Abstract
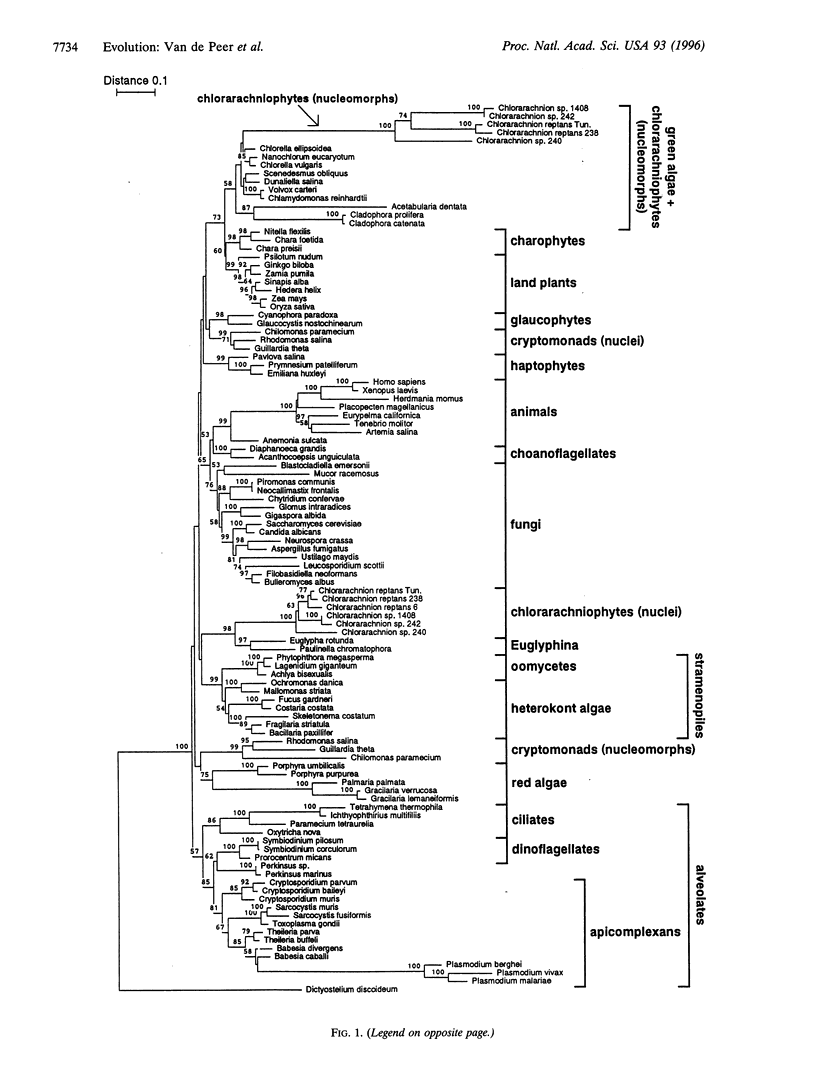
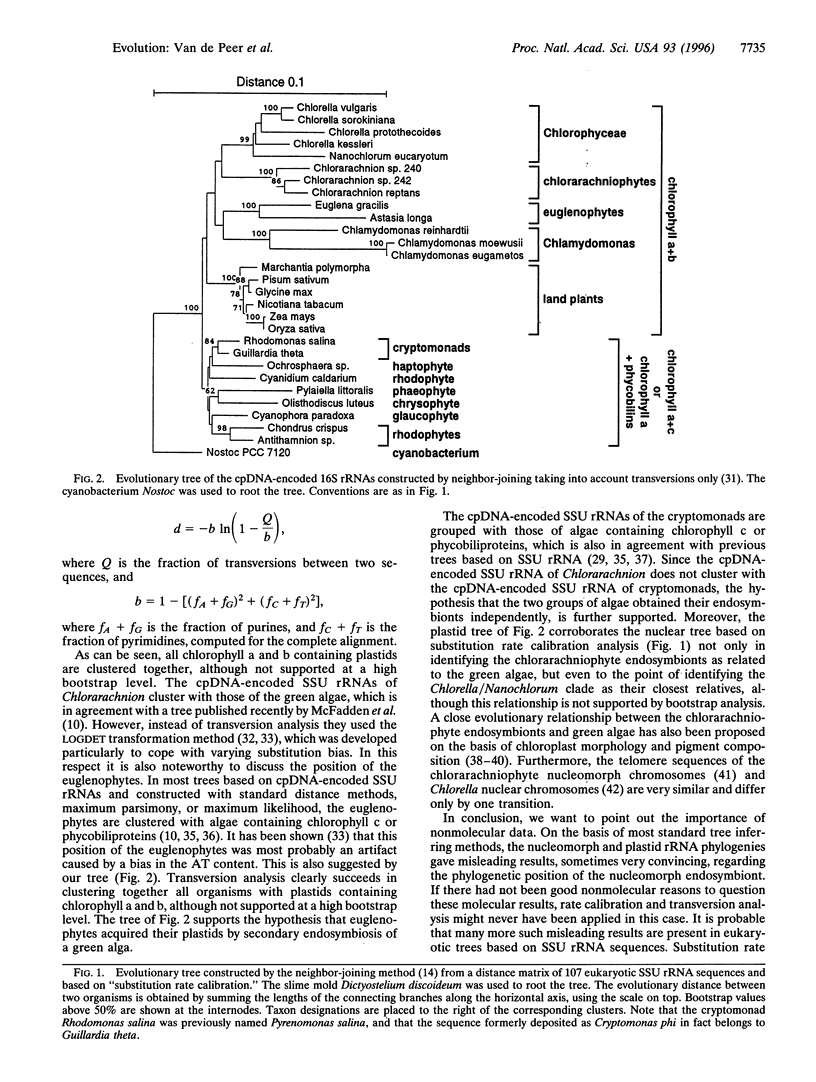
Chlorarachniophytes are amoeboid algae with chlorophyll a and b containing plastids that are surrounded by four membranes instead of two as in plants and green algae. These extra membranes form important support for the hypothesis that chlorarachniophytes have acquired their plastids by the ingestion of another eukaryotic plastid-containing alga. Chlorarachniophytes also contain a small nucleus-like structure called the nucleomorph situated between the two inner and the two outer membranes surrounding the plastid. This nucleomorph is a remnant of the endosymbiont's nucleus and encodes, among other molecules, small subunit ribosomal RNA. Previous phylogenetic analyses on the basis of this molecule provided unexpected and contradictory evidence for the origin of the chlorarachniophyte endosymbiont. We developed a new method for measuring the substitution rates of the individual nucleotides of small subunit ribosomal RNA. From the resulting substitution rate distribution, we derived an equation that gives a more realistic relationship between sequence dissimilarity and evolutionary distance than equations previously available. Phylogenetic trees constructed on the basis of evolutionary distances computed by this new method clearly situate the chlorarachniophyte nucleomorphs among the green algae. Moreover, this relationship is confirmed by transversion analysis of the Chlorarachnion plastid small subunit ribosomal RNA.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhattacharya D., Helmchen T., Bibeau C., Melkonian M. Comparisons of nuclear-encoded small-subunit ribosomal RNAs reveal the evolutionary position of the Glaucocystophyta. Mol Biol Evol. 1995 May;12(3):415–420. doi: 10.1093/oxfordjournals.molbev.a040216. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D., Helmchen T., Melkonian M. Molecular evolutionary analyses of nuclear-encoded small subunit ribosomal RNA identify an independent rhizopod lineage containing the Euglyphina and the Chlorarachniophyta. J Eukaryot Microbiol. 1995 Jan-Feb;42(1):65–69. doi: 10.1111/j.1550-7408.1995.tb01541.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T., Allsopp M. T., Chao E. E. Chimeric conundra: are nucleomorphs and chromists monophyletic or polyphyletic? Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11368–11372. doi: 10.1073/pnas.91.24.11368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijk P., De Wachter R. DCSE, an interactive tool for sequence alignment and secondary structure research. Comput Appl Biosci. 1993 Dec;9(6):735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- Douglas S. E., Murphy C. A., Spencer D. F., Gray M. W. Cryptomonad algae are evolutionary chimaeras of two phylogenetically distinct unicellular eukaryotes. Nature. 1991 Mar 14;350(6314):148–151. doi: 10.1038/350148a0. [DOI] [PubMed] [Google Scholar]
- Douglas S. E., Turner S. Molecular evidence for the origin of plastids from a cyanobacterium-like ancestor. J Mol Evol. 1991 Sep;33(3):267–273. doi: 10.1007/BF02100678. [DOI] [PubMed] [Google Scholar]
- Escalante A. A., Ayala F. J. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci U S A. 1995 Jun 20;92(13):5793–5797. doi: 10.1073/pnas.92.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson P., McFadden G. I. The chlorarachniophyte: a cell with two different nuclei and two different telomeres. Chromosoma. 1995 May;103(9):635–641. doi: 10.1007/BF00357690. [DOI] [PubMed] [Google Scholar]
- Goggin C. L., Barker S. C. Phylogenetic position of the genus Perkinsus (Protista, Apicomplexa) based on small subunit ribosomal RNA. Mol Biochem Parasitol. 1993 Jul;60(1):65–70. doi: 10.1016/0166-6851(93)90029-w. [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Maki S., Yamada T. Molecular organization of Chlorella vulgaris chromosome I: presence of telomeric repeats that are conserved in higher plants. Mol Gen Genet. 1995 Jan 6;246(1):29–36. doi: 10.1007/BF00290130. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Knoll A. H. The early evolution of eukaryotes: a geological perspective. Science. 1992 May 1;256(5057):622–627. doi: 10.1126/science.1585174. [DOI] [PubMed] [Google Scholar]
- Kuma K., Nikoh N., Iwabe N., Miyata T. Phylogenetic position of Dictyostelium inferred from multiple protein data sets. J Mol Evol. 1995 Aug;41(2):238–246. doi: 10.1007/BF00170678. [DOI] [PubMed] [Google Scholar]
- Lockhart P. J., Beanland T. J., Howe C. J., Larkum A. W. Sequence of Prochloron didemni atpBE and the inference of chloroplast origins. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2742–2746. doi: 10.1073/pnas.89.7.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerz M., Wolters J., Hofmann C. J., Sitte P., Maier U. G. Plastid DNA from Pyrenomonas salina (Cryptophyceae): physical map, genes, and evolutionary implications. Curr Genet. 1992 Jan;21(1):73–81. doi: 10.1007/BF00318658. [DOI] [PubMed] [Google Scholar]
- Maier U. G., Hofmann C. J., Eschbach S., Wolters J., Igloi G. L. Demonstration of nucleomorph-encoded eukaryotic small subunit ribosomal RNA in cryptomonads. Mol Gen Genet. 1991 Nov;230(1-2):155–160. doi: 10.1007/BF00290663. [DOI] [PubMed] [Google Scholar]
- McFadden G. I., Gilson P. R., Douglas S. E. The photosynthetic endosymbiont in cryptomonad cells produces both chloroplast and cytoplasmic-type ribosomes. J Cell Sci. 1994 Feb;107(Pt 2):649–657. doi: 10.1242/jcs.107.2.649. [DOI] [PubMed] [Google Scholar]
- McFadden G. I., Gilson P. R., Hofmann C. J., Adcock G. J., Maier U. G. Evidence that an amoeba acquired a chloroplast by retaining part of an engulfed eukaryotic alga. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3690–3694. doi: 10.1073/pnas.91.9.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morden C. W., Delwiche C. F., Kuhsel M., Palmer J. D. Gene phylogenies and the endosymbiotic origin of plastids. Biosystems. 1992;28(1-3):75–90. doi: 10.1016/0303-2647(92)90010-v. [DOI] [PubMed] [Google Scholar]
- Nelissen B., Van de Peer Y., Wilmotte A., De Wachter R. An early origin of plastids within the cyanobacterial divergence is suggested by evolutionary trees based on complete 16S rRNA sequences. Mol Biol Evol. 1995 Nov;12(6):1166–1173. doi: 10.1093/oxfordjournals.molbev.a040289. [DOI] [PubMed] [Google Scholar]
- Olsen G. J. Earliest phylogenetic branchings: comparing rRNA-based evolutionary trees inferred with various techniques. Cold Spring Harb Symp Quant Biol. 1987;52:825–837. doi: 10.1101/sqb.1987.052.01.090. [DOI] [PubMed] [Google Scholar]
- Ragan M. A., Bird C. J., Rice E. L., Gutell R. R., Murphy C. A., Singh R. K. A molecular phylogeny of the marine red algae (Rhodophyta) based on the nuclear small-subunit rRNA gene. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7276–7280. doi: 10.1073/pnas.91.15.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994 Sep;10(5):569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Neefs J. M., De Rijk P., De Wachter R. Reconstructing evolution from eukaryotic small-ribosomal-subunit RNA sequences: calibration of the molecular clock. J Mol Evol. 1993 Aug;37(2):221–232. doi: 10.1007/BF02407359. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Nicolaï S., De Rijk P., De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1996 Jan 1;24(1):86–91. doi: 10.1093/nar/24.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Van der Auwera G., De Wachter R. The evolution of stramenopiles and alveolates as derived by "substitution rate calibration" of small ribosomal subunit RNA. J Mol Evol. 1996 Feb;42(2):201–210. doi: 10.1007/BF02198846. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Achenbach L., Rouviere P., Mandelco L. Archaeal phylogeny: reexamination of the phylogenetic position of Archaeoglobus fulgidus in light of certain composition-induced artifacts. Syst Appl Microbiol. 1991;14(4):364–371. doi: 10.1016/s0723-2020(11)80311-5. [DOI] [PubMed] [Google Scholar]



