Abstract
Botanically derived natural products have recently become an attractive source of new chemotherapeutic agents. To explore active anti-colorectal cancer compounds, we performed phytochemical studies on Alkanna tinctoria and isolated eight quinone compounds. Using different spectrum methods, compounds were identified as alkannin (1), acetylalkannin (2), angelylalkannin (3), 5-methoxyangenylalkannin (4), dimethylacryl alkannin (5), arnebifuranone (6), alkanfuranol (7), and alkandiol (8). Compounds 4, 7, and 8 are novel compounds. The structures of the three novel compounds were elucidated based on extensive spectroscopic evidence including high-resolution mass spectrometry and NMR spectra. The antiproliferative effects of these eight compounds on HCT-116 and SW-480 human colorectal cancer cells were determined by the MTS method. Cell cycle and apoptosis were determined using flow cytometry. Enzymatic activities of caspases were determined by colorimetric assay, and interactions of compound 4 and caspase 9 were explored by docking analysis. Among the eight compounds, alkannin (1), angelylalkannin (3), and 5-methoxyangenylalkannin (4) showed strong antiproliferative effects, while compound 4 showed the most potent effects. Compound 4 arrested cancer cells in the S and G2/M phases, and significantly induced cell apoptosis. The apoptotic effects of compound 4 were supported by caspase assay and docking analysis. The structural functional relationship assay suggested that to increase anticancer potential, future modifications on alkannin (1) should focus on the hydroxyl groups at C-5 and C-8.
Keywords: Alkanna tinctoria, naphthoquinones, alkannin derivatives, novel compounds, antiproliferation, cell cycle, apoptosis, caspase 9, docking analysis, colorectal cancer
Introduction
Alkanna tinctoria (L.) Tausch is widely distributed in Europe and Western Asia. Its root has been used as a botanical drug for ulcers, inflammation, and wounds [1]. Previous phytochemical studies on this plant have resulted in the isolation of a series of naphthoquinone pigments, including alkannin and its derivatives [2–5]. Some of these compounds exhibit biological properties like cytotoxic, antimicrobial, antileishmanial, and anti-inflammatory activities [6, 7]. In our previous study, we isolated two known naphthoquinone compounds from this plant, and observed their antiproliferative effects on human colorectal cancer cells [8].
In this study, we performed a systemic phytochemical isolation on A. tinctoria. Eight quinone compounds were isolated and their chemical structures were elucidated by different spectral methods. Among the eight compounds, three novel compounds were characterized. The antiproliferative effects of the eight compounds on human colorectal cancer cells were determined. The effects of these compounds on the cell cycle and apoptosis were evaluated. To further explore the potential mechanisms of apoptosis, a structure-based docking model was simulated to get an inside view of ligand–receptor interaction. One novel alkannin derivative showed the most potent antiproliferative effect, and the structural-functional relationship of these naphthoquinone compounds was explored.
Materials and methods
Chemicals and reagents
All solvents were of high-performance liquid chromatography grade (Wako Pure Chemicals, Osaka, Japan). Materials for column chromatography included silica gel 60 (230–400 mesh, Nacalai Tesque Inc., Kyoto, Japan), YMC ODS-A gel (50 μm, YMC Co. Ltd., Kyoto, Japan), and Sephadex LH-20 gel (Uppsala, Sweden). TLC was performed on Kieselgel 60 F254 (Merck, Damstadt, Germany) plates. Milli-Q water was supplied by a water purification system (US Filter, Palm Desert, CA, USA). All cell culture plasticware was obtained from Falcon Labware (Franklin Lakes, NJ, USA) and Techno Plastic Products (Trasadingen, Switzerland). Trypsin, McCoy's 5A and Leibovitz's L-15 media, and phosphate buffered saline were obtained from Mediatech, Inc. (Herndon, VA, USA). Penicillin and streptomycin were obtained from Sigma-Aldrich (St. Louis, MO, USA). An MTS assay kit, CellTiter 96 Aqueous Solution Cell Proliferation Assay, was obtained from Promega (Madison, WI, USA). PI/RNase staining buffer was obtained from BD Biosciences Pharmingen (San Diego, CA, USA). An annexin V-FITC apoptosis detection kit was obtained from BD Biosciences (Rockville, MD, USA). Caspase 3, 8, 9 kits were obtained from BioVison (Mountain View, CA, USA).
Instrument procedures for structure elucidation
Optical rotations were obtained using a DIP-360 digital polarimeter (JASCO, Easton, USA). NMR spectra were recorded on a JEOL ECX 400 NMR spectrometer (JEOL, Tokyo, Japan). ESI-TOFMS experiments utilized a JEOLAccuTOF™LC 1100 mass spectrometer (JEOL, Tokyo, Japan).
Plant materials
Dried roots of Alkanna tinctoria (L.) Tausch (Boraginaceae) were donated by Professor Tibor Wenger, Semmelweis University, Budapest, Hungary and authenticated by Professor Yukihiro Shoyama. A voucher specimen (Number 201112) was deposited at the Herbarium of Faculty of Pharmaceutical Sciences, Nagasaki International University, Nagasaki, Japan.
Extraction, isolation and structure elucidation
900 g of air-dried A. tinctoria root powder was extracted with 95% EtOH to obtain 35 g of EtOH extract. The EtOH extract was suspended in H2O and partitioned with CHCl3 to obtain 14 g of CHCl3-soluble pigment fraction. The pigment fraction was then fractionated on a silica gel column with a gradient of hexane–EtOAc (20:1→0:1, v/v) to obtain five fractions (fr.1.1–1.5). Fr.1.1 (2100 mg) was repeatedly chromatographed on a silica gel column with hexane–EtOAc (20:1), followed by a sephadex LH-20 column with MeOH as an eluent to obtain 3 (950 mg). Fr.1.2 (450 mg) was repeatedly chromatographed on a silica gel column with hexane–EtOAc (10:1, v/v), followed by a sephadex LH-20 column with MeOH as an eluent to obtain 2 (20 mg). Fr.1.3 (1700 mg) was subjected to a silica gel column with hexane–EtOAc (5:1, v/v), followed by a sephadex LH-20 column with MeOH as an eluent to yield 4 (15 mg) and 8 (20 mg). Similarly, fr.1.4 (610 mg) was loaded onto a silica gel column with hexane–EtOAc (5:1 and 4:1, v/v) and then followed by sephadex LH-20 chromatography with MeOH as an eluent to give 1 (25 mg), 6 (10 mg), and 7 (15 mg). Finally, compound 5 (8 mg) was isolated from fr. 1.5 (300 mg) by means of reverse-phase column chromatography with MeOH–H2O (3:1, v/v) and preparative TLC with hexane–EtOAc (2:1, v/v). Spectral data of novel compounds, 4, 7, and 8 are included in the following paragraphs. The amounts of purified compounds are 8–950 mg. Thus, the percentage of isolated compounds in relation to plant materials is between 0.0009% and 0.106%.
5-Methoxyangenylalkannin (4): red solid; [α]D25-18 (c=0.23, CHCl3); HR-ESI-MS m/z: 385.1653 [M + H]+ (calcd. for C22H25O6, 385.1651); 1H-NMR (CDCl3, 400 MHz) δ: 1.58 (3H, s, H-6′), 1.68 (3H, s, H-5′), 1.95 (3H, m, H-5″), 2.00 (3H, dq, J = 5.6, 1.6 Hz, H-4″), 2.48 (1H, m, H-2′), 2.65 (1H, m, H-2′), 3.99 (3H, s, 5-OCH3), 5.15 (1H, t, J = 6.8 Hz, H-3′), 6.02 (1H, dd, J = 7.2, 4.4 Hz, H-1′), 6.15 (1H, m, H-3″), 6.73 (1H, s, H-3), 7.29 (1H, d, J = 9.2 Hz, H-6), 7.36 (1H, d, J = 9.2 Hz, H-7); and 13C-NMR (CDCl3, 100 MHz) δ: 182.0 (C-1), 152.4 (C-2), 131.3 (C-3), 190.2 (C-4), 154.3 (C-5), 56.9 (5-OCH3), 123.3 (C-6), 127.3 (C-7), 156.4 (C-8), 114.7 (C-9), 117.8 (C-10), 69.9 (C-1′), 33.0 (C-2′), 118.2 (C-3′), 135.7 (C-4′), 25.8 (C-5′), 18.0 (C-6′), 166.4 (C-1″), 127.3 (C-2″), 139.6 (C-3″), 15.9 (C-4″), 20.6 (C-5″).
Alkanfuranol (7): purple solid; HR-ESI-MS m/z: 319.1549 [M + H]+ (calcd. for C18H23O5, 319.1546); 1H-NMR (CDCl3, 400 MHz) δ: 1.62 (3H, d, J = 1.2 Hz, H-16), 2.36 (2H, q, J = 7.2 Hz, H-10), 2.48 (2H, J = 7.2 Hz, H-11), 3.29 (2H, s, H-7), 3.89 (3H, s, 3-OCH3), 3.91 (3H, s, 4-OCH3), 5.36 (1H, t, J = 6.8 Hz, H-9), 6.28 (1H, br s, H-13), 6.40 (1H, s, H-6), 7.23 (1H, br s, H-15), 7.34 (1H, br s, H-14); and 13C-NMR (CDCl3, 100 MHz) δ: 121.1 (C-1), 140.5 (C-2), 137.4 (C-3), 60.8 (3-OCH3), 139.0 (C-4), 60.7 (4-OCH3), 141.7 (C-5), 109.9 (C-6), 31.1 (C-7), 133.8 (C-8), 126.3 (C-9), 28.6 (C-10), 25.1 (C-11), 124.9 (C-12), 111.1 (C-13), 142.6 (C-14), 138.9 (C-15), 23.5 (C-16).
Alkandiol (8): white amorphous powder; [α]D25-13 (c=0.20, CHCl3); HR-ESI-MS m/z: 257.1179 [M + H]+ (calcd. for C16H17O3, 257.1178); 1H-NMR (CDCl3, 400 MHz) δ: 0.90 (3H, s, H-16), 2.40 (1H, d, J = 16.0, H-14), 2.71 (1H, d, J = 6.4 Hz, H-8), 2.76 (1H, d, J = 16.0 Hz, H-14), 4.36 (2H, s, H-15), 4.97 (1H, d, J = 6.4 Hz, H-7), 5.04 (1H, dd, J = 5.6, 1.2 Hz, H-10), 5.10 (1H, dt, J = 10.4, 1.2 Hz, H-11), 5.99 (1H, dd, J = 17.6, 10.4 Hz, H-12), 6.53 (1H, d, J = 8.8 Hz, H-6), 6.62 (1H, d, J = 8.8 Hz, H-5); and 13C-NMR (CDCl3, 100 MHz) δ: 148.6 (C-1), 121.9 (C-2), 124.5 (C-3), 150.5 (C-4), 116.2 (C-5), 113.8 (C-6), 76.8 (C-7), 51.5 (C-8), 150.0 (C-9), 107.6 (C-10), 113.0 (C-11), 148.6 (C-12), 38.8 (C-13), 36.6 (C-14), 71.9 (C-15), 18.5 (C-16).
Cell culture
Human colorectal cancer cell lines HCT-116 (McCoy's 5A) and SW-480 (Leibovitz's L-15) were obtained from American Type Culture Collection (Manassas, VA, USA). Cells were grown in the indicated medium supplemented with 10% FBS and 50 IU penicillin/streptomycin in a humidified atmosphere with 5% CO2 at 37°C.
Cell proliferation analysis
Compounds 1–8 were dissolved in DMSO and were stored at −20°C before use. Cells were seeded in 96-well plates (1×104 cells/well). After 24 h, indicated concentrations of drugs were added to the wells. The final concentration of DMSO was 1%. Controls were exposed to culture medium containing 1% DMSO without drugs. All experiments were performed in triplicate. Following the indicated incubation period, cell proliferation was evaluated using an MTS assay according to the manufacturer's instructions. Briefly, the medium was replaced with 100 μl of fresh medium and 20 μl of MTS reagent (CellTiter 96 Aqueous Solution) in each well, and the plate was returned to the incubator for 1–2 h. A 60 μl aliquot of medium from each well was transferred to an ELISA 96-well plate and its absorbance at 490 nm was recorded. Since 1% DMSO did not influence the proliferation of the two cell lines, results were expressed as percent of control (DMSO vehicle set at 100%).
Cell cycle assay
Cells were seeded into 24-well tissue culture plates. On the second day, the medium was changed and cells were treated with test compounds. Cells were incubated for 48 h before they were harvested. These cells were fixed gently with 80% ethanol in a freezer for 2 hr and were then treated with 0.25% Triton X-100 for 5 min in an ice bath. Cells were resuspended in 300 μl of PBS containing 40 μg/ml propidium iodide (PI) and 0.1 mg/ml RNase. Then the cells were incubated in a dark room for 20 min at room temperature, and cell cycle analysis was performed using a FACScan flow cytometer (Becton Dickinson, Mountain View, CA, USA) and FlowJo 7.1.0 software (Tree Star, Ashland, OR, USA). For each measurement, at least 10,000 cells were counted.
Apoptotic analysis
Cell apoptosis was assayed by flow cytometry following a previously described procedure [9]. Briefly, after treatment for 48 h, cells floating in the medium were collected. The adherent cells were detached with trypsin. Then culture medium containing 10% FBS (and floating cells) was added to inactivate trypsin. After being pipetted gently, the cells were centrifuged for 5 min at 1500 g. The supernatant was removed and cells were stained with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) according to the manufacturer's instructions. Untreated cells served as a control. The cells were analyzed immediately after staining using flow cytometer. For each measurement, at least 20,000 cells were counted.
Caspase 3 and 9 assays
SW-480 cells were seeded in 6-well tissue culture plates. After 24 h, the medium was changed and compound 4 was added. After treatment for 24 h, cell lysates were collected. Expression levels of caspases 3 and 9 were determined by the colorimetric method according to the manufacturer's instructions. Briefly, cell lysates were diluted with 50 μl of 2× reaction buffer (containing 10 mM DTT) to a protein concentration of 0.5 mg/ml in an ELISA 96-well plate. Then, 5 μl of colorimetric tetrapeptide substrate (DEVD-pNA for caspase 3 and LEHDpNA for caspase 9) and cell lysate were added, and the plate was incubated at 37°C for 24 h. Absorbance was recorded at 405 nm. The change in caspase activity was calculated as absorbance of Compound 4 treated cells/absorbance of untreated controls.
Western blot analysis
SW-480 cells were collected 24 h after exposure to 1–15 μM of compound 4 or vehicle and then lysed in cold radio immunoprecipitation assay (RIPA) buffer supplemented with 1% (v/v) protease inhibitor cocktail, and stored in aliquots at −80°C for later analysis. The protein concentration of the samples was determined by a BCA protein assay kit (Pierce, Rockford, IL). Lysates (40 μg protein) were denatured by heating at 95°C for 10 min and loaded on 4–15% Mini-PROTEAN TGX precast gels (Bio-Rad, Hercules, CA) for electrophoresis. Then, the proteins were transferred to a PVDF membrane (Bio-Rad, Hercules, CA) and blocked in PBST buffer (PBS with 0.05% Tween 20) containing 5% nonfat dried milk. The blots were incubated with the indicated primary antibodies (p21waf1/cip1, cdc2, cyclin B1, cdk2, caspase 3, caspase 9 and β-actin, from Cell Signaling, Danvers, MA) overnight at 4°C, followed by incubation for 1 h with the appropriate secondary antibodies conjugated with horseradish peroxidase in blocking buffer. β-actin was measured as the loading control. Immunocomplexes were detected using SuperSignal West Pico Substrate (Thermo-pierce, Rockford, IL) following the manufacturer's directions, and quantified by densitometry using Image J software. Intensities were normalized to vehicle-treated samples after adjusting for β-actin loading.
Receptor docking analysis
The possible binding modes of compound 4 at the catalytic domains of human caspase 9 were predicted using the docking program Surflex-Dock (Tripos, St. Louis, MO, USA). The structure of compound 4 was generated (through Ligand model in Sybyl), and the protein crystal structure of caspase 9 was obtained (PDB code 2AR9). For the preparation for the docking analysis, the protein structure was prepared by adding hydrogen atoms and missing side chain atoms and removing water molecules. Intermolecular interaction between compound 4 and caspase 9 was analyzed, and the key pharmacophore in the ligand was identified [10, 11].
Statistical analysis
Data are presented as mean ± standard error (SD). A one-way ANOVA was employed to determine the statistical significance of the results. In some cases, Student's t-test was used to compare two groups. The level of statistical significance was set at p < 0.05.
Results
Isolation and identification of the compounds
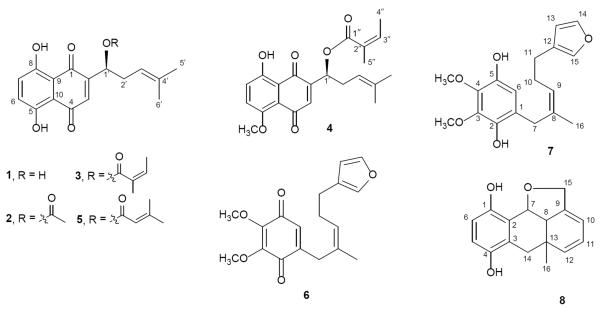
Alkannin (1) and angelylalkannin (3) were previously reported from the pigment fraction of A. tinctoria roots by partition between CHCl3 and water [8], and their structures were confirmed by their spectral data and compared with those previously reported [12, 13]. Other fractions were subjected to a series of column chromatographic separation to yield three novel compounds, named 5-methoxyangenylalkannin (4), alkanfuranol (7), and alkandiol (8), together with three known compounds, acetylalkannin (2) [11], dimethylacrylalkannin (5) [14], and arnebifuranone (6) [15], respectively (Figure 1). The structures of the three novel compounds were elucidated based on extensive spectroscopic evidence including high-resolution mass spectrometry and NMR spectra (1H-, 13C-, HMQC, HMBC, and COSY spectra). The detailed 1H and 13C data of the new compounds are provided in the Methods section.
Figure 1.
Chemical structures of the eight compounds isolated from Alkanna tinctoria.
Compound 4, which was obtained as a red solid, gave a quasimolecular ion peak at m/z 385.1653 [M + H]+ in the high-resolution electrospray-ionization time-of-flight mass spectrometry (HRESITOFMS). The 1H- and 13C-NMR spectra of 4 exhibited proton signals of two ortho-coupled and one singlet aromatic protons and two conjugated carbonyl groups, indicating 4 belongs to the 1,4-naphthoquinone derivatives like alkannin (1). In addition, signals of an aromatic methoxy (5-OCH3) were observed. Further analyses of 1H and 13C-NMR data suggested the presence of a 4-methylpent-3-en-1-ol side chain and an angenoyl group, which are similar to angelylalkannin (3) [14]. Furthermore, the overall structure of 4, especially linkage sites, was assigned by 1H-1H COSY, HMQC, and HMBC. Having established the structure of 4, like alkannin (1), the absolute configuration of C-1′ was assigned to be S using the optical rotation value [14]. Hence, the structure of 4 was identified as 5-methoxyangenylalkannin (1-[8-hydroxy-5-methoxy-1,4-dioxo-1,4-dihydronaphthalen-2-yl]-4-methylpent-3-enyl angenyloate).
Alkanfuranol (7) was obtained as a purple solid and had the molecular formula C18H22O5 based on HR-MS measurement. The 1H NMR spectrum of 7 showed five aromatic/olefinic signals (δ 5.36 [t, J = 6.8 Hz, H-9], 6.28 [br s, H-13], 6.40 [s, H-6], 7.23 [br s, H-15], and 7.34 [br s, H-14]), three methylene groups (δ 2.36 [q, J = 7.2 Hz, H-10], 2.48 [t, J = 7.2 Hz, H-11], and 3.29 [s, H-7]], one vinyl methyl [δ 1.62 [d, J = 1.2 Hz, H-16]), and two methoxy groups (δ 3.89 [s, 3-OCH3] and 3.91 [s, 4-OCH3]). The 13C NMR spectrum showed sixteen signals of a typical 3,4-dimethoxy-benzodihydroxyquinone (δ 121.1 [C-1], 140.5 [C-2], 137.4 [C-3], 139.0 [C-4], 141.7 [C-5], and 109.9 [C-6]), a mono-substituted furan ring (δ 124.9 [C-12], 111.1 [C-13], 142.6 [C-14], and 138.9 [C-15]), and isoheptenyl moiety. The 1H and 13C NMR data of 7 were similar to those of arnebifuranone (6) apart from the 3,4-dimethoxy-benzodihydroxyquinone moiety [15]. Furthermore, the structure of 7 was confirmed by the COSY and HMBC spectra. Consequently, the structure of alkanfuranol (7) was unambiguously established as depicted in Fig. 1.
Compound 8, a white amorphous powder, had the molecular formula C16H16O3 based on HRESIMS. The 1H NMR spectrum of 8 showed signals of two aromatic protons (δ 6.62 [d, J = 8.8 Hz, H-5], 6.53 [d, J = 8.8 Hz, H-6]) typical of 2,3-disubstituted benzodihydroxyquinone, three conjugated olefinic protons (δ 5.99 [dd, J = 17.6, 10.4 Hz, H-11], 5.10 [dt, J = 10.4, 1.2 Hz, H-12], and 5.04 [dd, J = 5.6, 1.2 Hz, H-10]), one oxymethine proton (δ 4.97 [d, J = 6.4 Hz, H-7]), one oxymethylene proton (δ 4.36 [s, H-1]), and one methyl group (δ 0.90 [s, H-16]). The 13C NMR spectrum showed sixteen signals including six aromatic carbons of a benzodihydroxyquinone (δ 148.6 [C-1], 121.9 [C-2], 124.5 [C-3], 150.5 [C-4], 116.2 [C-5], and 113.8 [C-6]), four conjugated olefinic carbons (δ 150.0 [C-9], 107.6 [C-10], 113.0 [C-11], and 148.6 [C-12]) and two oxygenated carbons (δ 76.8 [C-7] and 71.9 [C-15]). The 1H and 13C NMR data of 8 were quite similar to those of rhizonone, which was isolated from Lithospermum erythrorhizon (Boraginaceae), apart from the benzodihydroxyquinone moiety [16]. Furthermore, the planar structure of 8 was confirmed by COSY and HMBC experiments. The 1H-1H COSY experiment on 8 indicated the presence of partial structures written in bold lines; and in the HMBC spectrum, the long-range correlations were observed between the following protons and carbons: H-5 and C-1,3; H-6 and C-2,4; H-7 and C-1,3,9; H-15 and C-8,10; H-16 and C-8,12,14. Hence, the planar structure of alkandiol (8) was established as depicted in Fig. 1. The absolute stereochemistry of 8 has not been achieved yet, although its NOESY spectrum was recorded and the cross-peaks of H-7/H-8 and H-7/H-16 were observed. The occurrence of alkandiol (8) in A. tinctoria, the diol form of rhizonone, which is a novel metabolite putatively derived from geranylhydroquinone, has further suggested the similar biosynthesis pathway between alkannin and shikonin derivatives [16–18].
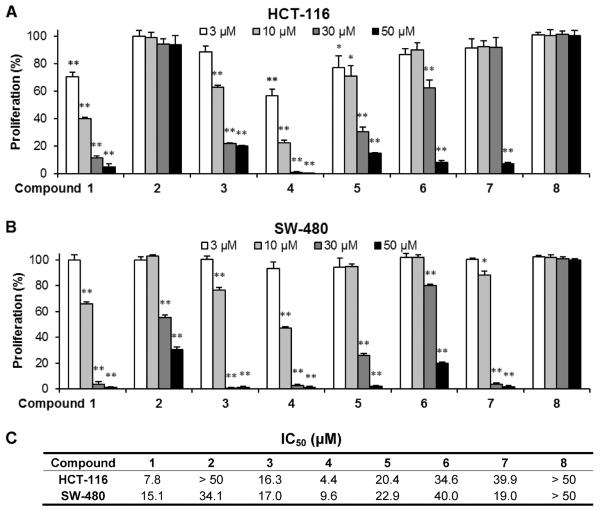
Antiproliferative effects of 8 compounds on HCT-116 and SW-480 cells
We evaluated the antiproliferative effects of eight compounds using two human colorectal cancer cell lines, HCT-116 and SW-480. As shown in Figure 2, the eight compounds showed different antiproliferative effects on the two cancer cell lines. At the tested concentrations (3–50 μM), compound 8 did not inhibit cancer cell growth on either of the two cancer cell lines. Compound 2 showed some antiproliferative effects on SW-480 cells, but such effects were not observed in HCT-116 cells. Compound 6 showed different potential for cancer cell growth inhibition.
Figure 2.
Antiproliferative effects of the eight compounds on human colorectal cancer cells. HCT-116 (A) and SW-480 (B) cells were treated with compounds at concentrations of 3, 10, 30, and 50 μM for 48 h. Results are expressed as percent of control (solvent vehicle set at 100%). Data are presented as mean ± SD of triplicate experiments. *P<0.05, **P<0.01 compared with control. (C). IC50 of compounds 1–8 on HCT-116 and SW-480 cells.
Regarding the effects on HCT-116 cells, Compounds 6 and 7 showed relatively weak antiproliferative effects, and at the concentration of 50 μM, HCT-116 cell growth was inhibited by 91.7% and 92.5%, respectively (both P<0.01 vs. control). Compounds 3 and 5 showed moderate effects; when treated with 30 μM the cancer cell growth was inhibited by 77.8% and 69.6%, respectively (both P<0.01). Compounds 1 and 4 showed strong antiproliferative effects. At concentrations of 3 μM and 10 μM, compound 1 inhibited cell growth by 29.7% (P<0.05) and 59.3% (P<0.01), while compound 4 inhibited cell growth by 43.1% and 77.4%, respectively (both P<0.01). Compared to the other seven compounds, compound 4 showed the strongest antiproliferative effects on HCT-116 cells (Figure 2A). The IC50 of compounds 1–8 on HCT-116 cells are: 7.8, >50, 16.3, 4.4, 20.4, 34.6, 39.9, >50 (μM), respectively (Figure 2C).
Isolated compounds showed different antiproliferative effects on SW-480 cells (Figure 2B). However, the sensitivity of certain compounds on SW-480 cells was not similar to HCT-116 cells. Compounds 2 and 6 showed relatively weak antiproliferative effects; compound 5 showed a moderate effect; compounds 1, 3, and 4 showed relatively strong effects, while compound 4 showed the strongest cancer cell inhibition effect on SW-480 cells (Figure 2B). The IC50 of compounds 1–8 on SW-480 cells are shown in Figure 2C. Data from both the HCT-116 and SW-480 cell lines suggested that the strongest antiproliferative effect was observed when using compound 4, a novel isolated compound.
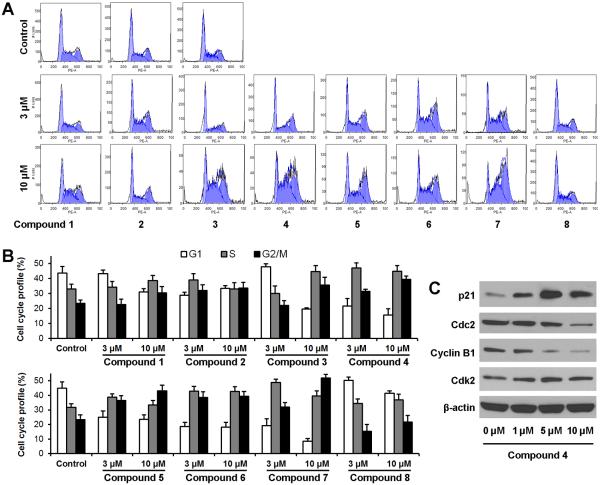
Effects of 8 compounds on cell cycle distribution of SW-480 cells
To examine whether proliferation in treated cells decreased because of cell cycle arrest at a specific phase, cell cycle profiles were determined using flow cytometry after staining with PI. As shown in Figure 3, besides compound 8, the other seven compounds induced cell cycle profile changes in different patterns. After treatment with 10 μM of compounds 5 and 7, the SW-480 cells were arrested mainly in the G2/M phase. Compounds 1, 2, 3, 4, and 6 decreased the percentage of cells in the G1 phase and increased cell proportions in both the S and G2/M phases. S phase cells became a major group, especially in treatment with higher concentrations. Among the 8 compounds, because compound 4 showed most potent antiproliferative effect, and also possessed marked activity on S and G2/M phase arrest, the expressions of proteins that control S and G2/M phase transition were evaluated. Compared to the control, expression of cyclin B1 and cdc2 were decreased in a dose-dependent manner. On the other hand, compound 4 significantly up-regulated p21, while slightly increasing cdk2 (Figure 3C). Our results were consistent with the role of these proteins in the regulation of the S and G2/M phase transition [19, 20]. Data from this study suggests that those seven compounds arrested SW-480 cells in the S and G2/M phases, and compound 4 induced S and G2/M arrest through the modulation of cell cycle-regulating proteins.
Figure 3.
Effects of the eight compounds on SW-480 cell cycle. (A) Representative histograms of DNA content in each experimental group. SW-480 cells were treated with 3 and 10 μM of compounds for 48 h. Cell cycle profile was determined using flow cytometry after staining with PI/RNase. (B). Percentage of each cell cycle phase with various treatments or with control. Data are presented as the mean ± SD of triplicate experiments. (C) Western blot analysis of cell cycle regulatory proteins from compound 4 treated SW-480 cells. Cells were treated with 1, 5 and 10 μM of compound 4 for 24 h.
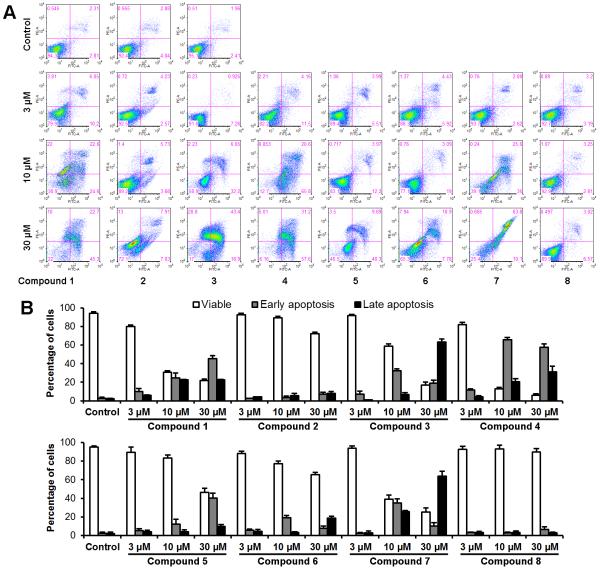
Apoptotic induction effects of 8 compounds on SW-480 cells
To explore the potential mechanism through which the tested compounds inhibit cell growth, cell apoptosis was assayed by flow cytometry after staining with double annexin V and PI in SW-480 cells (Figure 4). Annexin V can be detected in both the early and late stages of apoptosis. PI enters the cell in late apoptosis or necrosis. Viable cells were negative for both annexin V and PI (lower left quadrant); early apoptotic cells were positive for annexin V and negative for PI (lower right quadrant); late apoptotic or necrotic cells displayed both positive annexin V and PI (upper right quadrant); non-viable cells which underwent necrosis were positive for PI and negative for annexin V (upper left quadrant). After treatment for 48 h, except for compounds 2, 5, 6, and 8, the other four compounds significantly induced cancer cell apoptosis at concentrations of 10 and 30 μM (Figure 4A).
Figure 4.
Effects of the eight compounds on SW-480 cell apoptosis. SW-480 cells were treated with 3, 10, and 30 μM of compounds for 48 h. Apoptosis was quantified using flow cytometry after staining with annexin V/PI. (A) Representative scatter plots of PI (y-axis) vs. annexin V (x-axis). (B) Percentage of viable, early apoptotic, and late apoptotic cells. Data are presented as the mean ± SD of triplicate experiments.
Compared to the control (5.1%), the percentages of cells in both early and late apoptosis for each compound (treatment with 10 μM and 30 μM) were: Compound 1 (47.2%, 68.0%), compound 3 (38.9%, 83.2%), compound 4 (86.4%, 88.8%), and compound 7 (60.6%, 73.9%), respectively (all P<0.01) (Figure 4B). This result suggests that the novel compound 4 possesses the most potent apoptotic induction activity.
Activities of compound 4 on caspases 3 and 9
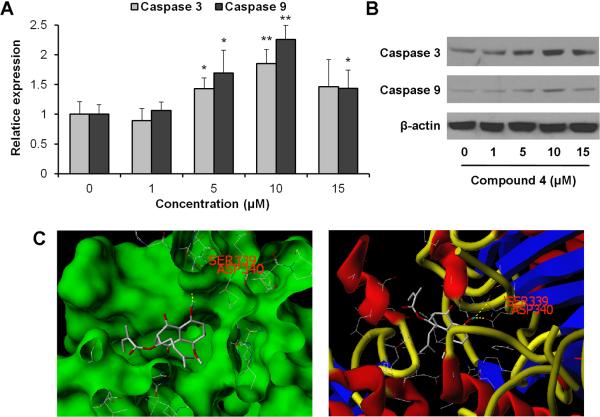
Caspase 3 and caspase 9 are two key proteins of the caspase family of proteases that are highly conserved in multicellular organisms and function as central regulators of apoptosis. They have been identified as playing a key role in the progression of apoptosis [21–23]. To further characterize the potential mechanism of the anticancer effect of compound 4, we conducted caspase assays. As shown in Figure 5A, treatment of SW-480 cells with 5 μM of compound 4 for 24 h increased the protease activities of caspases 3 and 9. These activities were further increased when treated with 10 μM of compound 4. At this treatment concentration, protease activity was increased by 85.2% for caspase 3, and 125.5% for caspase 9 (both P<0.01 vs. control). However, treatment with 15 μM did not increase their activities further (Figure 5A). The Western blot assay supported our results obtained by the protease activity assay (Figure 5B).
Figure 5.
Interactions of compound 4 with caspases 3 and 9. (A) Effects of compound 4 on caspases 3 and 9 activities in SW-480 cells. After treatment with 1–15 μM of compound 4 for 24 h, cell lysates were prepared and enzymatic activities of caspases 3 and 9 were measured by colorimetric assay. Results are normalized to each control and expressed as average ± SD of triplicate experiments (*P<0.05, **P<0.01 vs. control). (B) Western blot analysis of caspases 3 and 9. SW-480 cells were treated with 1–15 μM of compound 4 for 24 h. (C) Three-dimensional docking model of compound 4 at the binding site of human caspase 9 protein. Compound 4 docked with caspase 9 through hydrogen bond interactions with residues Asp-340 and Ser-339. Surface views are shown in the left panel and stick-ribbon models are shown in the right.
Since compound 4 significantly increased caspase 9 activity, to address if compound 4 might physically interact with caspase 9, we examined compound 4 docking for human caspase 9. The Surflex-Dock program was used to predict the binding sites of compound 4 to caspase 9. The energetically most favorable position for compound 4 interaction with caspase 9 is shown in Figure 5C. With in silico modeling, it is suggested that compound 4 forms hydrogen bonds with Asp-340 and Ser-339 at the active site of caspase 9 through its hydroxyl groups. In addition, compound 4 is predicted to show significant binding affinity for caspase 9 (CScore 3.82), suggesting that compound 4 directly interacts with caspase 9.
Discussion
Throughout the past two decades, the use of herbal medicines has been on the rise in cancer patients [24, 25]. As an alternative adjuvant to cancer chemotherapy, herbal medicines may act by increasing the sensitivity of neoplastic cells to chemotherapy and by reducing toxic and adverse effects induced by chemotherapeutic agents, without interfering with their tumoricidal effects [26, 27]. It is known that botanicals have been a significant resource to several of the currently used efficacious chemotherapeutic agents [28, 29]. There needs to be a continued effort to tap this resource and explore botanical or herbal active components with novel, potent, and distinct anti-cancer actions [30, 31].
Alkanna is a genus of herbaceous plants including about 60 species of the family Boraginaceae. The major bioactive compounds isolated from Alkanna plants are naphthoquinones [2]. Previous studies found that some naphthoquinone compounds showed anticancer activity [32, 33], but information on the structural-functional relationship of this type of compound was limited.
In our recent preliminary study, we isolated two naphthoquinone compounds, alkannin and angelylalkannin, from A. tinctoria, and observed significant anti-colorectal cancer cell proliferation activities. The antiproliferative potential of these two compounds was different, and the cancer cell inhibition of these two compounds was related to cell cycle arrest and induction of apoptosis. This promising data encouraged us to further isolate compounds from this plant. In this study, eight naphthoquinone compounds were isolated and purified from A. tinctoria. Structural elucidation confirmed that three of them are novel compounds.
The antiproliferative effects of isolated compounds were determined using human colorectal cancer cells. The two cell lines used in this study have varied p53 expression. HCT-116 is p53 wild type, while SW-480 cells contain a p53 mutation. Cancer cells with p53 mutations are resistant to many chemotherapeutic agents [34]. Interestingly, we observed that compounds 2 and 7 showed higher antiproliferative abilities in the p53 mutant cell line (SW-480) than in the p53 wild type line (HCT-116), suggesting that special structural modifications might prove useful in p53-mutated colon cancers. Moreover, we observed that compound 4, a novel naphthoquinone compound, showed the most potent antiproliferative effect, and the antiproliferative effect of compound 4 is stronger than that of fluorouracil [35], a commonly used chemotherapeutic agent.
Apoptosis is considered an important mechanism in the inhibition of cancer cells, and many cancer chemotherapeutic agents are strong apoptotic inducers against cancer cells [36, 37]. In this study, we assayed the effect of eight compounds on cancer cell apoptosis, and observed that the antiproliferation of potent compounds was related to their apoptotic response on colon cancer cells. To explore apoptotic induction mechanisms, we performed a caspase assay. We observed that compound 4 upregulated the expression of caspases 3 and 9, though this compound more significantly induced expression of caspase 9. Apoptosis occurs via a complex signaling cascade that is regulated at multiple points and involves many proteins. Caspase 9 is situated at critical points in apoptotic pathways and as a result plays a central role in apoptosis signal transduction [38]. Our docking analysis suggested the existence of an interaction site between compound 4 and caspase 9. Compound 4-induced apoptosis may occur in part through direct interactions with this protein.
Based on chemical structures and observed biological activities, we explored the structure-activity relationships of these compounds. Of the eight compounds, six are alkannin derivatives. Among the six derivatives, alkannin (1) showed strong antiproliferative effects. When alkannin (1) was acetylated to form acetylalkannin (2), its antiproliferative effect was almost eliminated, in which IC50 increased from 7.8 μM (1) to >50 μM (2) (for HCT-116 cells, same as follows). Continued modification on the 1'-acetyl group led to the increase of the antiproliferative effect. Specifically, IC50 decreased from >50 μM (acetylalkannin [2]) to 20.4 μM (dimethylacryl alkannin [5]), then to 17.0 μM (angelylalkannin [3]), but its antiproliferative potential is still less than that of alkannin (1). Furthermore, when 5-OH of angelylalkannin (3) was methylated to form the novel Compound 4 (5-Methoxyangenylalkannin), antiproliferative activities were significantly increased, with an IC50 of 4.4 μM. These data suggested that 1'-OH modification decreased antiproliferative effect, while modification on 5-OH increased its antiproliferative activity. Future structural modification should focus on 5-OH or 8-OH.
It is known that clinical drug development is a very long process and its success rate is low. However, many available anti-cancer drugs were originated from natural products. Our reported data are only an initial step for possible chemopreventive drug development. Compound 4 could be a good chemopreventive potential with possible clinical implications since the compound inhibited human colon cancer growth. Future in vivo pharmacological observations, as well as PK and toxicity studies will be conducted.
In conclusion, we isolated eight compounds from Alkanna tinctoria, with three novel compounds (compounds 4, 7, and 8). Among the eight compounds, six are alkannin derivatives, including compound 4. The antiproliferative activities of the eight compounds on different human colorectal cancer cell lines were determined. 5-Methoxyangenylalkannin (4) showed the most potent antiproliferative and apoptotic induction activities compared to alkannin (1) and its other derivatives. The analysis of the structural-functional relationship suggests that to improve anticancer activity, structural modification on alkannin should focus on hydroxyl groups at C-5 and C-8.
Acknowledgements
This work was supported in part by the NIH/NCCAM grants P01 AT004418 and K01 AT005362, and JICA-JST/NMIMR/CSRPM Medicinal Plants Research Project. We thank Professor Tiber Wenger, Semmelweis University, for plant donation.
Footnotes
Conflict of Interest The authors have no potential conflicts of interest.
References
- 1.Papageorgiou VP. Wound healing properties of naphthaquinone pigments from Alkanna tinctoria. Experientia. 1978;34:1499–1501. doi: 10.1007/BF01932375. [DOI] [PubMed] [Google Scholar]
- 2.Papageorgiou VP, Winkler A, Sagredos AN, Digenis GA. Studies on the relationship of structure to antimicrobial properties of naphthaquinones and other constituents of Alkanna tinctoria. Planta Med. 1979;35:56–60. doi: 10.1055/s-0028-1097184. [DOI] [PubMed] [Google Scholar]
- 3.Papageorgiou VP. New pigment of Alkanna-tinctoria having naphthaquinone structure. Planta Med. 1977;31:390–394. [Google Scholar]
- 4.Papageorgiou VP, Digenis GA. Isolation of 2 new alkannin esters from Alkanna-tinctoria. Planta Med. 1980;39:81–84. [Google Scholar]
- 5.Assimopoulou AN, Sturm S, Stuppner H, Papageorgiou VP. Preparative isolation and purification of alkannin/shikonin derivatives from natural products by high-speed counter-current chromatography. Biomed Chromatogr. 2009;23:182–198. doi: 10.1002/bmc.1101. [DOI] [PubMed] [Google Scholar]
- 6.Assimopoulu AN, Boskou D, Papageorgiou VP. Antioxidant activities of alkannin, shikonin and Alkanna tinctoria root extracts in oil substrates. Food Chem. 2004;87:433–438. [Google Scholar]
- 7.Ozer MS, Sarikurkcu C, Tepe B, Can S. Essential oil composition and antioxidant activities of alkanet (Alkanna tinctoria subsp. tinctoria) Food Sci Biotechnol. 2010;19:1177–1183. [Google Scholar]
- 8.Huu Tung N, Du GJ, Wang CZ, Yuan CS, Shoyama Y. Naphthoquinone components from Alkanna tinctoria (L.) Tausch show significant antiproliferative effects on human colorectal cancer cells. Phytother Res. 2013;27:66–70. doi: 10.1002/ptr.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang CZ, Luo X, Zhang B, Song WX, Ni M, Mehendale S, et al. Notoginseng enhances anti-cancer effect of 5-fluorouracil on human colorectal cancer cells. Cancer Chemother Pharmacol. 2007;60:69–79. doi: 10.1007/s00280-006-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain AN. Morphological similarity: a 3D molecular similarity method correlated with protein-ligand recognition. J Comput Aided Mol Des. 2000;14:199–213. doi: 10.1023/a:1008100132405. [DOI] [PubMed] [Google Scholar]
- 11.Giganti D, Guillemain H, Spadoni JL, Nilges M, Zagury JF, Montes M. Comparative evaluation of 3D virtual ligand screening methods: impact of the molecular alignment on enrichment. J Chem Inf Model. 2010;50:992–1004. doi: 10.1021/ci900507g. [DOI] [PubMed] [Google Scholar]
- 12.Papageorgiou VP. A new pigment of Alkanna tinctoria having naphthaquinone structure. Planta Med. 1977;31:390–394. [Google Scholar]
- 13.Papageorgiou VP, Digenis GA. Isolation of two new alkannin esters from Alkanna tinctoria. Planta Med. 1980;39:81–84. [Google Scholar]
- 14.Sevimli-Gur C, Akgun IH, Deliloglu-Gurhan I, Korkmaz KS, Bedir E. Cytotoxic naphthoquinones from Alkanna cappadocica (perpendicular) J Nat Prod. 2010;73:860–864. doi: 10.1021/np900778j. [DOI] [PubMed] [Google Scholar]
- 15.Yao XS, Ebizuka Y, Noguchi H, Kiuchi F, Shibuya M, Iitaka Y, et al. Biologically active constituents of Arnebia euchroma: structures of new monoterpenylbenzoquinones: arnebinone and arnebifuranone. Chem Pharm Bull. 1991;39:2962–2964. doi: 10.1248/cpb.39.2962. [DOI] [PubMed] [Google Scholar]
- 16.Fukui H, Hasan AFMF, Kyo M. Formation and secretion of a unique quinone by hairy root cultures of Lithospermum erythrorhizon. Phytochemistry. 1999;51:511–515. [Google Scholar]
- 17.Papageorgiou VP, Assimopoulou AN, Couladouros EA, Hepworth D, Nicolaou KC. The chemistry and biology of alkannin, shikonin, and related naphthazarin natural products. Angew Chem Int Ed Engl. 1999;38:271–300. doi: 10.1002/(SICI)1521-3773(19990201)38:3<270::AID-ANIE270>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Inouye H, Ueda S, Inoue K, Matsumura H. Quinones and related compounds in higher plants. Part 8. Biosynthesis of shikonin in callus cultures of Lithospermum erythrorhizon. Phytochemistry. 1979;18:1301–1308. [Google Scholar]
- 19.Agarwal C, Singh RP, Dhanalakshmi S, Tyagi AK, Tecklenburg M, Sclafani RA, et al. Silibinin upregulates the expression of cyclin-dependent kinase inhibitors and causes cell cycle arrest and apoptosis in human colon carcinoma HT-29 cells. Oncogene. 2003;22:8271–8282. doi: 10.1038/sj.onc.1207158. [DOI] [PubMed] [Google Scholar]
- 20.Hsu JD, Kao SH, Ou TT, Chen YJ, Li YJ, Wang CJ. Gallic acid induces G2/M phase arrest of breast cancer cell MCF-7 through stabilization of p27(Kip1) attributed to disruption of p27(Kip1)/Skp2 complex. J Agric Food Chem. 2011;59:1996–2003. doi: 10.1021/jf103656v. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson DW, Thornberry NA. Apoptosis. Life and death decisions. Science. 2003;299:214–215. doi: 10.1126/science.1081274. [DOI] [PubMed] [Google Scholar]
- 22.Creagh EM, Martin SJ. Caspases: cellular demolition experts. Biochem Soc Trans. 2001;29:696–702. doi: 10.1042/0300-5127:0290696. [DOI] [PubMed] [Google Scholar]
- 23.D'Amelio M, Cavallucci V, Cecconi F. Neuronal caspase-3 signaling: not only cell death. Cell Death Differ. 2010;17:1104–1114. doi: 10.1038/cdd.2009.180. [DOI] [PubMed] [Google Scholar]
- 24.Olaku O, White JD. Herbal therapy use by cancer patients: a literature review on case reports. Eur J Cancer. 2011;47:508–514. doi: 10.1016/j.ejca.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang CZ, He H, Wang X, Yuan CS. Trends in scientific publications of chinese medicine. Am J Chin Med. 2012;40:1099–1108. doi: 10.1142/S0192415X12500814. [DOI] [PubMed] [Google Scholar]
- 26.Wang HY, Qi LW, Wang CZ, Li P. Bioactivity enhancement of herbal supplements by intestinal microbiota focusing on ginsenosides. Am J Chin Med. 2011;39:1103–1115. doi: 10.1142/S0192415X11009433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang CZ, Calway T, Yuan CS. Herbal medicines as adjuvants for cancer therapeutics. Am J Chin Med. 2012;40:657–669. doi: 10.1142/S0192415X12500498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann J. Natural products in cancer chemotherapy: past, present and future. Nat Rev Cancer. 2002;2:143–148. doi: 10.1038/nrc723. [DOI] [PubMed] [Google Scholar]
- 29.Balunas MJ, Kinghorn AD. Drug discovery from medicinal plants. Life Sci. 2005;78:431–441. doi: 10.1016/j.lfs.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Ernst E. The role of complementary and alternative medicine in cancer. Lancet Oncol. 2000;1:176–180. doi: 10.1016/s1470-2045(00)00031-0. [DOI] [PubMed] [Google Scholar]
- 31.Sampson W. Natural products and cancer. In: Yuan CS, Bieber EJ, Bauer BA, editors. A Textbook of Complementary and Alternative Medicine. 2nd edn Parthenon/CRC; London: 2006. pp. 645–654. [Google Scholar]
- 32.Kapadia GJ, Balasubramanian V, Tokuda H, Konoshima T, Takasaki M, Koyama J, et al. Anti-tumor promoting effects of naphthoquinone derivatives on short term Epstein-Barr early antigen activation assay and in mouse skin carcinogenesis. Cancer Lett. 1997;113:47–53. doi: 10.1016/s0304-3835(96)04582-x. [DOI] [PubMed] [Google Scholar]
- 33.Kim JA, Lee EK, Park SJ, Kim ND, Hyun DH, Lee CG, et al. Novel anti-cancer role of naphthazarin in human gastric cancer cells. Int J Oncol. 2012;40:157–162. doi: 10.3892/ijo.2011.1195. [DOI] [PubMed] [Google Scholar]
- 34.Papenfuss K, Cordier SM, Walczak H. Death receptors as targets for anti-cancer therapy. J Cell Mol Med. 2008;12:2566–2585. doi: 10.1111/j.1582-4934.2008.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang CZ, Li B, Wen XD, Zhang Z, Yu C, Calway TD, et al. Paraptosis and NF-kappaB activation are associated with protopanaxadiol-induced cancer chemoprevention. BMC Complement Altern Med. 2013;13:2. doi: 10.1186/1472-6882-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z, Chen X, Fu S, Bao J, Dang Y, Huang M, et al. Dehydrocorydaline inhibits breast cancer cells proliferation by inducing apoptosis in MCF-7 cells. Am J Chin Med. 2012;40:177–185. doi: 10.1142/S0192415X12500140. [DOI] [PubMed] [Google Scholar]
- 37.Du GJ, Dai Q, Williams S, Wang CZ, Yuan CS. Synthesis of protopanaxadiol derivatives and evaluation of their anticancer activities. Anticancer Drugs. 2011;22:35–45. doi: 10.1097/CAD.0b013e32833fde29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wurstle ML, Laussmann MA, Rehm M. The central role of initiator caspase-9 in apoptosis signal transduction and the regulation of its activation and activity on the apoptosome. Exp Cell Res. 2012;318:1213–1220. doi: 10.1016/j.yexcr.2012.02.013. [DOI] [PubMed] [Google Scholar]