Abstract.
Understanding the mechanism of angiogenesis could help to decipher wound healing and embryonic development and to develop better treatment for diseases such as cancer. Microengineered devices were developed to reveal the mechanisms of angiogenesis, but monitoring the angiogenic process nondestructively in these devices is a challenge. In this study, we utilized a label-free imaging technique, ultrahigh-resolution optical coherence microscopy (OCM), to evaluate angiogenic sprouting in a microengineered device. The OCM system was capable of providing axial resolution and transverse resolution. Three-dimensional (3-D) distribution of the sprouting vessels in the microengineered device was imaged over , and details such as vessel lumens and branching points were clearly visualized. An algorithm based on stretching open active contours was developed for tracking and segmenting the sprouting vessels in 3-D-OCM images. The lengths for the first-, second-, and third-order vessels were measured as (), (), and (), respectively. The outer diameters for the first-, second-, and third-order vessels were , , and , respectively. These results demonstrate OCM as a promising tool for nondestructive and label-free evaluation of angiogenic sprouting in microengineered devices.
Keywords: optical coherence microscopy, microengineered device, angiogenesis, image segmentation
1. Introduction
Angiogenesis or the formation of new blood vessels from existing vasculature is a highly organized morphogenetic process including vessel initiation, formation, branching, maturation, and remodeling.1 It plays an essential role in many physiological conditions such as wound healing, embryonic development, and granulation formation. Dysregulation of angiogenesis contributes to numerous diseases such as cancer, ischemic disease, and infectious and immune disorders.1–3 Hence, understanding the fundamental morphogenetic processes of how cells organize to form new vessels and determining the function of various angiogenic factors that regulate angiogenesis are essential for developing novel therapeutic strategies. In order to decipher these processes, both in vitro and in vivo experimental systems have been developed.4 Living microvascular networks engineered in three-dimensional (3-D) tissue scaffolds have been successfully used for studying complex vascular phenomena including angiogenesis and thrombosis.5 The 3-D microengineered tissue constructs populated at physiologic cell densities have been made viable by using perfusable vascular networks that can meet its complex mass transport requirements.6 In a recent study, a microengineered organotypic model with endothelium-lined channels was used as a platform to recapitulate the morphogenetic processes of angiogenic sprouting in vitro.3 The success of these in vitro vascular models, which closely mimic in vivo conditions, points toward the potential of using this platform to investigate fundamental questions pertaining to angiogenesis and its role in disease progression.
Confocal microscopy and phase contrast microscopy have been used to characterize 3-D organotypic vascular models. Confocal microscopy has provided valuable insights into the morphogenetic processes central to angiogenesis such as endothelial invasion and sprout extension.3 It requires the specimens to be fixed and stained before imaging. However, these processes are destructive and not suitable for longitudinal studies using the same microvascular devices. Phase contrast microscopy has been utilized to observe dynamic events such as cell proliferation and migration in in vitro and in vivo assays.7 However, phase contrast microscopy lacks 3-D information, which is critical for evaluating angiogenic processes in organotypic models. Hence, a nondestructive imaging technique that can be used to monitor complex dynamic processes occurring in 3-D organotypic vascular models will be a valuable tool for investigating basic mechanisms in angiogenesis.
Optical coherence tomography (OCT) is a noninvasive optical imaging modality that can provide 3-D, high-resolution images of biological tissues without staining and processing.8,9 Optical coherence microscopy (OCM), which is an extension of OCT, combines the advantages of OCT and confocal microscopy using high-numerical aperture objectives to provide cellular-resolution images.10 OCM provides greater imaging depth compared with confocal microscopy. The OCM images can also be obtained using intrinsic scattering contrast and do not need fluorescent labeling or external contrast agents compared with confocal microscopy. In addition, OCM is a high-speed imaging modality that can acquire 3-D images of a specimen in seconds. OCT has been used for obtaining images of natural physiological blood vessels11 as well as engineered biological tissues.12–15 However, few studies have utilized OCM to evaluate microstructural organization and development in engineered tissues.12,16
In this study, we employ an ultrahigh-resolution OCM (UHR-OCM) system to evaluate angiogenic sprouting in a microengineered device. An algorithm based on stretching open-active contours (SOACs) was developed to track and segment the vessels in the OCM images for quantitative analysis of vessel diameter and length.17,18 The feasibility of using UHR-OCM for quantitative evaluation of morphologic features in 3-D organotypic vascular models is demonstrated. These results provide the basis for future label-free and longitudinal evaluation of microengineered devices using UHR-OCM.
2. Methods
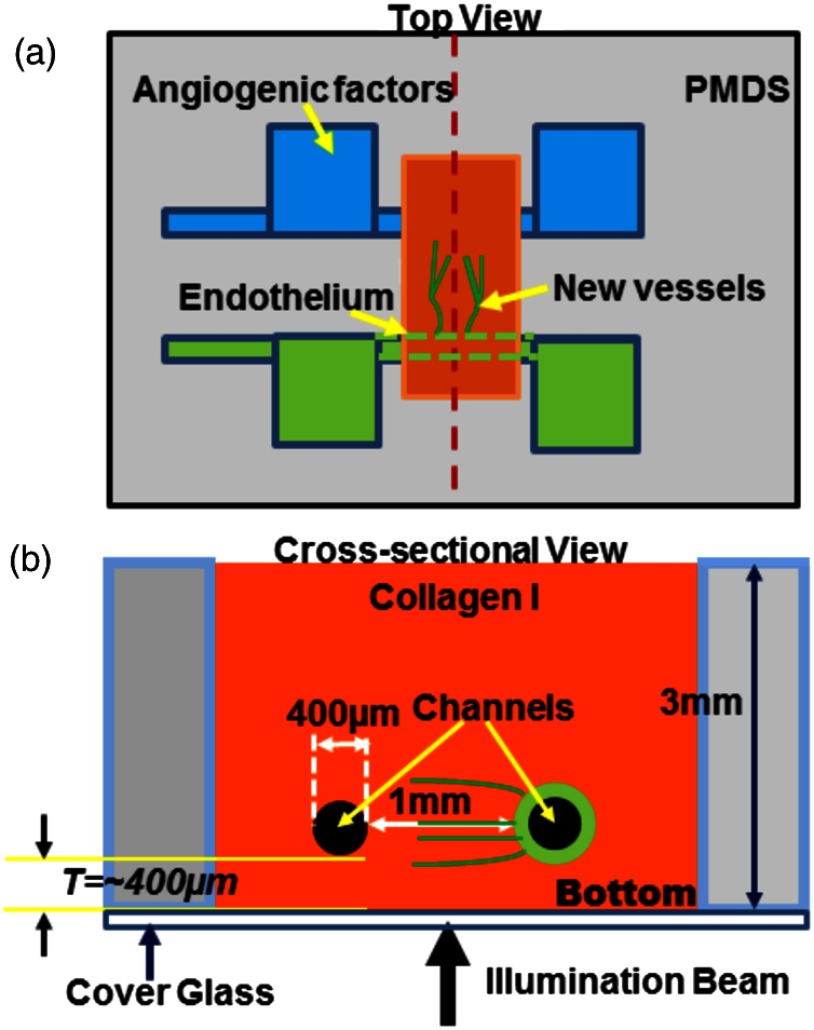
The in vitro organotypic model of angiogenic sprouting was developed by putting type-I collagen into a polydimethylsioxane mold/gasket shown in Fig. 1.3 Two needles were originally placed into the collagen matrix and extracted during the polymerization of the collagen to create two cylindrical channels. Endothelial cells were injected into one channel and adhered to form an endothelium. The other channel was filled with angiogenic factors to establish a gradient across the collagen matrix, which induced angiogenesis from the endothelium. The device was placed on a rocker to provide flow across the channels. Throughout the experiment, the device was placed in an incubator at 37°C, 5% , and 85% to 90% humidity at the University of Pennsylvania. Media and angiogenic factors in both channels were replenished daily, as previously described.3 After , the device was fixed with 3.7% formaldehyde and subsequently submerged in phosphate buffered saline before it was transported and imaged at Lehigh University with the UHR-OCM system.
Fig. 1.
Diagrams of the top (a) and cross-sectional (b) views of the microengineered device. The orange region shows the type-I collagen matrix. Sprouting vessels, shown as green lines, start from the channel lined with endothelial cells (green) toward the channel filled with angiogenic factors (blue). The two channels are marked as black circles in (b).
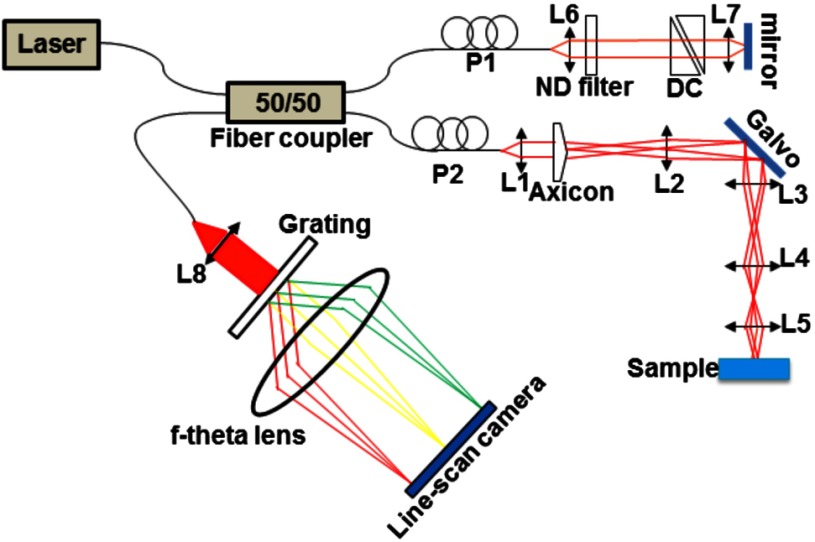
A schematic of the UHR-OCM system is shown in Fig. 2. The system was constructed with a supercontinuum light source (SC-400-4, Fianium Ltd., Southampton, UK), and a portion of the output spectrum with a center wavelength of and a spectral range of was used to provide axial resolution in tissue. The beam from the light source was split by a fiber coupler with about half of the input power directed toward the sample arm. A 175-deg conical lens19,20 was used in the sample arm to achieve an extended depth-of-field of and a transverse resolution of using a Olympus objective. The sample arm was configured as an inverted microscope, and the micro-engineered device was imaged from the bottom surface [Fig. 1(b)]. Backscattered light from the sample was interfered with the reference beam and was detected using a custom-built spectrometer, including a transmission grating (600 lpmm, Wasatch Photonics, Logan, Utah), an -theta lens (, Sill Optics, Wendelstein, Germany), and a 2048-pixel line-scan camera (AViiVA EM4, E2V Technologies Plc, Essex, UK). The line-scan camera was operated at 20,000 axial-scans/s. The sensitivity of the UHR-OCM system was measured to be with incident power on the sample. The OCM images of the region adjacent to the channel seeded with endothelial cells were acquired to map the distribution of neovessels. The data acquisition window was (). The region of interest was imaged 10 times repeatedly, and the datasets were averaged to reduce speckle noise and to improve vessel contrast in the OCM images. A segmentation algorithm based on SOACs17,18 was used to identify 3-D distribution of the vessels and locations of vessel junctions from the OCM images. The algorithm was written to track centers of the vessels in 3-D in order to extract the geometry and topology of the whole vascular network. Image acquisition and postprocessing were performed using custom-written software.
Fig. 2.
Schematic of the ultrahigh-resolution optical coherence microscopy (UHR-OCM) system. L1 to L8, lens; P1 and P2, polarization controller; DC, dispersion compensation glasses; and ND filter, neutral density filter.
3. Results
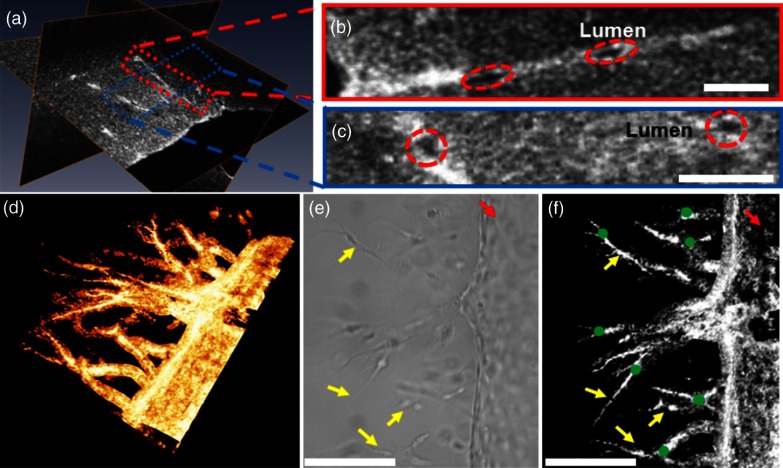
Orthogonal slices from a 3-D-OCM dataset are shown in Fig. 3(a), demonstrating angiogenic sprouting within the device. Magnified views of the regions marked in Fig. 3(a) are shown in Figs. 3(b) and 3(c), respectively. Sprouting vessels present as bright and connected lines in the OCM images. Lumens of the sprouting vessels can be clearly observed in Figs. 3(b) and 3(c). The lumens can be better appreciated from the 3-D imaging stack shown in Video 1, which corresponds to the same vessel shown in Fig. 3(b). Diameters of the highlighted lumens were measured to be to 7 μm. The segmentation algorithm was used to track the centers of the vessels and to remove the noisy background signal originated from the collagen matrix. As a result, rendering of the neovessels in 3-D is presented in Fig. 3(d) (see Video 2). All vessels sprouted from the channel seeded with endothelial cells and extended in the same direction toward the second channel filled with angiogenic factors. Figure 3(e) shows a white-light microscopy (Olympus CKX41, Tokyo, Japan) image of the sprouting vessels ( magnification). In comparison, Fig. 3(f) shows the en face maximum intensity projection from the 3-D-OCM dataset, corresponding to the same sample location as in Fig. 3(e). Vessel branches (yellow arrows) and branching points (green dots) can be identified more clearly in Fig. 3(f).
Fig. 3.
Figure 4(a) shows a cross-sectional projection of the sprouting vessels from the center region of the 3-D-OCM dataset, which cannot be obtained with traditional white-light microscopy. A vessel with second- and third-order branches [highlighted in Fig. 4(a)] is shown in Fig. 4(b). The lengths and diameters of the first-, second-, and third-order vessels were manually quantified in 3-D, and the results are compared in Figs. 4(c) and 4(d). The lengths for the first-, second-, and third-order vessels were measured to be (), (), and (), respectively [Fig. 4(c)]. The diameter of each vessel was obtained by averaging measurements obtained from several equidistant sites on that vessel. The outer diameters of the first-, second-, and third-order vessels were , , and , respectively [Fig. 4(d)]. As expected, significant decreases in vessel lengths and diameters were observed in higher order vessels compared with the first-order vessels (Student’s t-tests, ).
Fig. 4.
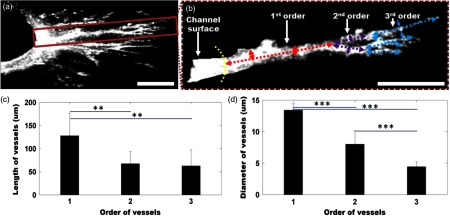
(a) Cross-sectional projection of the sprouting vessel. (b) The highlighted vessel in (a) is shown separately to illustrate the vessel orders and diameter and length measurements. The yellow dashed line marks the surface of the channel seeded with endothelial cells. The first-, second-, and third-order vessels are marked with red, purple, and blue dashed lines, respectively. (c, d) Quantitative comparison of vessel lengths and diameters for different vessel orders. Student’s t-tests were performed for statistically analysis (** and ***). Scale bar is 100 μm.
4. Discussion
In this study, we combined label-free and nondestructive UHR-OCM imaging and a segmentation algorithm based on SOACs in order to image and quantitatively characterize sprouting vessels in a microengineered device simulating the angiogenic process in biological tissues. The ultrahigh-imaging resolutions ( and for axial and transverse resolutions, respectively) provided by OCM made it possible to reveal fine structures in the vessels (e.g., lumens and branching points) and to accurately measure vessel diameters as thin as a few microns. Due to speckle noise and scattering signal originated from the background collagen matrix, the OCM image contrast was reduced. However, the segmentation algorithm successfully tracked the sprouting vessels in 3-D and enabled effective removal of background noise and signals originated from the collagen matrix. This turned out to be very useful for quantitative characterization of the vessel properties such as vessel lengths and diameters. The measured diameter for the third-order vessels is , which is smaller compared with well-developed capillaries observed in vivo. However, these third-order vessels were more likely to be vessel sprouts which were developing and not fully mature yet. Furthermore, our measurements were consistent with our previous observation from the same device model using confocal microscopy.3
Toxicity associated with sample staining is a limiting factor for confocal microscopy to perform longitudinal studies in order to monitor the dynamics of vessel growth over time.3 In comparison, OCM relies on intrinsic optical scattering contrast of the specimen and may be more suited for longitudinal imaging of a growing sample. The 3-D imaging capability of OCM also provides comprehensive information about the vessel distribution. Although the current study was performed using a fixed microengineered device, we demonstrated that OCM imaging has sufficient contrast and is capable of differentiating sprouting vessels from background collagen matrix without staining the specimen. Future experiments will be designed to monitor vessel growth over time using OCM and to quantitatively characterize sprouting speed, branching dynamics, and the influence of different angiogenic factors in microengineered devices.
5. Conclusions
In conclusion, we demonstrated UHR-OCM as a promising label-free and nondestructive imaging tool to evaluate angiogenic sprouting in microengineered devices. The algorithm based on SOACs was effective in tracking and segmenting the sprouting vessels in 3-D. Important parameters, such as vessel lengths and diameters, can be accurately quantified based on OCM images. The UHR-OCM can be used for longitudinal evaluation of microengineered devices in order to gain further insights about the mechanisms of angiogenesis.
Acknowledgments
The authors would like to thank Dr. Aneesh Alex for helpful discussion, Dr. Yevegeny Berdichevsky for providing Olympus CKX41 microscope, and Nicole Pirozzi for constructive feedback. This work was supported by the Lehigh University Start-up Fund, the National Institute of Health/National Institute of Biomedical Imaging and Bioengineering (NIH/NIBIB) Pathway to Independence Award (R00-EB010071 to C.Z.), the National Institute of Health/National Institute of General Medical Sciences (NIH/NIGMS R01GM098430) to X.H., and NIBIB R01EB00262 and R01EB08396 to C.C. D.T.N also acknowledges fellowship support from NHLBI (T32HL007954).
Biographies
Fengqiang Li received his BE degree in optoelectronics information engineering from Huazhong University of Science and Technology, Wuhan, China, in 2011. He is currently working toward his PhD degree in the Department of Electrical and Computer Engineering, Lehigh University, Bethlehem, PA. His research interest is biomedical imaging especially in optical coherence tomography and its applications in biological studies.
Ting Xu obtained his BE degree in electrical engineering in 2005, and MS degree in biomedical engineering in 2009 both from University of Science and Technology of China. He is currently a research assistant and a PhD candidate in the Department of Computer Science and Engineering at Lehigh University, where he is working on segmenting, tracking, and quantifying dynamic curvilinear networks in various kinds of biomedical imagery. His research interests include image analysis and computer vision.
Duc-Huy T. Nguyen received his BS degree in chemical engineering with honors from California Institute of Technology, in 2009. He is currently pursuing his PhD degree in chemical engineering at University of Pennsylvania. He is also affiliated with Boston University and the Wyss Institute for Biologically Inspired Engineering at Harvard University.
Xiaolei Huang earned her doctorate and master’s degrees in computer science from Rutgers University and her bachelor’s in computer science from Tsinghua University, China. She is currently an associate professor in the Computer Science and Engineering Department at Lehigh University, Bethlehem, PA. Her research involves the interfaces among medical imaging processing, computer vision, and machine learning. She has presented nationally and internationally and published several book chapters and numerous journal and conference proceeding articles.
Christopher S. Chen is a professor of biomedical engineering and director of the Tissue Microfabrication Laboratory at Boston University. He was previously the Skirkanich Professor of Innovation and founding director of the Center for Engineering Cells and Regeneration at the University of Pennsylvania. His group strives to identify the underlying mechanisms by which cells interact with materials and each other to build tissues and to apply this knowledge in the biology of stem cells, tissue vascularization, and cancer.
Chao Zhou obtained his MSc and PhD degrees in physics from the University of Pennsylvania and his BS degree in physics from Peking University, China. He is currently an assistant professor of electrical engineering and bioengineering at Lehigh University. He has published numerous journal articles and is a member of the International Society for Optical Engineering. His research group focuses on developing advanced optical imaging technologies and utilizing these technologies for various biomedical applications.
References
- 1.Carmeliet P., “Angiogenesis in health and disease,” Nat. Med. 9(6), 653–660 (2003). 10.1038/nm0603-653 [DOI] [PubMed] [Google Scholar]
- 2.Folkman J., “Angiogenesis in cancer, vascular, rheumatoid and other disease,” Nat. Med. 1(1), 27–31 (1995). 10.1038/nm0195-27 [DOI] [PubMed] [Google Scholar]
- 3.Nguyen D. H. T., et al. , “Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro,” Proc. Natl. Acad. Sci. U. S. A. 110(17), 6712–6717 (2013). 10.1073/pnas.1221526110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staton C. A., Reed M. W. R., Brown N. J., “A critical analysis of current in vitro and in vivo angiogenesis assays,” Int. J. Exp. Pathol. 90(3), 195–221 (2009). 10.1111/j.1365-2613.2008.00633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y., et al. , “In vitro microvessels for the study of angiogenesis and thrombosis,” Proc. Natl. Acad. Sci. U. S. A. 109(24), 9342–9347 (2012). 10.1073/pnas.1201240109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller J. S., et al. , “Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues,” Nat. Mater. 11(9), 768–774 (2012). 10.1038/nmat3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selbonne S., et al. , “In vitro and in vivo antiangiogenic properties of the serpin protease nexin-1,” Mol. Cell Biol. 32(8), 1496–1505 (2012). 10.1128/MCB.06554-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang D., et al. , “Optical coherence tomography,” Science 254(5035), 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drexler W., et al. , “In vivo ultrahigh-resolution optical coherence tomography,” Opt. Lett. 24(17), 1221–1223 (1999). 10.1364/OL.24.001221 [DOI] [PubMed] [Google Scholar]
- 10.Izatt J. A., et al. , “Optical coherence microscopy in scattering media,” Opt. Lett. 19(8), 590–592 (1994). 10.1364/OL.19.000590 [DOI] [PubMed] [Google Scholar]
- 11.Makita S., et al. , “Optical coherence angiography,” Opt. Express 14(17), 7821–7840 (2006). 10.1364/OE.14.007821 [DOI] [PubMed] [Google Scholar]
- 12.Yang Y., et al. , “Investigation of optical coherence tomography as an imaging modality in tissue engineering,” Phys. Med. Biol. 51(7), 1649–1659 (2006). 10.1088/0031-9155/51/7/001 [DOI] [PubMed] [Google Scholar]
- 13.Bagnaninchi P. O., et al. , “Chitosan microchannel scaffolds for tendon tissue engineering characterized using optical coherence tomography,” Tissue Eng. 13(2), 323–331 (2007). 10.1089/ten.2006.0168 [DOI] [PubMed] [Google Scholar]
- 14.Ko H. J., et al. , “Optical coherence elastography of engineered and developing tissue,” Tissue Eng. 12(1), 63–73 (2006). 10.1089/ten.2006.12.63 [DOI] [PubMed] [Google Scholar]
- 15.Liang X., Graf B. W., Boppart S. A., “Imaging engineered tissues using structural and functional optical coherence tomography,” J. Biophotonics 2(11), 643–655 (2009). 10.1002/jbio.v2:11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y. B., et al. , “Integrated multimodal optical microscopy for structural and functional imaging of engineered and natural skin,” J. Biophotonics 5(5–6), 437–448 (2012). 10.1002/jbio.v5.5/6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu T., et al. , “Delineating 3D angiogenic sprouting in OCT images via multiple active contours,” in Augmented Reality Environments for Medical Imaging and Computer-Assisted Interventions, Liao H., Linte C., Masamune K., Peters T., Zheng G., Eds., pp. 231–240, Springer, Berlin Heidelberg: (2013). [Google Scholar]
- 18.Xu T., Vavylonis D., Huang X., “3D actin network centerline extraction with multiple active contours,” Med. Image Anal. 18(2), 272–284 (2014). 10.1016/j.media.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leitgeb R. A., et al. , “Extended focus depth for Fourier domain optical coherence microscopy,” Opt. Lett. 31(16), 2450–2452 (2006). 10.1364/OL.31.002450 [DOI] [PubMed] [Google Scholar]
- 20.Bolmont T., et al. , “Label-free imaging of cerebral beta-amyloidosis with extended-focus optical coherence microscopy,” J. Neurosci. 32(42), 14548–14556 (2012). 10.1523/JNEUROSCI.0925-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.