SUMMARY
To characterize patient-derived xenografts (PDXs) for functional studies, we made whole-genome comparisons with originating breast cancers representative of the major intrinsic subtypes. Structural and copy number aberrations were found to be retained with high fidelity. However, at the single-nucleotide level, variable numbers of PDX-specific somatic events were documented, although they were only rarely functionally significant. Variant allele frequencies were often preserved in the PDXs, demonstrating that clonal representation can be transplantable. Estrogen-receptor-positive PDXs were associated with ESR1 ligand-binding-domain mutations, gene amplification, or an ESR1/YAP1 translocation. These events produced different endocrine-therapy-response phenotypes in human, cell line, and PDX endocrine-response studies. Hence, deeply sequenced PDX models are an important resource for the search for genome-forward treatment options and capture endocrine-drug-resistance etiologies that are not observed in standard cell lines. The originating tumor genome provides a benchmark for assessing genetic drift and clonal representation after transplantation.
INTRODUCTION
Many stage 3 breast cancers and effectively all stage 4 breast cancers are fatal, with annual worldwide deaths from the disease approaching one-half million (Youlden et al., 2012). Large-scale partial and whole-genome sequencing (WGS) was recently conducted on early-stage, treatment-naive breast cancer samples (Ellis and Perou, 2013). By contrast, the genomic landscape of advanced and treatment-resistant breast cancer is poorly documented. We therefore developed a panel of patient-derived xenografts (PDXs) from patients with poor-prognosis, treatment-resistant disease for genomic and functional studies, because early-passage PDX models reproduce gene expression patterns observed in the originating human tumors and recapitulate the chemotherapy response (DeRose et al., 2011; Fleming et al., 2010; Kabos et al., 2012; Marangoni et al., 2007; Zhang et al., 2013). However, it has remained unclear to what extent PDX models accurately represent the genomic characteristics of the originating tumor at a whole-genome level. The value of the PDX approach in the setting of estrogen-receptor-positive (ER+) breast cancer has also been questioned because very few breast cancer PDXs expressing ER have been reported.
WGS using massively parallel techniques is the gold standard for comparing an originating tumor with a counterpart PDX because partial-genome sequencing, which focuses on the coding sequence alone (i.e., exome sequencing), does not fully document all mutations, particularly structural variations (SVs) or other mutational events that occur in noncoding space (Ley et al., 2008). Promisingly, WGS of a single example of a breast cancer primary, a brain metastasis, and a PDX basal-like breast cancer “trio” demonstrated that the PDX model efficiently captures almost all of the genome-wide somatic mutations observed in the originating tumor, and displayed enrichment for mutations that were present in the metastatic sample even though they were derived from the primary tumor (Ding et al., 2010). Heterogeneity in mutation frequencies also has not been comparatively evaluated for PDX models and originating tumors, so a customized capture approach (Welch et al., 2012) was used to generate high depth at somatic variant positions genome wide, coupled with statistical analyses for this comparison. RNA sequencing (RNA-seq) was conducted to determine the expression level of individual mutations and to confirm gene fusion events (Iyer et al., 2011). Reverse phase protein array (RPPA) was employed to determine whether protein and phosphoprotein expression patterns were stable upon serial transplantation (Tabchy et al., 2011). We also successfully developed multiple ER+ PDXs from patients with endocrine-therapy-resistant disease, and our genomic and functional analyses revealed mechanistic insights into resistance that have not been achieved with conventional cell line approaches.
RESULTS
Derivation and RNA/Protein Expression Patterns in Xenografts from Advanced Stage Breast Cancer
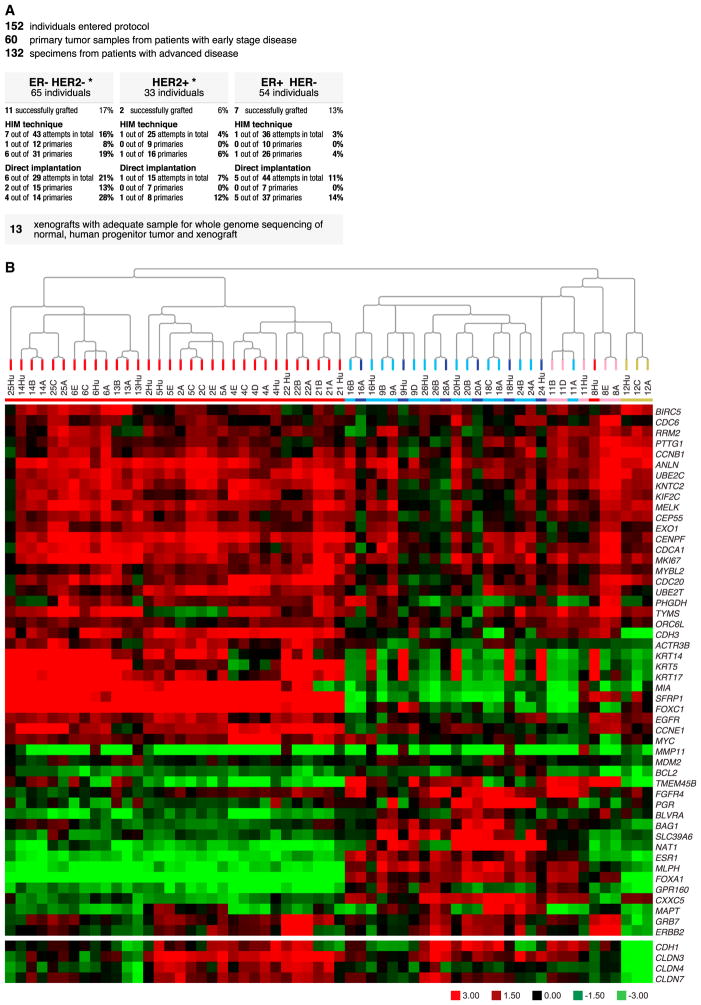
Samples were obtained from 152 patients (Figure 1A), which yielded 22 serially transplantable PDXs from 20 patients, for an engraftment rate of 13.1%. These PDXs are referred to as “Washington University Human in Mouse” (WHIM) lines and were mostly obtained from patients with advanced disease or larger primary tumors that rapidly developed lethal metastasis (Table S2A). Concordant ER and HER2 status was demonstrated at the mRNA level (Table 1), and PDX expression of ER and HER2 protein was confirmed by western blot (Figure S1). Mouse and human centromere-specific fluorescent in situ hybridization (FISH) assays were conducted to demonstrate that stromal elements were murine in origin, the malignant cells were human, and there was no evidence for interspecies cellular fusion events (Figures S2A–S2C). Agilent 44K Array-based mRNA expression data were generated from the originating tumors and from matched early- and late-passage PDX counterparts. We subjected the matched pairs to unsupervised hierarchical clustering after removing genes that were highly differentially expressed between progenitor and PDX models (FDR = 0) to ensure that this comparison was “tumor centric” and not confounded by differences in the hybridization properties of mRNA arising from human versus mouse stroma (Table S2B). In almost all cases, the originating tumor and WHIM lines derived from the same individual clustered adjacently (Figure 1B). Each sample was also classified into one of five intrinsic gene-expression subtypes; lumenal A (dark blue), lumenal B (light blue), HER2-enriched (pink), basal-like (red), and Claudin-low (yellow) using PAM50 (Parker et al., 2009) and the “Nine-Cell Line Claudin-low subtype predictor” (Prat et al., 2010; Table 1; Figure 1B). The PDX lines derived from ER+ clinical samples were all subtyped lumenal by PAM50 in both the human and mouse samples, with the exception of WHIM11, which was classified as HER2-E. Of note, the human lumenal originating tumors expressed high levels of cytokeratin 14 (CK14), CK5, and CK17 mRNA, but there was no evidence for expression of these CKs by the counterpart lumenal PDX. To investigate this discordance, we conducted immunohistochemistry for CK5 on the human lumenal tumor progenitor samples (derived from cutaneous metastases). This revealed normal-appearing CK5-positive epithelial cells arranged in ducts “trapped” among CK5-negative malignant lumenal epithelial cells, thereby “contaminating” the progenitor tumor samples with basal epithelial keratins (Figure S2D). To investigate the lumenal classification further, PDX mRNA was also profiled on a 244K customized UNC Agilent chip (Cancer Genome Atlas Network, 2012a) and the PDX data clustered with clinical breast cancer samples profiled on the same platform. In this analysis, all ER+ PDXs segregated with lumenal B tumors (Figure S3). The WHIM12 line was derived from a metaplastic carcinoma and showed a near-perfect correlation with the claudin-low signature (Figure S4). To address the stability of PDXs at the level of protein and phosphoprotein expression, multiple samples taken from the same passage and upon serial passage were assayed by RPPAs (Tabchy et al., 2011). Data from 110 antibodies for 68 samples harvested from 20 WHIM lines were clustered with the data from 386 primary breast cancers studied by The Cancer Genome Atlas (TCGA) research network (Cancer Genome Atlas Network, 2012a; Table S2C). In every case, the samples from each WHIM line clustered adjacently, including the two double isolates (WHIM2 and WHIM5, and WHIM20 and WHIM23; Figure S5). This suggests that the intra-PDX proteomic heterogeneity was considerably lower than the intertumoral heterogeneity in a large RPPA data set and was relatively stable over time and passage. The PDX samples were dispersed across the breast TCGA data, indicating that they are representative of the heterogeneous biology of breast cancer. An analysis was conducted to determine the relative rank of protein and phosphoprotein levels for each WHIM tumor with respect to the ranges in the TCGA data set (Table S2C). Here, the WHIM lines did not contain any extreme data outliers with respect to the phosphoprotein levels documented in the TCGA data. Phosphorylation of the phosphatidylinositol-3-kinase/AKT pathway proteins represented the highest-ranked pathway activation event (Table 1).
Figure 1. Generation of a Biologically Diverse Panel of PDX Models from Patients with Advanced Breast Cancer.
(A) Diagram indicating the genesis of the PDX models from patients with primary and advanced breast cancer, using two different implantation techniques (human in mouse [Kuperwasser et al., 2004] and simple orthotopic).
(B) Unsupervised hierarchical clustering of samples using all genes of the microarrays except the stromal-related genes. The colors of the array tree and the squares below the tree denote the subtype call of each sample. Red, basal-like; pink, HER2-enriched; dark blue, lumenal A; light blue, lumenal B; yellow, Claudin-low. Below the array tree and the subtype identification row, the heatmap of the 50 PAM50 genes as well as selected tight-junction-related genes (E-cadherin [CDH1], claudin 3 [CLDN3], CLDN4, and CLDN7) are shown. The stromal-related genes were identified after a two-class paired SAM was performed with an FDR of 0% between 18 paired progenitor human tumors and xenografts. The complete list of up- and down-regulated genes can be found in Table S2B.
See also Figures S3 and S4.
Table 1.
Summary of the WHIM Line Biomarker Status
| Intrinsic Subtype | P1/Late WHIM | HER2 RNAc | ER RNAc | P53 Mutation | PIK3CA Mutation | ESR1 Mutation Translocation | Highest Ranked | |
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
| WHIM Number | human | passage | human/P1 | human/P1 | human/P1 | human/P1 | amplification | phospho-protein by RPPA |
| 2a | basal | basal | 1.3 | 3.1 | S166S ins | WT | WT | p90RSK_pT359 |
|
|
|
|||||||
| Primary | basal | 1.0 | 1.0 | S166S ins | WT | WT | rank = 25.5 | |
|
| ||||||||
| 3 | N/A | Her2-E | N/A | N/A | N/A | N/A | WT | Src_pY527 |
|
|
|
|||||||
| Primary | Her2-E | 1.8 | 3.8 | WT | WT | WT | rank = 4.0 | |
|
| ||||||||
| 4 | basal | basal | −5.9 | −5.7 | R175H | WT | WT | p27_pT198 |
|
|
|
|||||||
| Skin met | basal | 2.2 | 2.2 | R175H | WT | WT | rank = 28.7 | |
|
| ||||||||
| 5a | basal | basal | 3.1 | 1.1 | S166S ins | WT | WT | p90RSK_pT359 |
|
|
|
|||||||
| Brain met | basal | 1.8 | −1.7 | S166S ins | WT | WT | rank = 62.3 | |
|
| ||||||||
| 6 | basal | basal | −1.5 | 3.1 | WT | WT | WT | PRAS40_pT246 |
|
|
|
|||||||
| Primary | basal | 1.2 | 3.6 | WT | WT | WT | rank = 59.2 | |
|
| ||||||||
| 8 | basal | HER2-E | 8.6 | 3.6 | Y234H | WT | WT | Shc_pY317 |
|
|
|
|||||||
| Skin met | HER2-E | 16.8 | 4.9 | Y234H | WT | WT | rank = 24.7 | |
|
| ||||||||
| 9 | LumA | LumB | 1.2 | 72.2 | WT | H1047R | WT | Akt_pS474 |
|
|
|
|||||||
| Skin met | LumB | 2.1 | 365.2 | WT | H1047R | WT | rank = 25.0 | |
|
| ||||||||
| 10 | N/A | basal | N/A | N/A | N/A | N/A | WT | N/A |
|
|
|
|||||||
| Skin met | basal | 1.6 | −2.1 | Y220C | WT | WT | ||
|
| ||||||||
| 11 | HER2-E | LumB | −1.6 | 31.8 | D184X ins | WT | WT | p70S6K_pT389 |
|
|
|
|||||||
| Primary | HER2-E | 1.2 | 112.1 | D184X ins | WT | WT | rank = 13.3 | |
|
| ||||||||
| 12 | CLDN low | CLDN low | −5.0 | 2.4 | R248Q | WT | WT | Akt_pT308 |
|
|
|
|||||||
| Primary | CLDN low | −5.0 | −1.2 | R248Q | WT | WT | rank = 10 | |
|
| ||||||||
| 13 | basal | basal | −4.8 | −2.1 | C238Y | WT | WT | Src_pY527 |
|
|
|
|||||||
| Skin met | basal | −1.5 | 1.0 | C238Y | WT | WT | rank = 14.3 | |
|
| ||||||||
| 14 | basal | basal | 1.0 | 5.3 | I195T | WT | WT | N/A |
|
|
|
|||||||
| Skin met | basal | 1.3 | 1.2 | I195T | WT | WT | ||
|
| ||||||||
| 16 | LumB | LumA | −2.9 | 40.1 | R248W | H1047R | AMP | 4E-BP1_pT37 |
|
|
|
|||||||
| Skin met | LumB | −1.2 | 171.5 | R248W | H1047R | AMP | rank = 13.7 | |
|
| ||||||||
| 17 | N/A | basal | N/A | N/A | N/A | N/A | WT | PKC-delta_pS664 |
|
|
|
|||||||
| Primary | basal | −22.8 | 4.1 | WT | WT | WT | rank = 2.5 | |
|
| ||||||||
| 18 | LumA | LumB | −3.9 | 65.5 | WT | E545K | ESR1 YAP1 | ER-alpha_pS118 |
|
|
|
|||||||
| Skin met | LumB | 3.1 | 19.9 | WT | E545K | ESR1 YAP1 | rank = 16.5 | |
|
| ||||||||
| 20 | LumB | LumA | 3.1 | 135.2 | C182X | E542K | Y537S | PDK1_pS241 |
|
|
|
|||||||
| Skin metb | LumB | 5.9 | 71.6 | C182X | E542K | Y537S | rank = 57.8 | |
|
| ||||||||
| 21 | basal | basal | −7.5 | 6.4 | P151H | WT | WT | Chk1_pS345 |
|
|
|
|||||||
| Primary | basal | −6.8 | −1.9 | P151H | WT | WT | rank = 16.7 | |
|
| ||||||||
| 22 | basal | basal | 2.5 | 1.2 | C275F | WT | WT | EGFR_pY1068 |
|
|
|
|||||||
| Skin met | basal | 7.7 | 2.6 | C275F | WT | WT | rank = 24.4 | |
|
| ||||||||
| 23 | N/A | LumA | N/A | N/A | N/A | N/A | N/A | Rb_pS807_S811 |
|
|
|
|||||||
| Node metb | LumB | −1.3 | 75.8 | C182X | E542K | Y537S | rank = 27.7 | |
|
| ||||||||
| 24 | LumA | LumB | −2.1 | 18.9 | WT | H1047K | WT | ER-alpha_pS118 |
|
|
|
|||||||
| Skin met | LumB | −1.3 | 719.1 | WT | H1047K | E380Q | rank = 7.0 | |
|
| ||||||||
| 25 | basal | basal | −3.9 | −1.1 | R273H | WT | WT | MAPK_pT202_Y204 |
|
|
|
|||||||
| Skin met | basal | 1.8 | −1.3 | R273H | WT | WT | rank = 20.3 | |
|
| ||||||||
| 26 | LumB | LumA | 1.9 | 59.6 | WT | WT | WT | mTOR_pS2448 |
|
|
|
|||||||
| Nodal met | LumB | 3.5 | 52.3 | WT | WT | WT | rank = 2.7 | |
The characteristics of 22 WHIM lines discussed in the text are summarized, including the site of the originating tumor; the PAM50 subtype of the originating tumor (“human”) and passage 1 (P1) and a later passage (late); HER2 and ESR1 mRNA levels normalized to WHIM2 P1, PIK3CA, TP53, and ESR1 mutation status and phosphoprotein ranking based on WHIM RPPA data within the context of 386 TCGA breast tumors (Table S2C; Figure S3).
See also Figures S5 and S12. WT, wild-type.
WHIM2 and WHIM5 from the same patient.
WHIM20 and WHIM23 from the same patient.
Reference WHIM2 early passage.
Genomic Fidelity of PDX Models
Using paired-end massively parallel sequencing, we sequenced 17 originating tumor, xenograft, and germ-line trio DNA samples to ≥ 30-fold average whole-genome coverage. For 13 trios, we subsequently validated each candidate mutation using solution-based hybridization capture followed by deep sequencing of the originating tumor, the paired WHIM line, and the normal DNA (for the somatic variants observed in WHIM4, WHIM24, WHIM25, and WHIM26, further validation was not conducted; see Table S2D for the coding region single-nucleotide variation [SNV] observed in these examples). In the 13 cases subjected to validation, a total of 59,189 genome-wide SNVs were confirmed (Table S2E). Of these, 1,056 (1.8%) were nonsilent protein coding mutations or in RNA genes (Table S2F). Across all WHIM lines, there were 241 (range 0–77) out of a total of 58,814 validated genome-wide SNVs that were unique to the originating tumor (0.4%). In contrast, a much higher number of sites (5,450, range 29–1,564, 9.3%) were PDX specific (Table S2G). Seventy-one mutations were detected in “significantly mutated genes” (SMGs) as defined by TCGA data (Cancer Genome Atlas Research Network, 2012b), lumenal tumors (Ellis et al., 2012), or triple-negative tumors (Shah et al., 2012; Table S2H). Each PDX harbored mutation(s) in at least one SMG (WHIM6) and up to 12 SMGs (WHIM14). A small number of potentially significant mutations were observed in the PDX but not in the originating tumor (WNK2 in WHIM8, PIK3R4 and KRAS in WHIM9, MAP4K2 in WHIM16, and CBLB in WHIM18). Thirty-four PDX-specific missense mutations were identified as potentially deleterious or functionally significant by a mutation impact assessment algorithm (Xi et al., 2004), and all examples contained at least one predicted deleterious PDX-specific SNV, except for WHIM 2 (Table S2I). The PDX data analysis pipeline removed sequence reads contributed by the murine genome, thereby “computationally purifying” the human tumor DNA (Ding et al., 2010). Biallelic deletions (e.g., in PTEN) were therefore revealed with clarity in the WHIM analysis (Figure S6) and amplified regions were often enhanced (Figures 2A, B, and S7–S9). Of the 5,336 copy-number variation (CNV) phenotypes that were detected in the 13 cases analyzed, 5,036 (94.4%) had the same call (amplified or deleted) in both the originating tumor and the counterpart PDX line (Table S2L). Remarkably, all SVs (translocations, large deletions, and inversions) were preserved upon transplantation, including regions characteristic of chromothripsis (Stephens et al., 2011; Figures 2A, 2B, and S7).
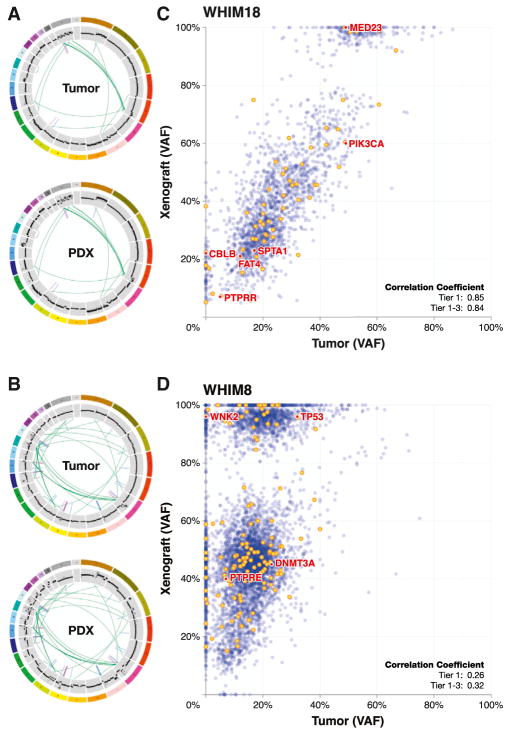
Figure 2. Pairwise Genome-Wide VAF, CNV, and SV Analyses.
(A and B) The circos plots for (A) WHIM18 and (B) WHIM8 show the closely matched SVs and CNVs in the tumor of origin and the paired WHIM line. To compare differences in mutant allele frequency between the originating tumor and the PDX counterpart, the read counts for each mutant and wild-type allele were expressed as a percentage of all reads at that position and analyzed by scatterplot and simple correlation coefficient.
(C) WHIM18 has a high correlation coefficient (0.84) in both the coding region (yellow) and noncoding region (blue). The VAF stability was maintained across all six SMG mutations.
(D) WHIM8 represented the opposite extreme with a low correlation coefficient (0.32) and a relatively large number of xenograft-specific mutations in the homozygous range of 80% or higher. Related to this figure are analyses for the other whole-genome sequenced originating tumor/PDX pairs that are displayed in Figures S7A–S9.
See also Figure S6.
Genomic Stability of PDX across Early and Late Passages
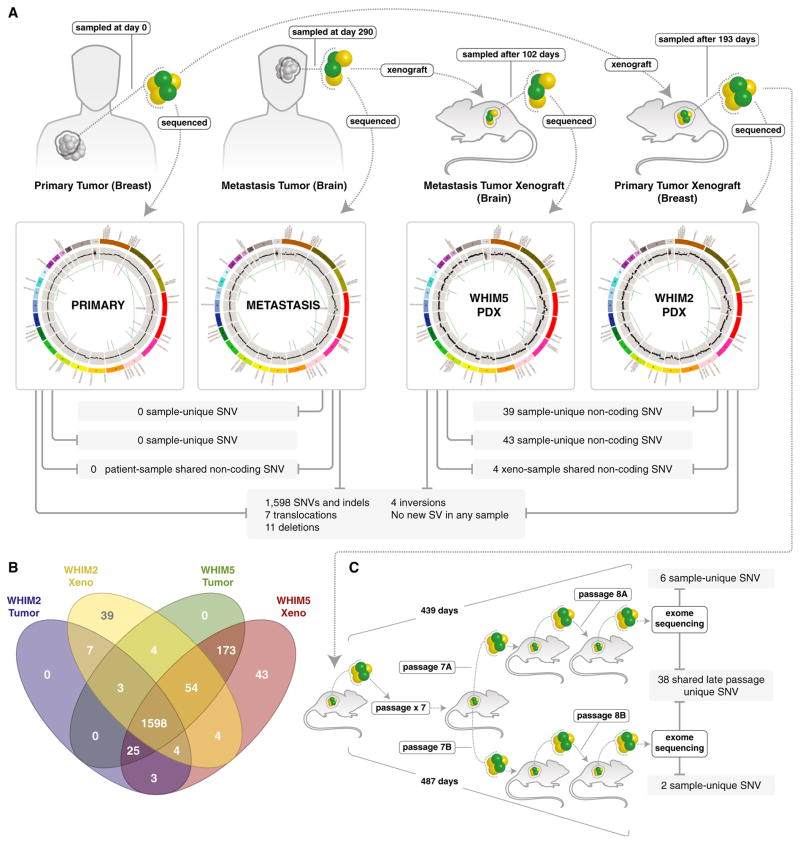
Subsequent to our earlier report on comparative WGS of a primary tumor, brain metastasis, and primary-derived PDX trio (Ding et al., 2010), a PDX model also was derived from the patient’s brain metastasis (WHIM5), enabling a deep genomic analysis of two xenografts from the same patient. The vast majority of the validated somatic SNVs and indels were shared by the four genomes (n = 1,598) as well as seven translocations, 11 deletions, and four inversions. The breast primary tumor and brain metastasis contained no single sample-unique SNV or SV (i.e., all of the SNVs were noted in at least one other sample; Figure 3A). However, in every comparison, more SNVs were observed in the tumor sample taken at a later time point when compared with a sample taken at an earlier time point (whether a PDX sample pair or a human sample pair). For example, in a comparison of the two human specimens, 13 SNVs were unique to the primary tumor, but 231 SNVs were unique to the metastasis. Additionally, both WHIM lines harbored additional sample-unique noncoding SNVs that become detectable after xenografting (39 in the case of WHIM2 and 43 in the case of WHIM5; Figure 3B). Since therapeutic experiments require extensive expansion of PDX models, we also conducted a “late-exome” study to characterize genomic drift in the WHIM2 genome, performing exome sequencing on two separate passage eight tumor grafts (Figure 3C). This experiment detected 38 additional variants in both specimens, although none of the SNVs were clearly damaging mutations in cancer-associated genes (Table S2K).
Figure 3. Whole-Genome Comparisons of Breast Primary Tumor and Brain Metastasis with their Counterpart PDX Model Xenografts from the Same Patient.
(A) The majority of the validated somatic SNVs were shared by the breast primary tumor, brain metastasis, and xenografts (1,598). In addition, seven translocations, 11 large deletions, and four inversions were present in all samples, without any SV detected or lost upon engraftment.
(B) The breast primary tumor and brain metastasis contained no sample-unique SNVs, i.e., all of the SNVs were noted in at least one other sample. However in every comparison, more SNVs were observed in the later time sample than in the earlier sample. In a comparison of the two human specimens, 13 were unique to the primary tumor and 231 were unique to the metastasis. Additionally, both WHIM lines harbored additional sample-unique noncoding SNVs (39 in the case of WHIM2 and 43 in the case of WHIM5).
(C) Exome sequencing of two separate DNA samples isolated from WHIM2 passage 8, after expansion for therapeutic studies. Mutations in coding space have accumulated, but a study of the 38 mutations observed in both samples suggests that most are passengers rather than biological drivers.
The Genome-Wide Variant Allele Frequency Is a Transplantable Phenotype
To compare mutant allele representation in the originating tumor isolates versus their corresponding PDX models, we obtained deep coverage through our capture-based validation approach and then calculated the proportion of sequencing reads that contained a mutant allele. This value was expressed as a percentage (variant allele frequency [VAF]) and analyzed by scatter-plot (Figures 2C and 2D; see Figures S7–S9 for the remaining examples). The genome-wide correlation coefficients across the 13 tumor/PDX pairs varied from 0.32 (WHIM8) to 0.86 (WHIM5; Table S2L and Figures S7–S9). In the majority of cases, there was statistical evidence for VAF stability genome wide, with nine out of the 13 comparisons showing correlation coefficients above 0.65. For example, WHIM18 (R = 0.85) displayed coding region (yellow) and noncoding region (blue) VAF stability, including all six SMG mutations (Figure 2C). Eight other pairs exhibited correlation coefficients above 0.65 (Table S1L), suggesting that VAF stability was the rule, not the exception. Clearly, differences in tumor purity biased the correlation, as the originating tumors were variably contaminated with DNA from normal stromal elements, whereas the PDX had been computationally purified. However, WHIM8 stood out from the other cases by exhibiting a low correlation coefficient (0.26) and a relatively large number of xenograft-specific mutations in the homozygous range of 80% or higher (Figure 2D). This pattern suggests the emergence of a clone that was below the detection limit in the originating tumor sample but had become a significant contributor to the PDX mutational repertoire.
Most PDX-Specific Mutations Are Not Expressed
The RNA-seq approach (Cancer Genome Atlas Research Network, 2012b) detected mRNA expression from 462 (44%) of the 1,056 validated, nonsilent SNVs identified by DNA sequencing of 13 PDX tumors (Table S2M). Expression was detected for only 39 of the 69 SMG mutations. Of the PDX-unique SMG mutations, only WNK2 in WHIM8, PIK3R4 and KRAS in WHIM9, and MAP4K2 in WHIM16 were detectable in the RNA-seq data. However, all TP53 mutations were expressed at high levels (75%–100% of reads; Table S2H). The RNA-seq data were also used to examine the expression of PDX-specific missense mutations predicted to be functionally significant by Polyphen (Xi et al., 2004). Of the 34 mutations in this class, only 11 were expressed according to the RNA-seq reads (Table S2I): Hist1H1E in WHIM6, ABCC1 in WHIM8, WDR81 in WHIM13, MAP4K2 in WHIM16, ZNF687 in WHIM21, and KRAS, SLC23A, LRRC58, MAPK9, KIF21B, and PIK3R4 in WHIM9. Since MAP3K1 mutations have not been previously reported in available cell lines, RNA-seq analysis was used to confirm that a splice site mutation in WHIM20 indeed generated a splice donor, leading to an out-of-frame MAP3K1 transcript (Figure S10).
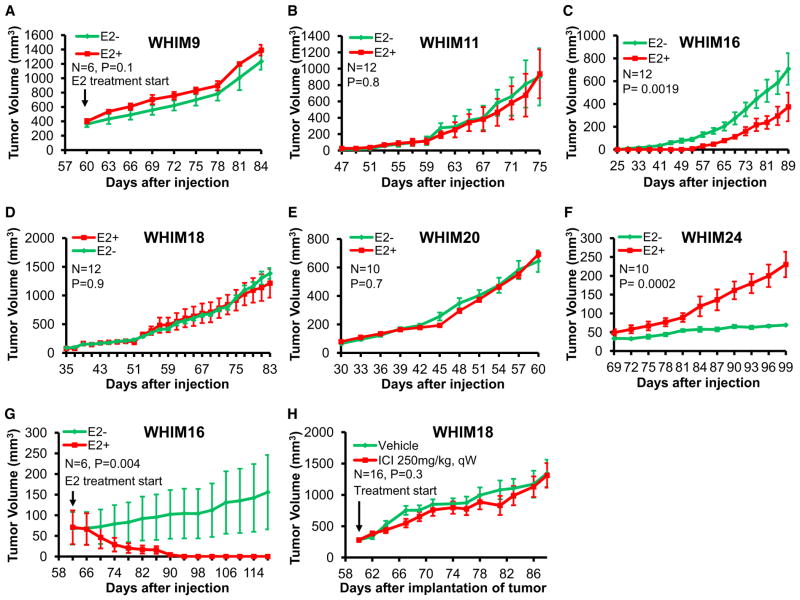
The Estradiol Response of ER+ PDX Mirrors the Clinical Phenotype of the Originating Tumor
The estradiol dependence of each ER+ PDX was studied by transplantation into oophorectomized mice with or without estradiol supplementation. Four lumenal PDX exhibited estradiol-independent growth (Figures 4A, 4B, 4D, and 4E) consistent with the fact these xenografted samples were accrued after the development of aromatase inhibitor resistance (Table S2A). WHIM24 was the only example that exhibited estradiol-dependent growth (Figure 4F); the patient who contributed this sample had a protracted clinical course and experienced a durable clinical response to tamoxifen after xenograft sample accrual. The growth of WHIM16 was delayed by estradiol (Figure 4C). Furthermore, established WHIM16 tumors exhibited marked regression in response to estradiol exposure (Figure 4G), modeling the paradoxical estradiol treatment of advanced breast cancer, which produced a modest response in the patient who contributed this sample (Ellis et al., 2009). The patient who donated WHIM18 had a particularly striking history of fulvestrant resistance (progression within 1 month of therapy; Table S2A), and WHIM18 proved to be just as fulvestrant unresponsive in the PDX setting (Figure 4H).
Figure 4. Estradiol Dependency and Tumor Doubling Times for the ER+ WHIM Lines.
(A and G) WHIM9 cells (A) or WHIM16 cells (G) were allowed to establish tumor nodules in ovariectomized nonobese diabetic (NOD)/severe combined immunodeficiency (SCID) mice and then treated with or without 17β-estradiol pellets.
(B–F) Tumor cells were subcutaneously injected into ovariectomized NOD/SCID mice and then immediately treated with 17β-estradiol pellets or observed.
(H) Fragments of WHIM18 tumor tissue were subcutaneously engrafted into female CB.17 SCID mice. Tumor-bearing mice were treated in the presence or absence of fulvestrant when tumor size reached 300 mm3. All data were analyzed in SAS using the PROC MIXED function.
ESR1 Translocation, Point Mutation, or Gene Amplification in ER+ PDX Models
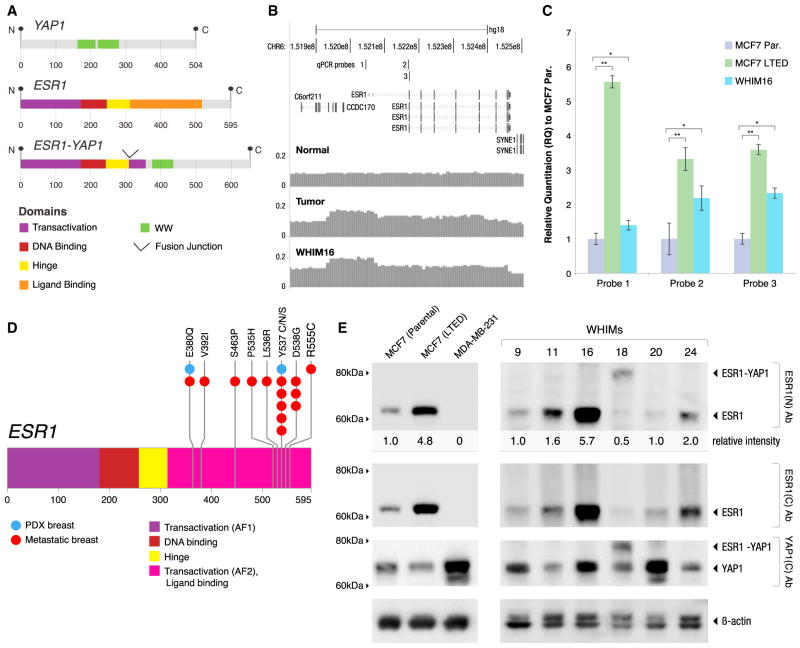
The RNA-seq data analysis identified five interchromosomal in-frame gene fusion events (Figure S11), including a balanced translocation between 6q and 11q in WHIM18 that created a transcript encoding the 5′ four exons of ESR1 (amino acids 1–365), fused to the C terminus of YAP1 (amino acids 230–504; Figure 5A). Western blots on WHIM18 extracts confirmed the presence of an appropriately sized ESR1/YAP1 fusion protein that was detected by an N-terminal ESR1 antibody and a YAP1 antibody, but not by a C-terminal ESR1 antibody (Figure 5E). Gene amplification across the ESR1 promoter and coding region was observed in WHIM16 (Figure 5B) and was associated with high levels of ESR1 protein (Figure 5D). To quantify and confirm the degree of amplification in WHIM16, we conducted quantitative PCR (qPCR) of the promoter region and two regions of the coding sequence of ESR1 (Figure 5C). In this experiment, MCF7 cell DNA was used as a nonamplified ESR1 control, and, unexpectedly, MCF7 cells that had been subjected to long-term estrogen deprivation (LTED) (Sanchez et al., 2011), but not parental MCF7 cells, showed marked ESR1 gene amplification (Figure 5C) with associated increased expression levels of ESR1 protein (Figure 5E). These data suggest that ESR1 amplification is an adaptation to estrogen deprivation in this well-studied model. WHIM20 expressed an ESR1-Y537S point mutation that was present in the majority of reads (96%) in the RNA-seq data (Table S2M). WHIM24 harbored an ESR1-E380Q mutation (Table S2D), which was not detected in the originating tumor but was present in 42 of 42 reads in the PDX (Figure S12). Low estradiol xenografting (i.e., no E2 supplementation) may therefore favor the growth of tumors with somatic variants in ESR1, since four out of seven ER+ PDXs analyzed by sequencing contained a mutation or a gene rearrangement. Interestingly, in a recent study by Piccart et al. (2013), ESR1 sequencing of advanced breast cancer samples from a clinical trial revealed both ESR1-Y537S and ESR1-E380Q. Piccart et al.’s report, in combination with our observations, clearly delineates a hot spot of ligand-binding-domain ESR1 mutations in advanced breast cancer (Figure 5D) that complement the initial example (Y537N) reported in the 1990s (Zhang et al., 1997).
Figure 5. ESR1 Gene Rearrangements and Point Mutations in Lumenal PDX Models.
(A) WHIM18 and the originating tumor harbored a balanced translocation between 6q and 11q in WHIM18 that created a fusion-transcript-defected mRNA-seq that encodes the 5′ four exons of ESR1 (amino acids 1–365, including the DNA-binding domain but not the steroid-binding domain) fused to the C terminus of YAP1 (amino acids 230–504), thereby excluding the TEAD domain and the first WW motif of YAP1, but retaining the second WW motif, the SH3 domain, the YES phosphorylation site, and the transactivation domain.
(B) WHIM16 and the originating tumor harbor amplification of the ESR1 gene that extends from the promoter region throughout the coding sequence that was mapped using read counts obtained during WGS.
(C) qPCR on genomic DNA using three separate probes was used to confirm gene amplification in WHIM16 cells. The negative control was MCF7 cells. In a screen for ESR1-gene-amplified cell lines, MCF7 cells that were adapted after LTED were found to have developed ESR1 gene amplification. qPCR results were normalized relative to parental MCF7 (Par.). The positions of probes 1, 2, and 3 are displayed in (B). Error bars are ±1 SD of the mean relative quantification (RQ); *p < 0.05, ** is p < 0.01.
(D) WHIM20 cells harbored and expressed a mutation in ESR1-Y537S, and WHIM24 harbors ESR1-E380Q (indicated in blue). The finding of ESR1-V537S and ESR1-E380Q in these PDX lines complements a recent report on ESR1 sequencing of advanced disease samples in which multiple mutations in the AF2/ligand-binding domain (in pink) were observed (Piccart et al., 2013; mutation positions from this report are indicated in red).
(E) Tumor lysates from six ESR1+ WHIM lines (WHIM9, WHIM11, WHIM16, WHIM18, WHIM20, and WHIM24) were analyzed by western blot using antibodies targeting the N terminus or C terminus of ESR1 or the C terminus of YAP1. In parallel, lysates from three breast cancer cell lines (parental MCF7, LTED MCF7, and MDA-MB-231) were analyzed as controls. All blots were replicated four times. ESR1 intensity detected by the N-terminal ER antibody was quantified and normalized against the actin level. For WHIM lines, the normalized ESR1 levels were averaged from four replicate blots and expressed as relative intensities using WHIM9 as the internal reference (arbitrarily set at one). For cell lines, ESR1 levels were similarly normalized against actin and expressed as relative values using parental MCF7 as the internal reference. Lysates from cell lines and WHIM tumors were analyzed in the same blot, but the images displayed reflect different exposure times.
See also Figure S15.
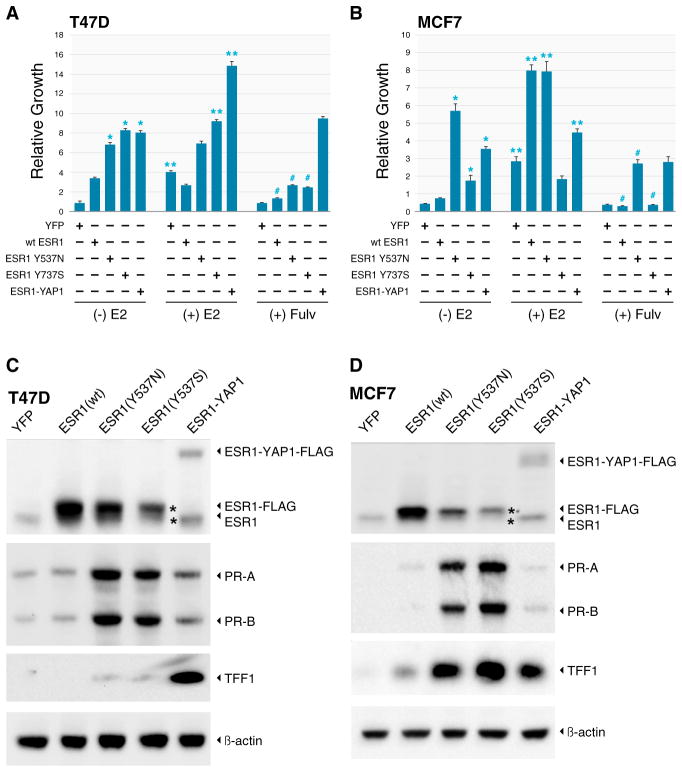
ESR1/YAP1 and ESR1-Y537S Induce Estradiol-Independent Growth
ESR1-Y537S and ESR1-Y537N are known to induce estradiol-independent transcriptional activity (Weis et al., 1996; Zhang et al., 1997). To compare the properties of ESR1 point mutations affecting the Y537 residue with the ESR1/YAP1 fusion gene product, MCF7 and T47D lines were engineered to overexpress wild-type ESR1, ESR1YAP1, ESR1-Y537S, and ESR1-Y537N proteins (YFP provided the control). Under low-estrogen conditions, all three mutant ESR1 constructs increased proliferation in T47D and MCF7 cells compared with the YFP control, and the three mutant ESR1 constructs were all more active than wild-type ESR1 (Figure 6A). In T47D cells, the proliferation of cells harboring ESR1-Y537N or ESR1-Y537S was largely estradiol independent, although some estradiol responsiveness was retained for ESR-Y537N in MCF7 cells. ESR1/YAP1 was as active as the two point mutants in inducing estradiol-independent growth, but E2 was able to further stimulate growth in ESR1/YAP1-expressing cells in T47D cells. This indicates that ESR1/YAP1 does not obviously function to inhibit the function of endogenous ESR1 (i.e., it was not dominant negative). Fulvestrant significantly inhibited the growth of cells expressing ESR1-Y537S, ESR1-Y537N, and wild-type ESR1, and induced downregulation of wild-type and mutant ER protein expression (Figure S13). However, growth suppression was incomplete for the two point mutants, suggesting partial resistance to fulvestrant. Cells expressing the ESR1/YAP1 fusion were clearly insensitive to fulvestrant and the fusion protein was not downregulated, since the ligand-binding domain of ESR1 is absent from this chimeric protein (Figure S13). Similar overall findings regarding growth induction under low-estrogen conditions by ESR1 mutants and the ESR1/YAP1 fusion were made in MCF7 cells (Figure 6B). Of note, however, cells expressing ESR1-Y537S grew poorly relative to cells expressing other ESR1 mutant constructs and exhibited signs of cell death (data not shown). Gain-of-function ESR1 point mutants may therefore require a cellular background that is tolerant of the extreme properties of these constitutively active ESR1 mutants. In accord with this hypothesis, ectopic expression levels for the Y537S mutant were lower than wild-type ER in both T47D and MCF7 (Figures 6C and 6D), and were extremely low in WHIM20, which naturally expresses the ESR1-Y537S mutant (Figure 5E). Lysates from cells grown in charcoal-stripped serum (CSS) media were analyzed for progesterone receptor (PR) and trefoil factor 1 (TFF1) expression by western blot. In both cell lines, the ESR1 point mutations strongly induced PR expression in a hormone-independent fashion relative to wild-type ESR1 and YFP control lines, whereas ESR1/YAP1 had a more modest effect. For TFF1, the two ESR1 point mutants induced expression in MCF7 cells in low-estradiol conditions, but caused less induction in the T47D cells. ESR1/YAP1 strongly induced TFF1 expression relative to wild-type in both cell lines. In the WHIM tumors that expressed these mutations naturally, high PR expression was associated with the lines expressing the mutations or gene rearrangements (WHIM16, WHIM18, WHIM20, and WHIM24). In contrast, WHIM lines with a wild-type ESR1 locus (WHIM9 and WHIM11) but estradiol-independent growth showed very low levels of PR expression (Figure S14).
Figure 6. Point Mutations and a Translocation in ESR1 Induce Estradiol-Independent Growth.
(A and B) T47D(A) and MCF7 (B) cells stably transduced with lentiviral vectors expressing the YFP control gene (YFP), wild-type ESR1 (WT ESR1), ESR1 point mutants (ESR1-Y537N and ESR1-Y537S), and the ESR1-YAP1 fusion gene (ESR1-YAP1) were grown in CSS medium for at least 2 weeks. Cells were then plated in CSS medium containing no supplemental estrogen (−E2), 10 nM estradiol (+E2), or medium without estrogen +500 nM fulvestrant (+ Fulv), and growth was measured after 10 days by Alamar blue assay. Mean results, with standard SEM as error bars, are shown for four experiments (T47D) and three experiments (MCF7), with each experiment conducted in quadruplicate. Cell growth in each line was normalized to baseline values obtained the day after the cells were plated, prior to the beginning of treatment. Expression of the ESR1 point mutants and ESR1-YAP1 fusion significantly promoted the growth of estrogen-deprived cells compared with WT ESR1 or YFP control (*p < 0.05 indicates significant growth stimulation versus YFP or WT ER). The effect of estradiol was then assessed for each lentivirus construct (**p < 0.05 indicates a significant stimulatory effect for each construct with and without estradiol). In T47D cells, estradiol stimulated the growth of YFP, ESR1-Y537S (minimally), and ESR1-YAP1, but not WT-ESR1 or ESR1-Y537N. In contrast, in MCF7 cells, estradiol promoted the growth of WT-ESR1, ESR1-Y537N, and to a much lesser extent ESR1-YAP1, but not ESR1-Y537S. Treatment with fulvestrant significantly inhibited estrogen-independent growth of cells expressing WT ER and ER point mutants (#p < 0.05), but not the ER-YAP1 fusion.
(C and D) T47D (C) and MCF7(D) cells were cultured for 8 days in CSS medium, followed by western blot for the expression of endogenous and exogenous ESR1 using an N-terminal antibody and two direct ESR1 downstream targets (progesterone receptor [PR-A and PR-B] and TFF1) with an actin loading control. Due to the substantially lower basal TFF1 expression in T47D cells compared with MCF7 cells, the T47D TFF1 blot was intentionally exposed for a longer time for visualization.
See also Figures S13 and S14.
DISCUSSION
In this study, we assessed the degree to which breast cancer PDX models are genomic replicas of human tumors based on genome-wide analysis, including translocations, insertions, deletions, point mutations, and amplification events. The stability of SVs was striking, suggesting that these genomic features may stabilize early in pathogenesis, perhaps during telomere crisis (Chin et al., 2004). Regarding SNVs, almost all mutations detected in the originating tumor were present in the PDX, but a variable number of SNVs were PDX unique (Table S2I). However, RNA-seq data indicated that most PDX-unique SNVs have expression levels below the detection limit and therefore may be passengers. PDX-unique mutations could arise in the mouse after xenografting or may represent a rare subclone that existed below the detection limit in the originating tumor but increased into the detectable range during growth in the mouse. Deep analysis of a “quartet” of a primary and metastasis pair with their counterpart PDX lines WHIM2 and WHIM5 suggests that xenograft-specific mutations indeed arise over time. When we compared samples taken at a later time point with those obtained an earlier time point, whether in the human setting (primary tumor and brain metastasis) or xenograft setting, we found that the later samples contained multiple SNVs (Figure 3). These mutations may increase “tumor fitness” in the transplanted environment or may just be passengers in a constantly mutating tumor (most were in noncoding regions). PDX-specific SNVs may simply arise from serial population reductions during repeated xenografting events, which can select a passenger mutation at random due to cell attrition during transplantation (“population bottlenecking”; Gisselsson et al., 2010). This might explain the accumulation of seemingly nonfunctional mutations observed with late-passage exome sequencing (Table S2K). However, selection by increasing tumor fitness is a more likely explanation for cases in which the PDX-specific mutations were detectable at the mRNA level and were functionally linked to cancer biology, such as WNK2 in WHIM8 (Jun et al., 2009; Moniz et al., 2007), PIK3R4 (Huang et al., 2011; Shull et al., 2012) and KRAS (Santos et al., 1984) in WHIM9, MAP4K2 in WHIM16 (Lau et al., 2012), and ESR1-E380Q in WHIM24. Thus, we are not arguing that PDXs are perfect genomic replicas of the originating tumors; rather, we suggest that tracking the PDX genome, benchmarked against the originating tumor, is a way to assess the ongoing genomic integrity of the model during experimentation. This is something that has never been considered for conventional cell-line approaches, where the progenitor tumor genome is rarely available and analysis of ongoing genetic drift is not a routine aspect of experimental design.
An important issue that was not addressed by previous investigations is the effect of the xenografting process on the VAF, since each mutation can either be present in the founder clone (and therefore present in all cells) or arise later in a subclone and therefore occur with a lower frequency because it is present in only a subpopulation of cells. VAF analysis, perhaps surprisingly, showed that the VAF for many mutations was preserved in the PDX, even in the case of rare mutations. This implies that clonal representation can be transplantable, i.e., different clones maintain their relative prevalence in equilibrium. Since clonal prevalence is maintained despite growth in an immunocompromised host, immunoediting (differential immune responses against particular mutant proteins) is an unlikely explanation for relative clone abundance in this setting (DuPage et al., 2012; Matsushita et al., 2012). Our findings are compatible with other recent studies on the clonal diversity of epithelial cancers, which showed that minor clones are carried at low frequencies for many passages until a section event (e.g., therapeutic intervention or the process of adaption to growth in a new organ) increases the minor mutation prevalence (Ding et al., 2012; Kreso et al., 2013).
Genomic analysis of each ER+ PDX raised tumor-unique hypotheses to explain endocrine-therapy resistance, underscoring the etiological heterogeneity of this common clinical problem. The WHIM11 line was isolated from a patient with a fulminant clinical course and little evidence for sensitivity to endocrine therapy (Table S2A). Despite the patient’s ER+ HER2− status, WHIM11 was classified as HER2-E by PAM50. This biomarker pattern predicts poor responsiveness to aromatase inhibition (Ellis et al., 2011). WHIM11 was a TP53 mutant and harbored a biallelic deletion in PTEN (Figure S6). RPPA data confirmed high levels of pS70S5K and 4EBP1 protein phosphorylation, indicating phosphoinositol-3-kinase pathway activation (Figure S3), which has been implicated in endocrine-therapy resistance (Sanchez et al., 2011). WHIM9 harbored monoallelic expression of an R515I mutation in SMAD4 (Table S2M). SMAD4 mutations were recently associated with genome instability in head and neck cancer (Bornstein et al., 2009), which could explain why this particular lumenal PDX had a high rate of PDX-specific mutations, although which mutation caused endocrine resistance in this line remains unclear.
WHIM16 exhibited paradoxical regression with estradiol, which is an effective but nonintuitive late-line endocrine therapy for some advanced ER+ breast cancers (Ellis et al., 2009). The ESR1 amplification and high-level ESR1 protein expression in WHIM16 therefore raise the hypothesis that ESR1 amplification may be a predictive marker for responsiveness to estradiol therapy in advanced disease. This suggestion is consistent with the finding (Figures 5B and 5C) that MCF7 cells develop ESR1 gene amplification after LTED in vitro, conditions under which estradiol is well known to induce apoptosis (Lewis et al., 2005; Song et al., 2001). In T47D cells, overexpression of wild-type ESR1 gene/protein increased growth in low-estradiol conditions, supporting the notion that by driving ESR1 overexpression, ESR1 gene amplification promotes adaptive resistance to estrogen deprivation (Figure 6B). This hypothesis is also compatible with clinical observations indicating that ESR1 amplification is associated with poor clinical outcome (Ejlertsen et al., 2012; Lin et al., 2013; Nielsen et al., 2011).
The ESR1-Y537S hormone-binding-domain mutation is clearly a potent cause of aromatase-inhibitor resistance. Expression of ESR1-Y537S produced greater growth than wild-type ESR1 under estrogen-deprived conditions and very strong induction of PR in the absence of estradiol in both cell lines tested (Figures 6C and 6D). Since ESR1-Y537S (or other mutations in this region of ESR1) was not observed in >500 exome sequencing experiments by the TCGA, it seems likely that mutations in the hormone-binding domain largely occur as an adaptation to endocrine treatment. Consistent with this conclusion, an ESR1 mutation hotspot in the ligand-binding-domain/AF2 region was observed in metastatic samples from a clinical trial for patients with nonsteroidal aromatase-inhibitor-resistant advanced breast cancer (Piccart et al., 2013). Our in vitro data indicate that ESR1-Y537S was responsive to fulvestrant, as protein expression was downregulated. However, suppression of growth was incomplete, indicating partial resistance (Figures 6 and S13). The patient whose tumor harbored ESR1-Y537S (WHIM20) experienced only 4 months of clinical benefit from fulvestrant, which is compatible with the hypothesis that ESR1-Y537S-positive tumors may be less responsive to this commonly used second-line endocrine intervention for advanced breast cancer. WHIM24, a PDX that was estradiol dependent (Figure 4) and associated with a tamoxifen clinical response (but resistance to aromatase inhibition) harbored an ESR1-E380Q mutation. This mutation has already been documented to be associated with estradiol hypersensitivity, increased DNA binding, and estradiol-independent activity (Pakdel et al., 1993). However, ESR1-E380Q was not detected in the relatively low-coverage WGS analysis of the originating tumor, so the link with the clinical phenotypes observed is uncertain (Figure S11).
The identification of the ESR1/YAP1 fusion gene in WHIM18 completes the mechanistic spectrum of gain-of-function mutations in ESR1 associated with endocrine-resistant breast cancers. YAP1 plays a central role in organ size and tumorigenesis through the Hippo pathway (Lin et al., 2013); however, the domains that are responsible for most of these biological properties are in the N terminus of YAP1 and therefore absent from the fusion gene. Analysis of TCGA breast cancer RNA-seq data revealed two other in-frame fusion genes that preserve at least the first four exons of ESR1 (preserving DNA binding). In one case, a fusion was detected with AKAP12, a putative tumor-suppressor gene (Gelman, 2012), and in the second case it was detected with POLH, a DNA polymerase associated with xeroderma pigmentosum (Ortega-Recalde et al., 2013; Figure S15). These findings indicate that the ESR1/YAP1 translocation documented in WHIM18 is not a private event, but is a member of a class of translocations that preserve the DNA-binding and AF1 domains of ESR1 with variable in-frame C-terminal partners that replace the ligand-binding and AF2 domains. Although these in-frame ESR1 translocations are likely rare, the denominator for breast cancer is so large that even low percentages of particular somatic events can represent clinically significant patient populations if the effect on the disease course is dramatic (i.e., in this instance, intrinsic and universal endocrine-therapy resistance).
In conclusion, PDX models validated through comparative whole-genome analysis against the originating tumor are a useful starting point for studies of the molecular pharmacology of advanced breast cancer. No actively growing cancer has a static genome, and genetic drift though cell attrition is inherent in the xenografting process. Furthermore, selection of mutations that increase tumor fitness in the murine environment is to be expected. However, unlike conventional cell lines, PDX-specific mutations can be monitored with reference to the genome of the originating human tumor, establishing a tumor “pedigree” that can be checked before and after each functional or pharmacological experiment. This continuous genomic annotation approach is illustrated by the late-passage exome sequencing conducted in WHIM2 (Figure 3).
The identification of endocrine-resistance-associated ESR1 gene rearrangements and point mutations has deep implications for the management of metastatic breast cancer. The choice of endocrine therapy in an advanced-disease setting could be based on the presence and class of ESR1 gene mutations and rearrangements if more were known about the correlations with outcomes. The detection of these mutations in the xenograft setting establishes the principle that the PDX approach captures genomic events that have been understudied in the past because they are not present in conventional ER+ cell lines even when experimentally selected for endocrine drug resistance in vitro. The availability of authentic PDX-based models of endocrine-therapy-resistant lumenal breast cancer will facilitate the testing of therapeutic interventions and perhaps particularly those designed to more effectively target mutant forms of ESR1.
EXPERIMENTAL PROCEDURES
Generation and Analysis of PDX Breast Cancer Models
All human tissues for these experiments were processed in compliance with NIH regulations and institutional guidelines, and approved by the institutional review board at Washington University. All animal procedures were reviewed and approved by the institutional animal care and use committee at Washington University in St. Louis. Detailed methods are provided in Extended Experimental Procedures. PDX models are available through the application to the Human and Mouse-Linked Evaluation of Tumors core at http://digitalcommons.wustl.edu/hamlet/.
WGS and Capture Validation
Seventeen patients with blood, tumor, and xenograft were selected for WGS. Detailed histories for these patients and xenografts are provided in Table S2A. Libraries were prepared using unamplified genomic DNA from blood (normal), tumor, and xenograft samples. Paired-end sequencing was performed on the Illumina platform as previously described (Ellis et al., 2012; Walter et al., 2012). Variant calling and validation of all mutations using liquid-phase hybridization capture were performed as previously described (Welch et al., 2012). All DNA have been deposited with dbGAP under accession number phs000611 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000611.v1.p1).
mRNA-Seq
mRNA-seq was performed as previously described (Cancer Genome Atlas Research Network, 2012b). Reads were mapped using the MapSplice algorithm (Wang et al., 2010). Expressed gene fusions were nominated using ChimeraScan v0.4.3 (Iyer et al., 2011) with default parameters. Gene fusion nominations were required to have at least two independent spanning junction reads. Sequences will be made available upon application to CGHub. TCGA mRNA-seq data can be accessed through the TCGA program (http://cancergenome.nih.gov). All PDX mRNA-seq data have also been deposited with dbGAP under accession number phs000611 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000611.v1.p1).
DNA Microarray-Based Gene-Expression Analysis
Agilent’s 4x44K Whole Human Gene expression microarray processing, data quality control and processing, and research use only PAM50 subtype classification were previously described (Ellis et al., 2011). The stromal-related genes were identified after a two-class paired significance analysis of microarrays (SAM) with an FDR of 0% between 18 paired progenitor human tumors and xenografts (Table S2B). The GEO accession number for the chip-based gene expression data reported in this paper is GSM41685. PDX tumors were also analyzed by 244K UNC customized Agilent chips for clustering with data with unmatched primary tumors (GEO accession number GSE46604).
Quantification of ESR1 Amplification
A real-time PCR system (Life Technologies) was run for ESR1 amplifications using control genes FAM38B and ASXL2 as described previously (Reis-Filho et al., 2008).
RPPA, In Vitro Growth Assays, Lentivirus Gene Transduction, and Western Blots
Standard methods were used for RPPA (Tabchy et al., 2011); see Extended Experimental Procedures for other standard protein-analysis approaches.
Statistical Methods
Hierarchical clustering was applied with a distance metric of one minus the Pearson correlation coefficient and using the average linkage method. Clustering results were visualized as dendrograms in heatmaps. Pearson and Spearman rank-based correlation coefficients were separately calculated to demonstrate VAF stability between a PDX and its patient origin.
Supplementary Material
Acknowledgments
This work was funded by grants to M.J.E. by Susan G. Komen for the Cure (BCTR0707808, KG090422, and PG12220321). The DNA sequencing and analysis were performed by the Genome Institute at Washington University School of Medicine and supported by grants to R.K.W. from the National Human Genome Research Institute (NHGRI U54 HG003079). The Tissue Procurement Core was supported by NCI 3P50 CA68438. The HAMLET Core was supported by CTSA grant UL1 RR024992. M.J.E. is also supported as a Susan G. Komen Scholar and by the Breast Cancer Research Fund, the Barnes Jewish Hospital Foundation, the Theresa Harpole Foundation for Metastatic Breast Cancer, and a gift from the Mary Speak family. The RPPA analysis was supported by NCI grants PO1CA099031, U54CA112970, KG081694, and P30 CA16672 to G.M. The RNA-seq was supported by grants from the TCGA Project (U24-CA143848), the NCI Breast SPORE Program (P50-CA58223-09A1), and the Breast Cancer Research Foundation to C.M.P. M.J.E. and C.M.P. own shares and have leadership positions in Bioclassifier and University Genomics, which has licensing income from patents related to the ProSigna breast cancer intrinsic subtype test.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ACCESSION NUMBERS
The dbGAP accession number for the DNA and PDX mRNA sequences reported in this paper is phs000611. The GEO accession number for the gene-expression data used in Figure 1B is GSM41685. PDX tumors were also analyzed by 244K UNC customized Agilent chips for clustering with data from primary tumors (GEO accession number GSE46604; Figure S3).
Supplemental Information includes Extended Experimental Procedures, 15 figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.08.022.
WEB RESOURCES
The URLs for data presented herein are as follows:
The Cancer Genome Atlas, http://cancergenome.nih.gov
Digital Commons, http://digitalcommons.wustl.edu/hamlet/
Index of OOMPA, http://bioinformatics.mdanderson.org/OOMPA
NCBI, http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000611.v1.p1
UNC Microarray Database, https://genome.unc.edu/
References
- Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, Reh D, Andersen P, Gross N, Olson S, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–3419. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012a;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012b;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo WL, Ljung BM, Chew K, Myambo K, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, Factor R, Matsen C, Milash BA, Nelson E, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejlertsen B, Aldridge J, Nielsen KV, Regan MM, Henriksen KL, Lykkesfeldt AE, Müller S, Gelber RD, Price KN, Rasmussen BB, et al. Prognostic and predictive role of ESR1 status for postmenopausal patients with endocrine-responsive early breast cancer in the Danish cohort of the BIG 1-98 trial. Ann Oncol. 2012;23:1138–1144. doi: 10.1093/annonc/mdr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Perou CM. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov. 2013;3:27–34. doi: 10.1158/2159-8290.CD-12-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Gao F, Dehdashti F, Jeffe DB, Marcom PK, Carey LA, Dickler MN, Silverman P, Fleming GF, Kommareddy A, et al. Lower-dose vs high-dose oral estradiol therapy of hormone receptor-positive, aromatase inhibitor-resistant advanced breast cancer: a phase 2 randomized study. JAMA. 2009;302:774–780. doi: 10.1001/jama.2009.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Suman VJ, Hoog J, Lin L, Snider J, Prat A, Parker JS, Luo J, DeSchryver K, Allred DC, et al. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype—ACOSOG Z1031. J Clin Oncol. 2011;29:2342–2349. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JM, Miller TC, Meyer MJ, Ginsburg E, Vonderhaar BK. Local regulation of human breast xenograft models. J Cell Physiol. 2010;224:795–806. doi: 10.1002/jcp.22190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman IH. Suppression of tumor and metastasis progression through the scaffolding functions of SSeCKS/Gravin/AKAP12. Cancer Metastasis Rev. 2012;31:493–500. doi: 10.1007/s10555-012-9360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Lindgren D, Mengelbier LH, Øra I, Yeger H. Genetic bottlenecks and the hazardous game of population reduction in cell line based research. Exp Cell Res. 2010;316:3379–3386. doi: 10.1016/j.yexcr.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang L, Greshock J, Colligon TA, Wang Y, Ward R, Katsaros D, Lassus H, Butzow R, Godwin AK, et al. Frequent genetic abnormalities of the PI3K/AKT pathway in primary ovarian cancer predict patient outcome. Genes Chromosomes Cancer. 2011;50:606–618. doi: 10.1002/gcc.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun P, Hong C, Lal A, Wong JM, McDermott MW, Bollen AW, Plass C, Held WA, Smiraglia DJ, Costello JF. Epigenetic silencing of the kinase tumor suppressor WNK2 is tumor-type and tumor-grade specific. Neuro-oncol. 2009;11:414–422. doi: 10.1215/15228517-2008-096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabos P, Finlay-Schultz J, Li C, Kline E, Finlayson C, Wisell J, Manuel CA, Edgerton SM, Harrell JC, Elias A, Sartorius CA. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res Treat. 2012;135:415–432. doi: 10.1007/s10549-012-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, O’Brien CA, van Galen P, Gan OI, Notta F, Brown AM, Ng K, Ma J, Wienholds E, Dunant C, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci USA. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KS, Zhang T, Kendall KR, Lauffenburger D, Gray NS, Haigis KM. BAY61-3606 affects the viability of colon cancer cells in a genotype-directed manner. PLoS ONE. 2012;7:e41343. doi: 10.1371/journal.pone.0041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JS, Meeke K, Osipo C, Ross EA, Kidawi N, Li T, Bell E, Chandel NS, Jordan VC. Intrinsic mechanism of estradiol-induced apoptosis in breast cancer cells resistant to estrogen deprivation. J Natl Cancer Inst. 2005;97:1746–1759. doi: 10.1093/jnci/dji400. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JI, Poon CL, Harvey KF. The Hippo size control pathway—ever expanding. Sci Signal. 2013;6:pe4. doi: 10.1126/scisignal.2003813. [DOI] [PubMed] [Google Scholar]
- Marangoni E, Vincent-Salomon A, Auger N, Degeorges A, Assayag F, de Cremoux P, de Plater L, Guyader C, De Pinieux G, Judde JG, et al. A new model of patient tumor-derived breast cancer xenografts for pre-clinical assays. Clin Cancer Res. 2007;13:3989–3998. doi: 10.1158/1078-0432.CCR-07-0078. [DOI] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz S, Veríssimo F, Matos P, Brazão R, Silva E, Kotelevets L, Chastre E, Gespach C, Jordan P. Protein kinase WNK2 inhibits cell proliferation by negatively modulating the activation of MEK1/ERK1/2. Oncogene. 2007;26:6071–6081. doi: 10.1038/sj.onc.1210706. [DOI] [PubMed] [Google Scholar]
- Nielsen KV, Ejlertsen B, Müller S, Møller S, Rasmussen BB, Balslev E, Lænkholm AV, Christiansen P, Mouridsen HT. Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res Treat. 2011;127:345–355. doi: 10.1007/s10549-010-0984-y. [DOI] [PubMed] [Google Scholar]
- Ortega-Recalde O, Vergara JI, Fonseca DJ, Ríos X, Mosquera H, Bermúdez OM, Medina CL, Vargas CI, Pallares AE, Restrepo CM, Laissue P. Whole-exome sequencing enables rapid determination of xeroderma pigmentosum molecular etiology. PLoS ONE. 2013;8:e64692. doi: 10.1371/journal.pone.0064692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakdel F, Reese JC, Katzenellenbogen BS. Identification of charged residues in an N-terminal portion of the hormone-binding domain of the human estrogen receptor important in transcriptional activity of the receptor. Mol Endocrinol. 1993;7:1408–1417. doi: 10.1210/mend.7.11.8114756. [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccart M, Rugo H, Chen D, Campone M, Burris AH, Taran T, Sahmoud T, Deleu I, Hortobagyi GN, Baselga J. Assessment of genetic alterations in postmenopausal women with hormone receptor-positive, HER2-negative advanced breast cancer from the BOLERO-2 trial by next-generation sequencing. Ann Oncol. 2013;24:iii25–iii26. [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho JS, Drury S, Lambros MB, Marchio C, Johnson N, Natrajan R, Salter J, Levey P, Fletcher O, Peto J, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet. 2008;40:809–810. doi: 10.1038/ng0708-809b. author reply 810–812. [DOI] [PubMed] [Google Scholar]
- Sanchez CG, Ma CX, Crowder RJ, Guintoli T, Phommaly C, Gao F, Lin L, Ellis MJ. Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer. Breast Cancer Res. 2011;13:R21. doi: 10.1186/bcr2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos E, Martin-Zanca D, Reddy EP, Pierotti MA, Della Porta G, Barbacid M. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull AY, Latham-Schwark A, Ramasamy P, Leskoske K, Oroian D, Birtwistle MR, Buckhaults PJ. Novel somatic mutations to PI3K pathway genes in metastatic melanoma. PLoS ONE. 2012;7:e43369. doi: 10.1371/journal.pone.0043369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RX, Mor G, Naftolin F, McPherson RA, Song J, Zhang Z, Yue W, Wang J, Santen RJ. Effect of long-term estrogen deprivation on apoptotic responses of breast cancer cells to 17beta-estradiol. J Natl Cancer Inst. 2001;93:1714–1723. doi: 10.1093/jnci/93.22.1714. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabchy A, Hennessy BT, Gonzalez-Angulo AM, Bernstam FM, Lu Y, Mills GB. Quantitative proteomic analysis in breast cancer. Drugs Today (Barc) 2011;47:169–182. doi: 10.1358/dot.2011.47.2.1576695. [DOI] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis KE, Ekena K, Thomas JA, Lazennec G, Katzenellenbogen BS. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol Endocrinol. 1996;10:1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi T, Jones IM, Mohrenweiser HW. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004;83:970–979. doi: 10.1016/j.ygeno.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Youlden DR, Cramb SM, Dunn NA, Muller JM, Pyke CM, Baade PD. The descriptive epidemiology of female breast cancer: an international comparison of screening, incidence, survival and mortality. Cancer Epidemiol. 2012;36:237–248. doi: 10.1016/j.canep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang QX, Borg A, Wolf DM, Oesterreich S, Fuqua SA. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 1997;57:1244–1249. [PubMed] [Google Scholar]
- Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, Landis MD, Wiechmann L, Schiff R, Giuliano M, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.