Abstract
Higher plants assimilate nitrogen in the form of ammonia through the concerted activity of glutamine synthetase (GS) and glutamate synthase (GOGAT). The GS enzyme is either located in the cytoplasm (GS1) or in the chloroplast (GS2). To understand how modulation of GS activity affects plant performance, Lotus japonicus L. plants were transformed with an alfalfa GS1 gene driven by the CaMV 35S promoter. The transformants showed increased GS activity and an increase in GS1 polypeptide level in all the organs tested. GS was analyzed by non-denaturing gel electrophoresis and ion-exchange chromatography. The results showed the presence of multiple GS isoenzymes in the different organs and the presence of a novel isoform in the transgenic plants. The distribution of GS in the different organs was analyzed by immunohistochemical localization. GS was localized in the mesophyll cells of the leaves and in the vasculature of the stem and roots of the transformants. Our results consistently showed higher soluble protein concentration, higher chlorophyll content and a higher biomass accumulation in the transgenic plants. The total amino acid content in the leaves and stems of the transgenic plants was 22–24% more than in the tissues of the non-transformed plants. The relative abundance of individual amino acid was similar except for aspartate/asparagine and proline, which were higher in the transformants.
Keywords: Glutamine synthetase, Lotus, Over-expression, Transformation
Introduction
Nitrogen is one of the most limiting factors for plant growth. Plants obtain their nitrogen in the form of nitrate or ammonia. The nitrate is reduced to ammonia, which is then assimilated through the concerted activity of glutamine synthetase (GS; EC 6.3.1.2) and glutamate synthase (GOGAT; EC 1.4.7.1 and EC 1.4.1.14). Glutamine synthetase catalyzes the first step in the pathway, the synthesis of glutamine from ammonia and glutamate, while GOGAT is responsible for the synthesis of glutamate from 2-oxoglutarate and glutamine (Lea and Ireland 1999). The GS/GOGAT cycle sits at the interface of nitrogen (N) and carbon (C) metabolism. Recent work has shown strict coordination between plant N and C metabolism as the assimilation of N demands C skeleton in the form 2-oxoglutarate and considerable ATP and reductant (Oliviera and Coruzzi 1999; Lancien et al. 2000). Higher-plant GS is an octameric enzyme that occurs as different isoforms of Mr 320–360 kDa. The individual GS isoforms appear to have specific non-redundant physiological roles in ammonia assimilation (Lancien et al. 2000). GS enzyme is either located in the cytoplasm (GS1) or in the chloroplast (GS2). GS2 is predominantly expressed in green tissues. It has been proposed that GS2 plays an important role in the assimilation of ammonia from nitrate reduction, and it has also been demonstrated that GS2 is indispensable for re-assimilation of photorespiratory ammonia (Blackwell et al. 1987; Wallsgrove et al. 1987; Kozaki and Takeba 1996). GS1 is encoded by a small multigene family, and the different GS1 isoforms are differentially regulated in a tissue- and developmental stage-specific manner (Bennett et al. 1989; Peterman and Goodman 1991; Temple et al. 1995; Dubois et al. 1996; Morey et al. 2002). GS1 plays a major role in the assimilation of ammonia derived from N2 fixation in the nodules and also has an important role in the remobilization of nitrogen during senescence, bacterial infection, herbicide treatment and water stress (Bauer et al. 1997; Pérez-García et al. 1998; Brugiere et al. 2000; Masclaux et al. 2000). GS1 is the main isoform involved in the assimilation of external ammonia (Sukanya et al. 1994) and the ammonia derived from other sources of nitrogen, like the synthesis of secondary metabolites and the recycling of nitrogen from protein catabolism (Joy 1988). Root GS1 may also have a role in the assimilation of the ammonia derived from nitrate reduction in the roots (Lancien et al. 1999). GS1 is also present in the shoots where it is localized in the vascular tissue (Carvalho et al. 1992; Kamachi et al. 1992; Pereira et al. 1992; Dubois et al. 1996; Sakurai et al. 1996; Pérez-García et al. 1998). The role of GS1 in the vasculature, however, is not well defined but it has been suggested that it may be involved in the synthesis of glutamine for nitrogen transport (Edwards et al. 1990; Dubois et al. 1996).
Since GS activity plays a central role in utilizing carbon skeletons to produce amino acids and other nitrogen compounds, a question that is important to address is whether modulating GS activity levels would have any effect on plant metabolism and performance. Several attempts have been made to modulate the levels of the GS enzyme in different plants using genetic engineering tools, with rather mixed outcomes (Eckes et al. 1989; Hemon et al. 1990; Hirel et al. 1992, 1997; Temple et al. 1993, 1994, 1998; Su et al. 1995; Vincent et al. 1997; Brugiere et al. 1999; Gallardo et al. 1999; Limami et al. 1999; Ortega et al. 2001; Fuentes et al. 2001; Oliveira et al. 2002; Carvalho et al. 2003; Fei et al. 2003; Fu et al. 2003; Harrison et al. 2003). Tobacco plants transformed with different GS1 genes driven by the CaMV 35S promoter have shown an increase in GS activity and GS1 polypeptide level in the leaves (Eckes et al. 1989; Hemon et al. 1990; Hirel et al. 1992, 1997; Temple et al. 1993; Fuentes et al. 2001; Oliveira et al. 2002). These plants showed little change in growth or in leaf protein content under optimal growth conditions. However, under nitrogen deficiency or moderate light conditions the GS1 tobacco transformants showed improved plant performance compared to control plants and this was attributed to increased photosynthetic rates and increased rates of ammonia assimilation (Fuentes et al. 2001; Oliveira et al. 2002). Rice plants expressing a bacterial GS gene performed better than control plants under low nitrogen conditions (Su et al. 1995). Poplar trees expressing a conifer GS gene were taller than the non-transformed trees under low nitrogen conditions and showed altered leaf morphology (Gallardo et al. 1999; Fu et al. 2003). A 50–80% increase in GS activity was detected in the leaves of Lotus corniculatus transformed with a soybean GS1 gene driven by the CaMV 35S promoter (Vincent et al. 1997). Pea plants transformed with a soybean GS1 gene driven by the CaMV 35S promoter or a nodule-specific promoter, while showing increased levels of GS1, showed no increase in GS activity or in total nitrogen or protein content. Similarly, our attempts to over-express alfalfa and soybean GS1 genes in alfalfa in both a constitutive and nodule-specific manner have to date resulted in no increase in either the GS1 polypeptide level or GS activity (Ortega et al. 2001). There are some reports of successful antisense RNA technology-mediated down-regulation of GS1 in plants resulting in physiological and/or biochemical changes (Temple et al. 1994; Carvahlo et al. 2003; Harrison et al. 2003). Vasculature-specific down-regulation of GS1 in alfalfa, using the acidic chitinase promoter to drive the antisense GS1 construct, has produced nitrogen deficiency symptoms in the young leaves (Temple et al. 1994) while nodule-specific down-regulation of GS1 produced an increase in asparagine synthetase activity and asparagine content in the nodules of L. japonicus and Medicago truncatula (Carvahlo et al. 2003; Harrison et al. 2003).
The differences in the response to the introduction of a GS1 transgene reported in the different plants has prompted us to study how L. japonicus, a model legume, responds to the introduction of the GS1 transgene driven by the constitutive CaMV 35S promoter. Our results suggest that GS1 could represent a key component of nitrogen-use efficiency and yield in L. japonicus.
Materials and methods
GS recombinant DNA clones
Plasmid pMsGS100, which contains a constitutively expressed class of alfalfa (Medicago sativa L.) GS1 cDNA (Temple et al. 1995; Ms glnβ in this work), was kindly provided by Dr. H. M. Goodman (Dept. of Molecular Biology and Genetics, Harvard Medical School, MA, USA). This clone was isolated from a herbicide-resistant alfalfa cell culture (DasSarma et al. 1986). The construction of the plasmid pGS111 containing the Ms glnβ cDNA under the control of the CaMV 35S promoter and the NOS 3′ transcription terminator was previously reported (Temple et al. 1993). A DNA fragment that contains the 3′-untranslated region (UTR) of the Ms glnβ (Temple et al. 1995) was used to specifically monitor the expression of the transgene. Also, a DNA fragment that contain the 3′-UTR of the L. japonicus GS1 cDNA (pLj glnβ) was used to monitor the expression of an endogenous GS1 gene. Clone pLj glnβ was cloned from an L. japonicus nodule λZAP cDNA library, kindly provided by Dr. J. Stougaard (University of Aarhus, Denmark).
Plant material
The plasmid pGS111 containing the CaMV35S–Ms glnβ gene construct was introduced into L. japonicus through Agrobacterium tumefaciens-mediated transformation. Transformation and regeneration procedures have been previously reported (Handberg and Stoutgaard 1992). The presence of the transgene in the transformants was checked by PCR using primers in the GS1 coding region and the CaMV 35S promoter region. Two independent transgenic lines with high GS activity and increased accumulation of GS1 protein (M-4 and K-25) were selected for further characterization from the several independent primary transformants previously analyzed (Temple et al. 1994). These transformed lines were selfed and the seeds obtained were germinated to identify the plants with the highest GS activity and GS1 polypeptide accumulation. The plants with the highest relative level of GS activity and GS1 polypeptide were selected as the putative homozygous lines. The selfed progeny of the putative homozygous K-25 and M-4 lines was analyzed for kanamycin (100 µg ml−1) resistance. Lines whose progeny showed 100% kanamycin resistance were confirmed as the homozygous R1 lines. The progeny of these R1 lines were used for all the experiments. Seeds of the progeny from the homozygous transgenic lines and the control non-transformed plants were germinated on peat pellets and grown under constant light. Cool-white fluorescent lamps which produced a measured photon flux density of 47 µmol m−2 s−1 were used. Seven weeks after germination plant growth was measured in the transgenic and control L. japonicus plants as plant fresh weight accumulation, and tissues were harvested to measure total protein content and chlorophyll content. At this stage, control and transformed plants were transferred to soil and established in the greenhouse. Plants were inoculated with Rhizobium loti. Plants were watered alternatively with water and 0.25×Hoagland solution. The typical photon flux density in the greenhouse was 382 µmol m−2 s−1 at midday. Tissues from control and transgenic plants were harvested at different times after planting and used for RNA, protein, enzyme activity and amino acid analysis. Nodules were usually obtained 4–5 weeks after inoculation. All experiments described in this paper were performed at least three times with consistent results and only representative experiments are shown here.
RNA isolation and analysis
Total RNA was isolated using an RNA isolation kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s procedure. Total RNA was fractionated on 1.35% agarose/formaldehyde gels, stained with SYBR Gold nucleic acid stain (Molecular Probes, Eugene OR, USA) and blotted onto nitrocellulose. Filters were pre-hybridized overnight and hybridized to 32P-labeled DNA probes for 20–24 h in hybridization buffer [50% formamide, 5×SSC, 5×Denhardt’s solution, 50 mM sodium phosphate (pH 7.0), 0.1% SDS, 0.1 mg ml−1 denatured salmon sperm DNA] at 42°C. Following hybridization, the filters were washed 4 times with 2×SSC, 0.5% SDS at 54°C for 20 min each and exposed to X-ray film.
Protein and chlorophyll extraction, enzyme assay
L. japonicus tissues were ground in liquid nitrogen and homogenized with 2 (roots, stems) or 5 (leaves, nodules) volumes of cold extraction buffer [50 mM Tris –HCl (pH 8.0), 20% glycerol, 3% insoluble polyvinylpolyactivity is reportedpyrrolidone (PVPP)] containing a protease inhibitor cocktail used according to the manufacturer (Roche Applied Science, Germany). The homogenate was centrifuged for 15 min at 12,500 g, and the supernatant was used for enzyme activity and protein analysis. For chlorophyll extraction, the insoluble pellet was re-extracted with cold acetone and clarified by centrifugation. Chlorophyll content was determined in the acetone fraction by measuring the absorbance at 425 nm. Protein concentration was measured in the soluble fraction by a Bradford protein assay (BioRad, Hercules, CA, USA), using BSA as protein standard. GS activity was measured spectrophotometrically at 500 nm by the transferase assay reported by Ferguson and Sims (1971). GS activity is reported as µmol of γ-glutamyl hydroxamate produced per min, at 30°C. GS activity data presented are the average of not less than 3 independent experiments with at least 12 samples in each experiment.
GS protein analysis
GS protein was analyzed by western blots after polyacrylamide gel electrophoresis. Three different electrophoresis systems were employed:
-
–
(A) SDS–PAGE using 12% acrylamide large-format slab gels, prepared and run according to the manufacturer (Bio-Rad). Five µg of denatured protein was loaded per lane.
-
–
(B) Two-dimensional SDS–PAGE was performed as previously described (Ortega et al. 2001). An amount of protein equivalent to 0.05 µmol min−1 of transferase activity was loaded per sample.
-
–
(C) Non-denaturing (native) PAGE was performed in 7.5% acrylamide slab gels; the pH of the resolving gel was 8.15. For native gels, 10 µg of total soluble protein was loaded per lane and fractionated at 100 V for 16 h at 4°C.
For western analysis, proteins from the polyacrylamide gels were electroblotted onto nitrocellulose in 25 mM Tris, 192 mM glycine, 20% methanol (pH 8.3). The blots were blocked with 2% BSA in Tris-buffered saline containing 0.1% Tween 20 and incubated with an antibody against Phaseolus vulgaris nodule GS1 (kindly provided by Dr. J. Cullimore, Castanet-Tolosan, France). Reacting bands were made visible with an alkaline phosphatase-linked second antibody, using nitro blue tetrazolium and 5-bromo-4-chloro-3-indoyl-phosphate as substrates. GS immunoreactive bands were quantified with Kodak 1D image analysis software (Kodak Scientific Imaging Systems, Rochester NY, USA). Experiments were repeated not less than three times with the same results. Only representative experiments are presented.
Ion-exchange chromatography
Fractionation of GS from the leaves of control and transgenic L. japonicus was performed by ion-exchange chromatography in DEAE-Sephacel. Two grams of fresh leaf tissue, including the vascular tissues, were ground in liquid nitrogen with 0.5 g insoluble PVPP. Protein was extracted in 25 ml extraction buffer [20 mM Tris (pH 8.4), 1 mM EDTA, 0.5 mM magnesium acetate, 5% glycerol, 2.5% ethylene glycol] plus a mixture of protease inhibitors [1 mM phenylmethylsulfonyl fluoride (PMSF), 50 µg ml−1 antipain, 1 µg ml−1 cystatin, 10 µg ml−1 leupeptin]. Following centrifugation at 25,000 g, the crude extract was loaded on a 12.5-ml DEAE-Sephacel column equilibrated in the extraction buffer. After washing the column in the same buffer, proteins were eluted in a 100 ml of a 0–450 mM KC1 linear gradient in extraction buffer. One-ml fractions were collected and GS activity was assayed by the transferase reaction. Representative DEAE fractions were subjected to PAGE followed by western blot analysis.
Amino acid analysis
Total amino acid analysis in leaf, stem and root tissues of control and transgenic L. japonicus plants was performed at two different locations: at the Molecular Structure Facility, University of California, Davis, and at the Experimental Station Chemical Laboratories, University of Missouri, Columbia. Results obtained were similar. Data presented are the averages of at least four different samples. Tissue samples were soaked overnight in performic acid and the liquid-phase hydrolysis was performed in 6 N HC1/0.1% phenol at 110°C for 24 h; then samples were dried and suspended in buffer containing norleucine as an internal standard.
Histochemical localization of GS in L. japonicus tissues
Freshly cut leaves, stems, roots and nodules from non-transformed and transgenic L. japonicus plants were fixed in 4% paraformaldehyde in 50 mM phosphate buffer (pH 7.2). Tissues were washed in the same buffer and dehydrated in graded series of ethanol dilutions, followed by incubation in a graded series of xylene dilutions, then infiltrated and embedded in Paraffin (Paraplast; Oxford Labware, St. Louis, MO, USA). Samples were sectioned into 10-µm slices and mounted on slides. Mounted sections were deparaffinized and GS immunodetected with an anti-GS primary antibody in combination with an immunoperoxidase staining system, according to the manufacturer’s instructions (Vectastain Elite ABC; Novocastra laboratories, UK). Digital images were taken from a Zeiss axioplan microscope at 400× magnification.
Results
Identification of homozygous lines of L. japonicus transformed with the GS1 transgene
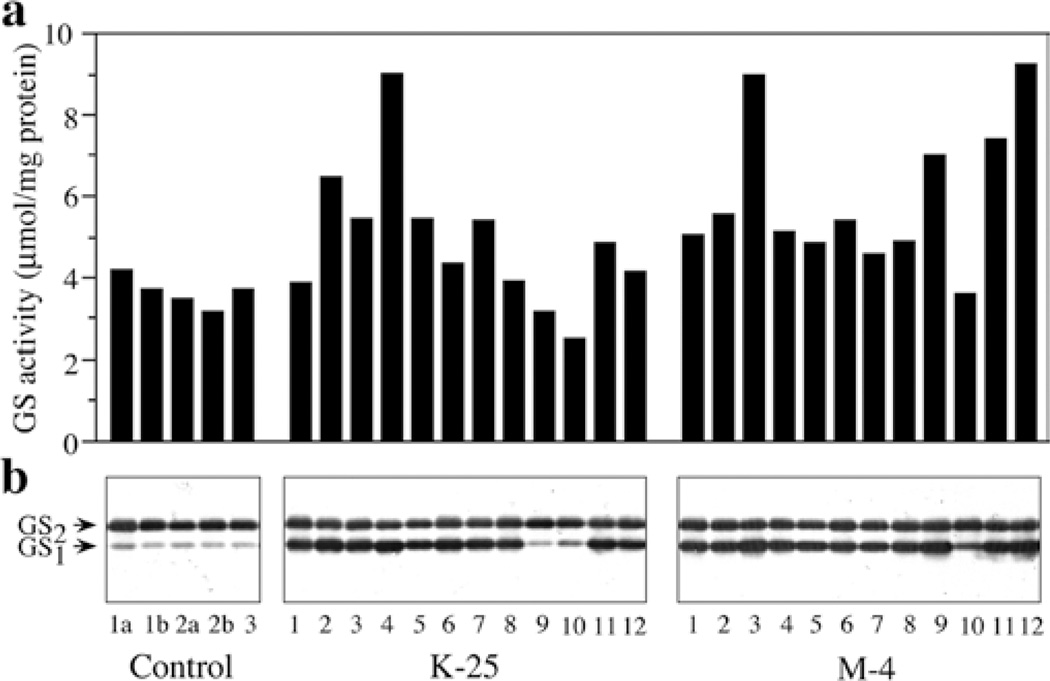
We introduced a cDNA that encodes the constitutive form of cytosolic GS from alfalfa (Ms glnβ DasSarma et al. 1986) behind the CaMV 35S promoter (CaMV35S–Ms glnβ) into L. japonicus, a model legume. Several independent L. japonicus primary transformants with the Ms glnβ were obtained (Temple et al. 1994). These transformants were tested positive for the integration of the transgene by genomic PCR. GS activity assays and GS1 protein analysis identified two independent transgenic lines (K-25 and M-4) with higher GS activity and increased accumulation of GS1 protein when compared to control non-transformed plants (Temple et al. 1994). These transformed lines were selfed and the seeds obtained were germinated on peat pellets, along with seeds from self-fertilized control non-transformed plants. The GS activity and GS1 polypeptide accumulation were analyzed in protein extracts from the leaves of these plants to identify the high expressors. A subset of the analysis is shown in Fig. 1. The plants with the highest relative level of GS activity and GS1 polypeptide were selected as putative homozygous lines. For confirmation of homozygosity for the transgene, the high expressors in the Rl generation were selfed and the seeds obtained were germinated in media containing kanamycin. All the plants selected for analysis showed progeny that was 100% kanamycin resistant and all showed a significantly higher level of GS1 polypeptide compared to control. The genetics of segregation confirmed that the R1 lines of K-25 and M-4 that were selected for further analysis were all homozygous.
Fig. 1.
a, b Analysis of GS polypeptide and GS activity in the leaves of the progeny of two independent Lotus japonicus transformants that over-express an alfalfa GS1 gene, a GS transferase activity was measured in leaf extracts from control plants and the progeny of two independent transformants following selfing. GS activity values plotted are µmol β-glutamyl hydroxamate produced per minute per mg of protein at 30°C. b Five µg of leaf protein from the same plants as those used for enzyme activity in a were subjected to SDS–PAGE followed by western blot hybridization using GS antibodies
Transgenic L. japonicus plants show accumulation of transcript corresponding to the transgene in the different organs
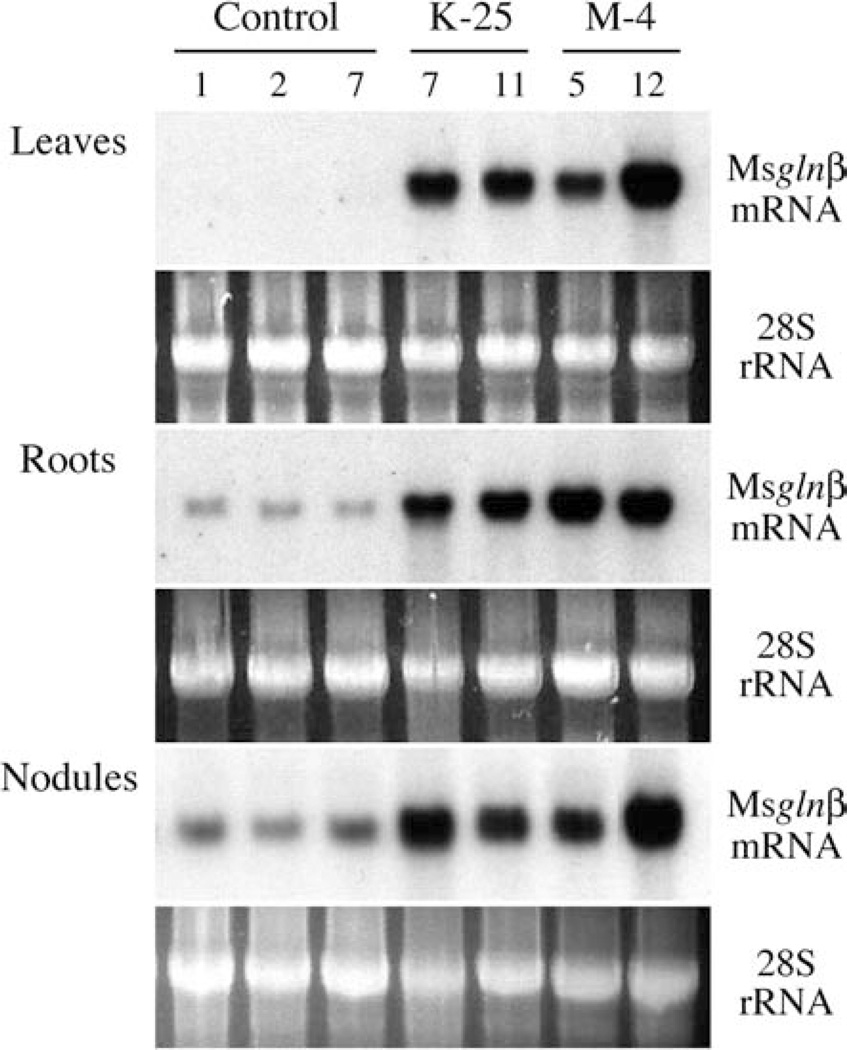
Alfalfa plants transformed with a GS1 gene driven by the CaMV 35S promoter showed a complete absence of the transgene transcript in the nodules (Ortega et al. 2001). To investigate if the Ms glnβ transgene was expressed in the nodules of transformed L. japonicus and to demonstrate that the increase in GS activity and GS1 protein in the M-4 and K-25 progeny was the result of the expression of the Ms glnβ transgene, total RNA was isolated from leaves, roots and nodules of control and transformed L. japonicus plants and following fractionation it was subjected to northern blot hybridization using the 3′-UTR of the Ms glnβ cDNA as the probe. The SYBR Gold-stained rRNA profile was used as a loading control. As seen in Fig. 2, compared to the controls the transformants showed a strong hybridization signal in all the organs including the nodules. The Ms glnβ 3′-UTR of the cDNA showed some degree of cross-hybridization to RNA from the roots and nodules of control non-transformed plants, suggesting cross-hybridization with the endogenous L. japonicus GS1 mRNA, and the level of hybridization probably reflects the abundance of the endogenous GS1 mRNA in these organs. The blots were also probed with a L. japonicus GS1 cDNA probe to monitor changes in the expression of endogenous GS1 genes. There was no significant difference in the accumulation of the endogenous LjGS1 mRNA in the non-transformed plants compared to the plants expressing the Ms glnβ transgene (data not shown).
Fig. 2.
Northern blot analysis of RNA from different tissues of control and Ms glnβ-transformed L. japonicus plants. Total RNA from different tissues (as indicated) of individual plants from control and homozygous lines of K-25 and M-4 transformants was subjected to northern blot analysis using a 32P-labeled probe for the 3′-UTR of the Ms glnβ gene. The SYBR Gold-stained pattern is shown as an internal standard for RNA loadings for each gel. Each lane represents an individual plant
Transgenic L. japonicus plants show accumulation of GS1 polypeptide and increased GS activity in the different organs
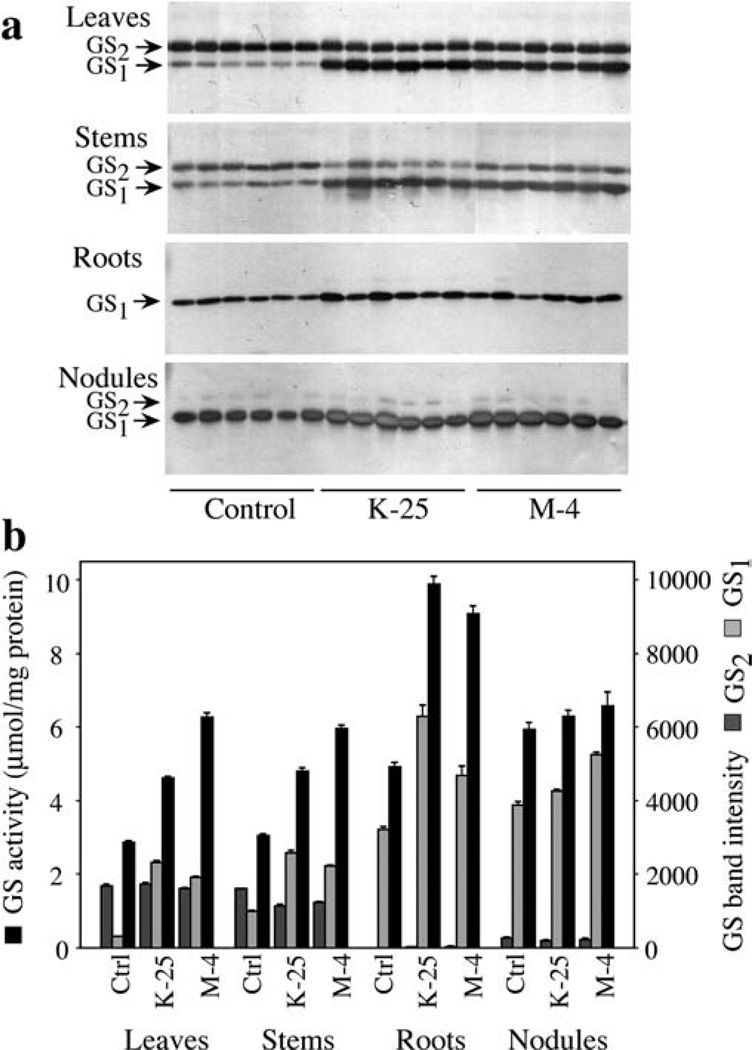
Proteins from leaves, stem, roots and nodules from randomly selected control plants and plants homozygous for the M-4 and K-25 transgenic lines were subjected to western blot analysis using an anti-GS antibody. A subset of the analysis is shown in Fig. 3a. The immunoreactive bands were quantified using the Kodak 1D image analysis software and the intensity values were plotted (Fig. 3b). Our results showed that, compared to control plants, the progeny of the plants transformed with the CaMV35S–Ms glnβ gene construct had an increased accumulation of the GS1 protein in all the organs tested; the leaves showed the highest level of increase (3- to 4-fold), the stem showed a 2- to 3-fold increase while the roots and nodules showed a smaller but significant increase. It is interesting to note that the immunoreactive band representing GS2 polypeptide showed a slightly lower level in the stem of the transformants compared to the control.
Fig. 3.
a, b GS activity and GS protein analysis in different tissues of control and Ms glnβ-transformed L. japonicus plants, a Five µg of protein extracted from leaves, stems, roots and nodules of control plants and plants from the homozygous lines (for the transgene), M-4 and K-25, were separated by SDS–PAGE, and GS was immunodetected in western blots with an anti-GS antibody, b The GS band intensities from the western blots in this experiment and other replicates of this experiment were quantified and the average band intensities (±SE) were plotted. GS1 and GS2 indicate the cytosolic and the chloroplastic GS, respectively. GS activity was determined by the transferase assay in extracts from different tissues of control non-transformed plants and plants from the progeny of the K-25 and M-4 transformants. GS activity is presented as µmol β-glutamyl hydroxamate produced per minute per mg of protein at 30°C. Values are the average (± SE) of at least three different experiments with no less than six individuals per experiment
Protein extracts from the different organs of K-25, M-4 and control plants were also analyzed for GS enzyme activity, and the average value for each of the lines was plotted. Our results showed that the increase in GS1 polypeptide accumulation in the different organs is accompanied by an increase in GS activity (Fig. 3b). The leaves and stems of the transformants showed a 1.5- to 2-fold increase in GS activity over control while the level of GS1 polypeptide accumulation was 3- to 5-fold higher. In absolute terms, the roots of the transformants showed the highest increase in GS activity, while the nodules showed a very slight increase in activity (Fig. 3b). There does not appear to be a direct correlation between the increase in GS1 polypeptide level and GS enzyme activity in the different organs, suggesting that there could be regulation at the level of holoenzyme stability.
Increased GS activity in the CaMV35S–Ms glnβ transformants can be attributed to the accumulation of a novel GS1 enzyme
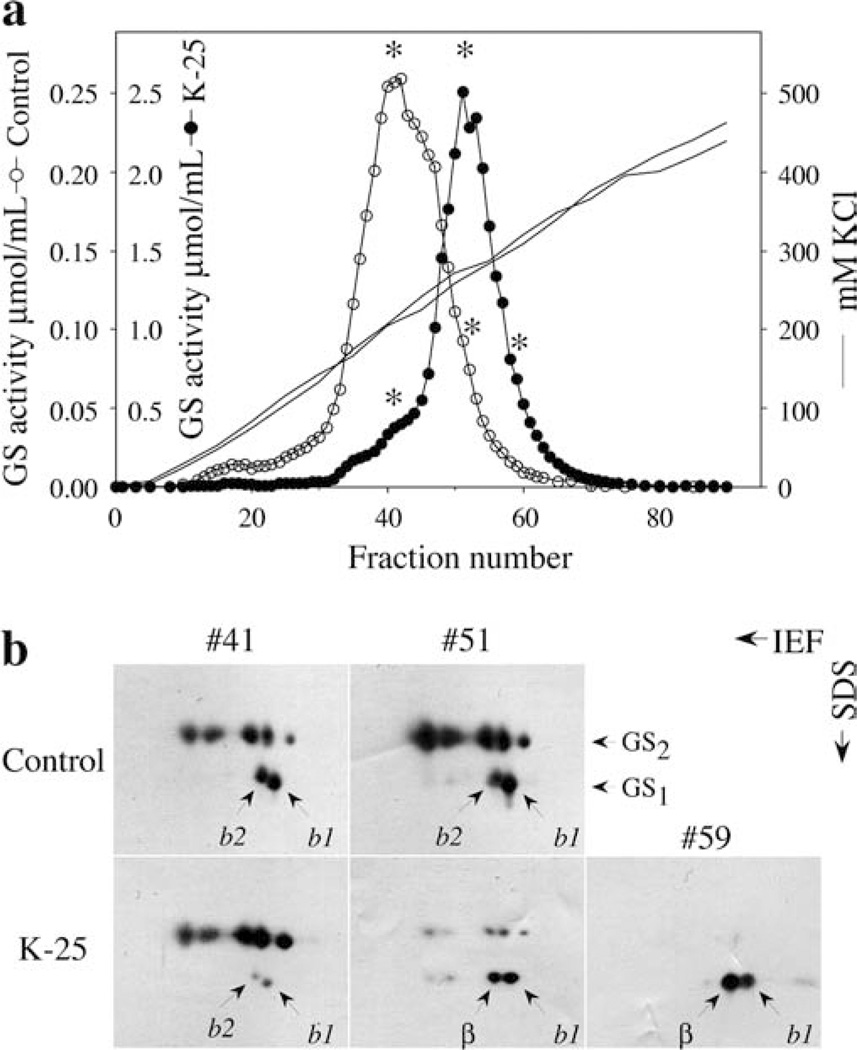
To check if the increase in GS activity in the transformants can be attributed to the accumulation of a GS1 encoded by the Ms glnβ gene, we analyzed the GS isoform profile of leaf extracts from a control plant and a K-25 plant by ion-exchange chromatography. Protein was extracted from equal amounts of leaf tissue, including the midribs, from the two plants and fractionated in a DEAE-Sephacel column. GS activity was eluted with increasing concentrations of KC1 and GS activity monitored by the transferase assay. The GS composition of the activity peaks was analyzed by two-dimensional PAGE, followed by western blot analysis with anti-GS antibodies. The leaves of the transformant (K-25) showed a significant increase in the total amount of GS activity on a fresh-weight basis (Fig. 4a; NB: the scale representing the GS activity profile for K-25 is 10-fold that for the control). Moreover, the peak of GS activity in the transgenic leaves eluted at a different position than the peak of GS activity from the leaves of the non-transformed plants. The protein fractions with the highest activity, and their counterpart fractions from control and transgenic (K-25) leaves, were analyzed by two-dimensional PAGE followed by western analysis in order to compare the GS polypeptide. (As a clarifying note: while the different panels cannot be compared quantitatively with one another because of variations due to blotting, they can be compared for qualitative and relative changes.) Two-dimensional PAGE analysis showed that the prevalent isoform in the activity peak of the transgenic K-25 leaves is the cytoplasmic GS1 isoform, while the peak of GS activity in the non-transformed L. japonicus leaves corresponded essentially to GS2 polypeptides (Fig. 4b). The fraction #41 from control plants, representing the peak activity in the control, and the corresponding fraction in the K-25 leaf extracts besides containing the GS2 polypeptides showed the presence of two spots, designated b1 and b2, corresponding in size to GS1. The b1 GS1 isoform appears to be the more abundant GS1 isoform as seen in the later fractions (fraction #51 of control in Fig. 4b). Fraction #41 of the control sample showed a significantly higher level of the GS1 polypeptides relative to GS2 when compared to fraction #41 of the K-25 sample. Fraction #51 from the K-25 plants representing the peak activity for the transformant showed mostly GS1 polypeptides consisting of the b1 subunit and a novel spot probably corresponding to the transgene product labeled as β. This polypeptide migrates very close to the endogenous GS1 polypeptide b2, but has a slightly different Mr. Fraction #51 of the control sample has relatively high levels of GS2 and there is definite drop in the amount of b1 polypeptide relative to the b2 subunit. The β polypeptide is the prevalent subunit in the later fractions of the GS activity peak from the K-25 extract. The last fractions of K-25 plants contained mainly the spot corresponding to β and to a lesser extent the spot corresponding to b1 (fraction #59 in Fig. 4b). The b2 subunit was not detected in fractions 51 and 59 from the K-25 extract. The results suggest that the majority of GS activity in the leaves of the transformant is due to a novel GS1 isozyme made up of the b1 subunit of L. japonicus and the transgene product, MsGSβ subunit.
Fig. 4.
a, b Fractionation of GS from the leaves of control and Ms glnβ-transformed L. japonicus plants by ion-exchange chromatography, a Protein extracted from 1 g of control (non-transformed) and transgenic L. japonicus (K-25) leaves, including the midribs, was loaded onto a 12.5-ml DEAE-Sephacel column equilibrated in extraction buffer and eluted with a 0–450 mM KCl linear gradient. GS activity was monitored in the eluate by the transferase assay and the activity for the different fractions as µmol β-glutamyl hydroxamate per minute per ml at 30°C was plotted on two different scales (0–0.25 µmol ml−1 for the control and 0–2.5 µmol ml−1 for K-25). The asterisks represent fractions that were further analyzed by two-dimensional gels, b Samples from selected DEAE fractions from control and transgenic (K-25) leaves (marked with asterisks in a) were separated by two-dimensional PAGE and subjected to western analysis. The loading in each gel was based on equal activity. While, the different panels cannot be compared quantitatively with one another because of variations due to blotting, they can be compared for qualitative and relative changes. The putative transgene GS product is marked as β. b1 and b2 indicate the L. japonicus GS1 products
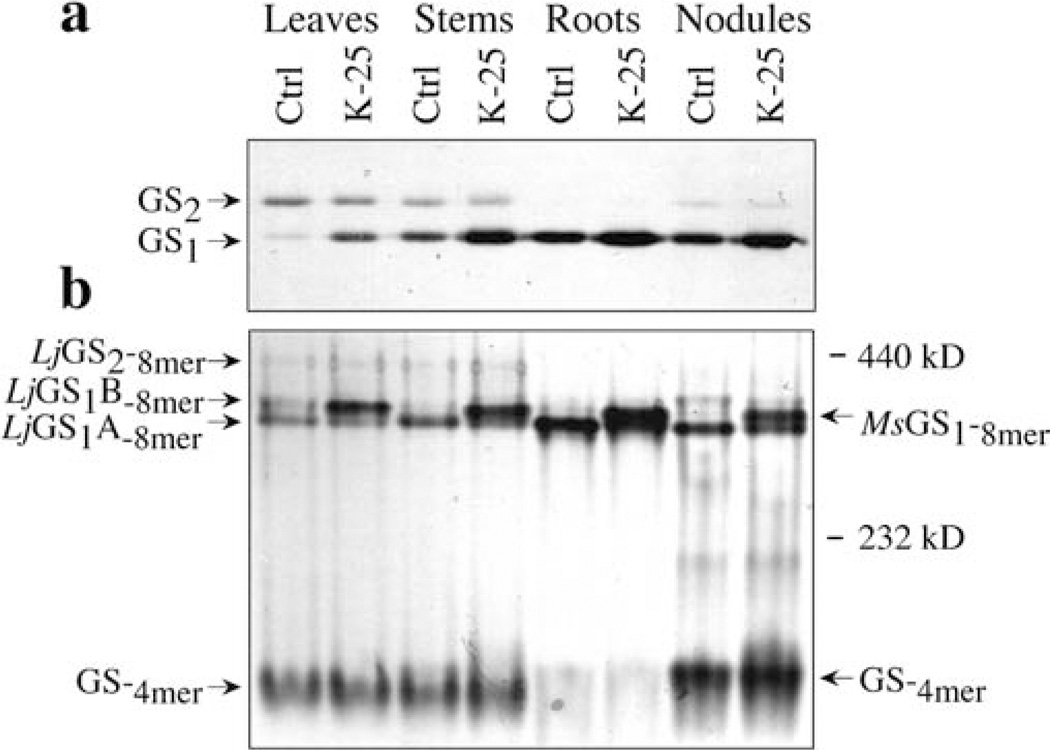
To further compare the GS isozyme profile between the transformant and the control plants, protein extracts from leaves, stems, roots and nodules of control and a K-25 transformant (homozygous for the transgene) were analyzed by non-denaturing gel electrophoresis followed by western blot analysis. The same samples were also subjected to SDS–PAGE followed by immunoblot analysis (Fig. 5a). Compared to the control samples, K-25 showed an increased level of GS1 polypeptide in all the organs. The native gel showed the presence of multiple GS immunoreactive bands with different mobilities on the gel (Fig. 5). The control organs each showed one major immunoreactive band (marked as LjGS1A-8mer), the band being more intense in the roots and nodules and with slightly faster mobility than the band in the leaves and stem. The difference in the mobility of this immunoreactive band between leaves/stem and nodules/roots would suggest differences in the subunit composition. The leaves, roots and nodules from the control plants showed a second slower migrating band (indicated as LjGS1B-8mer in Fig. 5). An immunoreactive band, marked as MsGS1-8mer, co-migrating with the LjGS1B-8mer is seen in all the organs of the transformant, the band being most intense in the roots followed by the nodules and stem. This MsGS1-8mer band may represent an octamer exclusively made up of the MsGSβ sub units or heteromers made up of L. japonicus GS1 subunits and the MsGSβ subunit. The leaves and stem of both control and transformed plants showed the presence of a slower migrating immunoreactive band, probably representing the GS2 isoform (indicated by LjGS2-8mer). DEAE chromatography coupled to non-denaturing and SDS–PAGE confirmed that this band corresponds to the GS2 isoform (data not shown). A fast migrating band that is observed at the bottom of the blot in Fig. 5 is prominent in leaves, stems and nodules from both control and transgenic plants, but not in the root extracts. Based on its migration pattern, as judged from the Mr standards included in the gel, we concluded that this faster migrating band corresponds to a tetrameric form of GS (GS-4mer in Fig. 5). The tetrameric form in the leaf and stem extracts appears to have slightly faster migration than the band from the nodules. Moreover, the tetrameric form appears to be more abundant in the nodules from the transformant compared to the control plant.
Fig. 5.
a, b Analysis of GS isoforms in the different organs of control and Ms glnβ-transformed L. japonicus a Five µg of protein from the different organs was subjected to SDS–PAGE followed by immunoblotting using the anti-GS antibodies. The cytosolic (GS1) and chloroplastic (GS2) subunits are indicated, b Ten-µg samples of the same proteins as in a were subjected to non-denaturing PAGE and GS isoforms were visualized by immunoblotting using anti-GS antibodies. The cytosolic (GS1) and the chloroplastic (GS2) isoforms are indicated. The GS octameric (8mer) and tetrameric (4mer) forms are identified by their MrA new GS isoform observed only in the transgenic tissues is represented as MsGS1 Native Mr standards were included in the gel and standard sizes are shown in kDa
The CaMV35S–Ms glnβ transformants exhibit major differences in the immunolocalization pattern for GS in all organs compared to control plants
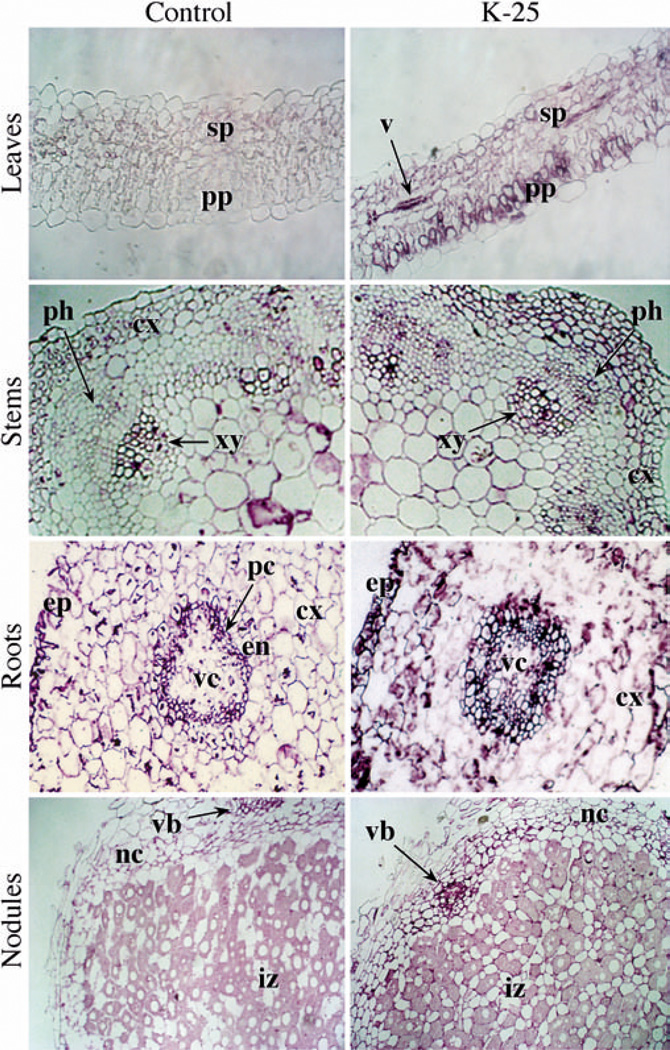
To determine the tissue distribution of the transgene product in the transformants, the different organs from control and transgenic plants were analyzed by immunolocalization, using GS antibodies. The mesophyll cells of leaves from control plants showed a very slight immunoreactive signal, suggesting that the antibodies used do not have affinity for chloroplastic GS in its native form. In contrast, the leaves of the transgenic plants showed a more widespread signal in all cell types—the palisade parenchyma, the spongy mesophyll, and in the vasculature (Fig. 6). Results also showed that GS is more abundant in the vasculature of the stem, roots and root nodules in both the control plants and in the transformants. However, there are some striking differences in the distribution of the GS between control and transgenic plants. The stems of the transgenic plants showed a generalized increase in the level of GS in comparison to the control stems. Whereas in the stems of control plants, the GS is mostly localized in the xylem (protoxylem) and the cortex, particularly in the collenchyma cells, in the transgenic stems the GS is expressed in the xylem, in all the cortex, and there is a distinctive accumulation in the phloem cells that is not seen in the control stems (Fig. 6). The transgenic roots showed an increase in the accumulation of the GS in the cortex and the epidermis, but the significant difference between transgenic and control roots is the accumulation of GS in the vascular cylinder of the transgenic plant. There is a slight increase in staining in the vascular tissue, the infected cells and the cortical cells in the nodules of the transformant compared to the nodules of the non-transformed plants. Taken together, the immunolocalization data suggest that constitutive over-expression of a GS1 gene construct results in higher accumulation of the protein in cells in which they are normally made and also in cells in which they normally do not accumulate.
Fig. 6.
Immunolocalization of GS in control and Ms glnβ-transformed L. japonicus plants. Cross-sections of leaves, stems, roots and nodules from non-transformed (Control) and MsGSβ-transformed L. japonicus plants (K-25) were stained with an anti-GS antibody in combination with a immunoperoxidase staining system (see Materials and methods). The tissues where the GS immuno-staining is comparatively more abundant in either control or transgenic plants are indicated: ex cortex, en endodermis, ep epidermis, iz infection zone, nc nodule cortex, pc pericycle, ph phloem, pp palisade parenchyma, sp spongy parenchyma, v vessel, vb vascular bundle, vc vascular cylinder, xy xylem. Magnification 400×
The CaMV35S–Ms glnβ transformants exhibit major physiological differences when compared to control plants
A physiological analysis to evaluate the effect of the constitutive over-expression of the Ms glnβ gene in L. japonicus included measurement of biomass accumulation (fresh weight and dry weight), protein content and chlorophyll content. In these experiments, the seeds from control and homozygous lines of K-25 and M-4 Ms glnβ transgenic plants were grown in peat pellets which were kept hydrated with water for the period of the experiment. Seven weeks after germination, tissues were harvested, fresh weight and dry weight in shoots were measured, and protein and chlorophyll concentrations were determined. The values from multiple experiments were used in preparing Table 1. There was a visible effect of the Ms glnβ over-expression on the growth of the transgenic L. japonicus plants. The significant gain in size of the transgenic L. japonicus plants corresponded to a higher accumulation of biomass measured as plant fresh weight (Table 1). Our results also consistently showed higher soluble protein concentration and higher chlorophyll content in the transgenic plants. These results show that the over-expression of a GS1 gene in a constitutive manner promotes growth and more protein and chlorophyll accumulation in L. japonicus plants, in comparison to control non-transformed plants.
Table 1.
Plant growth parameters in control and Ms glnβ-transformed Lotus japonicus plants. Seeds from non-transformed (Control) plants and the progeny of transformants K-25 and M-4 that contain a CaMV35S–Ms glnβ gene construct were planted in soil pellets. After 7 weeks, plant growth parameters were measured in the shoots. GS activity was measured by the transferase reaction (Ferguson and Sims 1971)
| Parameter | Control | K-25 | M-4 |
|---|---|---|---|
| Fresh weight (mg/plant ± SE) | 68.9±2.6 | 152.8 ±2.5 | 137.1 ±4.9 |
| Dry weight (mg/plant ± SE) | 22.9 ±1.7 | 39.6± 1.2 | 37.3 ±1.0 |
| Protein content (mg g W−1 ± SE) | 9.4±0.4 | 15.9±0.3 | 13.8±0.4 |
| Chlorophyll content (mg g FW−1 ± SE) | 4.0±0.3 | 8.5±0.9 | 5.9±0.6 |
| GS activity (µmol g FW−1 ± SE) | 26.9± 1.1 | 73.3± 1.4 | 86.7 ±2.8 |
The CaMV35S–Ms glnβ transformants show an increase in total amino acids and in the relative levels of Asp/Asn
The increase in plant biomass may be due to increased efficiency in nitrogen assimilation. To determine if GS over-expression parallels an increase in nitrogen assimilation, total amino acid analysis was carried out in leaves, stem and roots of control and transformed L. japonicus plants. The roots showed no significant difference in the total amino acid content or in any individual amino acid (data not shown). The total amino acid content in the leaves and stems of the transgenic plants showed a 20–21% average increase over the levels in control plants (Table 2). In comparison to the control tissues, the relative abundance of individual amino acids was the same in the transgenic tissues, except for a 39% and 55% increase in aspartate/asparagine in the leaves and stem, respectively. A significant increase in proline levels was detected in the leaves and stem of the transformants compared to control (27% and 31%, respectively) and a slight increase in the level of arginine (29%) was seen in the stem of the transformants (Table 2).
Table 2.
Total amino acid content in tissues of control and transgenic (K-25) L. japonicus plants that express the Ms glnβ gene (see Materials and methods). Data are the mean values in µmol g FW−1 ± SE (n = 4). The percent values that are significantly higher than the average increase are presented in bold. Asx represents a mixture of Asp/Asn, Glx represents a mixture of Glu/Gln
| Amino acid |
Leaf content (µmol g FW−1) | % Increase |
Stem content (µmol g FW−1) |
% Increase |
||
|---|---|---|---|---|---|---|
| Control | K-25 | Control | K-25 | |||
| Ala | 26.6±1.7 | 32.0±1.5 | 20 | 16.7±1.2 | 20.0 ±0.9 | 19 |
| Arg | 12.5±0.8 | 15.2±0.6 | 21 | 6.7 ±0.4 | 8.6±0.4 | 29 |
| Asx | 30.4±2.1 | 42.3 ±1.9 | 39 | 27.4 ±2.6 | 42.6 ±2.7 | 55 |
| Cys | 5.3 ±0.2 | 6.3±0.1 | 17 | 3.7±0.1 | 4.5±0.1 | 23 |
| Glx | 35.3±2.3 | 43.0 ±2.2 | 22 | 21.6±1.6 | 25.8± 1.1 | 19 |
| Gly | 28.6±1.7 | 34.3 ±1.4 | 20 | 17.9±1.2 | 21.6± 1.1 | 21 |
| His | 6.1 ±0.3 | 6.1±0.3 | 0 | 4.1 ±0.2 | 4.3±0.1 | 7 |
| Ile | 14.5±1.0 | 17.1 ±0.8 | 18 | 9.5 ±0.7 | 11.0±0.6 | 17 |
| Leu | 27.5±1.7 | 32.5±1.2 | 18 | 16.3 ±1.1 | 19.2±0.9 | 18 |
| Lys | 19.6±1.0 | 23.6 ±0.7 | 20 | 14.7± 1.0 | 17.0±0.7 | 16 |
| Met | 6.7 ±0.6 | 7.8 ±0.9 | 17 | 4.1 ±0.4 | 4.9 ±0.4 | 20 |
| Phe | 14.1 ±0.9 | 16.7±0.6 | 18 | 7.9 ±0.5 | 9.5±0.4 | 21 |
| Pro | 17.9±1.1 | 22.8 ±1.7 | 27 | 13.7± 1.2 | 18.0± 1.3 | 31 |
| Ser | 18.1 ±1.1 | 22.1 ±1.4 | 22 | 14.9± 1.1 | 17.6± 1.1 | 18 |
| Thr | 16.3±1.0 | 19.7±0.8 | 20 | 11.0±0.7 | 13.0±0.6 | 18 |
| Tyr | 0.4±0.1 | 0.5±0.2 | 12 | 1.9 ±0.2 | 1.8±0.3 | 0 |
| Val | 20.3±1.1 | 24.2 ±0.8 | 19 | 13.5±0.9 | 15.6±0.7 | 15 |
| Total | 300.3 ±18.4 | 365.9±15.8 | 22 | 205.6 ±14.8 | 255.2±12.5 | 24 |
Discussion
The data presented here show a strong correlation between the constitutive over-expression of GS1 and an increase in overall growth, fresh weight, chlorophyll and protein content in L. japonicus. The results suggest that in L. japonicus the incorporation of ammonium into organic compounds represents a rate-limiting step in biomass production. In the leaves of non-transformed plants GS2 is the major isoform, and it has been shown to function in the assimilation of nitrate and the reassimilation of photo respiratory ammonia. GS1 does not normally contribute to the re-assimilation of photorespiratory ammonia (Wallsgrove et al. 1987), probably because it is made only in the vasculature and is not found in the mesophyll cells where photorespiration occurs (Forde et al. 1989; Edwards et al. 1990). In this study, immunolocalization studies on the leaves of L. japonicus transformants have shown the presence of the GS1 protein in the mesophyll cells, which may function in providing a complementary or alternate route to GS2 for the re-assimilation of photorespiratory ammonia. As nitrogen flux through the photorespiratory pathway is 10-fold higher than primary N-assimilation (Keys et al. 1978), the enhanced re-assimilation of photorespiratory ammonium could lead to enhanced nitrogen-use efficiency. Oliviera et al. (2002) have attributed improved growth of GS1-expressing tobacco plants to increased assimilation of photorespiratory ammonia. However, the L. japonicus transformants in this study do not show an increase in the photorespiratory intermediates above the average increase in the amino acid content (Table 2). It is thus not likely that the improved performance of the L. japonicus transformants is due to improved assimilation of photorespiratory ammonia. GS1 has been shown to play a role in the recycling of nitrogen (Bauer et al. 1997; Perez-Garcia et al. 1998; Brugiere et al. 2000; Masclaux et al. 2000) and such increased GS1 activity could contribute to improved growth by increasing the recycling of proteins in the mature tissues followed by their mobilization into growing parts. The transformants also showed increased GS1 polypeptide in all the cells in the roots and specifically in the root vasculature and in the cortical cells. It is possible that increased GS1 activity in the roots positively influences the uptake of nitrate from the soil. Thus improved growth may be an attribute of the availability of nitrogen.
The L. japonicus transformants also show increased GS protein in the vasculature of the stem as shown by immunolocalization experiments. GS1 has been suggested to play a role in transport (Edwards et al. 1990; Dubois et al. 1996) and thus increased expression of GS1 in the vasculature may imply improved transport as the basis for increased growth of these transformants. Over-accumulation of GS1 in the vascular tissues may have a role in increasing the amount of amino acids available for transport and in increasing the recycling of nitrogen. This is supported by the fact that the stems of the transformants show a significant increase in the level of Asp/Asn (Table 2), amino acids with a functional role in transport (Joy 1988). The stems showed a significant increase in the proline levels and this agrees with the findings of Brugiere et al (1999), who showed a drop in proline levels in transgenic tobacco expressing an antisense GS1 construct in a phloem-specific manner. The importance of increased GS activity in the vasculature on nitrogen transport and plant performance is further emphasized by the fact that vascular-specific downregulation of GS1 genes in alfalfa produced a phenotype of nitrogen stress in the young leaves (Temple et al. 1994).
Comparison of L. corniculatus plants over-expressing a soybean GS1 gene driven by the CaMV 35S promoter (Hirel et al. 1997; Vincent et al. 1997) with L. japonicus plants over-expressing the alfalfa GS1 transgene showed some interesting similarities and dissimilarities. While both transformants showed an increase in biomass, the L. corniculatus transformants showed no increase in GS activity in the roots and nodules. In L. japonicus transformants, however, the roots showed the highest increase in GS activity compared to the control, and the nodules of the L. japonicus transformants also showed a significant increase in GS activity over the control. Moreover, the L. corniculatus transformants showed early senescence and premature flowering while the L. japonicus transformants showed no difference in the timing of senescence or flowering compared to the control plants. Since these are closely related plants, it is difficult for us to invoke some kind of mechanistic difference between the two plants at this stage.
While there are several reports in the literature where an increase in plant growth has been observed as a consequence of GS1 over-expression and the resulting increase in GS activity (Fuentes et al. 2001; Gallardo et al. 1999; Fu et al. 2003; Oliveira et al. 2002; Su et al. 1995), there are cases where the presence of a GS1 transgene is not accompanied by an increase in GS activity (Ortega et al. 2001; Fei et al. 2003). Since the GS1 transgene, in most of these studies, is being driven by the constitutive CaMV35S promoter, it would further appear that the regulation of the transgene is at a step(s) beyond transcription. This is further supported by the fact that the alfalfa transformants with the CaMV35S–GS1 gene construct, while accumulating significantly high levels of the transgene transcript in the leaves, showed no increase in GS1 polypeptide or GS activity (Ortega et al. 2001). Similarly, in pea plants transformed with different GS1 gene constructs, despite increased levels of GS1 polypeptide in the leaves and the nodules, no differences in GS activity in these tissues were detected between the wild-type plants and the transformants (Fei et al. 2003). One thing, however, to keep in mind in considering the different responses of plants to the presence of the GS1 transgene is that the transgene used in the different studies is not always the same. More recently we have shown that the 3′-UTR of the soybean GS1 gene plays a role in the turnover of the GS1 transcript (unpublished data).
While L. japonicus showed fewer constraints in the expression of the GS1 transgene compared to alfalfa and pea, translational/post-translational regulation in the expression of the GS1 transgene is also observed in L. japonicus. Thus, while the GS1 transgene transcript level appeared to be more or less similar in all the organs tested, the increase in the GS1 polypeptide level in the transformants over control varied considerably in the different organs. The leaves and stem showed an increase in the GS1 polypeptide level of between 3- and 5-fold, the roots showed an increase of between 1.5-and 2-fold while the nodules showed a much smaller increase. The GS activity correlates well with the intensity of the GS immunoreactive band on denaturing gels, except in the roots of the transgenic plants where the increase in activity exceeds the increase in the GS polypeptide (Fig. 3). The roots of the transformants, in absolute terms, showed the highest increase in GS activity and also showed the highest intensity in the novel immunoreactive band on native gels (Fig. 5). It is interesting to note that all organs other than the roots showed the presence of an immunoreactive band representing the tetrameric form. The nodules from the transformant showed an increase in the level of the tetramer compared to the control nodules. If the tetrameric form represents an intermediate in the pathway for GS holoenzyme turnover (Temple et al. 1996), it would appear that the GS holoenzyme is most stable in the roots of L. japonicus. A higher level of the tetramer in the nodules of the transformant compared to control would suggest that there is a higher rate of GS turnover in the transgenic nodules. Pea plants transformed with the soybean GS1 gene driven by the nodule-specific promoter while showing increased levels of GS1 protein in the nodules did not show any enhancement in GS activity compared to control plants (Fei et al. 2003). It is thus possible that the nodules have some mechanism to regulate the total level of GS activity. The stability of the GS enzyme in the roots of both control and transformed L. japonicus plants would suggest that, in L. japonicus, the main site of nitrogen assimilation may be the roots since the stability of the GS holoenzyme has been shown to be controlled by the availability of the enzyme substrates (Ortega et al. 1999; Suganuma et al. 1999).
The data presented in this paper clearly demonstrate the feasibility of increasing growth and improving performance in L. japonicus by the constitutive over-expression of GS1. Experiments are in progress to study the effect of tissue/organ-specific over-expression of GS1 in L. japonicus. These experiments should help determine the exact basis for improved performance of these GS1-over-expressing L. japonicus plants.
Acknowledgments
This work was supported by the National Institutes of Health (Grant Nos. GMO-8136-26, GMO-61222 and GMO-7667-25) and by the Agricultural Experiment Station at New Mexico State University.
Abbreviations
- GS
Glutamine synthetase
- UTR
Untranslated region
Contributor Information
Jose Luis Ortega, Department of Agronomy and Horticulture, New Mexico State University, Las Cruces, NM 88003, USA.
Stephen J. Temple, Department of Agronomy and Horticulture, New Mexico State University, Las Cruces, NM 88003, USA
Suman Bagga, Department of Agronomy and Horticulture, New Mexico State University, Las Cruces, NM 88003, USA.
Soumitra Ghoshroy, Department of Agronomy and Horticulture, New Mexico State University, Las Cruces, NM 88003, USA; Electron Microscopy Laboratory, New Mexico State University, Las Cruces, NM 88003, USA.
Champa Sengupta-Gopalan, Department of Agronomy and Horticulture, New Mexico State University, Las Cruces, NM 88003, USA.
References
- Bauer D, Biehler K, Fock H, Carrayol E, Hirel B, Migge A, Becker TW. A role for cytosolic glutamine synthetase in the remobilization of leaf nitrogen during water stress in tomato. Physiol Plant. 1997;99:241–248. [Google Scholar]
- Bennett MJ, Lightfoot DA, Cullimore JV. cDNA sequence and differential expression of the gene encoding the glutamine synthetase g polypeptide of Phaseolus vulgaris L. Plant Mol Biol. 1989;12:553–565. doi: 10.1007/BF00036969. [DOI] [PubMed] [Google Scholar]
- Blackwell RD, Murray AJS, Lea PJ. Inhibition of photosynthesis in barley with decreased levels of chloroplastic glutamine synthetase activity. J Exp Bot. 1987;38:1799–1809. [Google Scholar]
- Brugiere N, Dubois F, Limami AM, Lelandais M, Roux Y, Sangwan RS, Hirel B. Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell. 1999;11:1995–2011. doi: 10.1105/tpc.11.10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugiere N, Dubois F, Masclaux C, Sangwan RS, Hirel B. Immunolocalization of glutamine synthetase in senescing tobacco (Nicotiana tabacum L.) leaves suggests that ammonia assimilation is progressively shifted to the mesophyll cytosol. Planta. 2000;211:519–527. doi: 10.1007/s004250000309. [DOI] [PubMed] [Google Scholar]
- Carvalho H, Pereira S, Sunkel C, Salema R. Detection of cytosolic glutamine synthetase in leaves of Nicotiana tabacum L by immunocytochemical methods. Plant Physiol. 1992;100:1591–1594. doi: 10.1104/pp.100.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho HG, Lopes-Cardoso IA, Lima LM, Melo PM, Cullimore JV. Nodule-specific modulation of glutamine synthetase in transgenic Medicago truncatula leads to inverse alterations in asparagine synthetase expression. Plant Physiol. 2003;133:243–252. doi: 10.1104/pp.102.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma S, Tischer E, Goodman HM. Plant glutamine synthase complements a gln A mutant in Escherichia coli . Science. 1986;232:1242–1244. doi: 10.1126/science.2871626. [DOI] [PubMed] [Google Scholar]
- Dubois F, Brugiere N, Sangwan RS, Hirel B. Localization of tobacco cytosolic glutamine synthetase enzymes and the corresponding transcripts shows organ- and cell-specific patterns of protein synthesis and gene expression. Plant Mol Biol. 1996;31:807–817. doi: 10.1007/BF00019468. [DOI] [PubMed] [Google Scholar]
- Eckes P, Schmitt P, Daub W, Wengenmayer F. Overproduction of alfalfa glutamine synthetase in transgenic tobacco plants. Mol Gen Genet. 1989;217:263–268. doi: 10.1007/BF02464891. [DOI] [PubMed] [Google Scholar]
- Edwards JW, Walker EL, Coruzzi GM. Cell-specific expression in transgenic plants reveals nonoverlapping roles for chloroplast and cytosolic glutamine synthetase. Proc Natl Acad Sci USA. 1990;8:3459–3463. doi: 10.1073/pnas.87.9.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei H, Chaillou S, Hirel B, Mahon JD, Vessey KJ. Over-expression of a soybean cytosolic glutamine synthetase gene linked to organ-specific promoters in pea plants grown in different concentrations of nitrate. Planta. 2003;216:467–474. doi: 10.1007/s00425-002-0873-7. [DOI] [PubMed] [Google Scholar]
- Ferguson AR, Sims AP. Inactivation in vivo of glutamine synthetase and NAD-specific glutamate dehydrogenase. Its role in the regulation of glutamine synthetase in yeast. J Gen Microbiol. 1971;69:423–427. doi: 10.1099/00221287-69-3-423. [DOI] [PubMed] [Google Scholar]
- Forde BG, Day HM, Turton JF, Shen WJ, Cullimore JV, Oliver JE. Two glutamine synthetase genes from Phaseolus vulgaris L. display contrasting developmental and spatial patterns of expression in transgenic Lotus corniculatus plants. Plant Cell. 1989;1:391–401. doi: 10.1105/tpc.1.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Sampalo R, Gallardo F, Canovas FM, Kirby EG. Assembly of a cytosolic pine glutamine synthetase holoenzyme in leaves of transgenic poplar leads to enhanced vegetative growth in young plants. Plant Cell Environ. 2003;26:411–418. [Google Scholar]
- Fuentes SI, Allen DJ, Ortiz-Lopez A, Hernandez G. Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J Exp Bot. 2001;52:1071–1081. doi: 10.1093/jexbot/52.358.1071. [DOI] [PubMed] [Google Scholar]
- Gallardo F, Fu J, Canton FR, Garcia-Gutierrez A, Canovas FM, Kirby EG. Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta. 1999;210:19–26. doi: 10.1007/s004250050649. [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992;2:487–496. [Google Scholar]
- Harrison J, Crescenzo MP, Hirel B. Does lowering glutamine synthetase activity in nodules modify nitrogen metabolism and growth of Lotus japonicus L. Plant Physiol. 2003;133:253–262. doi: 10.1104/pp.102.016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemon P, Robbins MP, Cullimore JV. Targeting of glutamine synthetase to the mitochondria of transgenic tobacco. Plant Mol Biol. 1990;15:895–904. doi: 10.1007/BF00039428. [DOI] [PubMed] [Google Scholar]
- Hirel B, Marsolier MC, Hoarau A, Hoarau J, Brangeon J, Schafer R, Verma DPS. Forcing expression of a soybean root glutamine synthetase gene in tobacco leaves induces a native gene encoding cytosolic enzyme. Plant Mol Biol. 1992;20:207–218. doi: 10.1007/BF00014489. [DOI] [PubMed] [Google Scholar]
- Hirel B, Phillipson B, Murchie E, Suzuki A, Kunz C, Ferrario S, Limami A, Chaillou S, Deleens E, Brugiere N, Chaumont-Bonnet M, Foyer C, Morot-Gaudry J-F. Manipulating the pathway of ammonia assimilation in transgenic non-legumes and legumes. Z Pflanzeneraehr Bodenkd. 1997;160:283–290. [Google Scholar]
- Joy KW. Ammonia, glutamine and asparagine: a carbon nitrogen interface. Can J Bot. 1988;66:2103–2109. [Google Scholar]
- Kamachi K, Yamaya T, Hayakawa T, Mae T, Ojima K. Vascular bundle-specific localization of cytosolic glutamine synthetase in rice leaves. Plant Physiol. 1992;99:1481–1486. doi: 10.1104/pp.99.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys AJ, Bird IF, Cornelius MJ, Lea PJ, Wallsgrove RM, Miflin BJ. Photorespiratory nitrogen cycle. Nature. 1978;271:741–743. [Google Scholar]
- Kozaki A, Takeba G. Photorespiration protects C3 plants from photooxidation. Nature. 1996;384:557–560. [Google Scholar]
- Lancien M, Ferrario-Méry S, Roux Y, Bismuth E, Masclaux C, Hirel B, Gadal P, Hodges M. Simultaneous expression of NAD-dependent isocitrate dehydrogenase and other Krebs cycle genes after nitrate resupply to short-term nitrogen-starved tobacco. Plant Physiol. 1999;120:717–725. doi: 10.1104/pp.120.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancien M, Gadal P, Hodges M. Enzyme redundancy and the importance of 2-oxoglutarate in higher plant ammonium assimilation. Plant Physiol. 2000;123:817–824. doi: 10.1104/pp.123.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Ireland RJ. Nitrogen metabolism in higher plants. In: Singh BK, editor. Plant amino acids, biochemistry and biotechnology. New York: Dekker; 1999. pp. 1–47. [Google Scholar]
- Limami A, Phillipson B, Ameziane R, Perrollet N, Jiang Q, Roy R, Deleens E, Chaumont-Bonnet M, Gresshoff PM, Hirel B. Does root glutamine synthetase control plant biomass production in Lotus japonicus L. Planta. 1999;209:495–502. doi: 10.1007/s004250050753. [DOI] [PubMed] [Google Scholar]
- Masclaux C, Valadier MH, Brugiere N, Morot-Gaudry JF, Hirel B. Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta. 2000;211:510–518. doi: 10.1007/s004250000310. [DOI] [PubMed] [Google Scholar]
- Morey KJ, Ortega JL, Sengupta-Gopalan C. Cytosolic glutamine synthetase in soybean is encoded by a multigene family, and the members are regulated in an organ-specific and developmental manner. Plant Physiol. 2002;128:182–193. [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, Coruzzi GM. Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis . Plant Physiol. 1999;121:301–310. doi: 10.1104/pp.121.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM. Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 2002;129:1170–1180. doi: 10.1104/pp.020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JL, Roche D, Sengupta-Gopalan C. Oxidative turnover of soybean root glutamine synthetase. In vitro and in vivo studies. Plant Physiol. 1999;119:1483–1495. doi: 10.1104/pp.119.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JL, Temple SJ, Sengupta-Gopalan C. Constitutive overexpression of cytosolic glutamine synthetase (GS1) gene in transgenic alfalfa demonstrates that GS1 may be regulated at the level of RNA stability and protein turnover. Plant Physiol. 2001;126:109–121. doi: 10.1104/pp.126.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S, Carvalho H, Sukekl C, Salema R. Immunocytolo-calization of glutamine synthetase in mesophyll cells and phloem of leaves of Solarium tuberosum L. Protoplasma. 1992;167:66–73. [Google Scholar]
- Pérez-García A, Pereira S, Pissarra J, Garcia Gutierrez A, Cazorla FM, Salema R, de Vicente A, Canovas FM. Cytosolic localization in tomato mesophyll cells of a novel glutamine synthetase induced in response to bacterial infection or phos-phinotricin treatment. Planta. 1998;206:426–434. [Google Scholar]
- Peterman TM, Goodman HM. The glutamine synthetase gene family of Arabidopsis thaliana: light regulation and differential expression in leaves, roots and seeds. Mol Gen Genet. 1991;330:145–154. doi: 10.1007/BF00290662. [DOI] [PubMed] [Google Scholar]
- Sakurai N, Hayakawa T, Nakamura T, Yamaya T. Changes in the cellular localization of cytosolic glutamine synthetase protein in vascular bundles of rice leaves at various stages of development. Planta. 1996;200:306–311. [Google Scholar]
- Su J, Zhang XQ, Yan QS, Chen ZL, You CB. Construction of plant expression vectors carrying glnA gene encoding glutamine synthetase and regeneration of transgenic rice plants. Sci China Ser B. 1995;38:963–970. [Google Scholar]
- Suganuma N, Watanabe M, Yamada T, Izuhara T, Yamamoto K, Nishimura M, Toriyama K. Involvement of ammonia in maintenance of cytosolic glutamine synthetase activity in Pisum sativum nodules. Plant Cell Physiol. 1999;40:1053–1060. [Google Scholar]
- Sukanya R, Li M, Snustad DP. Root-specific and shoot-specific responses of individual glutamine-synthetase genes of maize to nitrate and ammonium. Plant Mol Biol. 1994;26:1935–1946. doi: 10.1007/BF00019504. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Knight TJ, Unkefer PJ, Sengupta-Gopalan C. Modulation of glutamine synthetase gene expression in tobacco by the introduction of an alfalfa glutamine synthetase gene in sense and antisense orientation: molecular and biochemical analysis. Mol Gen Genet. 1993;236:315–325. doi: 10.1007/BF00277128. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Bagga S, Sengupta-Gopalan C. Can glutamine synthetase activity levels be modulated in transgenic plants by the use of recombinant DNA technology? Biochem Soc Trans. 1994;22:915–920. doi: 10.1042/bst0220915. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Heard J, Ganter G, Dunn K, Sengupta-Gopalan C. Characterization of a nodule-enhanced glutamine synthetase from alfalfa: nucleotide sequence, in situ localization and transcript analysis. Mol Plant Microbe Interact. 1995;8:218–227. doi: 10.1094/mpmi-8-0218. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Kunjibettu S, Roche D, Sengupta-Gopalan C. Total glutamine synthetase activity during soybean nodule development is controlled at the level of transcription and holoprotein turnover. Plant Physiol. 1996;112:1723–1733. doi: 10.1104/pp.112.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Bagga S, Sengupta-Gopalan C. Down-regulation of specific members of the glutamine synthetase gene family in alfalfa by antisense RNA technology. Plant Mol Biol. 1998;37:535–547. doi: 10.1023/a:1006099512706. [DOI] [PubMed] [Google Scholar]
- Vincent R, Fraiser V, Chaillou S, Limami MA, Deleens E, Phillipson B, Douat C, Boutin J-P, Hirel B. Overexpression of a soybean gene encoding cytosolic glutamine synthetase in shoots of transgenic Lotus corniculatus L. plants triggers changes in ammonium assimilation and plant development. Planta. 1997;201:424–433. doi: 10.1007/s004250050085. [DOI] [PubMed] [Google Scholar]
- Wallsgrove RM, Turner JC, Hall NP, Kendall AC, Bright SJW. Barley mutants lacking chloroplast glutamine synthetase. Biochemical and genetic analysis. Plant Physiol. 1987;83:155–158. doi: 10.1104/pp.83.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]