Abstract
Background:
The CD1 family is less variable transmembrane antigen presenting molecules related to the MHC molecules. CD1a and CD1e genes are the most polymorphic ones associated with autoimmune diseases. The aim was to better clarify the map of CD1 genes in Southwest Iranian normal population for implications in vaccine design.
Methods:
In this study we investigated the polymorphism of CD1a, CD1d and CD1e in 311 healthy individuals from Fars Province in Southwest of Iran by PCR-SSP method.
Results:
Six of individuals had homozygote CD1a 01/01 genotype and 248 had homozygote CD1a 02/02 genotype. CD1d was found to be monomorphic with all tested individuals showing CD1d 01/01 genotype. Hundred and eleven individuals had homozygote CD1e 01/01 genotype and 48 had homozygote CD1e 02/02 genotype. The frequencies of CD1a 01 and CD1a 02 alleles were 11% and 89% while the frequencies of CD1e 01 and CD1e 02 alleles were 60.1% and 39.9%, respectively. Consistent with previous reports on other genes, a high degree of similarity in CD1a and CD1e allelic distribution was observed between Southwest Iranians and other Indo-European populations. However, the allelic frequency of the CD1a and CD1e alleles showed a significant difference from those of Chinese Han and She populations.
Conclusion:
These data are notable in the light of relatively recent genetic admixture along the Silk Road. Considering the significance of CD1 alleles in some autoimmune and infectious diseases and with the admixed nature of Iranian population, mapping the distribution of CD1e alleles in different regions of Iran can be useful in future designing of preventive and therapeutic vaccines.
Keywords: CD1, Normal population, PCR-SSP, Iran
Introduction
CD1 genes, located on human chromosome 1, comprise a less polymorphic lineage of antigen presenting molecules related to MHC class I glycoproteins (1, 2). The genes encoding the five isoforms of CD1 molecule, namely CD1a, CD1b, CD1c, CD1d and CD1e, are believed to have emerged as a result of MHC genes duplication before the divergence of birds and mammals in some 300 million years ago (3–5). Further translocation of the CD1 genes in mammals around 180 million years ago seems to have resulted in the translocation of CD1 locus from its ancestral position in the MHC locus on chromosome 6 (6–8). CD1 genes continued to expand and diverge among eutherians by virtue of duplication and diversification. This idea is supported by different types and copies of CD1 isoforms with a considerable degree of sequence homology in higher mammalian species such as rat, mice, cattle and human (9–11).
The functional importance of CD1 isoforms and their relative contribution to the immune response has been under investigation during the past two decades (12–17). While CD1 molecules are all considered as self and non-self lipid antigen presenting molecules, it is now more evident that CD1d and CD1c are directly involved in presentation of pathogens' lipids to NKT and T cells (2, 18, 19). Therefore, this ancient locus which presents limited variation could have been under some degree of selection pressure by infectious pathogens. CD1d, the most conserved molecule of all CD1 isoforms, has been saved from deletions that affected other CD1 family members in rat and mice (6, 9, 15). Limited allelic variation exists in CD1d gene compared to CD1a with two known polymorphisms at positions 13 and 51 and CD1e with 6 known alleles (20–23). The limited diversity of CD1 genes, even for the most polymorphic gene, i.e. CD1e, points to the critical role of these lipid presenting molecules in the defense against pathogens. In this regard, accumulating evidence imply the possible role of CD1-restricted T cells in infectious, inflammatory, autoimmune, and malignant disorders (2). Accordingly, the role of CD1 molecules in pathogenesis of multiple sclerosis (MS), rheumatoid arthritis (RA), atherosclerosis as well as human malignancies has been reported (2, 24–27). In addition, while an association between CD1a and CD1e alleles with multiple sclerosis is suggested (28), the association of these CD1 alleles with other neuropathies remains controversial (29–31). Despite the considerable conservation of CD1 genes, a high degree of difference in the distribution of CD1 alleles in different populations is reported (28, 32–34). In a previous study we did not observe any difference between CD1a and CD1d alleles in cancer patients compared to their healthy sex-matched controls (34). In our data, we found a dominant frequency of CD1D*01 allele similar to other populations but a somewhat different frequency of CD1a alleles from South-western part of Iran compared to the other Asian populations.
In a curious attempt to better clarify the map of CD1 genes in our population, we studied the frequency of CD1a, CD1d and CD1e alleles in a group of randomly recruited healthy blood donors.
Materials and Methods
Subjects
In a cross-sectional study from 2008–2010, 311 healthy blood donors including 194 men and 117 women (mean age 46 ± 10; range= 17–65 years) from Fars Province in Southwest of Iran were chosen randomly. All of these individuals had no history of autoimmune diseases, diabetes, malignancies or any familial diseases among their relatives.
DNA Extraction
Approximately 10 ml of fresh PBMCs were taken in EDTA 1%. Genomic DNA was extracted using salting out method (35). DNA concentration was measured by specterophotometeric analysis and optimal DNA concentration (0.3 ng/ml) was prepared for PCR reaction.
PCR Reaction
Using PCR-SSP method, three SNPs in CD1a (622 T/C), CD1d (354 A/T) and CD1e (6129 A/G) genes were amplified. For each sample, the PCR reaction was carried out in two separate tubes each containing one specific primer and a common primer which was common in both tubes. The primer sequences for each gene are depicted in Table 1. Two forward and reverse primers of β-globin gene were used as the internal control.
Table 1:
The primer sequences used for amplification of CD1a, d, e and β-globin genes
| Genes | Primer sequences |
|---|---|
| CD1A (T622C) | F Primer: CcTcTcTccTTccATGTcAt |
| R Primer: CcTCTcTccTTccATGTcAc | |
| Common primer: TTCAAACTGCAATTCATGGGC | |
| CD1D (A354T) | F Primer: GCTTCAGAGAGCGGACGGA |
| R Primer: GCTTCAGAGAGCGGACGGT | |
| Common primer: TGAAGTCCCGCAAAGGCTTT | |
| CD1E (A6129G) | F Primer: CCAGTTATACTTCCTAGTTTTATCCA |
| R Primer: CCAGTTATACTTCCTAGTTTTATCCG | |
| Common primer: GTGTATATGGTGGAGTGTGGG | |
| β-glubin | F primer: ACACAACTGTGTTCACTAGC |
| R primer: CAACTTCATCCACGT TCA CC |
PCR Condition
The PCR reaction was performed in a total volume of 25 μl including 13.5 μL DW, 2.5 μl of PCR buffer 10X, 0.75 μl MgCl2 (50 mM), 0.75 μl dNTP (10 mM), 1 μl of each primer (20 pm), 1 μL of each internal control primers, 1 μl of genomic DNA (0.3 ng/ml), 0.5 μl Tween-20 and 2 μl Taq polymerase (1U). The PCR program was a touch-down method and included an initial denaturation at 94°C for 3 min followed by three loops; loop one included 5 cycles of 94°C for 25 sec, 70°C for 60 sec and 72°C for 60 sec, loop 2 included 21 cycles of 94°C for 25 sec, 64°C for 70 sec and 72 °C for 70 sec and loop 3 consisted 94°C for 25 sec, 55°C for 60 sec and 72°C for 120 sec and a final extension at 72°C for 5 min for CD1a gene and an initial denaturation at 94°C for 6 min followed by three loops; loop one included a 5 cycles of 94°C for 30 sec, 70°C for 90 sec and 72°C for 60 sec, loop 2 included 26 cycles of 94°C for 30 sec, 63.5°C for 35 sec and 72°C for 60 sec and loop 3 consisted of 94°C for 30 sec, 55°C for 50 sec and 72°C for 60 sec and a final extension at 72°C for 5 min for CD1d and CD1e genes. Then, 8 μl of each PCR product was run in ethidium bromide-stained agarose gel and visualized under UV light.
Statistical Analysis
Data were analyzed by SPSS 11.5 and Epi-info 2002 software. The Hardy-Weinberg equilibrium was checked using chi-square t-test. P-values less than 0.05 were considered significant.
Results
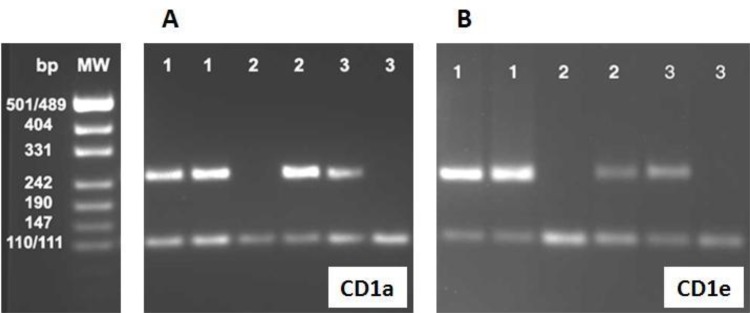
The electrophoresis results of the PCR products of CD1a (255 bp) and CD1e (241 bp) are shown in Fig. 1. The frequency of CD1a 01 and CD1a 02 alleles were 11% and 89% while the frequency of CD1e 01 and CD1e 02 alleles were 60.1% and 39.9%, respectively. Table 2 illustrates the genotype and allele frequencies of CD1a, CD1d and CD1e genes in the current study.
Fig. 1:
Gel electrophoresis of PCR products of CD1a and CD1e alleles. A. CD1a alleles amplification: The result of amplification for Isoleucine/Threonine (CT) heterozygote (lanes 1 and 1), Threonine (37) homozygote (lanes 2 and 2) genotypes and Isoleucine (TT) homozygote (lanes 3 and 3) are indicated. The reactions in both lanes 1 (Isoleucine/Threonine) indicate that the first individual is heterozygote, while lack of reaction in the first lanes 2 (Isoleucine) and the existence of PCR-amplified Threonine band in the second lane indicate that individual 2 is Threonine homozygote. The band in the first lane 3 (Isoleucine) and lack of reaction in the second lane 3 indicates that individual 3 is Isoleucine homozygote. B. CD1e alleles amplification: The result of amplification for Glutamine/Arginine heterozygote (lanes 1 and 1), Glutamine (52) homozygote (lanes 2 and 2) genotypes and Arginine (GG) homozygote (lanes 3 and 3) are indicated. The reactions in both lanes 1 (Glutamine/Arginine) indicate that the first individual is heterozygote, while lack of reaction in the first lanes 2 (Arginine) and the existence of PCR-amplified Glutamine band in the second lane indicate that individual 2 is Glutamine homozygote. The band in the first lane 3 (Arginine) and lack of reaction in the second lane 3 indicates that individual 3 is Arginine homozygote.
Table 2:
Genotype and allele frequency of CD1a, d and e genes in Iranian normal population
| Genes | Genotypes | Frequency | Alleles | Frequency |
|---|---|---|---|---|
| CD1a | 01/01 | 6 (1.9%) | 01 | 69 (11%) |
| 01/02 | 57 (18.3%) | 02 | 553 (89 %) | |
| 02/02 | 248 (79.8%) | |||
| CD1d | 01/01 | 311 (100%) | 01 | 622 (100%) |
| 01/02 | 0 (0%) | 02 | 0 (0%) | |
| 02/02 | 0 (0%) | |||
| CD1e | 01/01 | 111 (35.7%) | 01 | 374 (60.1%) |
| 01/02 | 152 (48.9%) | 02 | 248 (39.9%) | |
| 02/02 | 48 (15.4%) | |||
| Total | 311 (100%) |
A comparison of the CD1 allele frequencies between different populations is shown in Table 3. As indicated, the frequencies of CD1a and CD1e alleles were significantly different from that of Chinese Han (P=0.002) and Italian Abbruzzo (P=0.03) populations (33, 37). It is worth mentioning that all the selected individuals were in Hardy-Weinberg equilibrium for the three polymorphisms.
Table 3:
Comparison of CD1a, d and e allele frequency in different populations
| Alleles | Frequency in our population (n=311) | Italian population (n=100)(36) | Italian population (n=132)(30) | North American diverse ethnic group (n=110)(20) | Chinese Han (n=160) and She (n=260) population (51) | Dutch population (n=212)(31) | UK population (n=342)(38) | Italian Abbruzzo population (n=114)(37) |
|---|---|---|---|---|---|---|---|---|
| CD1A01 | 11% | 11% | 11% | 13% | 0% | 6.4% | 5% | 1% |
| CD1A02 | 89% | 89% | 89% | 87% | 100% | 93.6% | 95% | 99% |
| P-value | 1 | 1 | 0.6 | - | 0.2 | 0.1 | *0.002 | |
| CD1D01 | 100% | 100% | - | 99% | 100% | - | 100% | - |
| CD1D02 | 0% | 0% | - | 1% | 0% | - | 0% | - |
| P-value | ||||||||
| CD1E01 | 60.1% | 61% | 62% | 49% | 38% | 66.5% | 67% | 74% |
| CD1E02 | 39.9% | 39% | 38% | 51% | 62% | 33.5% | 33% | 26% |
| P-value | 0.9 | 0.8 | 0.1 | *0.002 | 0.3 | 0.3 | *0.03 |
Discussion
In present study, we investigated the polymorphisms in CD1a, CD1d and CD1e genes in 311 healthy individuals from Southwest of Iran. The results on CD1a and CD1d were very similar to the frequencies we obtained from another group of healthy individuals who were recruited and selected based on specific criteria in a previous study (34). Investigation of CD1e gene polymorphism in the current study revealed that the allelic frequency of these genes in our population was similar to some Italian, Dutch and British normal populations but far from that of Chinese Han population (P= 0.002, Table 3) (20, 30, 31, 33, 36, 37). The frequency of CD1a alleles in our population and in some Italian, Dutch and British populations were also found to be similar (Table 3) (31, 37, 38). Thus, it seems that in addition to CD1a, the CD1e alleles are also distributed very closely in our population and Italian population except Italian Abbruzzo and Chinese Han and She populations (Table 3) (37). These results are in accordance with the previous studies which showed a high degree of genetic relation between South-western Iranians and Italian population (39–41). Our findings also indicate that the CD1a and CD1e gene polymorphisms in exon 2 are largely different among various ethnic groups in Asia.
Current Iranian population is living in one of the oldest areas inhabited by modern humans (Homo sapiens sapiens). Despite the various ethnic groups and multiple languages spoken in Iran, investigation of the genetic structure of Iranian population is quite recent. The general consensus is that the Aryan tribes who first inhabited the South-western of Iran made a huge contribution to the genetic pool of current Iranians (42). However, a degree of genetic admixture has happened over the centuries of population movements and migrations from eastern and western neighboring lands (43). As such, the war and trade-related population movements along the Silk Road may have increased the diversity of the genetic pool in Iran (44–46). Previous studies have shown that despite this relatively recent genetic admixture, the footprints of Indo-Aryan alleles and mitochondrial DNA markers are still well preserved among Iranians (39, 40, 42). The extent of this preservation, however, varies in different areas of Iran owing to the extent of admixture with different populations and also inside-ethnicity breeding (42, 47).
It is suggested that the extent of genetic admixture can be studied by alleles that show a high frequency difference in different populations (48, 49). The high level of difference in allelic frequency allows scientists to estimate the extent of contribution of ancestral alleles in the genetic pool of an admixed population. This is especially important in the view of the associations between some alleles and diseases. Two recent reports with relatively large sample sizes from Iran and Switzerland point to the association of CD1a and CD1e alleles with susceptibility to multiple sclerosis (MS) (28, 50). Therefore, considering the relatively high difference in the frequency of CD1e and CD1a in our population and those from eastern Asian populations, investigation of other predisposing genes and environmental factors in Iranian and other Asian MS patients could be interesting (28, 33, 34). Moreover, given the importance of CD1 alleles in autoimmune diseases and neuropathies, tracing CD1a and CD1e alleles in admixed populations can be used in preventive and therapeutic planning, especially in designing efficient lipid and glycolipid vaccines. In this regard, the polymorphism of CD1 gene in other ethnic groups along with the functional consequences of such genetic variation should also be taken into account.
Ethical considerations
Ethical issues (Including plagiarism, Informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc) have been completely observed by the authors.
Acknowledgments
This work was financially supported by a grant (no. 1752) from Shiraz University of Medical Sciences and was performed in Shiraz Institute for Cancer Research. The authors declare that there is no conflict of interests.
References
- 1.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929–41. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 2.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–90. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 3.Salomonsen J, Sorensen MR, Marston DA, Rogers SL, Collen T, van Hateren A, Smith AL, Beal RK, Skjodt K, Kaufman J. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc Natl Acad Sci U S A. 2005;102:8668–73. doi: 10.1073/pnas.0409213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maruoka T, Tanabe H, Chiba M, Kasahara M. Chicken CD1 genes are located in the MHC: CD1 and endothelial protein C receptor genes constitute a distinct subfamily of class-I-like genes that predates the emergence of mammals. Immunogenetics. 2005;57:590–600. doi: 10.1007/s00251-005-0016-y. [DOI] [PubMed] [Google Scholar]
- 5.Miller MM, Wang C, Parisini E, Coletta RD, Goto RM, Lee SY, Barral DC, Townes M, Roura-Mir C, Ford HL, Brenner MB, Dascher CC. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc Natl Acad Sci U S A. 2005;102:8674–9. doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker ML, Miller RD. Evolution of mammalian CD1: marsupial CD1 is not orthologous to the eutherian isoforms and is a pseudogene in the opossum Monodelphis domestica. Immunology. 2007;121:113–21. doi: 10.1111/j.1365-2567.2007.02545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belov K, Harrison GA, Miller RD, Cooper DW. Molecular cloning of four lambda light chain cDNAs from the Australian brushtail possum Trichosurus vulpecula. Eur J Immunogenet. 2002;29:95–9. doi: 10.1046/j.1365-2370.2002.00286.x. [DOI] [PubMed] [Google Scholar]
- 8.Belov K, Deakin JE, Papenfuss AT, Baker ML, Melman SD, Siddle HV, Gouin N, Goode DL, Sargeant TJ, Robinson MD, Wakefield MJ, Mahony S, Cross JG, Benos PV, Samollow PB, Speed TP, Graves JA, Miller RD. Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex. PLoS Biol. 2006;4:e46. doi: 10.1371/journal.pbio.0040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 2003;24:412–8. doi: 10.1016/s1471-4906(03)00179-0. [DOI] [PubMed] [Google Scholar]
- 10.Ichimiya S, Kikuchi K, Matsuura A. Structural analysis of the rat homologue of CD1. Evidence for evolutionary conservation of the CD1D class and widespread transcription by rat cells. J Immunol. 1994;153:1112–23. [PubMed] [Google Scholar]
- 11.Bradbury A, Belt KT, Neri TM, Milstein C, Calabi F. Mouse CD1 is distinct from and co-exists with TL in the same thymus. Embo J. 1988;7:3081–6. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin LH, Calabi F, Lefebvre FA, Bilsland CA, Milstein C. Structure and expression of the human thymocyte antigens CD1a, CD1b, and CD1c. Proc Natl Acad Sci U S A. 1987;84:9189–93. doi: 10.1073/pnas.84.24.9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amiot M, Dastot H, Schmid M, Bernard A, Boumsell L. Analysis of CD1 molecules on thymus cells and leukemic T lymphoblasts identifies discrete phenotypes and reveals that CD1 intermolecular complexes are observed only on normal cells. Blood. 1987;70:676–85. [PubMed] [Google Scholar]
- 14.Cheroutre H, Holcombe HR, Tangri S, Castano AR, Teitell M, Miller JE, Cardell S, Benoist C, Mathis D, Huse WD, et al. Antigen-presenting function of the TL antigen and mouse CD1 molecules. Immunol Rev. 1995;147:31–52. doi: 10.1111/j.1600-065x.1995.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 15.Brossay L, Kronenberg M. Highly conserved antigen-presenting function of CD1d molecules. Immunogenetics. 1999;50:146–51. doi: 10.1007/s002510050590. [DOI] [PubMed] [Google Scholar]
- 16.Laskarin G, Redzovic A, Rubesa Z, Mantovani A, Allavena P, Haller H, Vlastelic I, Rukavina D. Decidual natural killer cell tuning by autologous dendritic cells. Am J Reprod Immunol. 2008;59:433–45. doi: 10.1111/j.1600-0897.2008.00599.x. [DOI] [PubMed] [Google Scholar]
- 17.Kasmar A, Van Rhijn I, Moody DB. The evolved functions of CD1 during infection. Curr Opin Immunol. 2009;21:397–403. doi: 10.1016/j.coi.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol. 1997;15:535–62. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 19.Porcelli SA. Bird genes give new insights into the origins of lipid antigen presentation. Proc Natl Acad Sci U S A. 2005;102:8399–400. doi: 10.1073/pnas.0503313102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han M, Hannick LI, DiBrino M, Robinson MA. Polymorphism of human CD1 genes. Tissue Antigens. 1999;54:122–7. doi: 10.1034/j.1399-0039.1999.540202.x. [DOI] [PubMed] [Google Scholar]
- 21.Oteo M, Parra JF, Mirones I, Gimenez LI, Setien F, Martinez-Naves E. Single strand conformational polymorphism analysis of human CD1 genes in different ethnic groups. Tissue Antigens. 1999;53:545–50. doi: 10.1034/j.1399-0039.1999.530604.x. [DOI] [PubMed] [Google Scholar]
- 22.Mirones I, Oteo M, Parra-Cuadrado JF, Martinez-Naves E. Identification of two novel human CD1E alleles. Tissue Antigens. 2000;56:159–61. doi: 10.1034/j.1399-0039.2000.560208.x. [DOI] [PubMed] [Google Scholar]
- 23.Tamouza R, Sghiri R, Ramasawmy R, Neonato MG, Mombo LE, Poirier JC, Schaeffer V, Fortier C, Labie D, Girot R, Toubert A, Krishnamoorthy R, Charron D. Two novel CD1 E alleles identified in black African individuals. Tissue Antigens. 2002;59:417–20. doi: 10.1034/j.1399-0039.2002.590509.x. [DOI] [PubMed] [Google Scholar]
- 24.Bell D, Chomarat P, Broyles D, Netto G, Harb GM, Lebecque S, Valladeau J, Davoust J, Palucka KA, Banchereau J. In breast carcinoma tissue, immature dendritic cells reside within the tumor, whereas mature dendritic cells are located in peritumoral areas. J Exp Med. 1999;190:1417–26. doi: 10.1084/jem.190.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coventry BJ. CD1a positive putative tumour infiltrating dendritic cells in human breast cancer. Anticancer Res. 1999;19:3183–7. [PubMed] [Google Scholar]
- 26.Iwamoto M, Shinohara H, Miyamoto A, Okuzawa M, Mabuchi H, Nohara T, Gon G, Toyoda M, Tanigawa N. Prognostic value of tumor-infiltrating dendritic cells expressing CD83 in human breast carcinomas. Int J Cancer. 2003;104:92–7. doi: 10.1002/ijc.10915. [DOI] [PubMed] [Google Scholar]
- 27.Melian A, Geng YJ, Sukhova GK, Libby P, Porcelli SA. CD1 expression in human atherosclerosis. A potential mechanism for T cell activation by foam cells. Am J Pathol. 1999;155:775–86. doi: 10.1016/S0002-9440(10)65176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamshidian A, Nikseresht AR, Vessal M, Kamali-Sarvestani E. Association of CD1A +622 T/C, +737 G/C and CD1E +6129 A/G genes polymorphisms with multiple sclerosis. Immunol Invest. 2010;39:874–89. doi: 10.3109/08820139.2010.503768. [DOI] [PubMed] [Google Scholar]
- 29.Uncini A, Notturno F, Pace M, Caporale CM. Polymorphism of CD1 and SH2D2A genes in inflammatory neuropathies. J Peripher Nerv Syst. 2011;16(Suppl 1):48–51. doi: 10.1111/j.1529-8027.2011.00307.x. [DOI] [PubMed] [Google Scholar]
- 30.De Angelis MV, Notturno F, Caporale CM, Pace M, Uncini A. Polymorphisms of CD1 genes in chronic dysimmune neuropathies. J Neuroimmunol. 2007;186:161–3. doi: 10.1016/j.jneuroim.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Kuijf ML, Geleijns K, Ennaji N, van Rijs W, van Doorn PA, Jacobs BC. Susceptibility to Guillain-Barre syndrome is not associated with CD1A and CD1E gene polymorphisms. J Neuroimmunol. 2008;205:110–2. doi: 10.1016/j.jneuroim.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Oteo M, Arribas P, Setien F, Parra JF, Mirones I, Gomez del Moral M, Martinez-Naves E. Structural characterization of two CD1A allelic variants. Hum Immunol. 2001;62:1137–41. doi: 10.1016/s0198-8859(01)00314-7. [DOI] [PubMed] [Google Scholar]
- 33.Gan LH, Pan YQ, Xu DP, Li M, Lin A, Yan WH. Polymorphism of human CD1a, CD1d, and CD1e in exon 2 in Chinese Han and She ethnic populations. Tissue Antigens. 2010;75:691–5. doi: 10.1111/j.1399-0039.2010.01443.x. [DOI] [PubMed] [Google Scholar]
- 34.Golmoghaddam H, Pezeshki AM, Ghaderi A, Doroudchi M. CD1a and CD1d genes polymorphisms in breast, colorectal and lung cancers. Pathol Oncol Res. 2011;17:669–75. doi: 10.1007/s12253-011-9367-x. [DOI] [PubMed] [Google Scholar]
- 35.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic acids research. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporale CM, Papola F, Fioroni MA, Aureli A, Giovannini A, Notturno F, Adorno D, Caporale V, Uncini A. Susceptibility to Guillain-Barre syndrome is associated to polymorphisms of CD1 genes. J Neuroimmunol. 2006;177:112–8. doi: 10.1016/j.jneuroim.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Aureli A, Fontecchio G, Altobelli E, Azzarone R, Del Beato T, Fioroni MA, Caporale CM, Adorno D, Papola F. CD1a and CD1e allele frequencies in an Italian population from the Abruzzo region. International journal of immunopathology and pharmacology. 2007;20:415–9. doi: 10.1177/039463200702000225. [DOI] [PubMed] [Google Scholar]
- 38.Jones DC, Gelder CM, Ahmad T, Campbell IA, Barnardo MC, Welsh KI, Marshall SE, Bunce M. CD1 genotyping of patients with Mycobacterium malmoense pulmonary disease. Tissue antigens. 2001;58:19–23. doi: 10.1034/j.1399-0039.2001.580103.x. [DOI] [PubMed] [Google Scholar]
- 39.Amirzargar A, Mytilineos J, Farjadian S, Doroudchi M, Scherer S, Opelz G, Ghaderi A. Human leukocyte antigen class II allele frequencies and haplotype association in Iranian normal population. Hum Immunol. 2001;62:1234–8. doi: 10.1016/s0198-8859(01)00320-2. [DOI] [PubMed] [Google Scholar]
- 40.Senemar S, Doroudchi M, Pezeshki AM, Bazrgar M, Torab-Jahromi A, Ghaderi A. Frequency of cystathionine beta-synthase 844INS68 polymorphism in Southern Iran. Mol Biol Rep. 2009;36:353–6. doi: 10.1007/s11033-007-9186-z. [DOI] [PubMed] [Google Scholar]
- 41.Cavalli-Sforza L, Menozzi P, Piazza A. In: The History and Geography of Human Genes. Princeton University Press, editor. 41 William Street, Princeton, New Jersey: 1994. [Google Scholar]
- 42.Farjadian S, Sazzini M, Tofanelli S, Castri L, Taglioli L, Pettener D, Ghaderi A, Romeo G, Luiselli D. Discordant patterns of mtDNA and ethno-linguistic variation in 14 Iranian Ethnic groups. Hum Hered. 2011;72:73–84. doi: 10.1159/000330166. [DOI] [PubMed] [Google Scholar]
- 43.Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A, Ayub Q, Mohyuddin A, Tyler-Smith C, Qasim Mehdi S, Torroni A, McElreavey K. Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet. 2004;74:827–45. doi: 10.1086/383236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao TM, Lee TD. Gm and Km allotypes in 74 Chinese populations: a hypothesis of the origin of the Chinese nation. Hum Genet. 1989;83:101–10. doi: 10.1007/BF00286699. [DOI] [PubMed] [Google Scholar]
- 45.Yao YG, Lu XM, Luo HR, Li WH, Zhang YP. Gene admixture in the silk road region of China: evidence from mtDNA and melanocortin 1 receptor polymorphism. Genes Genet Syst. 2000;75:173–8. doi: 10.1266/ggs.75.173. [DOI] [PubMed] [Google Scholar]
- 46.Yao YG, Kong QP, Wang CY, Zhu CL, Zhang YP. Different matrilineal contributions to genetic structure of ethnic groups in the silk road region in china. Mol Biol Evol. 2004;21:2265–80. doi: 10.1093/molbev/msh238. [DOI] [PubMed] [Google Scholar]
- 47.Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariev A, Bodmer WF. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci U S A. 2001;98:10244–9. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shriver MD, Smith MW, Jin L, Marcini A, Akey JM, Deka R, Ferrell RE. Ethnic-affiliation estimation by use of population-specific DNA markers. Am J Hum Genet. 1997;60:957–64. [PMC free article] [PubMed] [Google Scholar]
- 49.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporale CM, Notturno F, Pace M, Aureli A, Di Tommaso V, De Luca G, Farina D, Giovannini A, Uncini A. CD1A and CD1E gene polymorphisms are associated with susceptibility to multiple sclerosis. Int J Immunopathol Pharmacol. 2011;24:175–83. doi: 10.1177/039463201102400120. [DOI] [PubMed] [Google Scholar]