Abstract
In mass spectrometry based proteomics, data-independent acquisition (DIA) strategies have the ability to acquire a single dataset useful for identification and quantification of detectable peptides in a complex mixture. Despite this, DIA is often overlooked due to noisier data resulting from a typical five to ten fold reduction in precursor selectivity compared to data dependent acquisition or selected reaction monitoring. We demonstrate a multiplexing technique which improves precursor selectivity five-fold.
Keywords: Data Independent Acquisition, Q-Exactive, Multiplexing, Targeted Proteomics, Shotgun Proteomics
In discovery proteomics experiments, tandem mass spectrometry (MS/MS) data are usually collected on peptides by automated data-dependent acquisition (DDA). Using DDA, mass information on intact peptides in a previous MS spectrum is used to decide which subset of peptides will be targeted for acquisition of fragmentation (MS/MS) spectra necessary for sequence identification1. Although DDA is a powerful and versatile strategy, it suffers from several fundamental limitations.
When using DDA, the number of peptides sampled is limited by the MS/MS sampling speed despite the dynamic range and peak capacity of the mass analyzer. A single MS spectrum can contain over a hundred molecular species, of which only a handful are analyzed by MS/MS prior to the next full scan. This sampling of peptides for MS/MS analysis follows a random sampling model2 biased towards high-abundance peptides. Thus, in a complex protein digest, as many as 84% of peptides remain unsampled3 and as many as 30% of the sampled peptides can vary between replicate analyses of the same sample2.
DDA may compromise the sensitivity of MS/MS, because the full-scan mass spectrum used for the selection of the precursor ions typically contains greater chemical background interference than an MS/MS spectrum. A peptide could have abundance above the MS/MS detection limit but go unselected for fragmentation because the precursor is masked by background interference. Additional challenges are that 1) the MS/MS spectra acquired by DDA are rarely sampled at the optimal portion of the peptide elution profile4, and 2) as many as 15 – 20% of the sampled MS/MS spectra from a complex mixture are chimeric (i.e. contain two or more coeluting molecular species within the isolation window)5. An alternative to DDA is data-independent acquisition (DIA). In DIA, MS/MS scans are collected systematically and independent of precursor information. This approach has seen many variations such as collecting fragmentation data without precursor ion selection6, using ion mobility spectrometry-CID-time of flight mass spectrometry7, using wide isolation windows8, and using narrow isolation windows combined with many injections9. However, until recently, all of these methods have used a database searching strategy to assign peptide sequence to the fragmentation spectra. Because of the increased complexity of these MS/MS spectra, DIA methods have struggled to compete with the collection of MS/MS spectra using a traditional DDA strategy combined with database searching.
Recently, alternative strategies in which target peptides are queried against DIA data as opposed to trying to assign peptide sequences to every chimeric MS/MS spectrum acquired were reported10, 11. These strategies are similar to a targeted analysis using selected reaction monitoring (SRM) and fundamentally incomparable with a discovery experiment that uses a database search engine to qualitatively profile the peptide content in a mixture. The appeal of the approach is that any peptide precursor and product ion data within the limit of detection of the instrument can be extracted from the data. The relative fragment ion intensities, peptide precursor isotope peaks, and retention time of the extracted ion chromatograms are used to confirm the identity of the target molecular species just as with a targeted SRM experiment. Unlike SRM, different hypotheses can be tested on the data without having to perform another mass spectrometry experiment. However, with current mass spectrometers approaching acquisition speeds of 10 Hz, a 20 m/z wide precursor isolation window is required to sample a 400 m/z range every 2 seconds. This wide isolation width is undesirable due to substantially increased fragment ion interference (Supplementary Figs. 1–3). For example, peptides and their modified forms (e.g. oxidized methionine) are difficult to distinguish if they are isolated in the same window due to similar fragmentation patterns (Supplementary Fig. 1).
We present a multiplexing strategy (MSX) where five separate 4 m/z isolation windows are analyzed per spectrum. These spectra are demultiplexed into the five separate 4 m/z isolation windows using a novel strategy with similarities to Hadamard multiplexing12 resulting in data with the sampling frequency of a DIA approach using 20 × 20 m/z wide windows but the selectivity of an approach using 100 × 4 m/z wide windows. Demultiplexing improves precursor selectivity by 1) narrowing down the range of potential precursors for an MS/MS spectrum from a 20 to 4 m/z window (Supplementary Figs. 4 and 5), and 2) generating the unmixed fragment ion spectrum with signal from only the 4 m/z window. We implement this de-multiplexing approach within the popular open source Skyline software tool13 which provides a useful interface for the visualization and analysis of these data. For a detailed discussion on precursor selectivity, the design of MSX methods, and comparisons to other data acquisition techniques refer to Supplementary Note.
We collected MSX LC-MS/MS data on a S. cerevisiae lysate using five 4-m/z wide isolation windows per scan on a Q-Exactive (Thermo Fisher Scientific) mass spectrometer. Due to the multiple fills per mass analysis, this multiplexing technique is best suited for instrumentation where isolation and collisional activation of peptides is fast relative to mass analysis. We randomly selected five of the 100 possible 4 m/z isolation windows in the range of 500–900 m/z to be analyzed in each multiplexed scan (Fig. 1, Methods). To analyze these spectra, we modified Skyline to detect MSX spectra and de-multiplex them automatically on import (available in v. 1.3). We generated a Skyline document containing peptides with spectra in the NIST S. cerevisiae QTOF and Ion Trap spectral libraries (5/24/2011 builds)14. We analyzed the spectra with and without de-multiplexing.
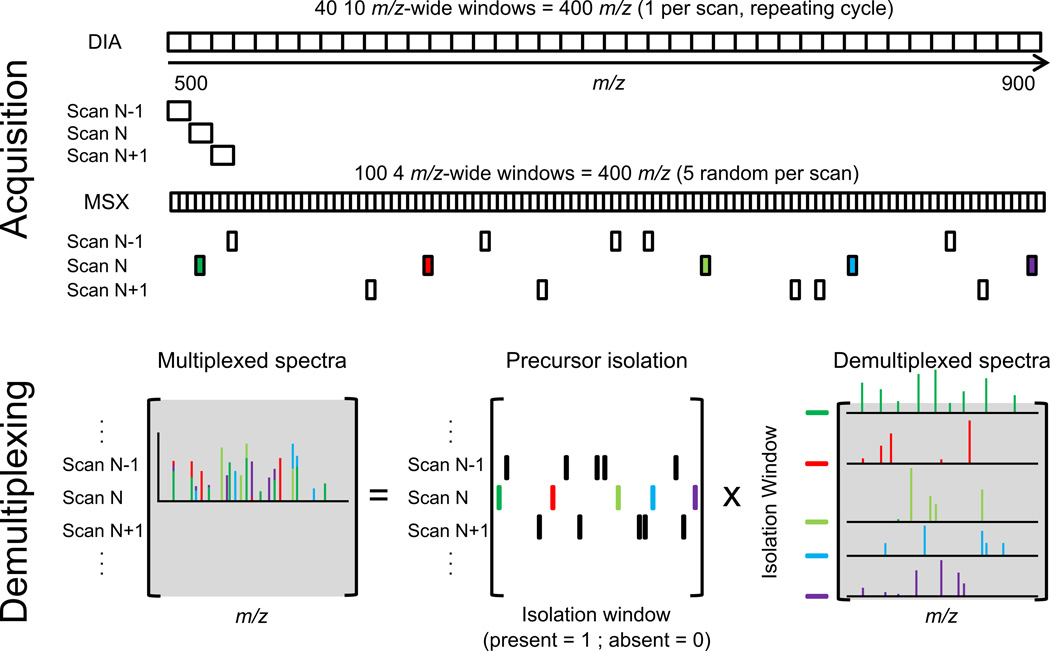
Figure 1. Multiplexed Data Independent Acquisition (MSX).
A common implementation of data independent acquisition (DIA) is to use a repeated cycle of wide isolation window MS/MS scans to cover a mass range. In this example, the 500–900 m/z range is covered with 40 scans each sampling a single 10 m/z wide window. In multiplexed DIA (MSX), each scan isolates five 4 m/z wide windows prior to fragment ion mass analysis. The five windows isolated in each scan are chosen randomly from the set of 100 possible non-overlapping windows covering the 500–900 m/z range. Each mixed MSX spectrum is de-multiplexed into the five component spectra corresponding to each isolated window.
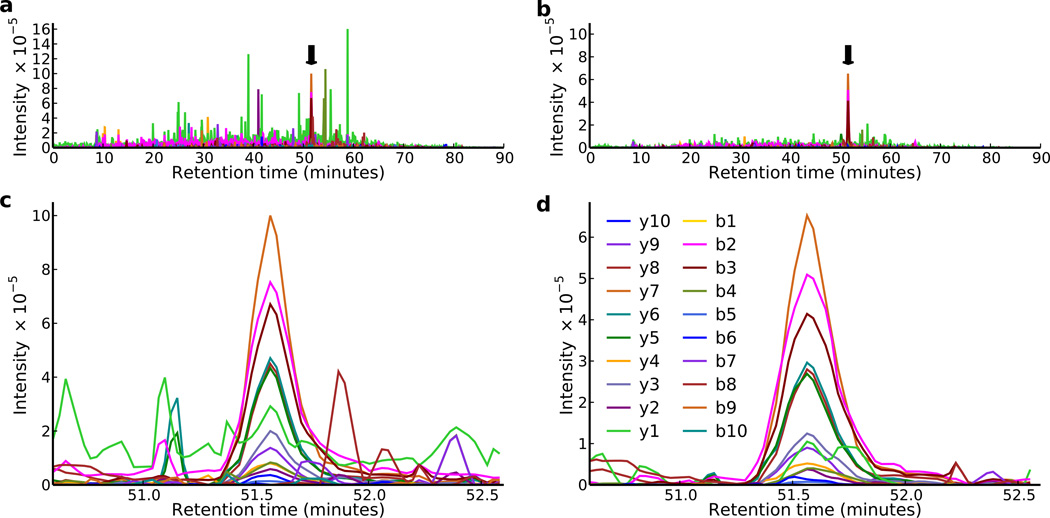
Extracted data for the peptide GPLVLEYETYR without de-multiplexing contained many intense fragment ion peaks present throughout the gradient which were fragments from other peptides (Fig. 2a). De-multiplexing removed the majority of these interfering peaks because they did not originate from precursors in the same isolation window as GPLVLEYETYR (Fig. 2b). There was also interference in many of the fragment ion chromatograms that overlapped in elution time with GPLVLEYETYR (Fig. 2c). This interference had a characteristic “spike” shape because it originated from precursors in different isolation windows than the target peptide. Due to the random sampling of isolation windows for each scan (Methods, Fig. 1, Supplementary Video), the window containing the interfering precursor was not isolated in consecutive scans containing the target isolation window. De-multiplexing removed the interfering signal while retaining the signal originating from the target isolation window (Fig. 2d), resulting in a higher dot-product similarity (0.96 vs. 0.94) to a DDA spectrum for GPLVLEYETYR acquired with a 2 m/z wide isolation window.
Figure 2. Demultiplexing Reduces Chemical Noise and Improves Selectivity.
The full b- and y- ion series for the peptide GPLVLEYETYR are plotted from an MSX experiment on the soluble fraction of S. cerevisiae lysate prior to (a, c) and after (b, d) demultiplexing. Prior to demultiplexing, there are many other peaks of similar or greater intensity than the peak for GPLVLEYETYR (indicated with an arrow in (a)). After demultiplexing, peaks from other precursors are effectively removed and the true peak is by far the most intense (b). Additionally, the demultiplexed peak (d) contains far less interference than the unprocessed peak (c).
To test the performance of this method for peptide quantification, we spiked a commercial six bovine protein digest (Bruker-Michrom) into a complex matrix (S. cerevisiae lysate, soluble fraction) in amounts ranging from 50 attomoles to 100 femtomoles on-column. MS1 and MSX data were collected simultaneously on each spike-in sample by acquiring MSX scans (R.P. 17,500) with MS1 scans (R.P. 35,000) interlaced every ten scans. We quantified five of the proteins using a total of 36 peptides (Fig. 3, Supplementary Data, Supplementary Table 1).
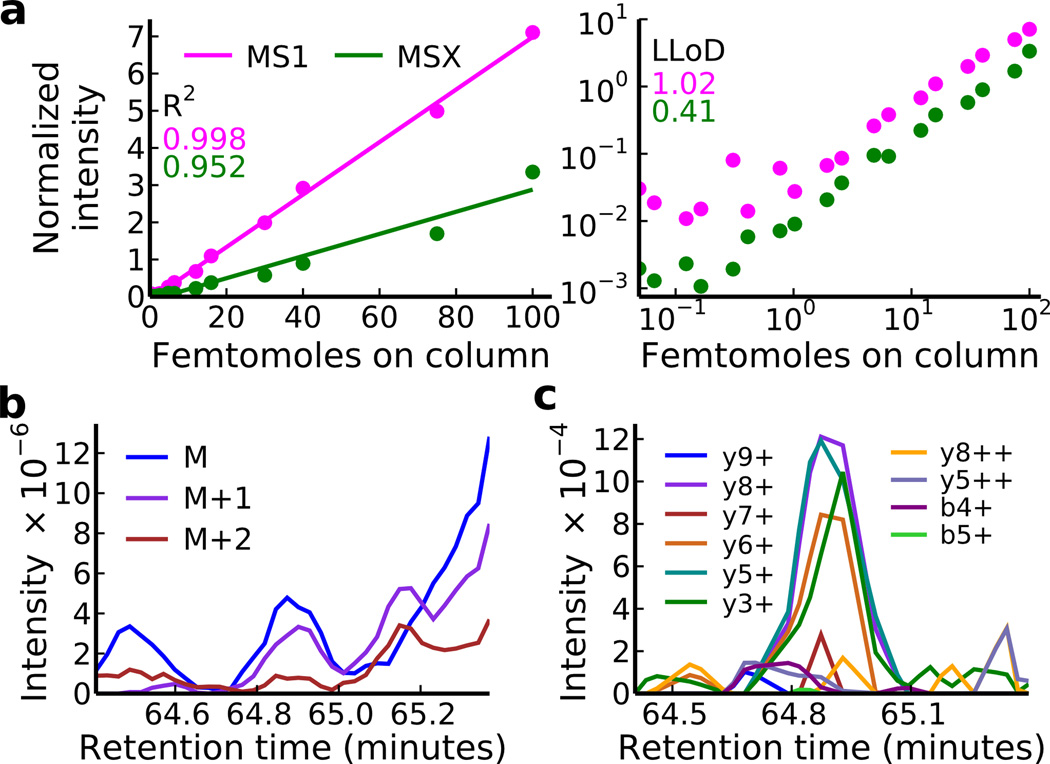
Figure 3. Quantitation of the LVNELTEFAK Peptide by MSX and MS1.
A commercial six protein digest was spiked into a complex matrix (soluble S. cerevisiae lysate) at amounts ranging from 50 attomoles – 100 femtomoles on column. MSX data were acquired with an MS1 scan interleaved every ten scans. The signal intensity for each spike in point (normalized to two background peptides) for the peptide LVNELTEFAK++ is plotted in (a) with (right pane) and without (left pane) log scaling of the x and y axes. The lower limit of detection is 0.41 femtomoles and 1.02 femtomoles for MSX and MS1-based quantitation respectively. The slopes of the regression lines were 0.030 ± 0.004 (95% confidence interval) and 0.071 ± 0.002 for MSX and MS1 respectively. (b) and (c) show the MS1 and MSX signal at 1.02 femtomoles. Although on average MS1 quantitation is more sensitive than MSX, chemical noise hinders quantitation of this peptide by MS1 and MSX is more sensitive due to greater selectivity.
The lower limit of detection for the 36 peptides quantified averaged 8.66 and 4.98 femtomoles for MSX and MS1 respectively. All peptides showed a linear response above the limit of detection with R2 values of the regression lines averaging 0.95 and 0.98 for MSX and MS1 respectively. Although the MSX method was less sensitive than the MS1 method on average (P = 0.016, paired t-test, n = 37), the results are impressive given that the MSX data provide structural selectivity information in addition to quantification. The slight reduction in sensitivity of MSX relative to MS1 is not unexpected in the absence of chemical noise. MS1 is expected to be more sensitive in a simple mixture because in MS/MS the ion beam is split into multiple fragmented products, each a fraction of the original intensity15. 7 of the 36 peptides suffered from interference in the MS1 signal, resulting in an average 3.4-fold improvement in sensitivity ranging from 1.3–8.3 fold by MSX quantification. Acquiring MS1 and MSX data simultaneously combines the high sensitivity of MS1 with the structural selectivity of MSX while theoretically improving quantitative precision by combining measurements from the precursor (MS1) and fragment ion peaks (MSX) for quantitation. The improved selectivity of MSX resulted in greater sensitivity (LOD of 0.41 femtomoles) compared to MS1 (LOD of 1.02 femtomoles) for the peptide LVNELTEFAK++ (Fig. 3, Supplementary Fig. 6).
We have demonstrated the efficacy of multiplexed DIA in addressing a major pitfall preventing wider adoption of these methods, low precursor selectivity. With wide isolation windows, there is an increased likelihood of relying on fragment ion data alone to discern between peptides with very similar fragmentation patterns (e.g. modified forms, or similar sequence). Even if a peptide can be unambiguously identified in these data, quantitation is negatively impacted by fragment ion interference. With these obstacles addressed, other proven advantages of DIA – including increased dynamic range, and more complete and reproducible MS/MS sampling across technical replicates – may be employed. This multiplexing technique, coupled with recent improvements in the interpretation of DIA data10, 11, make this a practical technique for global, highly-specific, and reproducible relative quantitation of peptides in a shotgun experiment.
Methods
Implementation of Multiplexed MS/MS on a Q-Exactive Mass Spectrometer
In a multiplexed MS/MS (MSX) scan, fragments from multiple precursor isolation windows are stored together prior to mass analysis in the Orbitrap. All of the MSX data in this manuscript were collected using five 4 m/z-wide isolation windows per MS/MS scan, although different combinations of isolation windows per scan and isolation width can be used. The five windows isolated for each MS/MS scan are chosen randomly from a list of all 4 m/z isolation windows to be analyzed in the experiment. In these experiments, the list is all 100 non-overlapping 4 m/z – wide windows between 500 and 900 m/z (Supplementary Table 2). The window width and position are shifted slightly from their integer values to reduce the likelihood of placing window edges where peptides are most likely to occur (Supplementary Fig. 7). For example, the first window is centered at 502.4783 m/z rather than 502 m/z and has a width of 4.002 m/z. This optimization is due to the fact that peptide masses are distributed across the mass range every ~1.0005 m/z in “allowable regions” interspersed with “forbidden zones”16 where peptide masses do not occur. Because the allowable regions are spaced apart by 1.00045475 m/z on average17, the isolation window width is set to be a multiple of this number. Additionally, windows are shifted so the edges fall in forbidden zones. All window edges are calculated using the equation 0.25 + 1.00045475 × N where N is an integer. The offset value of 0.25 was determined to be optimal based on analysis of the distribution of peptide precursor masses (+1, +2, +3 charged) in the Bibliospec18 spectral library. For window edges to be optimally placed, they must be a member of the series 0.25 + 1.00045475 × N. For these data, the edges of the windows are N × 4 × 1.00045475 + 0.25 where N is every integer in the range (inclusive) of 125 – 225.
Each scan is built by removing five random isolation windows from the isolation list. This process is repeated for each scan until the isolation window list is empty (20 scans in this case) at which point the list is repopulated. By sampling from the list without replacement, each isolation window is sampled every 20 scans (~3.5 seconds) on average and is guaranteed to be sampled at least every 40 scans. Since these data have been collected, an additional optimization avoids the selection of the same pairs of isolation windows in nearby scans. If selecting five isolation windows randomly from the isolation list results in pairing two windows together that have been paired in the same scan recently, the random selection is repeated until this is no longer the case or too much time has passed. With the 5 × 4 m/z multiplexing scheme, it is typical to not see the same two isolation window pairs observed within 200 scans (~35 seconds) of each other.
Scans are defined prior to an experiment using this method in Skyline (v1.3) and exported as an inclusion list containing a sequence of 5,000 isolation windows (1,000 scans). During analysis, the Q-Exactive (Thermo Fisher Scientific) loops through this list sequentially choosing five isolation windows for each MSX scan. This capability is implemented in a firmware modification which keeps the mass list from being sorted by the instrument computer prior to data acquisition.
When an MSX scan is acquired, each isolation window is isolated, fragmented, and stored serially-in-time prior to mass analysis in the Orbitrap. The fill time for all isolation windows in a given scan are the same. If the fill times for each window were different, it would be impossible to calculate the ion current (charges/second) for a fragment ion population because the fill time for the precursor(s) of that fragment ion population would be unknown.
To implement automated gain control (AGC), an MS1 scan is acquired every tenth scan. For each MSX scan, the most recent MS1 scan is analyzed to determine the summed ion current for all five precursor isolation windows in the scan. The summed ion current is determined by summing up the total MS1 extracted ion current for each of the five isolation windows. The target number of ions is divided by this summed ion current to determine the fill time (the same for each window). Using this approach, the total number of ions in the trap is fixed, but the number of ions from any of the five windows will vary. The maximum fill time for an isolation window is set to 20 milliseconds, meaning the maximum total fill time for a scan is 100 ms (5 windows × 20 ms). MS1 and MSX scans are acquired with resolving power 35,000 and 17,500 respectively. The AGC target for MS1 scans is set to 106 ions and 105 ions per isolation window for MS1 and MSX scans respectively.
De-Multiplexing by Non-Negative Least Squares Optimization
Due to the random selection of isolation windows for each scan, any two spectra with an isolation window in common will not share any other isolation windows. This characteristic helps when de-multiplexing these spectra because overlapping fragment ion information is more likely to come from the single isolation window the spectra have in common than any of the others. In other words, spectra can be de-multiplexed by leveraging information (the isolation windows used and fragment ion intensities) from neighboring spectra. To do this, a system of equations is built describing each observed multiplexed spectrum as a linear combination of multiple unobserved single-precursor component spectra (Fig. 1).
The system of equations is represented by matrix multiplication:
B = A * X
Matrix B contains the fragment ion intensities for the multiplexed spectra: each column is a fragment ion m/z, each row is a spectrum. Matrix A contains the isolation window pattern for each spectrum, each column is an isolation window, and each row is a spectrum. Isolation windows have the value 1 if they are present in a spectrum and 0 if they are absent. The matrix X is an unknown matrix containing the de-multiplexed component spectra. Each row is a de-multiplexed spectrum for an isolation window, each column is a de-multiplexed fragment ion intensity. The system of equations is solved by non-negative least squares19. For each spectrum to be de-multiplexed, this system of equations is built containing 140 consecutive MSX spectra with the spectrum to be de-multiplexed at the center of this window. 140 spectra are used for the 5 isolation window experiment, but for other methods, the number of spectra in the window is (# isolation windows total / isolation windows per scan) * (2 + isolation windows per scan). Setting the number of spectra this way guarantees that the system of equations is not underdetermined. Using neighboring spectra to de-multiplex a spectrum has the negative consequence of reducing signal intensity due to the averaging of the de-multiplexed spectra with nearby spectra similar to a boxcar smooth. To minimize this, each row of matrix B and A are multiplied by the Savitzky-Golay smoothing coefficient (5 wide, second order) for that row. Finally, each de-multiplexed peak is normalized such that the sum of the intensities from each de-multiplexed component is equivalent to the intensity of the original observed peak. This de-multiplexing happens automatically on import into Skyline.
Evaluation of De-Multiplexing
A soluble S. cerevisiae lysate was reduced, alkylated, digested for an hour with trypsin, and cleaned by dual mode solid phase extraction (Oasis MCX cartridges, Waters Corporation). LC-MSX data were acquired on the Q-Exactive as described above on 1.2 µg of sample per injection. A 40 cm 75 µM fused silica column packed with reversed-phase C12 Jupiter resin (Phenomenex) was used to separate the sample across a 90-minute linear acetonitrile gradient from 0 to 25% Buffer B. Chromatography was performed using a EASY-nLC II (Thermo Fisher Scientific) system set to a flow rate of 250 nL/min. Buffer A was 2% ACN, 0.1% formic acid, and 97.9% water. Buffer B was 99.9% ACN and 0.1% formic acid. The data were analyzed using a modified version of Skyline which processed the data with and without de-multiplexing enabled.
Spike-in Experiment to Evaluate Quantitative Performance
An equimolar six protein digest (Bruker-Michrom) was spiked into a complex matrix (soluble S. cerevisiae lysate digest) over four orders of magnitude to test the quantitative performance of MSX. The yeast sample was reduced, alkylated, digested for an hour with trypsin, and cleaned by dual mode solid phase extraction (Oasis MCX cartridges, Waters Corporation) after digestion. Prior to the spike-in experiments, the complex matrix was run four times to condition the liquid chromatography column. Spike-in experiments with 0.6 µg of the complex matrix loaded on column with 0.05, 0.12, 0.31, 0.77, 1.92, 4.8, 12, 30, and 75 femtomoles of the bovine protein digest spiked in were run followed by blanks of the complex matrix and buffer A, followed by 0.07, 0.16, 0.41, 1.02, 2.6, 6.4, 16, 40, and 100 fmol spike-ins. Samples were run using the same chromatography conditions and column type as in the previous section “Evaluation of De-Multiplexing, except using C18 Aqua resin (Phenomenex) instead of the C12 Jupiter reversed-phase resin.
36 peptides from 5 of the spiked-in proteins and not present in the background matrix were quantified (Fig. 3, Supplementary Data, and Supplementary Table 1). Peptides were quantified using the area under the curve of the M, M+1, and M+2 peaks for MS1, and a manually curated subset of the b- and y- ion series for MSX (Supplementary Table 1). The signal from each peptide was normalized by the signal from two highly-abundant peptides (DNSQVFGVAR++ and ESTLHLVLR++) from the background matrix. The lower limit of detection was determined for each peptide by manual inspection. For MSX data, the lower limit of detection was the lowest abundance where at least three transitions co-elute and have similar intensity ratios to that seen at higher abundance. For MS1 data, the lower limit of detection was the lowest abundance where at least the M and M+1 isotope peaks co-elute, and have the same rank-order as at higher abundances. The sensitivity of the two techniques was compared using a paired t-test on the log-transformed lower limits of detection for the 36 peptides (37 precursors) quantified. Regression lines were fit to the MS1 and MSX data for each peptide using all points at or above the lower limit of detection.
Supplementary Material
Acknowledgements
The authors would like to acknowledge financial support from National Institutes of Health Grants R01 GM103551, P41 GM103533, and F31 AG037265.
Competing financial interests:
The authors received financial support from ThermoFisher Scientific.
Abbreviations
- RP
Resolving power
- AGC
Automated Gain Control
- DDA
Data Dependent Acquisition
- DIA
Data Independent Acquisition
- MSX
Multiplexed Data Independent Acquisition
- SRM
Selected Reaction Monitoring
Footnotes
Author Contributions:
A.K., C.C.W, J.D.C., J.D.E., M.J.M, M.K., V.Z., and Y.S.T. designed experiments.
A.K., C.C.W., J.D.E., M.J.M., M.K., N.W.B., and V.Z. interpreted results.
J.D.E. and M.J.M wrote the manuscript.
A.K. created a firmware modification for the Q-Exactive.
A.K. and M.K. provided preliminary data.
G.E.M., J.D.E., and N.W.B. performed experiments
B.X.M., D.M.M., and J.D.E. wrote software.
References
- 1.Stahl DC, Swiderek KM, Davis MT, Lee TD. Journal of the American Society for Mass Spectrometry. 1996;7:532–540. doi: 10.1016/1044-0305(96)00057-8. [DOI] [PubMed] [Google Scholar]
- 2.Liu H, Sadygov RG, Yates JR., 3rd Analytical chemistry. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 3.Michalski A, Cox J, Mann M. Journal of proteome research. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 4.Wenner BR, Lynn BC. Journal of the American Society for Mass Spectrometry. 2004;15:150–157. doi: 10.1016/j.jasms.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Hoopmann MR, Finney GL, MacCoss MJ. Analytical chemistry. 2007;79:5620–5632. doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purvine S, Eppel JT, Yi EC, Goodlett DR. Proteomics. 2003;3:847–850. doi: 10.1002/pmic.200300362. [DOI] [PubMed] [Google Scholar]
- 7.Myung S, et al. Analytical chemistry. 2003;75:5137–5145. doi: 10.1021/ac030107f. [DOI] [PubMed] [Google Scholar]
- 8.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Nature methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 9.Panchaud A, et al. Analytical chemistry. 2009;81:6481–6488. doi: 10.1021/ac900888s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisbrod CR, Eng JK, Hoopmann MR, Baker T, Bruce JE. Journal of proteome research. 2012;11:1621–1632. doi: 10.1021/pr2008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillet LC, et al. Molecular & cellular proteomics : MCP. 2012;11 doi: 10.1074/mcp.O111.016717. O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams ER, Loh SY, McLafferty FW, Cody RB. Analytical chemistry. 1990;62:698–703. doi: 10.1021/ac00206a010. [DOI] [PubMed] [Google Scholar]
- 13.MacLean B, et al. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein SE, Rudnick PA. NIST Peptide Tandem Mass Spectral Libraries. Gaithersburg MD: National Institute of Standards and Technology; p. 20899. [Google Scholar]
- 15.Yost RA, Enke CG. Analytical chemistry. 1979;51:1251–1264. doi: 10.1021/ac50048a002. [DOI] [PubMed] [Google Scholar]
- 16.Frahm JL, Howard BE, Heber S, Muddiman DC. Journal of mass spectrometry : JMS. 2006;41:281–288. doi: 10.1002/jms.1024. [DOI] [PubMed] [Google Scholar]
- 17.Gay S, Binz PA, Hochstrasser DF, Appel RD. Electrophoresis. 1999;20:3527–3534. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3527::AID-ELPS3527>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Frewen BE, Merrihew GE, Wu CC, Noble WS, MacCoss MJ. Analytical chemistry. 2006;78:5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
- 19.Lawson CL, Hanson RJ Society for Industrial and Applied Mathematics. Classics in applied mathematics 15. 1 electronic text. Philadelphia, Pa: Society for Industrial and Applied Mathematics (SIAM, 3600 Market Street, Floor 6, Philadelphia, PA 19104); 1995. p. xii.p. 337. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.