Abstract
A set of 120 simple sequence repeats (SSRs) was developed from the newly assembled pear sequence and evaluated for polymorphisms in seven genotypes of pear from different genetic backgrounds. Of these, 67 (55.8 %) primer pairs produced polymorphic amplifications. Together, the 67 SSRs detected 277 alleles with an average of 4.13 per locus. Sequencing of the amplification products from randomly picked loci NAUPy31a and NAUpy53a verified the presence of the SSR loci. When the 67 primer pairs were tested on 96 individual members of eight species in the Rosaceae family, 61.2 % (41/67) of the tested SSRs successfully amplified a PCR product in at least one of the Rosaceae genera. The transferability from pear to different species varied from 58.2 % (apple) to 11.9 % (cherry). The ratio of transferability also reflected the closer relationships within Maloideae over Prunoideae. Two pear SSR markers, NAUpy43c and NAUpy55k, could distinguish the 20 different apple genotypes thoroughly, and UPGMA cluster analysis grouped them into three groups at the similarity level of 0.56. The high level of polymorphism and good transferability of pear SSRs to Rosaceae species indicate their promise for application to future molecular screening, map construction, and comparative genomic studies among pears and other Rosaceae species.
Electronic supplementary material
The online version of this article (doi:10.1007/s11105-013-0586-z) contains supplementary material, which is available to authorized users.
Keywords: Simple sequence repeat (SSR), Pear, Rosaceae, Transferability
Introduction
Pear (Pyrus spp.), one of the oldest fruit crops in the world, belongs to the genus Pyrus, subfamily Maloideae (Pomoideae), in the family Rosaceae. Economically, pear is the third most important temperate fruit species after grape and apple, and the recent genome sequencing of the diploid P. bretschneideri Rehd. cv. Dangshansuli (Wu et al. 2013) has allowed ready access to the DNA sequence of pear. Simple sequence repeats (SSRs) or microsatellite markers are widely used due to their multi-allelic nature, reproducibility, high polymorphism, and codominant inheritance (Campoy et al. 2011; Baraket et al. 2011; Swapna et al. 2011). Markers derived from the pear genome would be significant and useful in linkage map construction, genetic polymorphism evaluation, cultivar identification, traditional crossbreeding programs, and studies of genetic variability and the diversification process (Ferreira dos Santos et al. 2011).
Rosaceae is a large plant family containing more than 3,000 species, many of which are economically important fruit trees such as apple, apricot, cherry, peach, pear and plum as well as soft fruit crops like strawberry. In addition to pear, the whole genome sequences of both apple (Velasco et al. 2010) and strawberry (Shulaev et al. 2011) have been completed, making massive genomic data of pear, apple, and strawberry in terms of molecular marker maps, expressed sequence tags (EST), and gDNA sequences readily available. However, there is rather little genomic information available for other valuable fruit tree members of the big Rosaceae family. Due to their codominant and usually single-locus nature, SSR loci can be identified, and their alleles can be recognized in different genotypes of the same species and often in those of other close relatives. This means that a specific set of SSRs can be used in different sets of genotypes or mapping populations, making them particularly useful for variability analysis, fingerprinting, molecular marker development, marker-assisted selection (MAS), map construction, and comparative studies (Mnejja et al. 2010). Therefore, it is highly valuable to investigate the transferability of SSR markers from pear, where they are easily developed and evaluated, to other Rosaceae species. Transferability to relatives contributes to the wide applicability of SSRs developed based on the pear genome. Recently, there have been several reports on the transferability of SSR markers in or across genera among Rosaceae fruit crops (Decroocq et al. 2003; Gasic et al. 2009; Gisbert et al. 2009; Sargent et al. 2009, 2007; Yamamoto et al. 2004; Yao et al. 2010; Wünsch 2009; Mnejja et al. 2010; He et al. 2011; Bouvier et al. 2012).
Comparative mapping within many plant families has been well studied, for example, in Brassicaceae, Leguminosae, Poaceae, or Solanaceae (Devos and Gale 2000; Doganlar et al. 2002; Kalo et al. 2004; Lukens et al. 2003) and allows the identification of a marker framework for map-based prediction of the location of candidate genes linked with agriculturally important traits within different species of the same family. Recently, efforts have been undertaken for genome comparisons within Rosaceae (Shulaev et al. 2008). Molecular markers have been widely applied to various aspects of research in apple and pear (Celton et al. 2009; Yamamoto et al. 2007), and SSR markers with good transferability can be applied to comparative mapping and genomic synteny between these two important fruits. Bouvier et al. (2012) discovered that a new pear scab-resistant gene, Rvp1, is located close to microsatellite marker CH02b10 from the European pear cultivar Navara maps in a genomic region syntenic to an apple scab-resistant gene cluster on linkage group 2. This genomic region is known to carry a cluster of scab-resistant genes in apple, indicating the first functional synteny for scab resistance between apple and pear. Molecular marker linkage maps of apple and Prunus have been shown to have a high level of macro-synteny (Dirlewanger et al. 2004).
The study of transferability of SSRs and EST-SSRs among Rosaceae is significant. He et al. (2011) evaluated 71 apple SSR markers distributed across 17 linkage groups and identified 39 SSRs transferable to loquats. Mnejja et al. (2010) studied a total of 145 microsatellite primer pairs from Prunus DNA sequences for transferability in a set of eight cultivars from nine rosaceous species (almond, peach, apricot, Japanese plum, European plum, cherry, apple, pear, and strawberry). Gasic et al. (2009) investigated the level of transferability of 68 apple EST-SSRs in 50 individual members of the Rosaceae family, representing three genera and 14 species. Wünsch (2009) studied 13 Prunus SSRs known to be transferable to certain Prunus species, tested them in 10 Prunus, and found that three were transferable to all and eight to all but one species. Apple EST-SSR primer pairs (94) were tested on four accessions of Pyrus to evaluate transferability, and 40 of 72 functional SSRs produced polymorphic amplicons. From these, eight SSR randomly selected loci were used to analyze genetic diversity and relationships among a collection of Pyrus and displayed reliable amplification and considerable polymorphism in both Malus and Pyrus (Yao et al. 2010). Despite this abundance of research, most previous studies about transferability are related to apple genomic SSRs or EST-SSRs, with little research on pear SSR transferability.
In this study, we present 150 pear SSRs developed from publicly available Pyrus genome sequences (Wu et al. 2013). In order to ensure the quality of the markers, all SSRs have been evaluated for polymorphisms in seven pear genotypes from different genetic backgrounds, and their transferability to 96 individual members of eight species of the Rosaceae family was studied.
Materials and Methods
Plant Material
The seven pear genotypes used in the primary evaluation of pear primers have broad genetic variation. They are Doyenne du Comice from France (P. communis L.); Housui from Japan (P. pyrifolia Nakai); Huobali from Yunnan province of China (P. pyrifolia Nakai), which is a semi-cultivated species; Kuerlexiangli from Xinjiang province of China (P. sinkiangensis Yü); Yali from Hebei province of China (P. bretschneideri Rehd.); Jingbaili from Beijing, China (P. ussuriensis Maxim.); and Bean Pear (P. calleryana) often used as rootstock. A total of 96 Rosaceae genotypes (Table 1) of eight species from seven genera and three subfamilies were used for transferability exploration; these cultivars were chosen largely based on their economic value. All the pear leaves were collected from trees located at the Pukou District pomology farm of Nanjing Agriculture University; apple, plum, peach, cherry, and apricot samples were collected from trees located at the production demonstration bases of Shangdong Institute of Pomology; strawberry leaves were picked in the greenhouse of Shangdong Agriculture University; loquat leaf tissues were collected in the loquat germplasm nursery of Suzhou Polytechnic Institute of Agriculture; and Japanese apricot leaves were collected in the Garden of Fujiabian Agricultural Technology Park in Nanjing of China.
Table 1.
Plant material used for cross-species transferability in Rosaceae
| Sample no. | Individuals tested | Origin | Sample no. | Individuals tested | Origin | Sample no. | Individuals tested | Origin |
|---|---|---|---|---|---|---|---|---|
| Prunoideae Japanese apricot (Prunus mume) | Prunoideae Cherry (Cerasus pseudocerasus) | Prunoideae Apricot (Prunus armeniaca) | ||||||
| 1 | Bungo | Japan | 33 | Brooks | USA | 64 | Zhenzhuyouxing | China |
| 2 | Xiaoyezhugan | China | 34 | Nanyo | Japan | 65 | Hongguang | China |
| 3 | Gessekai | Japan | 35 | Van | Canada | 66 | Honghebao | China |
| 4 | Shirokaga | Japan | 36 | Hong Deng | China | 67 | Katy | USA |
| 5 | Xiaoqing | China | 37 | Ruby | USA | 68 | Mono | USA |
| 6 | Ruantiaohongmei | China | Maloideae Apple (Malus x domestica) | 69 | Jinkaite | China | ||
| 7 | Nankou | Japan | 38 | Sekaiichi | Japan | 70 | Sweet Production | Unkown |
| 8 | Xiyeqing | China | 39 | Huaguan | China | 71 | Red Mingtand | Unkown |
| 9 | Oushuku | Japan | 40 | Zaohong | China | 72 | Sungold | USA |
| 10 | Nanhong | China | 41 | Delicious | USA | 73 | Yubadan | China |
| Prunoideae Plum (Prunus salicina) | 42 | Braeburn | New Zealand | Prunoideae Peach (Prunus persica) | ||||
| 11 | Eldorda | Australia | 43 | Fuji | Japan | 74 | Beijing 9 | China |
| 12 | Grandora | USA | 44 | Golden Delicious | USA | 75 | Datuanmilu | China |
| 13 | Black Amber | USA | 45 | Mikilifi | Japan | 76 | Kurakatawase | Japan |
| 14 | Bluebyrd | USA | 46 | Longfeng | China | 77 | Huayu | China |
| 15 | Methley | South Africa | 47 | Starking | USA | 78 | Hanmilu | China |
| 16 | Angeleno | USA | 48 | Dailv | China | 79 | Hakuto | Japan |
| 17 | Autumn Giant | Japan | 49 | Gala | New Zealand | 80 | Yumyong | Korea |
| 18 | Catalina | USA | 50 | Qinyang | China | 81 | Fudaotaowang | China |
| 19 | Weikeseng | China | 51 | HoKuto | Japan | 82 | Zhaoyanghong | China |
| 20 | Red Beauty | USA | 52 | Jonagold | USA | 83 | Shuguang | China |
| 21 | June Rosa | Uknown | 53 | Orin | Japan | 84 | Zaofengwang | China |
| 22 | Early Beauty | USA | 54 | Lujia 1 | China | 85 | Hongjuhua | China |
| 23 | Bilvhongxin | China | 55 | Alps Otome | Japan | Maloideae Loquat (Eriobotrya japonica) | ||
| 24 | Early Beauty | USA | 56 | Hanfu | China | 86 | Zhaozhong | China |
| Prunoideae Cherry (Cerasus pseudocerasus) | 57 | Xiushui | China | 87 | Qingzhong | China | ||
| 25 | Kosmytchekaja | Ukraine | Rosoideae Strawberry (Fragaria × ananassa) | 88 | Guanyu | China | ||
| 26 | Summit | Canada | 58 | Meiho | Japan | 89 | Gaoliangjiang | China |
| 27 | Druichba | Ukraine | 59 | Toyonoka | Japan | 90 | Jidanbai | China |
| 28 | Early Ruby | Ukraine | 60 | Nyoho | Japan | 91 | Tianzhong | China |
| 29 | Tieton | USA | 61 | Benihoppe | Japan | 92 | Qianxipeiyu | China |
| 30 | Rainier | USA | 62 | Gongben | Japan | 93 | Subai 1 | China |
| 31 | Earty Rilbmond | China | 63 | Sweet Charlie | USA | 94 | Shiromogi | Japan |
| 32 | Lapins | Canada | 95 | Dahongpao | China | |||
| 96 | Dazhong | China | ||||||
DNA Extraction and Quality Control
Total genomic DNA of all the plants was extracted from young leaves based on the improved CTAB method (Pan et al. 2006). Subsequently, 5-μl DNA solution was loaded on 1.0 % agarose gel to check the quality.
SSR Markers of Pear Genome
A total of 120 primers (Supplemental Table 1) were developed from the pear genome sequence. All SSR markers were obtained by scanning the published pear whole genome sequencing scaffolds (Wu et al. 2013; http://peargenome.njau.edu.cn) and were assessed for their polymorphism levels in the seven pear genotypes mentioned. The SSR polymorphisms in pears were used to investigate transferability to 96 Rosaceae cultivars.
PCR and Electrophoresis
All polymerase chain reactions (PCRs) were performed in a total volume of 10 μl containing: 1 μl of 30 ng/μl genomic DNA template, 1 μl of 10 × PCR buffer (without MgCl2), 1 μl of 2.5 mM dNTP mixture, 0.6 μl of 25 mM MgCl2, 0.1 μl each of forward and reverse primer (10 pmol/μl), and 0.1 μl of 5 U/μl Taq polymerase (Takara Biotechnology Company, Dalian). The reactions were performed with the following conditions: 94 °C for 3 min, then 39 cycles of 94 °C for 40 s, 55 °C for 50 s, and 72 °C for 1 min, and a final step at 72 °C for 10 min. PCR products (10 μl) were mixed with 2.5 μl formamide loading buffer (98 % formamide, 10 mM EDTA, 0.25 % bromophenol blue, 0.25 % xylenecyanol, pH 8.0), and 1.5 μl of each mixture and a molecular size marker of 100-bp ladder, Trans DNA 2 K Marker, or pBR322 DNA-MspI Digest (Beijing BLKW Biotechnology Co., Ltd) were loaded onto an 8 % non-denaturing polyacrylamide gel in 1× TBE buffer (Tris-borate, EDTA, pH 8.0), running under a voltage of 200 V and visualized using the silver-staining protocol described by Bassam et al. (1991).
Sequencing of Three SSR Loci in Pyrus Accessions
In order to verify the presence of SSR and prove the allelic variation of the SSR loci, amplification products from two randomly picked loci (NAUPy31a and NAUpy53a) were isolated with a DNA Gel Extraction kit AxyPrep™ (Axygen Inc.), and the fragments were cloned into the pMD18-T vector. The fragments were sequenced by Invitrogen, Shanghai, China.
Allele Data and Statistical Analysis
Allele sizes were estimated by comparison to a 100-bp ladder marker, Trans DNA 2 K Marker, and a pBR322 DNA-MspI Digest (Beijing BLKW Biotechnology Co., Ltd). SSRs were scored as dominant markers: presence of a band was recorded as “1” and absence as “0.” All accessions were analyzed as a single population. The observed number of alleles (Na), effective number of alleles (Ne), number of alleles, Nei’s gene diversity (h), and Shannon’s information index (I) were calculated for each locus by the program POPGENE, version 1.31 (Yang and Yeh 1993). Only reproducible bands were used to calculate the Dice’s similarity coefficients (Nei and Li 1979). The chi-square test for Hardy–Weinberg equilibrium (Phw) of primers was calculated using the POPGENE program (version 1.32), with 0.01 significance and 10,000 times simulation. An unweighted pair group method using arithmetic average (UPGMA) cluster analysis was performed based on the similarity matrix for 20 apple individuals using the NTSYS-pc program (Version 2.2j) (Rohlf 1998).
Results and Discussion
Polymorphism of SSR Markers
In total, 67 of 120 (55.8 %) SSRs produced amplifications in the seven pear genotypes (Doyenne du Comice, Housui, Huobali, Kuerlexiangli, Bean Pear, Yali, and Jingbaili). In general, the SSR markers produced high-quality banding patterns (Fig. 1), most of them giving product sizes ranging from 90 to 280 bp (Table 2). The 67 SSRs detected 294 alleles with an average of 4.1 per locus; the observed number of alleles (Na) ranged from 1.57 to 2.00, with an average value of 1.89; the effective number of alleles (Ne) ranged from 1.19 to 1.88, with a mean level of 1.51; Nei’s gene diversity (h) ranged from 0.12 to 0.46, with a mean level of 0.31; and Shannon’s information index (I) varied from 0.17 to 0.65, with an average value of 0.46. High-quality NTSYS analysis of the genetic distance among the seven genotypes according to the 67 SSRs gave distance values ranging from 0.2426 (Huobali and Kuerlexiangli) to 1.0119 (Jingbaili and Bean Pear), with an average of 0.69, indicating a high level of genetic diversity among the seven pear individuals, suggesting a good applicability of these markers to germplasm diversity research. Consequently, we picked these 67 pear SSRs for further transferability exploration across Rosaceae. Excepting their high quality, these 67 SSRs were chosen because almost all primers produced clear bands in all seven pear genotypes representing different pear classes, suggesting that primer sites in the genome are well conserved. This indicates the larger possibility of successful transferability to other Rosaceae species.
Fig. 1.
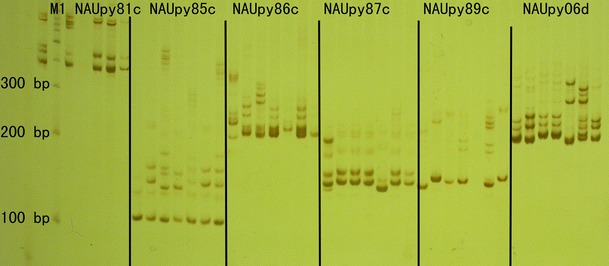
Amplification of six SSRs, namely, NAUpy81c, NAUpy85c, NAUpy86c, NAUpy87c, NAUpy89c, and NAUpy06d, in seven pear genotypes. Lanes: M 1 100-bp ladder DNA marker, 1 Doyenne du Comice, 2 Housui, 3 Huobali, 4 Kuerlexiangli, 5 Bean Pear, 6 Yali, 7 Jingbaili
Table 2.
Parameter values of 67 SSR polymorphisms in seven pear genotypes
| SSR locus | Size ranged in pears (bp) | Number of alleles | na | ne | h | I |
|---|---|---|---|---|---|---|
| NAUpy35c | 130–160 | 6 | 1.86 | 1.37 | 0.25 | 0.40 |
| NAUpy91b | 130–160 | 4 | 1.86 | 1.67 | 0.36 | 0.53 |
| NAUpy74v | 110–115 | 5 | 1.29 | 1.20 | 0.12 | 0.17 |
| NAUpy23f | 147–230 | 4 | 2.00 | 1.44 | 0.29 | 0.46 |
| NAUpy74f | 110–125 | 3 | 2.00 | 1.44 | 0.28 | 0.44 |
| NAUpy39h | 100–110 | 5 | 2.00 | 1.19 | 0.16 | 0.30 |
| NAUpy92a | 125–190 | 4 | 2.00 | 1.70 | 0.40 | 0.59 |
| NAUpy46k | 125–190 | 5 | 2.00 | 1.19 | 0.16 | 0.30 |
| NAUpy55k | 150–166 | 3 | 2.00 | 1.51 | 0.32 | 0.50 |
| NAUpy92e | 200–210 | 5 | 2.00 | 1.31 | 0.23 | 0.38 |
| NAUpy22c | 210–280 | 2 | 2.00 | 1.80 | 0.43 | 0.62 |
| NAUpy24c | 180–230 | 5 | 2.00 | 1.83 | 0.45 | 0.64 |
| NAUpy53a | 180–200 | 3 | 1.86 | 1.45 | 0.28 | 0.43 |
| NAUpy86n | 220–260 | 4 | 2.00 | 1.44 | 0.29 | 0.46 |
| NAUpy32c | 150–180 | 4 | 2.00 | 1.63 | 0.37 | 0.56 |
| NAUpy38c | 120–160 | 5 | 2.00 | 1.41 | 0.27 | 0.44 |
| NAUpy27c | 100–200 | 4 | 1.86 | 1.37 | 0.25 | 0.40 |
| NAUpy31a | 180–240 | 4 | 1.71 | 1.34 | 0.22 | 0.34 |
| NAUpy38e | 90–120 | 2 | 1.86 | 1.38 | 0.25 | 0.39 |
| NAUpy86c | 200–210 | 2 | 2.00 | 1.66 | 0.37 | 0.55 |
| NAUpy87c | 150–160 | 5 | 2.00 | 1.50 | 0.33 | 0.51 |
| NAUpy85d | 140–200 | 4 | 1.71 | 1.51 | 0.28 | 0.41 |
| NAUpy68n | 110–160 | 3 | 2.00 | 1.81 | 0.44 | 0.63 |
| NAUpy02z | 160–190 | 3 | 1.57 | 1.40 | 0.24 | 0.35 |
| NAUpy80k | 120–200 | 4 | 2.00 | 1.63 | 0.38 | 0.57 |
| NAUpy87d | 190–210 | 5 | 1.71 | 1.37 | 0.23 | 0.35 |
| NAUpy43c | 140–160 | 5 | 1.43 | 1.41 | 0.21 | 0.29 |
| NAUpy60e | 100–110 | 6 | 1.57 | 1.40 | 0.24 | 0.35 |
| NAUpy75t | 95–180 | 4 | 2.00 | 1.73 | 0.41 | 0.60 |
| NAUpy91t | 110–150 | 4 | 2.00 | 1.69 | 0.40 | 0.59 |
| NAUpy88x | 120–200 | 2 | 1.86 | 1.62 | 0.35 | 0.50 |
| NAUpy67t | 90–120 | 3 | 2.00 | 1.52 | 0.32 | 0.50 |
| NAUpy98x | 100–150 | 3 | 2.00 | 1.88 | 0.46 | 0.65 |
| NAUpy36t | 100–150 | 3 | 1.71 | 1.46 | 0.27 | 0.40 |
| NAUpy30u | 180–260 | 4 | 1.86 | 1.59 | 0.34 | 0.50 |
| NAUpy62u | 180–220 | 4 | 2.00 | 1.49 | 0.33 | 0.51 |
| NAUpy43w | 90–110 | 4 | 1.71 | 1.61 | 0.32 | 0.46 |
| NAUpy40c | 220–250 | 4 | 2.00 | 1.66 | 0.37 | 0.55 |
| NAUpy85c | 110–170 | 4 | 2.00 | 1.70 | 0.39 | 0.57 |
| NAUpy89c | 150–160 | 4 | 1.86 | 1.37 | 0.26 | 0.41 |
| NAUpy94x | 160–180 | 3 | 1.86 | 1.53 | 0.32 | 0.47 |
| NAUpy23d | 160–180 | 2 | 1.86 | 1.53 | 0.32 | 0.47 |
| NAUpy07u | 180–210 | 4 | 2.00 | 1.59 | 0.36 | 0.54 |
| NAUpy35v | 130–150 | 4 | 2.00 | 1.48 | 0.31 | 0.48 |
| NAUpy92x | 110–120 | 4 | 1.71 | 1.39 | 0.24 | 0.37 |
| NAUpy81m | 140–150 | 4 | 1.57 | 1.40 | 0.24 | 0.35 |
| NAUpy71v | 120–135 | 4 | 2.00 | 1.63 | 0.37 | 0.56 |
| NAUpy48t | 130–160 | 4 | 2.00 | 1.50 | 0.33 | 0.51 |
| NAUpy29u | 130–160 | 4 | 1.57 | 1.47 | 0.25 | 0.36 |
| NAUpy41v | 130–150 | 4 | 1.86 | 1.43 | 0.28 | 0.43 |
| NAUpy33v | 110–140 | 3 | 2.00 | 1.49 | 0.33 | 0.51 |
| NAUpy13d | 120–140 | 4 | 1.86 | 1.37 | 0.25 | 0.40 |
| NAUpy85x | 120–155 | 6 | 1.86 | 1.57 | 0.34 | 0.50 |
| NAUpy98s | 160–200 | 6 | 2.00 | 1.75 | 0.43 | 0.62 |
| NAUpy28d | 170–210 | 4 | 1.71 | 1.51 | 0.30 | 0.43 |
| NAUpy31t | 100–150 | 6 | 1.71 | 1.51 | 0.30 | 0.43 |
| NAUpy66w | 100–120 | 4 | 1.71 | 1.46 | 0.27 | 0.40 |
| NAUpy30d | 200–220 | 6 | 2.00 | 1.51 | 0.33 | 0.51 |
| NAUpy90w | 180–240 | 6 | 2.00 | 1.31 | 0.23 | 0.38 |
| NAUpy73m | 110–130 | 4 | 2.00 | 1.36 | 0.26 | 0.42 |
| NAUpy35d | 150–160 | 3 | 2.00 | 1.31 | 0.23 | 0.38 |
| NAUpy30c | 220–250 | 6 | 2.00 | 1.48 | 0.31 | 0.47 |
| NAUpy06z | 120–200 | 4 | 2.00 | 1.66 | 0.38 | 0.56 |
| NAUpy06d | 200–220 | 6 | 2.00 | 1.54 | 0.34 | 0.52 |
| NAUpy58t | 130–150 | 6 | 2.00 | 1.48 | 0.31 | 0.48 |
| NAUpy45f | 120–170 | 5 | 2.00 | 1.44 | 0.29 | 0.46 |
| NAUpy56h | 170–200 | 4 | 2.00 | 1.51 | 0.32 | 0.50 |
| Average | 4.14 | 1.89 | 1.51 | 0.31 | 0.46 |
Observed sizes—size range on 8 % high-resolution non-denaturing polyacrylamide gel. Number of alleles—total numbers observed in seven pear genotypes
na observed number of alleles, ne effective number of alleles [Kimura and Crow (1964)], h Nei’s (1973) gene diversity, I Shannon’s information index [Lewontin (1972)]
Sequencing Results of Two SSR Loci in Pyrus Accessions
The sequencing and alignment of SSR alleles derived from pears verified the presence of the SSR loci and revealed a high degree of conservation of SSR flanking regions in two loci (Fig. 2). The sequencing results showed a diversity of alleles in both loci. TA-repeats of allelic variation were detected for NAUPy31, and CT-repeats of allelic variation were detected for NAUpy53a (Fig. 2). The allelic diversity was mainly due to length variations of the microsatellite repeats, combined with point mutation events within the flanking regions. These variations may contribute to the formation of pear germplasm diversity.
Fig. 2.
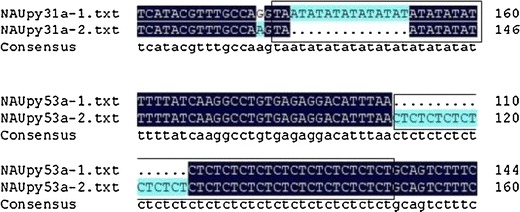
DNAMAN alignment of sequences obtained from selected PCR bands amplified by primer pairs NAUPy31a and NAUpy53a in pears. 1–2 bands of different sizes; TA-repeat and CT-repeat differences within the fragments are shown highlighted in blue
Transferability to Other Species in Rosaceae
A set of 67 selected SSR (Table 2) polymorphisms in seven pear genotypes was evaluated using genomic DNA of 96 genotypes belonging to seven genera in Rosaceae, including Prunus (10 Prunus mume and 14 P. salicina), Armeniaca (8 accessions), Cerasus (13 accessions), Amygdalus (12 accessions), Fragaria (6 accessions), Malus (20 accessions), and Eriobotrya (11 accessions). Prunus, Armeniaca, Cerasus, and Amygdalus belong to Prunoideae, Fragaria belongs to Rosoideae, and Malus and Eriobotrya belong to Maloideae (Table 1). Overall, 61.2 % (41/67) of the tested SSRs successfully amplified at least one PCR product of the approximate size expected for a homologous gene in at least one of the Rosaceae genera screened (Table 3; Fig. 3). This result (61.2 %) was lower than the 75 % achieved from apple EST-SSRs to other Rosaceae (Gasic et al. 2009). The most likely reason was that, compared to genomic SSRs, in theory, EST-SSRs have higher transferability because the EST-derived microsatellites are within transcribed regions of the DNA and are expected to be more conserved and less polymorphic than genomic SSR markers. However, the apple EST-SSRs to other Rosaceae had a lack of polymorphism, which is in agreement with the theory that genomic SSRs have a lower transferability but a better polymorphism. The good polymorphism characteristics of the transferable pear genomic SSRs in this study would be more valuable in application to Rosaceae genomic studies.
Table 3.
Transferability of pear SSRs to other Rosaceae fruit crops
| SSR locus | Number of accessions in which an SSR was amplified | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Japanese apricot | Plum | Cherry | Apple | Strawberry | Apricot | Peach | Loquat | ||
| NAUpy22c | 3/10 | 0/14 | 0/13 | 16/20 | 0/6 | 0/10 | 0/12 | 7/11 | 26/96 |
| NAUpy24c | 0/10 | 0/14 | 4/13 | 20/20 | 0/6 | 2/10 | 8/12 | 4/11 | 38/96 |
| NAUpy32c | 0/10 | 0/14 | 0/13 | 1/20 | 0/6 | 0/10 | 0/12 | 0/11 | 1/96 |
| NAUpy38c | 0/10 | 0/14 | 0/13 | 3/20 | 0/6 | 0/10 | 0/12 | 0/11 | 3/96 |
| NAUpy40c | 0/10 | 1/14 | 0/13 | 7/20 | 0/6 | 0/10 | 0/12 | 0/11 | 8/96 |
| NAUpy43c | 0/10 | 0/14 | 0/13 | 15/20 | 0/6 | 0/10 | 0/12 | 4/11 | 19/96 |
| NAUpy86c | 0/10 | 0/14 | 0/13 | 7/20 | 0/6 | 0/10 | 0/12 | 0/11 | 7/96 |
| NAUpy87c | 6/10 | 5/14 | 7/13 | 2/20 | 0/6 | 0/10 | 0/12 | 0/11 | 20/96 |
| NAUpy89c | 10/10 | 11/14 | 10/13 | 15/20 | 0/6 | 0/10 | 0/12 | 0/11 | 46/96 |
| NAUpy27c | 0/10 | 0/14 | 0/13 | 11/20 | 0/6 | 0/10 | 0/12 | 1/11 | 12/96 |
| NAUpy91b | 0/10 | 0/14 | 0/13 | 13/20 | 0/6 | 0/10 | 0/12 | 0/11 | 13/96 |
| NAUpy85d | 1/10 | 0/14 | 0/13 | 10/20 | 0/6 | 1/10 | 0/12 | 1/11 | 13/96 |
| NAUpy87d | 3/10 | 4/14 | 3/13 | 4/20 | 0/6 | 0/10 | 0/12 | 0/11 | 14/96 |
| NAUpy46k | 0/10 | 1/14 | 0/13 | 6/20 | 0/6 | 0/10 | 0/12 | 0/11 | 7/96 |
| NAUpy55k | 1/10 | 1/14 | 1/13 | 16/20 | 0/6 | 0/10 | 0/12 | 5/11 | 24/96 |
| NAUpy80k | 0/10 | 0/14 | 0/13 | 11/20 | 0/6 | 1/10 | 1/12 | 7/11 | 20/96 |
| NAUpy81m | 0/10 | 1/14 | 0/13 | 8/20 | 0/6 | 0/10 | 0/12 | 0/11 | 9/96 |
| NAUpy86n | 0/10 | 0/14 | 0/13 | 7/20 | 0/6 | 5/10 | 3/12 | 1/11 | 16/96 |
| NAUpy98s | 0/10 | 0/14 | 0/13 | 9/20 | 0/6 | 3/10 | 1/12 | 4/11 | 17/96 |
| NAUpy36t | 0/10 | 0/14 | 0/13 | 8/20 | 0/6 | 0/10 | 0/12 | 3/11 | 11/96 |
| NAUpy48t | 1/10 | 0/14 | 0/13 | 7/20 | 0/6 | 0/10 | 0/12 | 0/11 | 8/96 |
| NAUpy58t | 2/10 | 0/14 | 0/13 | 4/20 | 0/6 | 0/10 | 0/12 | 0/11 | 6/96 |
| NAUpy67t | 2/10 | 2/14 | 5/13 | 20/20 | 3/6 | 10/10 | 11/12 | 8/11 | 61/96 |
| NAUpy91t | 0/10 | 0/14 | 0/13 | 10/20 | 0/6 | 5/10 | 5/12 | 1/11 | 21/96 |
| NAUpy07u | 0/10 | 7/14 | 4/13 | 20/20 | 0/6 | 0/10 | 0/12 | 0/11 | 31/96 |
| NAUpy30u | 0/10 | 0/14 | 0/13 | 0/20 | 0/6 | 0/10 | 0/12 | 7/11 | 7/96 |
| NAUpy62u | 0/10 | 0/14 | 0/13 | 7/20 | 0/6 | 0/10 | 0/12 | 2/11 | 9/96 |
| NAUpy33v | 0/10 | 0/14 | 0/13 | 0/20 | 0/6 | 0/10 | 0/12 | 3/11 | 3/96 |
| NAUpy35v | 3/10 | 5/14 | 1/13 | 12/20 | 0/6 | 0/10 | 0/12 | 0/11 | 21/96 |
| NAUpy41v | 0/10 | 0/14 | 0/13 | 3/20 | 0/6 | 0/10 | 0/12 | 0/11 | 3/96 |
| NAUpy74v | 0/10 | 0/14 | 0/13 | 10/20 | 0/6 | 0/10 | 0/12 | 8/11 | 18/96 |
| NAUpy43w | 0/10 | 0/14 | 0/13 | 7/20 | 0/6 | 0/10 | 0/12 | 0/11 | 7/96 |
| NAUpy94x | 0/10 | 0/14 | 0/13 | 3/20 | 0/6 | 0/10 | 0/12 | 0/11 | 3/96 |
| NAUpy92a | 0/10 | 0/14 | 0/13 | 10/20 | 0/6 | 0/10 | 3/12 | 0/11 | 13/96 |
| NAUpy31a | 0/10 | 0/14 | 0/13 | 2/20 | 0/6 | 0/10 | 0/12 | 0/11 | 2/96 |
| NAUpy74f | 0/10 | 0/14 | 0/13 | 4/20 | 0/6 | 0/10 | 0/12 | 0/11 | 4/96 |
| NAUpy23f | 3/10 | 0/14 | 0/13 | 6/20 | 0/6 | 5/10 | 5/12 | 4/11 | 23/96 |
| NAUpy45f | 0/10 | 0/14 | 0/13 | 10/20 | 0/6 | 0/10 | 0/12 | 3/11 | 13/96 |
| NAUpy39h | 0/10 | 0/14 | 0/13 | 4/20 | 0/6 | 0/10 | 0/12 | 0/11 | 4/96 |
| NAUpy56h | 0/10 | 0/14 | 0/13 | 19/20 | 0/6 | 0/10 | 0/12 | 0/11 | 19/96 |
| NAUpy38e | 0/10 | 0/14 | 0/13 | 10/20 | 0/6 | 4/10 | 6/12 | 2/11 | 22/96 |
| Number of transferable SSRs | 11/67 | 10/67 | 8/67 | 39/67 | 1/67 | 9/67 | 9/67 | 22/67 | 41/67 |
| Percentage (%) | 16.4 | 14.9 | 11.9 | 58.2 | 1.5 | 13.4 | 13.4 | 32.8 | 61.2 |
Number of transferable SSRs—number of SSR markers that successfully amplified in at least one Rosaceae cultivar; the SSRs whose products amplified in Rosaceae crops ranged from 90 to 280 bp were considered to be transferable, and those beyond this range were not recorded in this table
Fig. 3.

Amplification of pear SSR marker NAUpy98s in Rosacea accessions; numbers in lanes are the same as Table 1; 1–10 Japanese apricot, 11–24 plum, 25–37 cherry, 38–57 apple, 58–63 strawberry, 64–73 apricot, 74–85 peach, 86–96 loquat, M 3 Trans DNA 2 K Marker
The highest transferability, 58.2 % (Table 3), was observed in the closely related apple (Malus domestica), in which the majority of pear SSRs are polymorphic (Supplemental Fig. 1) and had amplification patterns similar to those observed in pear. This indicates that primer binding sites between these two closely related rosaceous genera, Malus and Pyrus, are fairly well conserved (Table 3; Supplemental Fig. 1). This high level of transferability of SSRs was consistent with genome comparison of pear and apple (Wu et al. 2013) and was also similar to previous findings where apple SSRs have been reported capable of identifying polymorphism and detecting genetic diversity in pear (Yamamoto et al. 2001, 2004). Besides, Gasic et al. (2009) reported high transferability (59 %) of apple EST-SSRs to pear, and both Pieratoni et al. (2004) and Yamamoto et al. (2004) have reported amplification of apple SSRs in pear populations. Yao et al. (2010) tested 94 primer pairs on four accessions of Pyrus to evaluate the transferability of the markers, and 40 of 72 functional SSRs produced polymorphic amplicons. All of the above indicates that pear has close synteny with apple.
Two SSR loci, NAUpy43c and NAUpy55k, were then employed to analyze genetic diversity and relationship among a collection of 20 genotypes of apple. The two SSR loci could distinguish the 20 apple individuals and have detected 9 alleles (Table 4), and UPGMA cluster analysis grouped these individuals into three groups at the similarity level of 0.56 (Fig. 4). Based on the polymorphism revealed by the two SSR markers NAUpy43c and NAUpy55k, the cluster results largely agree with the traditional taxonomy based on pedigrees or geographic origins. The first group consisted of Longfeng and Xiushui: Longfeng was the progeny of Jinhong and Bailong, and Xiushui was a seedling of Ralls. The second group included Braeburn, Orin, Dailv, Qinyang, Lujia 1, Golden Delicious, and Starking, also called the Golden Delicious class. Among them, Dailv, a seedling of Golden Delicious, and Orin, whose female parent was Golden Delicious, were well known, and they belong to the Golden Delicious class. Lujia 1 has a close relationship with Starking, and Braeburn originated from New Zealand and was the progeny of Lady Hamilton and Granny Smith. Sekaiichi, Huaguan, Zaohong, Delicious, Fuji, Mikilifi, Gala, HoKuto, Jonagold, Alps Otome, and Hanfu formed the third group, also called the Delicious class. According to the pedigrees, Jonagold belongs to the Jonathan lineage, Mikilifi belongs to the Summer Pearmain lineage, and Sekaiichi, Huaguan, Zaohong, Fuji, Gala, HoKuto, Alps Otome, and Hanfu all have relationships with Delicious. In the third group, Fuji is the parent of HoKuto, Alps Otome, Hanfu, and Huaguan. Excepting Alps Otome, the other three cultivars grouped with Fuji together with Mikilifi with a similarity coefficient value of 0.78. These results were consistent with previous classifications (Zhang et al. 2012; Potts et al. 2012; Reim et al. 2012), indicating the good quality and usefulness of the two pear SSR loci in apple germplasm evaluation. In brief, SSR markers developed from pear showed high transferability in apple and can be valuable for application in germplasm evaluation and genetic relationship analysis of apple, as well as comparative mapping between pear and apple.
Table 4.
Data of the SSR fingerprint bands with two pear primers amplified among 20 apple genotypes
| SSR locus | Band no. | Apple cultivars (sample no.) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
| NAUpy43c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | |
| NAUpy55k | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 |
| 2 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | |
| 3 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | |
| 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
1 presence of band, 0 absence of band; sample no.: 1 Sekaiichi, 2 Huaguan, 3 Zaohong, 4 Delicious, 5 Braeburn, 6 Fuji, 7 Golden Delicious, 8 Mikilifi, 9 Longfeng, 10 Starking, 11 Dailv, 12 Gala, 13 Qinyang, 14 HoKuto, 15 Jonagold, 16 Orin, 17 Lujia 1, 18 Alps Otome, 19 Hanfu, 20 Xiushui
Fig. 4.
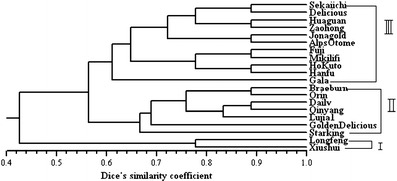
UPGMA dendrogram of Malus accessions based on Dice’s similarity coefficient (Nei and Li 1979) according to pear SSR markers NAUpy43c and NAUpy55k
Loquat (Eriobotrya japonica), a subtropical evergreen fruit tree belonging to the Rosaceae subfamily Maloideae that also contains apple and pear, ranked in second place with a transferability of 32.8 %. The pedigrees of the majority of loquat cultivars are historically unknown, and the current system used for cultivar classification gives little information on genetic identity and variability, as it is mainly based on morphological traits linked to ecotype, flesh color, fruit shape, usage, and ripening time (Martínez-Calvo et al. 2008). Genetic diversity and the relationships among different cultivars of loquat are of great importance for the conservation of genetic resources, breeding initiatives, and national and international exchanges of materials (He et al. 2011). The loquat genome has not been sequenced, and only a small number of molecular markers of loquat are available, but this number can be increased by identifying pear SSRs that are transferable to loquat cultivars/accessions. In this study, 22 SSRs from pear were successfully transferred to loquat (Table 3; Fig. 3), which will provide new insight into the level of genetic diversity within loquat and synteny with pear. In previous research, apple SSRs had been transferred and applied in loquat. Soriano et al. (2005) used 30 SSRs from apple to assay the genetic relationships in loquat, 13 of which amplified polymorphic products and distinguished 34 of the 40 loquat accessions originating from different European countries. He et al. (2011) also identified 39 SSRs from apple that could be transferred to loquat. The pear SSRs transferred to loquat could be applied as new SSR markers in loquat for genetic research, such as map construction and comparison, resource assessment, and molecular breeding.
Transferability of pear SSRs to the Prunoideae members was at an analogous level: Japanese apricot 18.3 %, plum 16.9 %, cherry 15.5 %, apricot 12.7 %, and peach 14.1 %, indicating that these Prunoideae stone fruits have homology regions with the pear genome. In addition, the Prunoideae species shared most of the same amplified markers (Table 3), indicating that they have a close genetic relationship within themselves, which largely agrees with the traditional taxonomy. The SSRs transferred to Prunoideae successfully were polymorphic and produced special band patterns in the Prunoideae (Fig. 3), suggesting application of these SSRs as valuable tools for comparative mapping and genetic diversity analysis. Gasic et al. (2009) surveyed the transferability of apple EST-SSRs to Prunoideae and showed that the frequency of transferability ranged from 25 % in the subgenus Armeniaca to 38 % in the subgenus Amygdalus. Apple EST-SSRs were successfully amplified in 14 members of the subgenus Prunophora, represented by apricot and European and Japanese plums, with an average of 40 %, with the highest frequency of transferability (35 %) observed for Japanese plum. Substantial transferability of apple EST-SSR to peach, apricot, and European plum was also noted at 37, 25, and 29 %, respectively. The value of apple EST-SSR to Prunoideae members was much higher than that observed in this study, likely suggesting the higher conservation of EST regions used for marker development. Vendramin et al. (2007) studied a set of EST-SSRs isolated from the peach fruit transcriptome and their transportability across Prunus species. Apricot genomic SSRs showed considerable transferability, 20 %, in all Prunus species, but failed to amplify in apple (Messina et al. 2004). Mnejja et al. (2010) studied the transferability of 145 Prunus microsatellite markers across a set of eight cultivars from nine rosaceous species (almond, peach, apricot, Japanese plum, European plum, cherry, apple, pear, and strawberry). Of these, 16.6 % were transferable in pear, and the polymorphism of Prunus microsatellites was also detected at a low level. In this study, the transferability of pear SSRs to Prunus, 38 % (27/71) in total (Table 3), is higher than the above, and some displayed variable band types (Fig. 3); thus, they can play an important role in the genetic study of Prunus or related species. In contrast, Wünsch (2009) studied 13 Prunus SSRs known to be transferable to certain Prunus species, tested them in 10 Prunus, and found that three were transferable to all of them and eight to all but one species. Prunus SSRs (63.9 %) were transferable to other Prunus crops (Mnejja et al. 2010), overall showing a higher level of transferability to close species. Most pear SSRs transferred to Prunus detected only a single band and had a lack of polymorphism (Fig. 3), but their transferability indicates some extent of genomic collineation between pear and Prunus.
Only a few SSR primers were amplified (1.4 %) among six strawberry cultivars. Unlike other Rosaceae family crops such as apple and peach, the strawberry is considered to be non-climacteric because the flesh does not ripen in response to ethylene. Genomically, strawberry genus has a small basic (x = 7) genome size of ~240 Mb, while the cultivated F. ×ananassa is among the most complex of crop plants, harboring eight sets of chromosomes (2n = 8x = 56) derived from as many as four different diploid ancestors (Shulaev et al. 2011). Other research about transferability to strawberry found that only 12 of 145 Prunus SSR loci (8.3 %) tested were polymorphic in strawberry (Mnejja et al. 2010). This indicates that the strawberry has a comparatively more distant relationship with pear than other Rosaceae fruit trees, which is consistent with the divergent relationship of pear, apple, and strawberry based on whole genome comparison (Wu et al. 2013). However, Gasic et al. (2009) reported good transferability of EST-SSRs (49 %) from apple to strawberry. In addition, Zorrilla-Fontanesithe et al. (2011) investigated the transferability of 174 Fragaria SSRs to rose and raspberry, with the data ranging from 28.7 % for genic SSRs in rose to 16.1 % for genomic SSRs in raspberry. Collinearity analysis between pear and two other sequenced rosaceous species, apple and strawberry, has revealed that they share the same ancestor, but unlike pear and apple, strawberry did not have a whole genome duplication event (Wu et al. 2013). So, the higher transferability of EST-SSR over genomic SSRs within species also reveals the common ancestral origins with differential genome evolution. In addition, the level of polymorphism of an SSR marker seems to greatly depend on the germplasm on which it is tested. Thus, more exploration such as sequence comparison should be done to study the genetic relationship between pear and strawberry.
Conclusion
The SSR markers in this study displayed a high level of polymorphism in pears, suggesting valuable applications of these markers in future pear linkage map construction, genetic polymorphism evaluation, cultivar identification, and MAS. As useful PCR-based genetic markers, the pear SSRs can be applied to these various aspects of genetic research not only for Pyrus, but also for other members of Rosaceae. Our results reveal a relatively high level of transferability between pear and several other Rosaceae species, which means an increased number of SSR markers available for Rosaceae crops, which is particularly of value for those species with little genomic information. Most of the pear SSRs presented diversity when assessed in other Rosaceae species, implying that they will be significant for genetic research. Besides this, when mapped, these markers can be used for conducting macro-synteny studies among Rosaceae species to better understand genome organization and evolutionary relationships in this important fruit family. The transferability to each Rosaceae species was largely related to their consanguinity with pear, demonstrating the applicability of pear SSRs in phylogenetic and evolutionary studies of the Rosaceae family. Overall, these findings suggest that the high level of polymorphism and good transferability of pear SSRs result in their promise for wide use in QTL mapping, molecular breeding, investigation of population genetic diversity, comparative mapping, and evolutionary studies among pears and other Rosaceae species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 647 kb)
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31171928), the National Public Benefit (Agricultural) Research Foundation of China (200903044), and the Earmarked Fund for China Agriculture Research System (CARS-29).
References
- Baraket G, Chatti K, Saddoud O, Abdelkarim A, Mars M, Trifi M, Hannachi A. Comparative assessment of SSR and AFLP markers for evaluation of genetic diversity and conservation of fig, Ficus carica L., genetic resources in Tunisia. Plant Mol Biol Rep. 2011;29:171–184. doi: 10.1007/s11105-010-0217-x. [DOI] [Google Scholar]
- Bassam BJ, Caetano-Anollés G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-I. [DOI] [PubMed] [Google Scholar]
- Bouvier L, Bourcy M, Boulay M, Tellier M, Guérif P, Denancé C, Durel CE, Lespinasse Y. A new pear scab resistance gene Rvp1 from the European pear cultivar ‘Navara’ maps in a genomic region syntenic to an apple scab resistance gene cluster on linkage group 2. Tree Genet Genomes. 2012;8:53–60. doi: 10.1007/s11295-011-0419-x. [DOI] [Google Scholar]
- Campoy J, Ruiz D, Egea J, Rees D, Celton J, Martínez-Gómez P. Inheritance of flowering time in apricot (Prunus armeniaca L.) and analysis of linked quantitative trait loci (QTLs) using simple sequence repeat (SSR) markers. Plant Mol Biol Rep. 2011;29:404–410. doi: 10.1007/s11105-010-0242-9. [DOI] [Google Scholar]
- Celton JM, Tustin DS, Chagné D, Gardiner SE. Construction of a dense genetic linkage map for apple rootstocks using SSRs developed from Malus ESTs and Pyrus genomic sequences. Tree Genet Genomes. 2009;2:86–97. [Google Scholar]
- Decroocq V, Fave MG, Hagen L, Bordenave L, Decroocq S. Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet. 2003;106:912–922. doi: 10.1007/s00122-002-1158-z. [DOI] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Genome relationships: the grass model in current research. Plant Cell. 2000;12:637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirlewanger E, Graziano E, Joobeur T, Garriga-Calderé F, Cosson P, Howad W, Arús P. Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci. 2004;101:9891–9896. doi: 10.1073/pnas.0307937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD. A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics. 2002;161:1697–1711. doi: 10.1093/genetics/161.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira dos Santos A, Ramos-Cabrer A, Díaz-Hernández M, Pereira-Lorenzo S. Genetic variability and diversification process in local pear cultivars from northwestern Spain using microsatellites. Tree Genet Genomes. 2011;7:1041–1056. doi: 10.1007/s11295-011-0393-3. [DOI] [Google Scholar]
- Gasic K, Han Y, Kertbundit S, Shulaev V, Iezzoni AF, Stover EW, Bell RL, Wisniewski ME, Korban SS. Characteristics and transferability of new apple EST-derived SSRs to other Rosaceae species. Mol Breed. 2009;23:397–411. doi: 10.1007/s11032-008-9243-x. [DOI] [Google Scholar]
- Gisbert AD, Martínez-Calvo J, Llácer G, Badenes ML, Romero C. Development of two loquat [Eriobotrya japonica (Thunb.) Lindl.] linkage maps based on AFLPs and SSR markers from different Rosaceae species. Mol Breed. 2009;23:523–538. doi: 10.1007/s11032-008-9253-8. [DOI] [Google Scholar]
- He Q, Li X, Liang G, Ji K, Guo Q, Yuan W, Zhou G, Chen K, Weg WE, Gao Z. Genetic diversity and identity of Chinese loquat cultivars/accessions (Eriobotrya japonica) using apple SSR markers. Plant Mol Biol Rep. 2011;29:197–208. doi: 10.1007/s11105-010-0218-9. [DOI] [Google Scholar]
- Kalo P, Seres A, Taylor SA, Jakab J, Kevei Z, Kereszt A, Endre G, Ellis THN, Kiss GB. Comparative mapping between Medicago sativa and Pisum sativum. Mol Genet Genomics. 2004;272:235–246. doi: 10.1007/s00438-004-1055-z. [DOI] [PubMed] [Google Scholar]
- Kimura, Crow The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. The apportionment of human diversity. Evol Biol. 1972;6:381–398. doi: 10.1007/978-1-4684-9063-3_14. [DOI] [Google Scholar]
- Lukens L, Zou F, Lydiate D, Parkin I, Osborn T. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics. 2003;164:359–372. doi: 10.1093/genetics/164.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Calvo JM, Gisbert AD, Alamar MC, Hernandorena R, Romero C, Llácer G, Badenes ML. Study of a germplasm collection of loquat (Eriobotrya japonica Lindl.) by multivariate analysis. Genet Resour Crop Evol. 2008;55:695–703. doi: 10.1007/s10722-007-9276-8. [DOI] [Google Scholar]
- Messina R, Laino O, Marrazzo MT, Cipriani G, Testolin R. New set of microsatellite loci isolated in apricot. Mol Ecol Notes. 2004;4:432–434. doi: 10.1111/j.1471-8286.2004.00674.x. [DOI] [Google Scholar]
- Mnejja M, Garcia-Mas J, Audergon JM, Arús P. Prunus microsatellite marker transferability across rosaceous crops. Tree Genet Genomes. 2010;6:689–700. doi: 10.1007/s11295-010-0284-z. [DOI] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Yang C, Wei Z, Jiang J. DNA extraction of birch leaves by improved CTAB method and optimization of its ISSR system. J For Res. 2006;4:298–300. doi: 10.1007/s11676-006-0068-3. [DOI] [Google Scholar]
- Pieratoni L, Cho KH, Shin IS, Chiodini R, Tartarini S, Dondini L, Kang SJ, Sansavini S. Characterization and transferability of apple SSRs to two European pear F1 populations. Theor Appl Genet. 2004;109:1519–1524. doi: 10.1007/s00122-004-1775-9. [DOI] [PubMed] [Google Scholar]
- Potts SM, Han Y, Khan MA, Kushad MM, Rayburn AL, Korban SS. Genetic diversity and characterization of a core collection of Malus germplasm using simple sequence repeats (SSRs) Plant Mol Biol Rep. 2012;30:827–837. doi: 10.1007/s11105-011-0399-x. [DOI] [Google Scholar]
- Rohlf FJ. Numerical taxonomy and multivariate analysis system. Version 2.0. Setauket: Exeter Software; 1998. [Google Scholar]
- Reim S, Höltken A, Höfer M. Diversity of the European indigenous wild apple (Malus sylvestris (L.) Mill.) in the East Ore Mountains (Osterzgebirge), Germany: II. Genetic characterization. Genet Resour Crop Evol. 2012 [Google Scholar]
- Sargent DJ, Marchese A, Simpson DW, Howad W, Fernandez-Fernandez F, Monfort A, Arus P, Evans KM, Tobutt KR. Development of “universal” gene-specific markers from Malus spp. cDNA sequences, their mapping and use in synteny studies within Rosaceae. Tree Genet Genome. 2009;5:133–145. doi: 10.1007/s11295-008-0178-5. [DOI] [Google Scholar]
- Sargent D, Rys A, Nier S, Simpson D, Tobutt K. The development and mapping of functional markers in Fragaria and their transferability and potential for mapping in other genera. Theor Appl Genet. 2007;114:373–384. doi: 10.1007/s00122-006-0441-9. [DOI] [PubMed] [Google Scholar]
- Shulaev V, Korban SS, Sosinski B, Abbott AG, Aldwinckle HS, Folta KM, Iezzoni A, Main D, Arus P, Dandekar AM, Lewers K, Brown SK, Davis TM, Gardiner SE, Potter D, Veilleux RE. Multiple models for Rosaceae genomics. Plant Physiol. 2008;147:985–1003. doi: 10.1104/pp.107.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, Burns P, Davis TM, Slovin JP, Bassil N, Hellens RP, Evans C, Harkins T, Kodira C, Desany B, Crasta OR, Jensen RV, Allan AC, Michael TP, Setubal JC, Celton J-M, Rees DJG, Williams KP, Holt SH, Rojas JJR, Chatterjee M, Liu B, Silva H, Meisel L, Adato A, Filichkin SA, Troggio M, Viola R, Ashman T-L, Wang H, Dharmawardhana P, Elser J, Raja R, Priest HD, Bryant DW, Fox SE, Givan SA, Wilhelm LJ, Naithani S, Christoffels A, Salama DY, Carter J, Girona EL, Zdepski A, Wang W, Kerstetter RA, Schwab W, Korban SS, Davik J, Monfort A, Denoyes-Rothan B, Arus P, Mittler R, Flinn B, Aharoni A, Bennetzen JL, Salzberg SL, Dickerman AW, Velasco R, Borodovsky M, Veilleux RE, Folta KM. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano JM, Romero C, Vilanova S, Llácer G, Badenes ML. Genetic diversity of loquat germplasm (Eriobotrya japonica (Thunb) Lindl) assessed by SSR markers. Genome. 2005;48:108–114. doi: 10.1139/g04-101. [DOI] [PubMed] [Google Scholar]
- Swapna M, Sivaraju K, Sharma RK, Singh NK, Mohapatra T. Single-strand conformational polymorphism of EST-SSRs: a potential tool for diversity analysis and varietal identification in sugarcane. Plant Mol Biol Rep. 2011;29:505–513. doi: 10.1007/s11105-010-0254-5. [DOI] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagne D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouze P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel C-E, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R. The genome of the domesticated apple (Malus × domestica Borkh.) Nat Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- Vendramin E, Dettori MT, Giovinazzi J, Micali S, Quarta R, Verde I. A set of EST-SSRs isolated from peach fruit transcriptome and their transportability across Prunus species. Mol Ecol Notes. 2007;7:307–310. doi: 10.1111/j.1471-8286.2006.01590.x. [DOI] [Google Scholar]
- Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, Chen NJ, Nishio T, Xu X, Cong L, Qi K, Huang X, Wang Y, Zhao X, Wu J, Deng C, Gou C, Zhou W, Yin H, Qin G, Sha Y, Tao Y, Chen H, Yang Y, Song Y, Zhan D, Wang J, Li L, Dai M, Gu C, Wang Y, Shi D, Wang X, Zhang H, Zeng L, Zheng D, Wang C, Chen M, Wang G, Xie L, Sovero V, Sha S, Huang W, Zhang S, Zhang M, Sun J, Xu L, Li Y, Liu X, Li Q, Shen J, Wang J, Paull RE, Bennetzen JL, Wang J, Zhang S. The genome of pear (Pyrus bretschneideri Rehd.) Genome Res. 2013;23:396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wünsch A. Cross-transferable polymorphic SSR loci in Prunus species. Sci Hortic. 2009;120:348–352. doi: 10.1016/j.scienta.2008.11.012. [DOI] [Google Scholar]
- Yamamoto T, Kimura T, Sawamura Y, Kotobuki K, Ban Y, Hayashi T, Matsuta N. SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor Appl Genet. 2001;102:865–870. doi: 10.1007/s001220000524. [DOI] [Google Scholar]
- Yamamoto T, Kimura T, Saito T, Kotobuki K, Matsuta N, Liebhard R, Gessler C, van de Weg WE, Hayashi T. Genetic linkage maps of Japanese and European pears aligned to the apple consensus map. Acta Hortic. 2004;663:51–56. [Google Scholar]
- Yamamoto T, Kimura T, Terakami S, Nishitani C, Sawamura Y, Saito T, Kotabuki K, Hayashi T. Integrated reference genetic linkage maps of pear based on SSR and AFLP markers. Breed Sci. 2007;57:321–329. doi: 10.1270/jsbbs.57.321. [DOI] [Google Scholar]
- Yang RC, Yeh FC. Multilocus structure in Pinus contorta Dougl. Theor Appl Genet. 1993;87:568–576. doi: 10.1007/BF00221880. [DOI] [PubMed] [Google Scholar]
- Yao L, Zheng X, Cai D, Gao Y, Wang K, Cao Y, Teng Y. Exploitation of Malus EST-SSRs and the utility in evaluation of genetic diversity in Malus and Pyrus. Genet Resour Crop Evol. 2010;57:841–851. doi: 10.1007/s10722-009-9524-1. [DOI] [Google Scholar]
- Zhang Q, Li J, Zhao Y, Korban SS, Han Y. Evaluation of genetic diversity in Chinese wild apple species along with apple cultivars using SSR markers. Plant Mol Biol Rep. 2012;30:539–546. doi: 10.1007/s11105-011-0366-6. [DOI] [Google Scholar]
- Zorrilla-Fontanesi Y, Cabeza A, Torres AM, Botella MA, Valpuesta V, Monfort A, Sánchez-Sevilla J, Amaya I. Development and bin mapping of strawberry genic-SSRs in diploid Fragaria and their transferability across the Rosoideae subfamily. Mol Breed. 2011;27:137–156. doi: 10.1007/s11032-010-9417-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 647 kb)


