Abstract
Objective
This study aimed to investigate the expression of the MSH2 DNA repair protein in head and neck squamous cell carcinoma (HNSCC) in order to analyze its association with clinicopathologic factors and overall survival of patients.
Material and Methods
Clinical data and primary lesions of HNSSC were collected from 55 patients who underwent surgical resection with postoperative radiotherapy in Montes Claros, state of Minas Gerais, Brazil, between 2000 and 2008. Immunohistochemical reactions were performed to analyze MSH2 protein expression.
Results
Bivariate analysis showed no significant correlation or association between MSH2 expression and clinicopathologic parameters by Mann-Whitney and Kruskal-Wallis tests. Patients with locoregional metastatic disease (OR=4.949, p<0.001) and lower MSH2 immunohistochemical expressions (OR=2.943, p=0.032) presented poorer survival for HNSCC by Cox regression models.
Conclusions
Our data demonstrated that lower MSH2 expression might contribute to a higher clinic aggressiveness of HNSCC by promoting an unfavorable outcome.
Keywords: Squamous cell carcinoma, DNA repair, MutS homolog 2 protein, Immunohistochemistry, Prognosis
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the most prevalent malignant neoplasm arising from the epithelial lining of the upper aerodigestive tract mucosa. It represents the 6th most common type of human cancer, and it is responsible for high death rates worldwide every year4,10. A plethora of sociodemographic, economic, and cultural factors are associated with the development of HNSCC, along with a background of molecular disturbances, both genetic and epigenetic, that influence the course and incidence of the disease1,8.
Mismatch repair (MMR) is responsible for mismatch base substitution and insertion-deletion mismatches generated during DNA replication. MMR also recognizes DNA damage formed by chemical compounds, such as alkylating agents. Inactivation of MMR genes and proteins can have profound biological consequences on cells such as an increased tendency of mutations in genes associated with maintenance and replication of genome14,20. A crucial part of this system is the MutS-homologue 2 (MSH2) gene, which is located at chromosome 2p22. MSH2 protein recognizes DNA mismatches by forming two functional heterodimers: MSH2-MSH6, and MSH2-MSH3. MSH2-MSH6 recognizes single-base mismatches and short insertion-deletion loops, and MSH2-MSH3 which recognizes larger loops that occur during DNA replication6,22.
Disorders in MMR results in microsatellite instability and the accumulation of mutations in proto-oncogenes or tumor suppressor genes which can lead to cancer development. Moreover, it has been observed that tumors with loss of MSH2 expression showed increased frequency of microsatellite instability23,24. This mechanism is characterized by small insertions or deletions within short tandem repeats in tumor DNA when compared with the corresponding normal DNA2. Tumors with MSI frequently show somatic alterations in repetitive DNA tracts of several genes involved in cell growth control, apoptosis, and DNA repair7,27,29.
Molecular disturbances in the repair mechanisms mediated by the MSH2 repair confer susceptibility on tumor cells to present somatic mutations throughout the genome. Furthermore, these alterations in expression of MSH2 have been associated with chromosomal or genomic instability which, at a molecular level, might predispose to HNSCC7,29. However, little is known about the role of MSH2 in prognostic of patients with HNSCC. The aim of this study was to investigate the expression of the MSH2 DNA repair protein in the carcinoma cells of HNSCC in order to analyze its association with clinicopathologic factors and overall survival of patients.
MATERIAL AND METHODS
Tissue specimens, patients, and ethical aspects
This retrospective analytical study was performed using archived tissue blocks from 55 surgically resected primary lesions from patients with HNSCC, confirmed by morphological analysis (male-to-female ratio, 4.5:1; mean age, 61.2±12.9 years). All selected patients underwent surgical resection followed by postoperative radiotherapy. Clinical and outcome data from the HNSCC patients were obtained from the health records at reference centers for diagnosis and oncology treatment between 2000 and 2008 in Montes Claros, Minas Gerais, Brazil. Ethical approval for this study was obtained from the relevant local ethics committees (Unimontes: 1564/2008).
Clinical staging
HNSCC tumors were classified according to the International Union Against Cancer TNM Classification of Malignant Tumors and the International Classification of Diseases for Oncology26. TNM staging resulted in the following distribution: stage I, 1 patient (1.8%); stage II, 4 (7.3%); stage III, 23 (41.8%); and stage IV, 27 (49.1%). The anatomical site of HNSCC lesions were distributed in oral cavity (n=19, 34.5%), oropharynx (n=23, 41,8%), hypopharynx (n=5, 9.1%), and larynx (n=8, 14.5%). The lesions located in the oral cavity were considered as the anterior group and those located in the oropharynx-hypopharynx-larynx as the posterior group for characterization of the anatomical site variable. Clinical factors were classified as follows: local recurrence: absence (n=38, 69.1%) and presence (n=17, 30.9%); tumor size: small lesions (T1/T2, n=8, 14.5%) and large lesions (T3/T4, n=47, 85.5%); locoregional metastasis: absent (n=24, 43.6%) and present (n=31, 56.4%); distant metastasis: absent (n=51, 92.7%) and present (n=4, 2.3%).
Morphological staging
Paraffin-embedded archival tissue from primary HNSCC were cut in 5 µm thick sections, deparaffinized, and stained with hematoxylin and eosin. Only specimens with clear morphological evidence of tumor infiltration into the connective tissue were included. Morphological analysis was carried out by an oral pathologist (AMB de Paula) with no prior knowledge of the epidemiological or clinical data of the selected patients. Kappa statistic was used to assess intra-examiner reproducibility (AMB de Paula) relating to morphological grades of OSCC. Kappa test revealed good concordance for OSCC groups (WHO, 1995, k=0.877, p=0.000; Invasive Front Grading, k=0.492, p=0.024). All HNSCC samples were reviewed and graded using the cell differentiation degrees of the World Health Organization (WHO)21 and Invasive Front Grading (IFG)3. In these morphological grading systems, high malignancy scores are considered to be representative of a poorly differentiated tumor and exhibited evidence of morphological severity, such as absence or decreased keratinization, high cell dissociation, and diffuse infiltrative pattern. The lesions were morphologically classified as follows: well/moderately differentiated (I/II) vs. poorly differentiated (III) (WHO grading) and accordingly with IFG, the grading was classified into three groups: F1 (4-8 points), F2 (9-12 points) and F3 (13-16 points). Higher scores for IFG have been associated with greater clinicopathologic aggressiveness of malignant disease.
Immunohistochemical (IHC) analysis
HNSCC sections were deparaffinized in xylene and rehydrated in a series of descending ethanol concentrations. Thereafter, the sections were treated in a pressure boiler with TRIS EDTA (pH=7.4) for 5 min at 121ºC. The sections were submitted to 5 min blocking with endogenous peroxidase and 0.03% hydrogen peroxide in 100% ethanol. After washing with 10 mM phosphate-buffered saline (PBS) (pH=7.6) for 5 min, the sections were incubated with Protein Block Serum-free (Dako, California, USA) prior to incubation with primary antibodies overnight at 4ºC. The primary antibody used was mouse monoclonal anti-MSH2 (dilution 1:80; clone 25D12, Abcam, Cambridge, USA). The sections were thereafter incubated with streptavidin-biotin labeled antibody (LSAB-Kit Plus Peroxidase; Dako, California, USA) for 30 min. Tissues were stained for 5 min with 3,3′-diaminobenzidine tetrahydrochloride freshly prepared in 10 mM PBS containing 0.1% hydrogen peroxide and then counterstained with Mayer's hematoxylin and mounted in Permount (Fisher Scientific, New Jersey, USA). Positive control was applied according to the manufacturer's instructions, and negative controls were employed by replacing the primary antibody with Universal Negative Control (Dako, California, USA).
Counting of immunostained samples
Samples were evaluated by one independent observer using an optical Olympus® BH2 microscope (model: CX31; RTSF, Miami, USA), with 10x ocular and 40x objective lenses and an ocular lattice (area: 0.092 mm2). Counting of 12 fields in the invasive front of HNSCC was performed to calculate the percentage of positive carcinoma cells. Only carcinoma cells with a distinct brown staining of the nucleus were considered positive (Figure 1). For statistical reasons, in order to analyze the overall survival of patients and clinicopathologic parameters, the mean values of the staining percentages of MSH2 were used in study.
Figure 1.
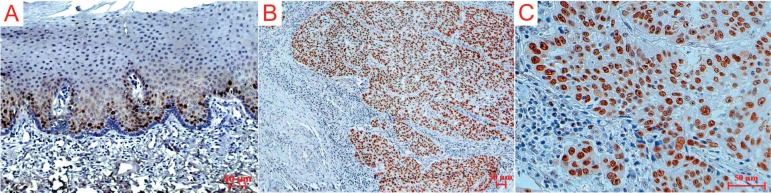
Immunohistochemical expression of MSH2 repair protein in healthy oral mucosa and carcinoma cells of the invasive front of head and neck squamous cell carcinoma (HNSCC). A: Low immunohistochemical expression in basal layer of oral healthy epithelium. B/C: High immunohistochemical expression cases of HNSCC. Slides were stained with diaminobenzidine and counterstained with Mayer’s haematoxylin
Statistical analysis
All data were transferred to SPSS 17.0 software (SPSS Inc., Chicago, Illinois, USA) and subjected to specific tests. The Kolmogorov-Smirnov test was used to determine whether the distribution of continuous variable was normal. The analysis of MSH2 protein expression in tissues assumed non-parametrical distribution, and comparisons between groups were performed using the Mann-Whitney test. The prognostic significance of all covariates (clinicopathologic factors and MSH2 protein immunoexpression) for overall survival data were evaluated using Kaplan-Meyer test and Cox regression models. Associations among variables were considered significant when the confidence level was >95% (p<0.05).
RESULTS
The immunohistochemical evaluation showed that MHS2 protein was detected only in the nucleus of malignant epithelial cells. In all HNSCC samples investigated, it was noted that the invasive fronts exhibited a higher number of cancer cells with a strong nuclear signal (Figure 1). Immunopositivity for MSH2 protein was noted in 96.4% of samples (mean±s.d. and median: 81.5%±21.5% and 90%, respectively). Table 1 shows bivariate comparisons between the clinicopathologic covariates and the mean values of MSH2 immunohistochemical expression. No clinicopathologic covariates exhibited a significant association with expression of the MSH2 protein.
Table 1.
Bivariate analysis between the clinicopathologic covariates and the mean values of MSH2 immunohistochemical expression in the head and neck squamous cell carcinoma samples
| DNA Repair Proteins Immunoexpression | ||
| Covariates | MSH2 | |
| Anatomical Site | Mean±SD | p |
| Anterior (n=20) | 80.77±22.10 | 0.523 |
| Posterior (n=35) | 81.92 ± 21.49 | |
| Recurrence | ||
| Absent (n=38) | 81.15±20.17 | 0.449 |
| Present (n=17) | 82.28±24.90 | |
| Tumor Size | ||
| Small (n=8) | 89.85±8.25 | 0.317 |
| Large (n=47) | 80.08±22.78 | |
| Locorregional Metastasis | ||
| Absent (n=24) | 79.90±26.56 | 0.946 |
| Present (n=31) | 82.74±16.99 | |
| WHO Grading | ||
| Well/Moderate differentiated (n=39) | 85.21±14.61 | 0.339 |
| Poorly differentiated (n=16) | 72.45±31.66 | |
| IFG | ||
| 4-8 (n=6) | 87.95±16.12 | 0.509 |
| 9-12 (n=22) | 77.85±25.36 | |
| 13-16 (n=27) | 83.04±19.20 | |
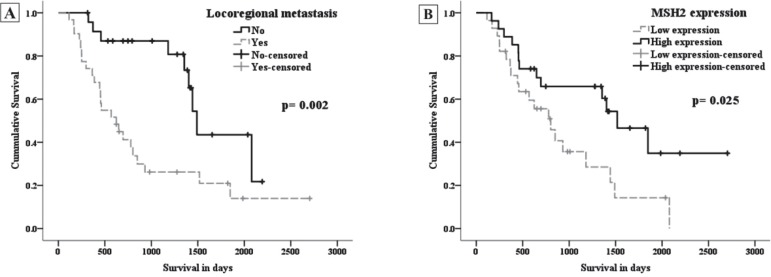
In overall survival analysis, HNSCC patients were followed for a period from 116 to 2704 days (mean: 929.1±622.8 days and median: 749 days). It was noted that 22 patients (40%) were alive and 33 patients (60%) died from disease at the end of the period. Patients who died without evidence of recurrence/metastasis were considered censored by the last clinical evaluation. In the bivariate analysis (Kaplan-Meyer curves), the covariates locoregional metastatic disease (p=0.002) and low expression of MSH2 protein (p=0.025) exhibited a significant association with poor overall survival (Figure 2). Subsequently, using Cox's regression multivariate models, significant differences were again noted when we tested the relationship between the covariates of locoregional metastasis (OR=4.949; CI 95%=2.045-11.979; p<0.001) and low expression of MSH2 (OR=2.943; CI 95%=1.212-7.142; p=0.032), with a worse overall survival (Table 2).
Figure 2.
Overall survival analysis in head and neck squamous cell carcinoma patients using Kaplan-Meier survival curves after 2704 days, according to locoregional metastatic disease (A) and MSH2 protein expression (B). Differences between groups were evaluated by the log-rank test
Table 2.
Overall survival for the head and neck squamous cell carcinoma patients of this study using Cox regression analysis
| Covariates | 95% CI | |||
| OR | Lower | Upper | p | |
| Anatomical Site | ||||
| Anterior | Referent | |||
| Posterior | 2.120 | 0.216 | 1.029 | 0.059 |
| Locoregional Metastasis | ||||
| No | Referent | |||
| Yes | 4.949 | 2.045 | 11.979 | 0.000* |
| MSH2 Expression | ||||
| High | Referent | |||
| Low | 2.943 | 1.212 | 7.142 | 0.032* |
DISCUSSION
The mismatch repair pathway is responsible for correcting base substitution mismatches and insertion-deletion mismatches generated during DNA replication or recognizing genetic damage14,20. MSH2 is the most abundantly expressed mismatch repair protein and its inactivation can have profound pathologic consequences on DNA, particularly in microsatellite sequences, rendering them dysfunctional due to instability. Importantly, the presence of this type of DNA damage is considered to be an established biomarker for loss of mismatch repair activities in cancer cells6,22. Notably, few data are available about the role of MSH2 gene status or tissue expression for both upper aerodigestive tract mucosa carcinogenesis and for the progression of HNSCC7,27,29. In this present study, similar with findings of other human cancers studies that showed a low frequency of MSH2 immunonegativity (microsatellite instability-high phenotype) in different human cancers5,18,19, we noted a down-regulation of MHS2 protein expression in approximately 9% of HNSCC samples. Different mechanisms may lead to loss of MSH2 protein expression, such as mutation in the MSH2 gene itself that leads to a lack of expression of this protein or to the expression of a truncated, non-functional form9. Another mechanism could be MSH2 gene promoter hypermethylation which can result in its transcriptional silencing15.
HNSCC patients frequently exhibit low rates of overall and disease-free survival4,10. In the present study, bivariate and multivariate analysis showed that HNSCC patients whose malignancy exhibited low protein expression of MSH2 and locoregional metastasis had a worse overall survival. Reduced MSH2 protein expression has also been shown to be of unfavorable prognostic value for prostate cancer30, soft tissue sarcoma28, and biliary tract carcinoma11. Unfortunately, in a variety of human cancers, data on the prognosis of down-regulation of MSH2 expression are still contradictory5,12,13,17,19,25. Taking into account HNSCC7, the prognostic value of loss of MSH2 protein expression is unclear, mainly because the few existing studies have presented an insufficient number of patients or perhaps because squamous cell carcinomas from the different anatomic sites of the upper aerodigestive tract seem to exhibit distinct risk factors (environmental and genetic/epigenetic factors), clinical presentations, and outcomes, which may influence the evaluation of potential prognostic markers. According to our findings, the down-regulation of MSH2 and consequent loss of mismatch repair activities in transformed cells could promote an increased tendency of the genome to acquire mutations in genes associated with maintenance and replication of the genome, inducing in cancer cells a more aggressive phenotype and resulting in a worse prognosis for HNSCC patients. Finally, we confirmed that HNSCC individuals presenting with locoregional metastatic disease also had a poor overall survival. This finding is consistent with fundamental concepts that lymph node status is the most important predictor of survival16, which is confirmed in our population of head and neck carcinoma patients.
Our study, however, has a number of limitations such as the small sample size and the typical difficulties of cross-sectional study design to establish causal inference. Unfortunately, it was not possible to investigate the participation of other genetic or epigenetic disturbances that could interfere with the expression and functional activity of MSH2. Otherwise, our findings show that the immunohistochemical expression of MSH2 protein seems to contribute to the identification of patients who had a poor prognosis. These findings might be useful for selecting HNSCC patients for adjuvant radiotherapy/chemotherapy protocols. However, future large clinical and molecular studies are needed to foster a better understanding about the reliability of MSH2 protein as an indicator of biological behavior of HNSCC or as a potential molecular therapeutic target for this type of cancer.
CONCLUSION
In conclusion, this study showed that reduced MSH2 expression in HNSCC tissues was associated with poor overall survival for patients. This low expression of MSH2 might contribute to a higher genomic instability and an increase in molecular disturbances resulting in a worse prognosis for patients.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful for support from Minas Gerais Foundation Research (FAPEMIG) and National Council for Scientific and Technological Development (CNPq). This study was supported by research grant (CNPq - 470331/2010-0).
Footnotes
Competing Interests: The authors declare no competing interests.
REFERENCES
- 1.Bansal S, Sircar K, Joshi SK, Singh S, Rastogi V. A comparative study of p53 expression in hyperplastic , dysplastic epithelium and oral squamous cell carcinoma. Braz J Oral Sci. 2010;9(2):85–88. [Google Scholar]
- 2.Baross-Francis A, Andrew SE, Penney JE, Jirik FR. Tumors of DNA mismatch repair-deficient hosts exhibit dramatic increases in genomic instability. Proc Natl Acad Sci U S A. 1998;95(15):8739–8743. doi: 10.1073/pnas.95.15.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryne M, Koppang HS, Lilleng R, Kjaerheim A. Malignancy grading of the deep invasive margins of oral squamous cell carcinomas has high prognostic value. J Pathol. 1992;166(4):375–381. doi: 10.1002/path.1711660409. [DOI] [PubMed] [Google Scholar]
- 4.Cooper JS, Porter K, Mallin K, Hoffman HT, Weber RS, Ang KK, et al. National Cancer Database report on cancer of the head and neck: 10-year update. Head Neck. 2009;31(6):748–758. doi: 10.1002/hed.21022. [DOI] [PubMed] [Google Scholar]
- 5.Cooper WA, Kohonen-Corish MR, Chan C, Kwun SY, McCaughan B, Kennedy C, et al. Prognostic significance of DNA repair proteins MLH1, MSH2 and MGMT expression in non-small-cell lung cancer and precursor lesions. Histopathology. 2008;52(5):613–622. doi: 10.1111/j.1365-2559.2008.02999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox EC, Degnen GE, Scheppe ML. Mutator gene studies in Escherichia coli: the mutS gene. Genetics. 1972;72(4):551–567. doi: 10.1093/genetics/72.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demokan S, Suoglu Y, Demir D, Gozeler M, Dalay N. Microsatellite instability and methylation of the DNA mismatch repair genes in head and neck cancer. Ann Oncol. 2006;17(6):995–999. doi: 10.1093/annonc/mdl048. [DOI] [PubMed] [Google Scholar]
- 8.Fuzikawa AK, Haddad LA, da-Cunha-Melo JR, Brasileiro-Filho G, Pena SD. Utilization of microsatellites for the analysis of genomic alterations in colorectal cancers in Brazil. Braz J Med Biol Res. 1997;30(8):915–921. doi: 10.1590/s0100-879x1997000800001. [DOI] [PubMed] [Google Scholar]
- 9.Jascur T, Boland CR. Structure and function of the components of the human DNA mismatch repair system. Int J Cancer. 2006;119(9):2030–2035. doi: 10.1002/ijc.22023. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 11.Kohya N, Miyazaki K, Matsukura S, Yakushiji H, Kitajima Y, Kitahara K, et al. Deficient expression of O(6)-methylguanine- DNA methyltransferase combined with mismatch-repair proteins hMLH1 and hMSH2 is related to poor prognosis in human biliary tract carcinoma. Ann Surg Oncol. 2002;9(4):371–379. doi: 10.1007/BF02573872. [DOI] [PubMed] [Google Scholar]
- 12.Kouso H, Yoshino I, Miura N, Takenaka T, Ohba T, Yohena T, et al. Expression of mismatch repair proteins, hMLH1/hMSH2, in non-small cell lung cancer tissues and its clinical significance. J Surg Oncol. 2008;98(5):377–383. doi: 10.1002/jso.21108. [DOI] [PubMed] [Google Scholar]
- 13.Kruschewski M, Noske A, Haier J, Runkel N, Anagnostopoulos Y, Buhr HJ. Is reduced expression of mismatch repair genes MLH1 and MSH2 in patients with sporadic colorectal cancer related to their prognosis? Clin Exp Metastasis. 2002;19(1):71–77. doi: 10.1023/a:1013853224644. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 15.Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1. Nat Genet. 2009;41(1):112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 16.Manikantan K, Khode S, Dwivedi RC, Palav R, Nutting CM, Rhys-Evans P, et al. Making sense of post-treatment surveillance in head and neck cancer: when and what of follow-up. Cancer Treat Rev. 2009;35(8):744–753. doi: 10.1016/j.ctrv.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Mylona E, Zarogiannos A, Nomikos A, Giannopoulou I, Nikolaou I, Zervas A, et al. Prognostic value of microsatellite instability determined by immunohistochemical staining of hMSH2 and hMSH6 in urothelial carcinoma of the bladder. APMIS. 2008;116(1):59–65. doi: 10.1111/j.1600-0463.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- 18.Nijhuis ER, Nijman HW, Oien KA, Bell A, ten Hoor KA, Reesink- Peters N, et al. Loss of MSH2 protein expression is a risk factor in early stage cervical cancer. J Clin Pathol. 2007;60(7):824–830. doi: 10.1136/jcp.2005.036038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohrling K, Edler D, Hallstrom M, Ragnhammar P. Mismatch repair protein expression is an independent prognostic factor in sporadic colorectal cancer. Acta Oncol. 2010;49(6):797–804. doi: 10.3109/02841861003705786. [DOI] [PubMed] [Google Scholar]
- 20.Peltomäki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21(6):1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 21.Pindborg JJ, Wahi PN. Histological typing of cancer and precancer of the oral mucosa. Berlin: Springer; 1997. [Google Scholar]
- 22.Plotz G, Piiper A, Wormek M, Zeuzem S, Raedle J. Analysis of the human MutLα.MutSα complex. Biochem Biophys Res Commun. 2006;340(3):852–859. doi: 10.1016/j.bbrc.2005.12.096. [DOI] [PubMed] [Google Scholar]
- 23.Ruszkiewicz A, Bennett G, Moore J, Manavis J, Rudzki B, Shen L, et al. Correlation of mismatch repair genes immunohistochemistry and microsatellite instability status in HNPCC-associated tumours. Pathology. 2002;34(6):541–547. doi: 10.1080/0031302021000035965-2. [DOI] [PubMed] [Google Scholar]
- 24.Shia J, Ellis NA, Klimstra DS. The utility of immunohistochemical detection of DNA mismatch repair gene proteins. Virchows Arch. 2004;445(5):431–441. doi: 10.1007/s00428-004-1090-5. [DOI] [PubMed] [Google Scholar]
- 25.Shih KK, Garg K, Levine DA, Kauff ND, Abu-Rustum NR, Soslow RA, et al. Clinicopathologic significance of DNA mismatch repair protein defects and endometrial cancer in women 40 years of age and younger. Gynecol Oncol. 2011;123(1):88–94. doi: 10.1016/j.ygyno.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Sobin LH. TNM: evolution and relation to other prognostic factors. Semin Surg Oncol. 2003;21(1):3–7. doi: 10.1002/ssu.10014. [DOI] [PubMed] [Google Scholar]
- 27.Souza LR, Fonseca-Silva T, Pereira CS, Santos EP, Lima LC, Carvalho HA, et al. Immunohistochemical analysis of p53, APE1, hMSH2 and ERCC1 proteins in actinic cheilitis and lip squamous cell carcinoma. Histopathology. 2011;58(3):352–360. doi: 10.1111/j.1365-2559.2011.03756.x. [DOI] [PubMed] [Google Scholar]
- 28.Taubert HW, Bartel F, Kappler M, Schuster K, Meye A, Lautenschläger C, et al. Reduced expression of hMSH2 protein is correlated to poor survival for soft tissue sarcoma patients. Cancer. 2003;97(9):2273–2278. doi: 10.1002/cncr.11326. [DOI] [PubMed] [Google Scholar]
- 29.Theocharis S, Klijanienko J, Giaginis C, Rodriguez J, Jouffroy T, Girod A, et al. Expression of DNA repair proteins, MSH2, MLH1 and MGMT in mobile tongue squamous cell carcinoma: associations with clinicopathological parameters and patients' survival. J Oral Pathol Medi. 2011;40(3):218–226. doi: 10.1111/j.1600-0714.2010.00945.x. [DOI] [PubMed] [Google Scholar]
- 30.Velasco A, Hewitt SM, Albert PS, Hossein M, Rosenberg H, Martinez C, et al. Differential expression of the mismatch repair gene hMSH2 in malignant prostate tissue is associated with cancer recurrence. Cancer. 2002;94(3):690–699. doi: 10.1002/cncr.10247. [DOI] [PubMed] [Google Scholar]