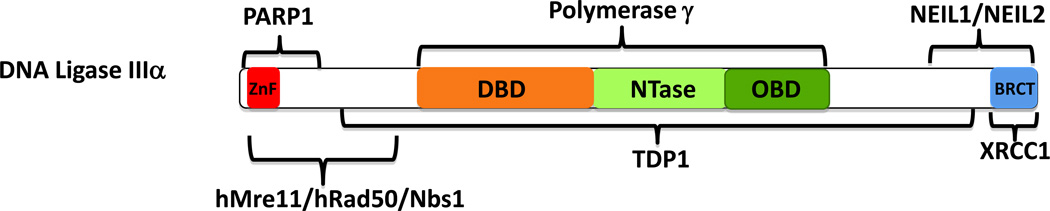
Proteins that bind to the DNA ligase IIIα subunit of the nuclear DNA ligase IIIα:XRCC1 complex have been identified (Fig. 4). For PARP1, these interactions involve both DNA ligase IIIα and XRCC1 (Caldecott et al., 1996; Masson et al., 1998; Leppard et al., 2003; Della-Maria et al., 2011). PARP1 is an abundant nuclear protein that has been implicated in many different DNA transactions, including the repair of single- and double-strand breaks (SSBs and DSBs) (Ame et al., 2004). During the repair of SSBs, PARP1 binds to the breaks via its N-terminal ZnFs. This activates the NAD-dependent polymerase activity of PARP1, resulting in the synthesis of ADP-ribose polymers on PARP1 itself and other chromatin proteins (Ame et al., 2004). Both XRCC1 and DNA ligase IIIα preferentially bind to poly (ADP-ribosylated) PARP1 in vitro and their recruitment to DNA damage sites in vivo is dependent upon poly (ADP-ribose) synthesis (Masson et al., 1998; Schreiber et al., 2002; Leppard et al., 2003; Okano et al., 2003; Okano et al., 2005). It has been suggested that the DNA ligase IIIα ZnF may enable DNA ligase IIIα:XRCC1 and associated proteins to find and engage SSBs in the presence of negatively charged poly (ADP-ribose) polymers generated by PARP1 (Leppard et al., 2003).
Figure 4. Protein partners of DNA ligase IIIα.
The regions of DNA ligase IIIα involved in interactions with hMre11/hRad50/Nbs1, NEIL1 and NEIL2, PARP1, TDP1, XRCC1 and mitochondrial DNA polymerase γ are indicated.
hMre11:hRad50:Nbs1
As with PARP-1, both DNA ligase IIIα and XRCC1 interact with the hMre11:hRad50:Nbs1 complex (Fig. 4) (Della-Maria et al., 2011) that plays a multifunctional role in the cellular response to DSBs, including end resection (D'Amours and Jackson, 2002; Paull and Lee, 2005; Haber, 2008). This generation of 3’ single strands is a key early step in hRad51-dependent recombinational repair and is also involved in a poorly defined, minor alternative non-homologous end-joining (NHEJ) pathway. hMre11:hRad50:Nbs1 and DNA ligase IIIα:XRCC1 associate in a DNA damage-dependent manner in cells that are deficient in the major DNA-dependent protein kinase (DNA PK)-dependent NHEJ pathway (Della-Maria et al., 2011). Furthermore, these complexes process and join DNA ends in vitro generating deletions and utilizing microhomologies at the repair sites, both of which are characteristics of the alternative NHEJ pathway (Nussenzweig and Nussenzweig, 2007; Della-Maria et al., 2011). Based on these results, it appears that hMre11:hRad50:Nbs1 and DNA ligase IIIα:XRCC1 act together in the alternative NHEJ pathway.
Tyrosyl phosphodiesterase 1 (TDP1)
This enzyme, which removes the covalently-linked topoisomerase I (topo I) peptide from aborted topo I-DNA complexes and participates in the repair of SSBs generated by ionizing radiation (Plo et al., 2003; El-Khamisy et al., 2005; El-Khamisy et al., 2007), appears to interact exclusively with DNA ligase IIIα (Fig. 4) (El-Khamisy et al., 2005). Protein complexes immunoprecipitated with Tdp1 antibody, which contain DNA ligase IIIα, XRCC1 and PNK, are active in the repair of 3’-tyrosyl DNA substrates (El-Khamisy et al., 2005). Although these complexes are presumably involved in the repair of the nuclear genome, a fraction of Tdp1 is localized in mitochondria and participates in mitochondrial base excision repair (Das et al., 2010), raising the possibility that the interaction between Tdp1 and DNA ligase IIIα contributes to repair in this organelle. Notably, mutations in the TDP1 gene are responsible for the inherited neurodegenerative disease, spinocerebellar ataxia with axonal neuropathy-1 (El-Khamisy et al., 2005). Thus, the nuclear and/or mitochondrial repair pathways involving DNA ligase IIIα and Tdp1 appear to play a key role in maintaining the viability of terminally differentiated neuronal cells. This is discussed in more detail in the section below.
NEIL1 and NEIL2
The NEIL DNA glycosylases, which also have AP lyase activity, remove oxidatively damaged DNA bases. These proteins also appear to interact exclusively with DNA ligase IIIα (Wiederhold et al., 2004; Das et al., 2006). The interaction of the C-terminal region of DNA ligase IIIα with either NEIL1 or NEIL2 (Fig. 4) is one in a network of protein:protein interactions that links DNA ligase IIIα:XRCC1 with NEIL1 or NEIL2, polynucleotide kinase 3’ phosphatase and DNA polymerase β in complexes that repair oxidative base lesions via an AP endonuclease-independent mechanism (Wiederhold et al., 2004; Das et al., 2006). As with Tdp1, NEIL1 and NEIL2 are present in mitochondria as well as nuclei (Hu et al., 2005; Mandal et al., 2012).
DNA polymerase γ
XRCC1 is absent from mammalian mitochondria suggesting that the mitochondrial version of DNA ligase IIIα has different protein partners in this organelle. Indeed, an interaction between DNA ligase IIIα and the mitochondrial DNA polymerase, Pol γ, has been identified (De and Campbell, 2007). This interaction, which involves the central catalytic region of DNA ligase IIIα (Fig. 4), presumably co-ordinates the activities of these enzymes during mitochondrial DNA replication and repair.
Cellular functions of DNA ligases encoded by the LIG3 gene
Following the identification of the interaction between DNA ligase IIIα and XRCC1 (Caldecott et al., 1994), it was assumed that DNA ligase IIIα participated in base excision repair and the repair of SSBs with XRCC1 and that mutational inactivation of the LIG3 gene would result in the same phenotype as xrcc1 mutant cells. However, expression of a mutant version of XRCC1, which did not interact with DNA ligase IIIα and, as a consequence, did not increase the steady state levels of nuclear DNA ligase IIIα, complemented the DNA damage sensitivity of cycling xrcc1 mutant cells (Taylor et al., 2000a). These studies demonstrated that XRCC1, which is present at higher levels than DNA ligase IIIα (Leppard et al., 2003), functions independently of DNA ligase IIIα in nuclear DNA repair. Furthermore, studies by the Campbell laboratory showed that, although XRCC1 was not detectable in mitochondria, the reduction of DNA ligase IIIα levels by siRNA disrupted mitochondrial function, indicating that the mitochondrial version of DNA ligase IIIα functions in mitochondrial DNA metabolism independently of XRCC1 (Lakshmipathy and Campbell, 2000; Lakshmipathy and Campbell, 2001).
DNA replication and repair of base lesions and SSBs
Recent conditional gene targeting approaches have shown that the LIG3 gene is essential because of the mitochondrial function of DNA ligase IIIα (Gao et al., 2011; Simsek et al., 2011b; Arakawa et al., 2012). Furthermore, cells lacking nuclear DNA ligase IIIα did not exhibit sensitivity to a variety of DNA agents that xrcc1 mutant cells are sensitive to (Gao et al., 2011; Simsek et al., 2011b). Using shRNA to knockdown expression of the other DNA ligases, it was concluded that DNA ligase I plays the predominant role in excision repair and the repair of SSBs in the nucleus (Gao et al., 2011). There are, however, conflicting reports regarding the contribution of DNA ligases I and IIIα to DNA repair. For example, mouse embryonic fibroblasts, either deficient in or lacking DNA ligase I, activity do not exhibit DNA damage sensitivity (Bentley et al., 1996; Bentley et al., 2002; Harrison et al., 2002) whereas human DNA ligase I-deficient fibroblasts are sensitive to DNA damage, in particular DNA alkylation (Teo et al., 1983a; Teo et al., 1983b; Barnes et al., 1992). Differences in DNA repair between mouse and human cells, the expression levels of DNA ligase I and DNA ligase IIIα and the degree of redundancy between DNA ligase I and DNA ligase IIIα in different cell types may underlie these apparently contradictory observations. In support of this idea, it has been shown recently that the relative stoichiometry of DNA ligases I and IIIα differed between the mouse striatum and cerebellum and that these differences correlated with BER efficiency and triplet repeat stability in these tissues (Goula et al., 2012).
Based on the compelling evidence linking DNA ligase I with DNA replication (Ellenberger and Tomkinson, 2008), it was surprising that lig1 null mouse cells were viable (Bentley et al., 1996; Bentley et al., 2002). Studies in chicken DT40 cells have shown that nuclear DNA ligase IIIα is essential for DNA replication in the absence of DNA ligase I and may even contribute to DNA replication in the presence of DNA ligase I (Arakawa et al., 2012). This appears to also be the case in mammalian cells as both DNA ligase IIIα and XRCC1 are required for proliferation in cells with reduced levels of DNA ligase I. In addition, there is increased association of DNA ligase IIIα and XRCC1 with chromatin and co-localization with replication in DNA ligase I-deficient cells (Chalony et al, 2012). At the present time, it is unclear how DNA ligase IIIα is targeted to replication forks. One possible mechanism is recruitment to single-strand interruptions in the lagging strand via an interaction of the DNA ligase IIIα:XRCC1 complex with poly (ADP-ribosylated) PARP1 (Masson et al., 1998; Schreiber et al., 2002; Leppard et al., 2003; Okano et al., 2003; Okano et al., 2005). Alternatively, the DNA ligase IIIα:XRCC1 complex may be recruited via an interaction between XRCC1 and PCNA (Fan et al., 2004). In base excision repair (BER), there are different subpathways involving either DNA ligase I or DNA ligase IIIα (Caldecott et al., 1996; Frosina et al., 1996; Cappelli et al., 1997; Levin et al., 2000). DNA ligase IIIα-dependent short-patch BER is thought to be a housekeeping repair pathway that acts on the entire genome whereas replication-associated BER appears to occur via DNA ligase I-dependent long-patch BER (Ellenberger and Tomkinson, 2008). Similarly, in nucleotide excision repair, there is a DNA ligase IIIα-dependent pathway that operates in cycling and non-cycling cells and a DNA ligase I-dependent pathway that operates in S phase cells (Moser et al., 2007). The results of recent studies are challenging this simple model, in which DNA ligase I-dependent excision repair is only active in proliferating cells. As mentioned previously, post-mitotic tissues with higher levels of DNA ligase I have increased BER activity and triplet repeat stability (Goula et al., 2012). Furthermore, DNA ligase I appears to be the predominant DNA ligase active in XRCC1-mediated DNA repair even in non-dividing cells (Gao et al., 2011; Katyal and McKinnon, 2011). It should, however, be noted that mouse embryonic fibroblasts, either lacking or having reduced DNA ligase I activity, do not exhibit increased sensitivity to DNA damaging agents (Bentley et al., 2002; Harrison et al., 2002). This suggests that DNA ligase IIIα may be able to effectively substitute for DNA ligase I in nuclear excision repair in certain cell types. At the present time, the identity of the DNA ligase(s) that completes DNA mismatch repair is not known.
Repair of DSBs
The majority of DSBs are repaired by the major NHEJ pathway involving DNA PK and DNA ligase IV (Ellenberger and Tomkinson, 2008). As mentioned above, there is evidence for a DNA ligase IIIα-dependent alternative NHEJ pathway that makes a minor contribution to DSB repair in cells with a functional DNA PK-dependent NHEJ pathway (Wang et al., 2005; Corneo et al., 2007; Yan et al., 2007; Xie et al., 2009; Simsek et al., 2011a). The alternative NHEJ pathway is predominantly responsible for chromosomal translocations (Simsek et al., 2011a) and, in accord with the DNA ligase IIIα ZnF being critical for intermolecular ligation in vitro (Taylor et al., 2000b; Cotner-Gohara et al., 2008; Cotner-Gohara et al., 2010), deletion of the DNA ligase IIIα ZnF significantly reduces the frequency of translocations (Simsek et al., 2011a). Interestingly, XRCC1 does not appear to be required for alternative NHEJ making this the first nuclear DNA repair pathway in which DNA ligase IIIα appears to function independently of XRCC1 (Boboila et al., 2012). DNA ligase IIIα is overexpressed in a significant fraction of cancer cell lines (Chen et al., 2008; Sallmyr et al., 2008). Notably, this overexpression is indicative of increased activity of the DNA ligase IIIα–dependent alternative NHEJ pathway and an increased dependence on this pathway for the repair of DSBs in these cells (Sallmyr et al., 2008; Tobin et al., 2012a; Tobin et al., 2012b). Cells with this DNA repair abnormality, which has been detected in samples from patients with therapy-resistant forms of chronic myeloid leukemia and breast cancer, can be selectively targeted by inhibiting both DNA ligase IIIα and PARP1 (Tobin et al., 2012a; Tobin et al., 2012b). These preclinical studies suggest that DNA ligase IIIα is a potential therapeutic target, in particular in cancers that have failed frontline therapy. There are, however, concerns about the potential toxic effects of DNA ligase III inhibitors on normal tissues and cells because of the essential role of this enzyme in mitochondria. At the present time, the identity of the DNA ligase(s) that completes recombinational repair of DSBs is not known.
Role of the LIG3 gene in neuronal cells
As mentioned above, defects in the DNA ligase IIIα-interacting protein TDP1 have been identified as the cause of the hereditary neurodegenerative disease, spinocerebellar ataxia with axonal neuropathy 1 (El-Khamisy et al., 2005). In addition, defects in two other DNA repair proteins, aprataxin and polynucleotide kinase phosphatase, that interact with the DNA ligase IIIα:XRCC1 complex have been identified as the causes of ataxia-oculomotor apraxia 1 (Moreira et al., 2001; Ahel et al., 2006) and a recently discovered disease characterized by microcephaly, early onset-intractable seizures and developmental delay (Shen et al.), respectively. TDP1, aprataxin and PNKP are all involved in the cleaning up of termini at SSBs both in nuclei and mitochondria (Rass et al., 2007; Das et al., 2010; Sykora et al., 2011; Mandal et al., 2012; Tahbaz et al., 2012). Aprataxin removes adenylate groups from 5’ phosphate DNA termini, generated by abortive ligation (Ahel et al., 2006) whereas PNKP phosphorylates 5’ hydroxyl termini and dephosphorylates 3’-phosphate termini (Karimi-Busheri and Weinfeld, 1997; Karimi-Busheri et al., 1998). These studies suggest that cells of the nervous system are more sensitive to the loss of proteins involved in the repair of single strand breaks than other cell types. This may be because terminally differentiated neuronal cells lack some of the DNA repair mechanisms that are active in proliferating cells and/or because of the high levels of active oxidative metabolism in the CNS that generate excessive DNA damage (Barzilai, 2007; Chen et al., 2007).
Interestingly, neural-specific inactivation of the XRCC1 and LIG3 genes have different effects on the developing nervous system (Lee et al., 2009; Katyal and McKinnon, 2011). Neuronal cells lacking XRCC1 are hypersensitive to DNA damaging agents that generate SSBs as are neuronal cells lacking either Tdp1 or aprataxin. However, the neuropathology resulting from the loss of XRCC1 function is more severe than that caused by loss of either Tdp1 or aprataxin. This presumably reflects the central role of XRCC1 in the repair of all nuclear SSBs whereas Tdp1 and aprataxin are only required for specific subsets of SSBs. These studies are consistent with the conclusion that DNA ligase I is the predominant activity in the XRCC1-dependent repair of SSBs in nuclear DNA (Gao et al., 2011). While neural inactivation of XRCC1 resulted in a seizure-like phenotype after about 3 months, neural inactivation of LIG3 had a more severe effect, with ataxia evident after two weeks and death within three weeks. The neuropathology induced by loss of DNA ligase IIIα function was markedly different than that induced by loss of XRCC1 with the neuronal cells lacking DNA ligase IIIα exhibiting mitochondrial defects. Thus, it appears that XRCC1-dependent repair of nuclear SSBs plays a critical neuroprotective role whereas neuropathologies associated with loss of DNA ligase IIIα function are due to mitochondrial dysfunction (Katyal and McKinnon, 2011). While the contribution of DNA ligase IIIα to nuclear DNA metabolism may vary depending on cell-type and growth status, it plays an essential and unique role in mitochondrial DNA metabolism. It is possible that the neuropathology resulting from defects in Tdp1, aprataxin or PNKP may be due, at least in part, to reduced DNA ligase IIIα-dependent repair of mitochondrial DNA.
Concluding Remarks
The mammalian LIG3 gene encodes distinct DNA ligase polypeptides that participate in nuclear and mitochondrial DNA metabolism. Recent studies have shown that the LIG3 gene is essential for cell viability because mitochondrial DNA ligase IIIα is required for mitochondrial function. There is a significant functional redundancy between DNA ligases I and IIIα in nuclear DNA replication and repair. Further studies are needed to characterize the role of DNA ligase IIIα in DNA replication in cells that are deficient in DNA ligase I activity. DNA ligase IIIα is the predominant activity involved in generating chromosomal translocation via its participation in an alternative NHEJ pathway. Notably, DNA ligase III is frequently overexpressed in a significant fraction of cancer cells, resulting in increased activity of the alternative NHEJ pathway. Initial studies with DNA ligase III inhibitors indicate that cancer cells with this DNA repair abnormality can be selectively targeted. Further studies are needed to elucidate the role of DNA ligase IIIβ, which is generated by an alternative splicing mechanism detected in male germs cells, in meiotic recombination and/or germ cell development.
Acknowledgements
Studies in the Tomkinson laboratory on DNA ligase III are supported by research grants from the National Institutes of Health (P01 CA92584 and ES12512 to AET), a grant from the V Foundation and the University of New Mexico Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- Ame J, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Arakawa H, Bednar T, Wang M, Paul K, Mladenov E, Bencsik-Theilen AA, Iliakis G. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res. 2012;40:2599–2610. doi: 10.1093/nar/gkr1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- Barzilai A. The contribution of the DNA damage response to neuronal viability. Antioxid Redox Signal. 2007;9:211–218. doi: 10.1089/ars.2007.9.211. [DOI] [PubMed] [Google Scholar]
- Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- Bentley DJ, Harrison C, Ketchen AM, Redhead NJ, Samuel K, Waterfall M, Ansell JD, Melton DW. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J Cell Sci. 2002;115:1551–1561. doi: 10.1242/jcs.115.7.1551. [DOI] [PubMed] [Google Scholar]
- Boboila C, Oksenych V, Gostissa M, Wang JH, Zha S, Zhang Y, Chai H, Lee CS, Jankovic M, Saez LM, Nussenzweig MC, McKinnon PJ, Alt FW, Schwer B. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1) Proc Natl Acad Sci U S A. 2012;109:2473–2478. doi: 10.1073/pnas.1121470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. Faseb J. 1997;11:68–76. [PubMed] [Google Scholar]
- Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular 'nick-sensor' in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- Chen J, Tomkinson AE, Ramos W, Mackey ZB, Danehower S, Walter CA, Schultz RA, Besterman JM, Husain I. Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination. Mol Cell Biol. 1995;15:5412–5422. doi: 10.1128/mcb.15.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lee HM, Greeley GH, Jr, Englander EW. Accumulation of oxidatively generated DNA damage in the brain: a mechanism of neurotoxicity. Free Radic Biol Med. 2007;42:385–393. doi: 10.1016/j.freeradbiomed.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson GM, MacKerrell J, A D, Tomkinson AE. Rational design of human DNA ligase inhbitors that target cellular DNA replication and repair. Cancer Res. 2008;68:3169–3177. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, Meek K, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- Cotner-Gohara E, Kim IK, Hammel M, Tainer JA, Tomkinson AE, Ellenberger T. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry. 2010;49:6165–6176. doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotner-Gohara E, Kim IK, Tomkinson AE, Ellenberger T. Two DNA-binding and nick recognition modules in human DNA ligase III. J Biol Chem. 2008;283:10764–10772. doi: 10.1074/jbc.M708175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuneo MJ, Gabel SA, Krahn JM, Ricker MA, London RE. The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes. Nucleic Acids Res. 2011;39:7816–7827. doi: 10.1093/nar/gkr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, Hazra TK. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair (Amst) 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BB, Dexheimer TS, Maddali K, Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci U S A. 2010;107:19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A, Campbell C. A novel interaction between DNA ligase III and DNA polymerase gamma plays an essential role in mitochondrial DNA stability. Biochem J. 2007;402:175–186. doi: 10.1042/BJ20061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Maria J, Zhou Y, Tsai MS, Kuhnlein J, Carney JP, Paull TT, Tomkinson AE. Human Mre11/Rad50/Nbs1 and DNA ligase III{alpha}/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair (Amst) 2007;6:1485–1495. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: Structural and functional insights. Annu. Rev. Biochem. 2008;7:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., 3rd XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goula AV, Pearson CE, Della Maria J, Trottier Y, Tomkinson AE, Wilson DM, 3rd, Merienne K. The nucleotide sequence, DNA damage location, and protein stoichiometry influence the base excision repair outcome at CAG/CTG repeats. Biochemistry. 2012;51:3919–3932. doi: 10.1021/bi300410d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Alternative endings. Proc Natl Acad Sci U S A. 2008;105:405–406. doi: 10.1073/pnas.0711334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C, Ketchen AM, Redhead NJ, O'Sullivan MJ, Melton DW. Replication failure, genome instability, and increased cancer susceptibility in mice with a point mutation in the DNA ligase I gene. Cancer Res. 2002;62:4065–4074. [PubMed] [Google Scholar]
- Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- Husain I, Tomkinson AE, Burkhart WA, Moyer MB, Ramos W, Mackey ZB, Besterman JM, Chen J. Purification and characterization of DNA ligase III from bovine testes. Homology with DNA ligase II and vaccinia DNA ligase. J Biol Chem. 1995;270:9683–9690. doi: 10.1074/jbc.270.16.9683. [DOI] [PubMed] [Google Scholar]
- Karimi-Busheri F, Lee J, Tomkinson AE, Weinfeld M. Repair of DNA strand gaps and nicks containing 3'-phosphate and 5'-hydroxyl termini by purified mammalian enzymes. Nucleic Acids Res. 1998;26:4395–4400. doi: 10.1093/nar/26.19.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi-Busheri F, Weinfeld M. Purification and substrate specificity of polydeoxyribonucleotide kinases isolated from calf thymus and rat liver. J Cell Biochem. 1997;64:258–272. [PubMed] [Google Scholar]
- Katyal S, McKinnon PJ. Disconnecting XRCC1 and DNA ligase III. Cell Cycle. 2011;10:2269–2275. doi: 10.4161/cc.10.14.16495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28:3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res. 2001;29:668–676. doi: 10.1093/nar/29.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chalony C, Hoffschir F, Gauthier LR, Gross J, Biard DS, Boussin FD, Pennaneach V. Partial Complementation of a DNA ligase I defciciency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells. Cell Mol Life Sci. 2012;69:2933–2949. doi: 10.1007/s00018-012-0975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Katyal S, Li Y, El-Khamisy SF, Russell HR, Caldecott KW, McKinnon PJ. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol Cell Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, Leng M, Mu JJ, Chen D, Songyang Z, Wang Y, Qin J. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol Cell Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey ZB, Niedergang C, Murcia JM, Leppard J, Au K, Chen J, de Murcia G, Tomkinson AE. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J Biol Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- Mackey ZB, Ramos W, Levin DS, Walter CA, McCarrey JR, Tomkinson AE. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol Cell Biol. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM, Sarkar PS, Hazra TK. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3'-phosphatase in maintenance of mitochondrial genome. J Biol Chem. 2012;287:2819–2829. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, Fousteri MI. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S, Lan L, Tomkinson AE, Yasui A. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res. 2005;33:422–429. doi: 10.1093/nar/gki190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JL, Dianova II, Finch D, Tait PS, Strom CE, Helleday T, Dianov GL. XRCC1 phosphorylation by CK2 is required for its stability and efficient DNA repair. 2010;9:835–841. doi: 10.1016/j.dnarep.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Paull TT, Lee JH. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle. 2005;4:737–740. doi: 10.4161/cc.4.6.1715. [DOI] [PubMed] [Google Scholar]
- Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst) 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Actions of aprataxin in multiple DNA repair pathways. J Biol Chem. 2007;282:9469–9474. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- Sallmyr A, Tomkinson AE, Rassool F. Up-regulation of WRN and DNA ligase IIIalpha in Chronic myeloid leukemia: Consequences for the repair of DNA double strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, Ganesh VS, Chang BS, Grix A, Hill RS, Topcu M, Caldecott KW, Barkovich AJ, Walsh CA. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011a;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011b;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Jasin M. DNA ligase III: a spotty presence in eukaryotes, but an essential function where tested. Cell Cycle. 2011;10:3636–3644. doi: 10.4161/cc.10.21.18094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderhall S, Lindahl T. Mammalian DNA ligases. Serological evidence for two separate enzymes. J Biol Chem. 1975;250:8438–8444. [PubMed] [Google Scholar]
- Sykora P, Croteau DL, Bohr VA, Wilson DM., 3rd Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc Natl Acad Sci U S A. 2011;108:7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahbaz N, Subedi S, Weinfeld M. Role of polynucleotide kinase/phosphatase in mitochondrial DNA repair. Nucleic Acids Res. 2012;40:3484–3495. doi: 10.1093/nar/gkr1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RM, Moore DJ, Whitehouse J, Johnson P, Caldecott KW. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol Cell Biol. 2000a;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RM, Whitehouse CJ, Caldecott KW. The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res. 2000b;28:3558–3563. doi: 10.1093/nar/28.18.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo IA, Arlett CF, Harcourt SA, Priestley A, Broughton BC. Multiple hypersensitivity to mutagens in a cell strain (46BR) derived from a patient with immuno-deficiencies. Mutat Res. 1983a;107:371–386. doi: 10.1016/0027-5107(83)90177-x. [DOI] [PubMed] [Google Scholar]
- Teo IA, Broughton BC, Day RS, James MR, Karran P, Mayne LV, Lehmann AR. A biochemical defect in the repair of alkylated DNA in cells from an immunodeficient patient (46BR) Carcinogenesis. 1983b;4:559–564. doi: 10.1093/carcin/4.5.559. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin LA, Robert C, Nagaria P, Chumsri S, Twaddell W, Ioffe OB, Greco GE, Brodie AH, Tomkinson AE, Rassool FV. Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol Cancer Res. 2012a;10:96–107. doi: 10.1158/1541-7786.MCR-11-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin LA, Robert C, Rapoport AP, Gojo I, Baer MR, Tomkinson AE, Rassool FV. Targeting abnormal DNA double strand break repair in tyrosine kinase inhbitor-resistant chronic meyloid leukemias. Oncogene. 2012b;32:1784–1793. doi: 10.1038/onc.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson AE, Roberts E, Daly G, Totty NF, Lindahl T. Three distinct DNA ligases in mammalian cells. J Biol Chem. 1991;266:21728–21735. [PubMed] [Google Scholar]
- Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- Wang YC, Burkhart WA, Mackey ZB, Moyer MB, Ramos W, Husain I, Chen J, Besterman JM, Tomkinson AE. Mammalian DNA ligase II is highly homologous with vaccinia DNA ligase. Identification of the DNA ligase II active site for enzyme-adenylate formation. J Biol Chem. 1994;269:31923–31928. [PubMed] [Google Scholar]
- Wei YF, Robins P, Carter K, Caldecott KW, Papin DJC, Yu G-L, Wang R-P, Shell BK, Nash RA, Schar P, Barnes DE, Haseltine WA, Lindahl T. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and genetic recombination. Mol. Cell. Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]